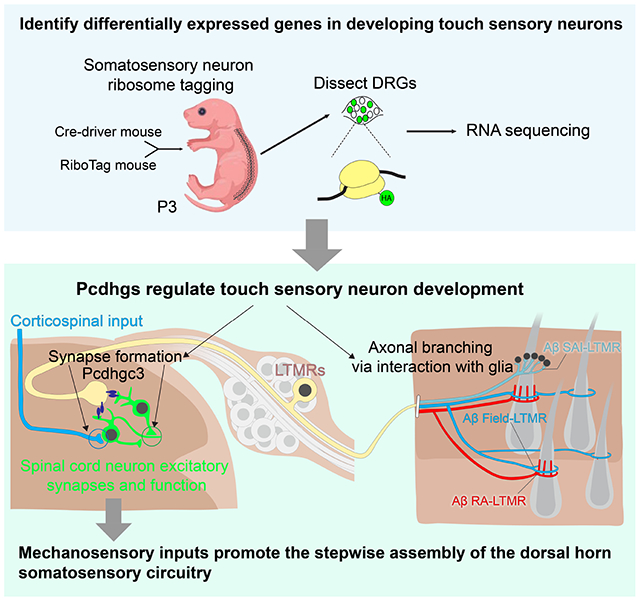
Summary
Light touch sensation begins with activation of low-threshold mechanoreceptor (LTMR) endings in the skin and propagation of their signals to the spinal cord and brainstem. We found that the clustered protocadherin gamma (Pcdhg) gene locus, which encodes 22 cell-surface homophilic binding proteins, is required in somatosensory neurons for normal behavioral reactivity to a range of tactile stimuli. Developmentally, distinct Pcdhg isoforms mediate LTMR synapse formation through neuron-neuron interactions and peripheral axonal branching through neuron-glia interactions. The Pcdhgc3 isoform mediates homophilic interactions between sensory axons and spinal cord neurons to promote synapse formation in vivo, and is sufficient to induce postsynaptic specializations in vitro. Moreover, loss of Pcdhgs and somatosensory synaptic inputs to the dorsal horn leads to fewer corticospinal synapses onto dorsal horn neurons. These findings reveal essential roles for Pcdhg isoform diversity in somatosensory neuron synapse formation, peripheral axonal branching, and step-wise assembly of central mechanosensory circuitry.
Graphical Abstract
eTOC Blurb
Meltzer et al. show that the clustered protocadherin gamma (Pcdhg) gene locus regulates somatosensory neuron synapse formation and peripheral axonal branching. Pcdhgc3 is the only isoform that promotes synapse formation between sensory neurons and spinal cord neurons. Further, they found that somatosensory inputs regulate the assembly of spinal cord circuitry.
Introduction
The skin, our largest organ, mediates our sense of touch, which is essential for fundamental tasks ranging from object recognition to social exchange. The sense of touch also contributes to brain development, such that deprivation or over-reactivity to touch during early stages of animal development can result in aberrant brain development and function.1-3 Light touch sensation begins with activation of low-threshold mechanoreceptor (LTMR) axonal endings in the skin4, and the relay of LTMR signals to the spinal cord and brainstem.4-7 Yet, of all the senses, touch remains one of the least understood at the developmental level.8-10
Based on their action potential conduction velocity, cutaneous LTMRs are classified as Aβ-, Aδ-, or C-LTMRs.11 LTMRs are further distinguished by the cutaneous end organs with which they associate, rates of adaptation to indentation of the skin (rapidly adapting (RA), intermediate adapting (IA), or slowly adapting (SA)), and preferred stimuli.4 In mouse hairy skin, guard hair follicles account for ~1% of the trunk hair follicles and are associated with Aβ RA-LTMRs, Aβ SAI-LTMRs, and Aβ field-LTMRs, which are differentially sensitive to hair deflection, skin indentation, and stroking.5,12,13 Awl/auchene and zigzag hair follicles, which together comprise ~99% of trunk hair follicles, are innervated by Aβ RA-LTMRs (Awl/auchene hair only), Aβ field-LTMRs, Aδ-LTMRs, and C-LTMRs.14,15 In glabrous skin, LTMRs also form end organs associated with Aβ RA-LTMRs and Aβ SA-LTMRs.16-18 Individual LTMR subtypes form a range of axonal branch numbers and patterns in the skin, which establishes their physiological receptive fields4,8,9. How LTMR branching patterns are controlled remains unknown.
