Abstract
Bacillus subtilis cwlD and dacB mutants produce spore peptidoglycan (PG) with increased cross-linking but with little change in spore core hydration compared to the wild type. A cwlD dacB double mutant produced spores with a two- to fourfold greater increase in PG cross-linking and novel muropeptides containing glycine residues but no significant changes in spore resistance or core hydration.
The peptidoglycan (PG) cortex of bacterial endospores is required for maintenance of spore core dehydration, heat resistance, and dormancy. This PG differs in structure from that of the vegetative cell walls of the corresponding species in that many of the PG peptide side chains are removed either partially, to leave single l-Ala side chains, or completely, with formation of muramic-δ-lactam (22, 24). The decrease in the number of side chains results in a very low degree of cross-linking of the PG strands in the spore cortex (1, 14, 22). This low degree of cross-linking should give the cortical PG a significant flexibility, which has been proposed to be required for achievement of spore core dehydration during spore formation (8, 21). Recently, it has been suggested that there is a gradient of cross-linking across the span of the cortical PG, with loose cross-linking on the inside and high cross-linking on the outside, which provides a directional flexibility to this structure that might participate in core dehydration (13). Recent advances in methods for rapid analysis of spore PG structure have allowed the identification of structural changes in several mutant strains. A Bacillus subtilis dacB mutant, which lacks the sporulation-specific d,d-carboxypeptidase penicillin-binding protein (PBP) 5* (4), has a fourfold increase in spore PG cross-linking compared to the wild type yet has normal spore core dehydration (1, 14) and only a slight loss of spore heat resistance due to an inability to maintain dehydration upon heating (16). A cwlD mutant produces spore PG with no muramic-δ-lactam and a twofold increase in cross-linking (1, 15), has almost-normal spore core dehydration and normal spore heat resistance (15), and is unable to degrade spore PG in order to complete germination (15, 19). The fact that major increases in the PG cross-linking in these mutant spores had little effect on spore core dehydration and heat resistance is suggestive that flexibility of the cortical PG may in fact be unimportant in achievement of spore core dehydration. However, it is not certain whether in these mutants the high amount of spore PG cross-linking is present during the process of spore core dehydration or is attained only after this process is complete.
Given the significant changes in PG structure in spores of cwlD and dacB mutants, we decided to determine if there were even greater changes in PG structure in spores of a cwlD dacB double mutant and whether this would have any significant effect on spore phenotype. We constructed a cwlD dacB double-mutant strain by transformation of strain PS2066 (ΔdacB) (16) with chromosomal DNA from PS2307 (cwlD::Cm) (15, 19) and selection for chloramphenicol resistance; the presence of each mutation in the resulting strain, PS2422, was verified by Southern blotting. The single mutants and the double mutant had vegetative growth rates in 2× SG medium (7) identical to that of a wild-type strain (PS832), and all produced equal numbers of spores per milliliter of culture based upon microscopic examination. As observed previously, the colony-forming ability of the cwlD spores was severely reduced (15, 19) (10−3 colonies/spore) and this was unaffected by the addition of the dacB mutation. When spore germination was assayed as described (15), the double mutant exhibited a phenotype identical to that previously found for the cwlD mutant (15, 19). Relative to wild-type and dacB spores, the cwlD and double-mutant spores lost optical density and released dipicolinic acid (DPA) a little more slowly following exposure to germinants. However, the extents of optical density loss and DPA release were similar for all spore preparations by 90 min after addition of germinants. Germinating wild-type and dacB spores released 45 and 19% of the total spore hexosamine as PG fragments within 90 min, respectively, while the cwlD and double-mutant spores released <5%. The latter severe defect is due to the inability of spores of strains with the cwlD mutation to hydrolyze their altered spore PG during germination (2, 15, 19). Spore heat resistance was determined by using a method (15) in which the mutant spores are subjected to a mild lysozyme treatment which allows them to complete germination and form colonies. As observed previously (15), the wild-type and cwlD spores had similar values for heat resistance (D85 values of 28 and 33 min, respectively; the D85 is the time at 85°C required to reduce spore viability tenfold), while the dacB spores had slightly reduced heat resistance (D85 of 16 min). The heat resistance of the double-mutant spores (D85 of 16 min) was identical to that of the dacB spores. These heat resistance values correlated well with spore core wet density values measured by metrizoic acid density gradient centrifugation of coat-permeabilized spores (9). The wild-type and dacB spores had identical spore core wet densities (two independent measurements [13, 16]), whereas the cwlD and double-mutant spores had a slightly reduced spore core wet density (reduced by 0.006 g/ml, three independent measurements). This slight increase in spore water content is not expected to produce a discernible change in heat resistance (3, 12). The decreased heat resistance of the dacB spores is due to the inability of these spores to maintain core dehydration upon heating (16), and this is presumably also the case for the cwlD dacB spores.
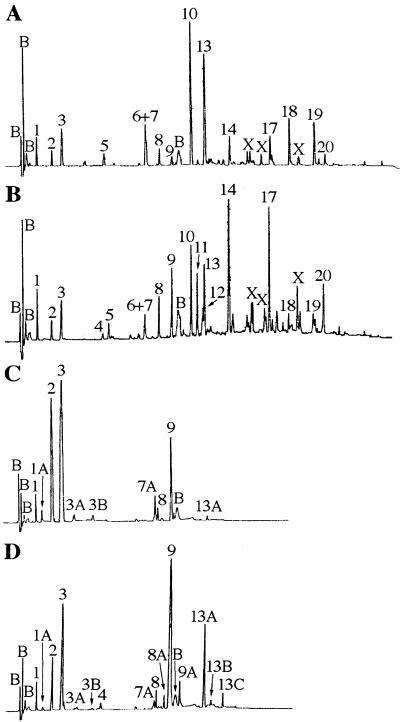
To determine if changes in spore cortex structure were correlated with changes in other properties of these spores, PG was purified from each of the spore preparations and digested with a muramidase (Mutanolysin; Sigma), and the digest was analyzed by reversed-phase high-performance liquid chromatography (HPLC) (Fig. 1) (1, 14). Most of the resulting muropeptides were identified by cochromatography with previously identified muropeptides (Table 1). Novel muropeptides were further purified by using a different reversed-phase HPLC gradient system and subjected to amino acid analysis and matrix-assisted laser desorption/ionization (MALDI) time-of-flight mass spectrometry (14) (Table 2). Newly identified muropeptides included two disaccharide (DS) pentapeptides, i.e., one containing a Gly residue (Fig. 1 and Table 2, peak 3A [DS-TP-Gly; TP is tetrapeptide]) and one containing an additional Ala residue (Fig. 1 and Table 2, peak 3B [DS-TP-Ala]). The presence of glycine in muropeptides from spore cortex had previously been seen in cwlD single-mutant spores (muropeptides 1A and 7A) (15) and in the PG of outgrowing spores (11), but the positions of these glycine residues in the peptides are uncertain (see below). Muropeptides 3A and 3B were present in small amounts in the cwlD mutant spores but had not been previously identified. Other newly identified muropeptides (Table 2) derived from the double-mutant spores included peak 8A, a cross-linked dimer of DS-TP-Gly and DS-TP; peak 9A, a cross-linked dimer of DS-TP-Ala and DS-TP; peak 13B, a cross-linked trimer of one DS-TP-Ala and two DS-TP; and peak 13C, a cross-linked tetramer of DS-TP.
FIG. 1.
HPLC analysis of spore PG muropeptides. PG purified from spores produced by strains PS832 (wild type) (A), PS2066 (ΔdacB) (B), PS2307 (cwlD::Cm) (C), and PS2422 (cwlD::CmΔdacB) (D) was muramidase digested and separated by using a methanol gradient as previously described (14). Peaks are numbered as previously described (14, 15), and the structures of the muropeptides are given in Table 1. Peaks labeled B are buffer components seen in control samples. Peaks labeled X are reduction products of muramic-δ-lactam-containing muropeptides; corrections for this reduction were performed in calculation of PG structural parameters (14). No additional peaks were observed upon longer elution of the samples in panels C and D.