In the spinal cord dorsal horn, LTMR terminals are organized in a somatotopic and highly overlapping manner in the LTMR-RZ, which spans lamina IIiv through V.6,15,19 During mouse embryonic development, beginning around E14, somatosensory neuron axons penetrate the spinal cord and establish synaptic connections with spinal cord neurons over the next several days to weeks.20,21 Mechanical stimuli evoked behavioral responses and physiological responses recorded in the dorsal horn are first observed at late gestational ages.22,23 The precise timing of the formation of synapses between LTMR subtypes and spinal cord neurons, the molecular signals that govern this process, and how LTMR inputs may affect the functional organization of other inputs to the dorsal horn remain poorly understood.
The clustered protocadherins encode a family of diverse cell-surface homophilic proteins and are tandemly arrayed in the genome in α-, β- and γ-subclusters, called Pcdha, Pcdhb and Pcdhg, respectively.24 In the Pcdha and Pcdhg subclusters, variable exons encoding extracellular, transmembrane, and juxtamembrane domains are spliced to three constant exons.25-28 Thus, each Pcdhg gene encodes a protein with a unique extracellular and transmembrane domain, a variable cytoplasmic domain, followed by a shared C-terminal domain.24,29-31 The clustered Pcdhs play critical roles in many aspects of nervous system development, including dendritic self-avoidance, dendrite arborization, neuronal survival, astrocyte-neuron interactions, and axonal tiling.32-39 Although Pcdhgs can be found at synapses, especially in postsynaptic density (PSD) fractions of purified synaptosomes,40-43 and they form two-dimensional structures between adjacent membranes,44-49 functional evidence demonstrating a direct role in synapse formation has remained elusive.50,51
Here, we determined the timing of synapse formation between LTMRs and spinal cord neurons of the LTMR-RZ and assessed LTMR gene expression profiles during the peak of postnatal synapse formation to identify candidate synaptogenic regulators. Our findings have revealed unique functions for distinct clustered protocadherin family members in somatosensory neuron synapse formation and peripheral axonal branching, and that sensory inputs to the spinal cord indirectly control the assembly of other components of spinal cord circuitry.
Results
Pcdhgs are expressed in the postnatal DRG and spinal cord when sensory neurons are forming synaptic connections
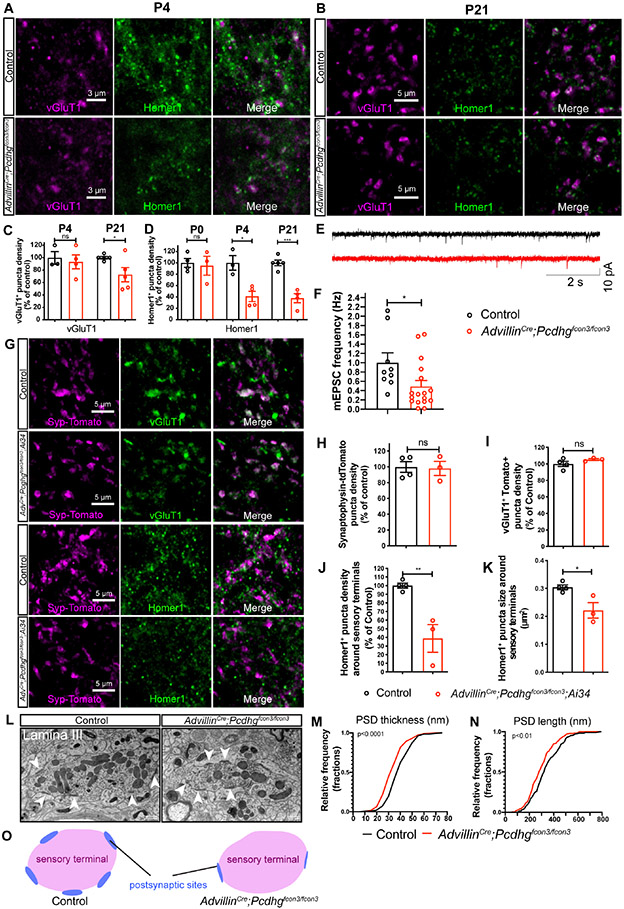
To define the postnatal period of synapse formation between mechanosensory neurons and spinal cord neurons, we used previously characterized genetic tools to label synapses of all primary somatosensory neuron subtypes (AdvillinCre)52 as well as synapses of two representative LTMR subtypes, Aδ-LTMRs (TrkBCreER)13 and Aβ RA-LTMRs (RetCreER).53 A Cre-dependent synaptophysin-tdTomato reporter line (Ai34) was used to visualize presynaptic terminals of these sensory neurons during postnatal development with a focus on synapses in the thoracic LTMR-RZ. The size of presynaptic sensory terminals increased between P0 and P21 for all three groups of sensory neurons (Figures 1A and 1B). To visualize postsynaptic specialization of these sensory neuron glutamatergic synapses, we stained for the postsynaptic scaffolding protein Homer1, which is present at postsynaptic densities (PSDs) of glutamatergic synapses within the dorsal horn.54,55 The number and size of Homer1+ puncta associated with sensory neuron terminals increased more than 2-fold between P0 and P21 (Figures 1C and S1A). Thus, somatosensory neuron synapses in the dorsal horn are formed before birth and throughout early postnatal development (Figure 1D).
Figure 1. Pcdhgs are expressed during postnatal development when somatosensory axons actively form synapses in the dorsal horn.
(A) IHC images of spinal cord dorsal horn lamina III from P0, P10 and P21 mice. Arrowheads point to some of the synapses made between sensory neuron terminals and spinal cord neurons. Scale bars represent 3 μm.
(B) Quantifications of the sizes of sensory terminals labeled by synaptophysin-tdTomato at P0, P3, P10 and P21. RetCreER;Ai34: n = 2 animals for each time point. TrkBCreER;Ai34: n = 4 animals for P0, 2 animals for P3, 3 animals for P10, and 3 animals for P21. AdvillinCre;Ai34: n = 3 animals for P0, 2 animals for P3, 2 animals for P10, and 4 animals for P21. Two-way ANOVA.
(C) Quantifications of the average numbers of Homer1+ puncta per sensory terminal labeled by synaptophysin-tdTomato at P0, P3, P10 and P21. Two-way ANOVA.
(D) Summary of the synapse formation surrounding sensory terminals in the LTMR recipient zone during postnatal development.
(E) Schematic of the RNA sequencing workflow.
(F) Genetic labeling strategies for each of DRG neuron groups.
(G) Heatmaps depicting expression patterns for differentially expressed genes encoding cell adhesion molecules or axon guidance proteins. Each column is one biological replicate, and each row shows the expression level for one gene. Prop., proprioceptors.
(H) Heatmap depicting expression patterns of Pcdhg genes in the DRGs at P3.
(I and J) RNAscope images (I) and quantification (J) for Pcdhga2, Pcdhga7, Pcdhgb1, Pcdhgc3, and Pcdhgc4 expression levels (n = 3 animals). SLC17A6(vGluT2) labels the excitatory neurons in the dorsal spinal cord. DAPI staining in blue labels cell nuclei. Student’s unpaired t test.
ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
To explore the molecular basis of LTMR synapse formation and other developmental processes, we performed deep RNA sequencing of major LTMR subtypes, proprioceptors, as well as all DRG neurons to obtain and compare their transcriptome profiles at a relevant developmental stage. P3 was chosen for our analysis because LTMRs exhibit both central synapse formation (Figures 1A-1C) and peripheral target innervation13,56-58 at this age. We used genetic tools that selectively label subsets of LTMRs and proprioceptors in combination with mice that express the epitope-tagged ribosomal protein RPL22HA (RiboTag mice), enabling isolation of mRNA transcripts in these populations using immunoprecipitated polyribosomes (Figures 1E, and 1F and S1B).59
To assess the validity of the transcriptome profiles, we examined expression patterns of several marker genes. While expression patterns of several genes in developing LTMRs were also detected in mature LTMRs, including Ntrk2, Calb2, Adtrp, and Cdh3 (Figures 1G and S1C and S1D),60-62 many subtype-uniquely enriched genes (SUEGs) were identified here for the first time (Figures S1C and S1D). We hypothesized that differentially expressed transmembrane molecules in developing LTMRs may regulate their synapse formation and peripheral innervation. Therefore, we examined expression patterns of cell adhesion molecules and axon guidance proteins previously implicated in neural circuit assembly.51,63,64 This analysis revealed several candidate differentially expressed genes encoding transmembrane proteins in P3 LTMR subtypes (Figure 1G).
Among the genes encoding transmembrane proteins implicated in developmental processes, Pcdhg subcluster genes stood out because they exhibited interesting differences in their expression levels of isoforms, with Pcdhga7 and Pcdhgc3 isoforms being expressed at the highest levels (Figure 1H). Moreover, at P0, at least 20 of the 22 Pcdhgs are expressed in DRGs.39 At P3, expression of Pcdhg isoforms in DRG neurons was confirmed with single-molecule RNA fluorescent in situ hybridization (smRNA-FISH) using probes against 5 representative isoforms (Figure S1E). While Pcdhgc3 is evenly and broadly expressed throughout the DRG, Pcdhga7 is enriched in a subset of DRG neurons (Figure S1E). In the spinal cord, Pcdhgc3 and Pcdhgc4 were detected throughout the spinal cord (Figures 1I and 1J). In addition, there is a significant decrease in the level of expression of three out of five isoforms between P3 and P21 (Figure 1J).
Pcdhgs function in primary sensory neurons for normal tactile behaviors and sensorimotor integration
To examine the role of Pcdhgs in LTMR development, we used a Pcdhg conditional allele (Pcdhgfcon3), thus bypassing the neonatal lethality associated with constitutive Pcdhgs deletion, and AdvillinCre to selectively delete all Pcdhgs in all DRG neurons during embryonic development.37,52,65 In the Pcdhgfcon3 allele, GFP is fused to the carboxy-terminal constant exon 3 shared by all isoforms and is flanked by loxP sites, such that loss of GFP expression indicates deletion of Cre-mediated excision of the constant exon 3 (Figures 2A and S2A).
Figure 2. Pcdhgs function in primary sensory neurons for normal tactile behaviors and sensorimotor integration.
(A) IHC images of DRG and spinal cord showing the GFP fused Pcdhg proteins in wildtype (negative control), Pcdhgfcon3/fcon3 (positive control) and AdvillinCre;Pcdhgfcon3/fcon3 mice. IB4 labels lamina IIi nonpeptidergic nociceptors in the DRG.
(B) Diagram for the tactile PPI behavior assay.
(C) Percentage of inhibition of startle response to 125 dB noise when the startle noise is preceded by a light air puff of 0.9 PSI (500 ms ISI). Student’s unpaired t test.
(D) Quantification of the average movement of the back in response to 1.0 PSI air puff applied to the back hairy skin of P5 control and AdvillinCre;Pcdhgfcon3/fcon3 pups. Student’s unpaired t test.
(E and F) Von Frey thresholds (E) and response rates (F) for littermate control and AdvillinCre;Pcdhgfcon3/fcon3 mice. Two-way ANOVA. Fisher's LSD post hoc test.
(G) Diagram for the rough floor aversion assay.
(H) Percentages of time spent on rough side for littermate control and AdvillinCre;Pcdhgfcon3/fcon3 mice. Student’s unpaired t test.