TABLE 1.
Muropeptide peaks derived from spore PG
| Peaka | Structureb | Reference for structure | Amino acids released during hydrazinolysis |
|---|---|---|---|
| 1 | DS-TriP | 14 | Dpm |
| 1A | DS-TriP-Gly | 15 | Gly |
| 2 | DS-Ala | 14 | Ala |
| 3 | DS-TP | 14 | Ala |
| 3A | DS-TP-Gly | Table 2 | Gly, Ala |
| 3B | DS-TP-Ala | Table 2 | Ala |
| 4 | DS-TP-TP | 14 | |
| 5 | TS-TP (open lactam) | 1, 14 | |
| 6 | TS-TP (reduced lactam) | 14 | |
| 7 | TS-Ala (reduced lactam) | 14 | |
| 7A | DS-TriP-Gly-DS-TP | 15 | Gly |
| 8 | DS-TriP-DS-TP | 14 | Dpm |
| 8A | DS-TP-Gly-DS-TP | Table 2 | Gly |
| 9 | DS-TP-DS-TP | 14 | Ala |
| 9A | DS-TP-Ala-DS-TP | Table 2 | Ala |
| 10 | TS-TP | 14 | |
| 11 | TS-TP-TP | 14 | |
| 12 | DS-TP-TS-TP (reduced lactam) | 14 | |
| 13 | TS-Ala | 14 | |
| 13A | DS-TP-DS-TP-DS-TP | 15 | |
| 13B | DS-TP-DS-TP-DS-TP-(Ala) | Table 2 | |
| 13C | DS-TP-DS-TP-DS-TP-DS-TP | Table 2 | |
| 14 | DS-TP-TS-TP | 14 | |
| 17 | TS-TP-TS-TP | 14 | |
| 18 | HS-TP | 14 | |
| 19 | HS-Ala | 14 | |
| 20 | TS-TP-HS-TP | 14 |
Peaks are numbered as shown in Fig. 1.
Abbreviations: HS, hexasaccharide; TS, tetrasaccharide. Where multiple saccharide moieties are indicated they are cross-linked via their peptide side chains with the first peptide providing the acceptor Dpm. The placement of an Ala residue in parentheses in muropeptide 13B indicates that this residue could be on either the first or second acceptor peptide.
TABLE 2.
Structural data for novel muropeptidesa
| Peak | Amt (nmol)
|
Predicted structure | (M-H)− calculated m/zb | (M-H)− observed m/zb | |||||
|---|---|---|---|---|---|---|---|---|---|
| N-acetylglucosamine | N-acetylmuramic acid | Ala | Glu | Dpm | Gly | ||||
| 3A | 1 (1) | 1.0 (1) | 2.1 (2) | 1.2 (1) | 1.2 (1) | 1.0 (1) | DS-TP-Gly | 999.0 | 998.7 |
| 3B | 1 (1) | 1.2 (1) | 2.9 (3) | 1.3 (1) | 1.2 (1) | 0.1 (0) | DS-TP-Ala | 1,036.0c | 1,034.5c |
| 8A | 1 (2) | 1.1 (2) | 2.0 (4) | 1.3 (2) | 1.1 (2) | 0.6 (1) | DS-TP-Gly-DS-TP | 1,921.9 | 1,921.5 |
| 9A | 1 (2) | 1.3 (2) | 3.2 (5) | 1.7 (2) | 1.6 (2) | 0.05 (0) | DS-TP-Ala-DS-TP | 1,936.0 | 1,935.0 |
| 13B | 1 (3) | 1.1 (3) | 2.1 (7) | 1.1 (3) | 1.3 (3) | 0.1 (0) | DS-TP-DS-TP-DS-TP-(Ala) | 2,859.9 | 2,859.1 |
| 13C | 1 (4) | 1.0 (4) | 2.0 (8) | 1.2 (4) | 1.2 (4) | 0.04 (0) | DS-TP-DS-TP-DS-TP-DS-TP | 3,712.7 | 3,712.5 |
Peaks are numbered as shown in Fig. 1. All nanomole amounts determined using amino acid and amino sugar analyses (14) are normalized to the amount of N-acetylglucosamine taken as 1. Values in parentheses are predicted numbers of moles of the residues within the molecule. The placement of an Ala residue in parentheses in muropeptide 13B indicates that this residue could be on either the first or second acceptor peptide.
(M-H)− is the deprotonated molecular ion observed in the negative-ion mode. Calculated m/z values are the mass-to-charge ratios predicted from the amino acid and amino sugar analyses. Average mass values are given. Observed m/z values are the values measured by MALDI time-of-flight mass spectrometry.
These are calculated and observed values for the (M+Na)+ ion.
The overall structure of the spore PG in the double mutant revealed a combination of the alterations seen in the two single mutants as well as newly apparent muropeptides (Fig. 1 and Tables 2 and 3). The complete lack of muramic-δ-lactam caused by the cwlD mutation (1, 15) was combined with the high amount of cross-linking caused by the dacB mutation (1, 14). The appearance of novel muropeptides 8A, 9A, 13B, and 13C in the double mutant is due to increased cross-linking of monomer muropeptides that are present in the single mutants. The presence of a small amount of ineffective cross-links, in which one of the peptides had been cleaved from the glycan strand, was previously seen in dacB spore PG (14). A very small amount of these ineffective cross-links was also found in the double-mutant spore PG (Fig. 1D, peak 4). The fact that there are fewer of these ineffective cross-links in the double-mutant spore PG than in the dacB spore PG suggests that while a CwlD muramoyl-l-alanine amidase activity may be responsible for some of this peptide cleavage there must also be an additional amidase functioning during spore PG synthesis. The percentage of muramic acid residues which retained peptides of any sort (87%) was increased above the level seen in the cwlD mutant (66%), and the increased cross-linking of these peptides may reduce the cleavage to produce single l-Ala side chains, as seen in the dacB mutant (Table 3) (1, 14). The degree of effective cross-linking (both peptides attached to glycans) of the spore PG in the double mutant (expressed as a percentage of muramic acid residues with cross-linked peptides) was ninefold, fourfold, and twofold greater than those of the PG from wild-type, cwlD, and dacB spores, respectively (Table 3). The degree of cross-linking of the PG from the cwlD dacB spores is similar to values obtained for B. subtilis vegetative cell wall PG (6, 17, 23).
TABLE 3.
Spore PG structural parametersa
| Strain (genotype) | % of muramic acid with side chain ofb:
|
% Peptides with Gly | % of muramic acid with a cross-linked peptidec | ||||
|---|---|---|---|---|---|---|---|
| Lactam | l-Ala | TP | TriP | TP-Ala | |||
| PS832 (wild type) | 50 | 24 ± 2 | 25 ± 2 | 2.1 ± 0.8 | 0 | 0 | 3.5 ± 0.6 |
| PS2066 (ΔdacB) | 46 ± 1 | 12 | 43 ± 2 | 2.2 ± 0.1 | 0 | 0 | 16.5 ± 1.3 (13.7) |
| PS2307 (cwlD::Cm) | 0 | 33 ± 1 | 58 ± 1 | 6.4 ± 0.4 | 1.2 ± 0.1 | 6.3 ± 0.7 | 7.9 ± 0.3 |
| PS2422 (cwlD::Cm ΔdacB) | 0 | 13 ± 3 | 80 ± 3 | 4.8 ± 0.4 | 2.4 ± 0.2 | 3.9 ± 1.0 | 32.4 ± 3.0 |
Data are averages of two (PS832, PS2066, and PS2307) or three (PS2422) analyses with errors of 1 standard deviation.
Lactam, muramic-δ-lactam. The values given for TP and TriP are the sum of TP and TP-Gly and the sum of TriP and TriP-Gly, respectively.
The value in parentheses is for effective cross-links; the remaining (ineffective) cross-links involved peptides which had been cleaved from the muramic acid (14). If no value is given in parentheses then there were not enough ineffective cross-links to alter the value.