(I) Distance traveled in an open field test. Student’s unpaired t test.
(J) Example images of the balance beam test.
(K and L) Total time it took for each animal to cross the beam (K) and average number of slips per trial (L) for littermate control and AdvillinCre;Pcdhgfcon3/fcon3 mice. Student’s unpaired t test.
Each dot represents an animal. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
To determine whether Pcdhgs in somatosensory neurons are essential for LTMR function and somatosensation, tactile sensitivity of AdvillinCre;Pcdhgfcon3/fcon3 mutant mice was tested. At weaning age, AdvillinCre;Pcdhgfcon3/fcon3 mutants exhibited reduced body weight, but quickly gained weight to a normal range by 6 weeks of age (Figure S2B). A tactile prepulse inhibition of the startle reflex assay (tactile PPI) was performed on mutant and littermate control mice66,67 to assess hairy skin sensitivity (Figures 2B). Adult AdvillinCre;Pcdhgfcon3/fcon3 mutants exhibited a reduction in tactile PPI (Figure 2C) but not the response to air puff alone (Figure S2C), suggesting a reduction in hairy skin mechanosensation. On the other hand, the mutants performed normally in an acoustic version of PPI (Figure S2D). Consistent with the adult tactile PPI findings, P5 AdvillinCre;Pcdhgfcon3/fcon3 mutants exhibited reduced reactivity to air puffs (1.0 PSI) applied to the back (Figure 2D).
We also asked whether glabrous skin sensitivity to mechanical stimuli is altered in AdvillinCre;Pcdhgfcon3/fcon3 mice, using von Frey filament stimulation of hindpaw glabrous skin.68 AdvillinCre;Pcdhgfcon3/fcon3 mutants displayed reduced hindpaw withdrawal responses (Figures 2E and 2F). The rough floor aversion test was performed as an additional measurement of glabrous skin sensitivity (Figure 2G)69. Here, AdvillinCre;Pcdhgfcon3/fcon3 mice exhibited decreased aversion to rough textured floors compared to littermate controls (Figure 2H).
Because somatosensory feedback contributes to the control of locomotion and corrective movements70-72, we also examined general locomotion in AdvillinCre;Pcdhgfcon3/fcon3 mice and their littermate controls using the open field assay. AdvillinCre;Pcdhgfcon3/fcon3 mice exhibited a decrease in both the total distance traveled (Figure 2I) and the percentage of time spent in the center of the chamber (Figure S2E). This decrease in the time spent in the center of the chamber may not represent an anxiety-like behavior (Figure S2F).73 When traversing a narrow elevated beam, AdvillinCre;Pcdhgfcon3/fcon3 mice spent more time crossing the beam (Figures 2J and 2K) and displayed more hindlimb slips than littermate control mice (Figures 2L), suggesting a disruption in their sensorimotor integration.
Pcdhgs function in primary somatosensory neurons for central synapse formation
The alterations in somatosensation in AdvillinCre;Pcdhgfcon3/fcon3 mice prompted us to ask whether Pcdhgs regulate synapse formation or other aspects of somatosensory neuron axon development. In the dorsal horn LTMR-RZ, ~60% of the vGluT1+ glutamatergic terminals are somatosensory neuron presynaptic terminals, while ~40% of vGluT1+ terminals are corticospinal presynaptic terminals.6 At P21, the density of both vGluT1+ and Homer1+ puncta was reduced in AdvillinCre;Pcdhgfcon3/fcon3 mutants (Figures 3A-3D). Consistent with this histological analysis, mutant spinal cord neurons displayed a ~50% reduction in mEPSC frequency, but no change in mEPSC amplitudes (Figures 3E, F and S3A). To determine when the reduction of synapse density emerges, excitatory synaptic densities during neonatal and postnatal times were examined. At P4 the density of vGluT1+ puncta appeared normal in AdvillinCre;Pcdhgfcon3/fcon3 mutants (Figures 3A and 3C). On the other hand, the density of Homer1+ puncta in the mutants appeared normal at P0, while at P4 it was decreased by 59.0 ± 15.1% compared to control littermates (Figure 3D). These findings suggest that loss of Pcdhgs in somatosensory neurons leads to a reduction of both vGluT1+ presynaptic terminals and Homer1+ postsynaptic puncta within the LTMR-RZ and a concomitant loss of functional synapses. This synapse reduction is unlikely caused by abnormal targeting of the sensory terminals, as the overall distribution of sensory axon terminals in the dorsal horn in AdvillinCre;Pcdhgfcon3/fcon3 mutants appeared normal (Figures 3B and S3B-G).