The fact that the extremely high level of PG cross-linking in cwlD dacB spores had no effect on attainment of relatively normal spore core dehydration suggests that either (i) the flexibility of the spore PG is irrelevant to core dehydration, (ii) high degree of cross-linking is achieved only after core dehydration has taken place, or (iii) the high degree of cross-linking is concentrated in the outer layers of the spore PG and the proposed gradient of cross-linking within this structure (13) is still present. Analysis of the PG structure in developing spores throughout sporulation should generate evidence for or against these various possibilities, and this work is currently in progress.
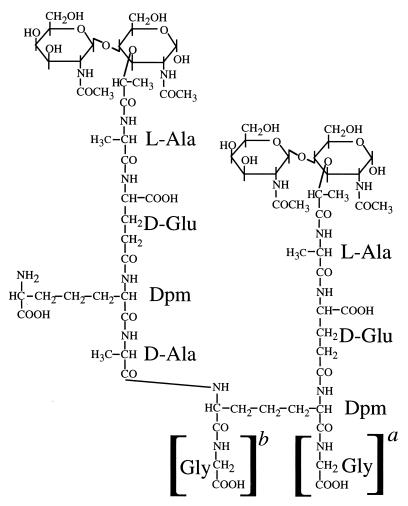
Since the observation of Gly in spore PG was rather novel, we attempted to determine the positions of the Gly residues in the appropriate muropeptides derived from cwlD and double-mutant spore PG. Treatment of the Gly-containing muropeptides with fluorodinitrobenzene (FDNB) never resulted in modification of the Gly residues, as judged by subsequent amino acid analysis, indicating that the Gly did not have a free amino group. However, FDNB treatment did result in modification of all diaminopimelic acid (Dpm) residues in monomer muropeptides (peaks 1, 1A, 3, 3A, and 3B) and 50% of the Dpm in cross-linked muropeptides (peaks 7A, 8, 8A, 9, 9A, and 13B) (the “acceptor” Dpm which is involved in cross-linking should not be modified). This indicates that the Gly in these muropeptides is not in a peptide bond to the ε-amino group of the Dpm, in the position of a Gly peptide cross bridge observed in the PG of some bacterial species (18). This is relevant to the observation that Lysostaphin, which cleaves glycyl-interpeptide cross bridges in PG, can exert some effect on the B. subtilis vegetative cell wall (20) despite the failure to identify such cross bridges in this species (23). Remaining possibilities for the position of the Gly are (i) the α-carboxyl group of the Dpm, (ii) the ω-carboxyl group of the Dpm, (iii) the α-carboxyl group of the d-Glu, and (iv) the carboxyl group of Ala in DS-TP-Gly. Hydrazinolysis of muropeptide 1A produced only Gly (Table 1), suggesting that the Gly is attached to Dpm (if the Gly were on the Glu then the Dpm should also be recovered as a C-terminal amino acid, as for muropeptide 1 [Table 1]). Hydrazinolysis of muropeptide 3A produced Gly and Ala. Two possible interpretations of this are (i) that the Gly and the Ala are peptide bonded to the α- and ω-carboxyls of the Dpm and (ii) that this is a mixture of two linear pentapeptides with the Gly in either the fourth or fifth position. The results for quantitative recovery of amino acids following hydrazinolysis were not consistent enough to differentiate between these two possibilities. Hydrazinolysis of muropeptide 3B released Ala. Again, either this additional amino acid could be on the ω-carboxyl of the Dpm or this could be a linear pentapeptide terminating in two Ala residues. However, we have found that this muropeptide is also present in the spore PG of a dacA mutant (10) that lacks the d,d-carboxypeptidase PBP5. Loss of this protein might be expected to result in the retention of the linear pentapeptide present in the PG precursors. Hydrazinolysis of muropeptide 8A produced only Gly. This indicates that the Gly must be the fifth residue of a linear acceptor pentapeptide. HPLC separation by using a different buffer system (14) resolved peak 7A into two muropeptides, each of which gave the identical results of amino acid analysis. Hydrazinolysis produced only Gly from each of these. Two potential explanations for this are (Fig. 2) (i) that these two muropeptides are DS-TriP-DS-TP (TriP is tripeptide) with an additional Gly attached to either the α- or the ω-carboxyl of the Dpm in the acceptor TriP and (ii) that these two muropeptides each have Gly attached to the same carboxyl of Dpm and that they differ in some other part of the molecule. Although MALDI postsource decay time-of-flight mass spectrometry has allowed us to determine the carbohydrate and amino acid sequences of some muramyl peptides (5), the signals from muropeptides 7A and 8A containing five free carboxyl groups were too low to permit fragment ion analysis.