Figure 3. Pcdhgs function in primary sensory neurons for postsynaptic specialization of synapses between sensory axon terminals and spinal cord neurons.
(A and B) IHC images of spinal cord dorsal horn lamina III from P4 (A) and P21 (B) littermate control and AdvillinCre;Pcdhgfcon3/fcon3 mice.
(C and D) Normalized densities of vGluT1+ (C) and Homer1+ (D) puncta from littermate control and AdvillinCre;Pcdhgfcon3/fcon3 mice. Student’s unpaired t test.
(E) Representative spontaneous mEPSC traces recorded in random spinal cord neurons in lamina III from P13-P16 littermate control (n = 3 animals) and AdvillinCre;Pcdhgfcon3/fcon3 mice (n = 4 animals).
(F) Presynaptic Pcdhg deletion decreases the frequency of spontaneous mEPSCs in the spinal cord neurons. Each dot is the mEPSC frequency of a spinal cord neuron. Mann-Whitney test.
(G) IHC images of spinal cord lamina III from P21 littermate control and AdvillinCre;Pcdhgfcon3/fcon3;Ai34 mice.
(H-K) Normalized densities of synaptophysin-tdTomato (Ai34) puncta density (H) and vGluT1+ Tomato+ double positive puncta (I). The size and normalized density of Homer1+ puncta around Tomato+ sensory terminals is quantified in (K) and (J), respectively. Each dot represents one animal. Student’s unpaired t test.
(L) EM images of synaptic glomeruli from lamina III in spinal cord dorsal horn from a control animal and a AdvillinCre;Pcdhgfcon3/fcon3 animal. Arrowheads point to the postsynaptic sites formed within the glomeruli. Scale bars represent 500 nm.
(M and N) Quantifications of PSD thickness (M, control 42.1 ± 0.6 nm, 304 synapses from 3 animals; AdvillinCre;Pcdhgfcon3/fcon3: 32.4 ± 0.6 nm, 286 synapses from 3 animals). Quantifications of PSD length (N, control 340.0 ± 7.1 nm; AdvillinCre;Pcdhgfcon3/fcon3: 299.3 ± 6.2 nm). Kolmogorov-Smirnov test.
(O) Summary of the synaptic formation phenotype in the AdvillinCre;Pcdhgfcon3/fcon3 mutants.
ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
Since vGluT1 labels both primary somatosensory neuron and corticospinal neuron terminals in the LTMR-RZ, we tested whether the disruption in vGluT1 labeling observed in the AdvillinCre;Pcdhgfcon3/fcon3 mutants is a reflection of altered somatosensory neuron synapses, corticospinal neuron synapses, or both. To selectively visualize primary somatosensory neuron terminals, we generated AdvillinCre;Pcdhgfcon3/fcon3;Ai34 animals, in which all somatosensory neurons express the presynaptic marker synaptophysin-tdTomato (Figure 3G). Interestingly, the density of primary sensory neuron synaptic terminals in the mutants was comparable to that of controls at P21 (Figures 3H and 3I), suggesting that the reduction in the number of vGluT1+ puncta in the mutants is caused by a decrease in corticospinal terminals (also see Figures 7H-K). On the other hand, the density and size of Homer1+ puncta surrounding tdTomato+ sensory terminals was reduced in AdvillinCre;Pcdhgfcon3/fcon3;Ai34 animals (Figures 3J and 3K). To address this further, electron microscopy was used to examine ultrastructural features of excitatory synapses associated with dorsal horn synaptic glomeruli, which are comprised of axon terminals of primary sensory afferents and synapses with spinal cord neurons (Figure 3L).74,75 Indeed, the thickness and length of PSDs associated with synaptic glomeruli in lamina III were reduced in AdvillinCre;Pcdhgfcon3/fcon3 mice (Figures 3M and 3N), while the overall morphology of glomeruli of mutants appeared normal. Similarly, AdvillinCre;Pcdhgfcon3/fcon3;Ai34 animals exhibited synapse formation deficits in laminae I, IIo, and IIi, which contain nociceptor, thermoreceptor, pruriceptor, and high-threshold mechanoreceptor synapses, and lamina IX, which contains proprioceptor synapses (Figures S3H-S3K). Deleting Pcdhgs in somatosensory neurons also led to a widespread reduction in the number of excitatory synapses in lamina I, II and IX (Figure S3L).