FIG. 2.
Proposed structure of muropeptide 7A. The DSs (N-acetylglucosamine–N-acetylmuramic acid) have the peptide side chains linked to the muramic acid residues. The peptides have the d-Glu residues linked through their γ-carboxyls to the α-amino groups of Dpm. The peptide cross-link is formed between the d-Ala carboxyl and the ε-amino group of the Dpm. The Gly residues in brackets are the positions where this residue can be linked to the α-carboxyl (a) and ω-carboxyl (b) of the Dpm in the acceptor peptide, respectively.
Our data indicates that Gly is present in some cases as the fourth amino acid in the peptide (muropeptide 1A and potentially 7A) and in some cases as the fifth (muropeptide 8A and potentially 3A). It is also possible that some Gly residues (and by analogy some extra Ala residues) are on the ω-carboxyl of the Dpm, a novel position for Gly in PG (18). The origin of these Gly residues is unclear, and they are seen only under unique circumstances—in strains with disrupted PG metabolism and in outgrowing spores (11). However, no Gly-containing peptides are seen in immature spore PG extracted from wild-type forespores (10). It is possible that these Gly residues are incorporated into PG due to some amino acid deficiency, which could arise during spore outgrowth in poor medium or due to a failure to cleave and recycle peptides during PG synthesis in sporulation of certain mutants. However, supplementation of cultures of the cwlD dacB mutant 2.5 h into sporulation with either 1 mM d-Ala or 1 mM l-Ala and analysis of the resulting spore PG structure showed that these medium additions had no effect on the incorporation of Gly residues onto PG (data not shown). An alternative possibility is that some Gly residues in mutant spore PG are a byproduct of an abnormal Dpm amidation reaction; indeed, Dpm in the vegetative wall of B. subtilis is normally amidated on the ω-carboxyl (23).
Acknowledgments
This work was supported by grants GM19698 (P.S.), GM56695 (D.L.P.), and RR10888 (C.E.C.) from the National Institutes of Health and by funds from Virginia Tech.
We thank John Helmann for helpful discussion.
REFERENCES
- 1.Atrih A, Zollner P, Allmaier G, Foster S J. Structural analysis of Bacillus subtilis 168 endospore peptidoglycan and its role during differentiation. J Bacteriol. 1996;178:6173–6183. doi: 10.1128/jb.178.21.6173-6183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atrih A, Zollner P, Allmaier G, Williamson M P, Foster S J. Peptidoglycan structural dynamics during germination of Bacillus subtilis 168 endospores. J Bacteriol. 1998;180:4603–4612. doi: 10.1128/jb.180.17.4603-4612.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaman T C, Gerhardt P. Heat resistance of bacterial spores correlated with protoplast dehydration, mineralization, and thermal adaptation. Appl Environ Microbiol. 1986;52:1242–1246. doi: 10.1128/aem.52.6.1242-1246.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchanan C E, Gustafson A. Mutagenesis and mapping of the gene for a sporulation-specific penicillin-binding protein in Bacillus subtilis. J Bacteriol. 1992;174:5430–5435. doi: 10.1128/jb.174.16.5430-5435.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costello C E. Time, life … and mass spectrometry—new techniques to address biological questions. Biophys Chem. 1997;68:173–188. doi: 10.1016/s0301-4622(97)00033-1. [DOI] [PubMed] [Google Scholar]
- 6.Forrest T M, Wilson G E, Pan Y, Schaefer J. Characterization of cross-linking of cell walls of Bacillus subtilis by a combination of magic-angle spinning NMR and gas chromatography-mass spectrometry of both intact and hydrolyzed 13C- and 15N-labeled cell-wall peptidoglycan. J Biol Chem. 1991;266:24485–24491. [PubMed] [Google Scholar]
- 7.Leighton T J, Doi R H. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem. 1971;254:3189–3195. [PubMed] [Google Scholar]
- 8.Lewis J C, Snell N S, Burr H K. Water permeability of bacterial spores and the concept of a contractile cortex. Science. 1960;132:544–545. doi: 10.1126/science.132.3426.544. [DOI] [PubMed] [Google Scholar]
- 9.Lindsay J A, Beaman T C, Gerhardt P. Protoplast water content of bacterial spores determined by buoyant density sedimentation. J Bacteriol. 1985;163:735–737. doi: 10.1128/jb.163.2.735-737.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meador-Parton, J., and D. L. Popham. 1999. Unpublished data.
- 11.Murray T, Popham D L, Pearson C B, Hand A R, Setlow P. Analysis of outgrowth of Bacillus subtilis spores lacking penicillin-binding protein 2a. J Bacteriol. 1998;180:6493–6502. doi: 10.1128/jb.180.24.6493-6502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakashio S, Gerhardt P. Protoplast dehydration correlated with heat resistance of bacterial spores. J Bacteriol. 1985;162:571–578. doi: 10.1128/jb.162.2.571-578.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popham D L, Gilmore M E, Setlow P. Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties. J Bacteriol. 1999;181:126–132. doi: 10.1128/jb.181.1.126-132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popham D L, Helin J, Costello C E, Setlow P. Analysis of the peptidoglycan structure of Bacillus subtilis endospores. J Bacteriol. 1996;178:6451–6458. doi: 10.1128/jb.178.22.6451-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popham D L, Helin J, Costello C E, Setlow P. Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc Natl Acad Sci USA. 1996;93:15405–15410. doi: 10.1073/pnas.93.26.15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popham D L, Illades-Aguiar B, Setlow P. The Bacillus subtilis dacB gene, encoding penicillin-binding protein 5*, is part of a three-gene operon required for proper spore cortex synthesis and spore core dehydration. J Bacteriol. 1995;177:4721–4729. doi: 10.1128/jb.177.16.4721-4729.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popham D L, Setlow P. The cortical peptidoglycan from spores of Bacillus megaterium and Bacillus subtilis is not highly cross-linked. J Bacteriol. 1993;175:2767–2769. doi: 10.1128/jb.175.9.2767-2769.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekiguchi J, Akeo K, Yamamoto H, Khasanov F K, Alonso J C, Kuroda A. Nucleotide sequence and regulation of a new putative cell wall hydrolase gene, cwlD, which affects germination in Bacillus subtilis. J Bacteriol. 1995;177:5582–5589. doi: 10.1128/jb.177.19.5582-5589.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulijasz A T, Grenader A, Weisblum B. A vancomycin-inducible lacZ reporter system in Bacillus subtilis: induction by antibiotics that inhibit cell wall synthesis and by lysozyme. J Bacteriol. 1996;178:6305–6309. doi: 10.1128/jb.178.21.6305-6309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warth A D. Mechanisms of heat resistance. In: Dring G J, Ellar D J, Gould G W, editors. Fundamental and applied aspects of bacterial spores. London, United Kingdom: Academic Press, Inc.; 1985. pp. 209–225. [Google Scholar]
- 22.Warth A D, Strominger J L. Structure of the peptidoglycan from spores of Bacillus subtilis. Biochemistry. 1972;11:1389–1396. doi: 10.1021/bi00758a010. [DOI] [PubMed] [Google Scholar]
- 23.Warth A D, Strominger J L. Structure of the peptidoglycan from vegetative cell walls of Bacillus subtilis. Biochemistry. 1971;10:4349–4358. doi: 10.1021/bi00800a001. [DOI] [PubMed] [Google Scholar]
- 24.Warth A D, Strominger J L. Structure of the peptidoglycan of bacterial spores: occurrence of the lactam of muramic acid. Proc Natl Acad Sci USA. 1969;64:528–535. doi: 10.1073/pnas.64.2.528. [DOI] [PMC free article] [PubMed] [Google Scholar]