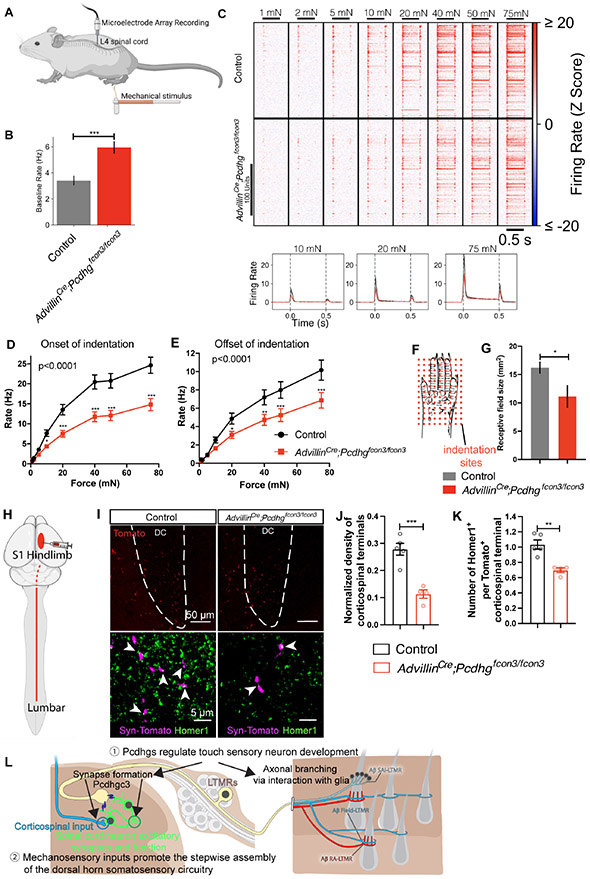
Figure 7. Aberrant physiological responses and corticospinal synaptic inputs in the dorsal horn of AdvillinCre; Pcdhgfcon3/fcon3 mice due to reduced functional mechanosensory synapses.
(A) MEA recordings from lumbar spinal cord dorsal horn in mice, while the glabrous skin from right hind paw is indented with a series of forces.
(B) Quantifications of the baseline firing rates in littermate control and AdvillinCre;Pcdhgfcon3/fcon3 mice. n = 3 animals per genotype. Student’s unpaired t test.
(C) Summary of representative units responding to indentation of the skin with different forces. The amount of force is labeled on the top. The indentations applied are indicated by the black bars below the force labels. Average firing rates in response to the indentation are shown on the bottom row. n = 187 neurons for 5 controls and n = 207 neurons for 5 mutants.
(D and E) Quantifications of the onset (D) and offset (E) responses in littermate control and AdvillinCre;Pcdhgfcon3/fcon3 mice. Two-way ANOVA.
(F) Diagram for mapping the functional receptive fields of the dorsal horn neurons.
(G) Quantifications of the receptive field sizes in littermate control and AdvillinCre;Pcdhgfcon3/fcon3 mice.
(H) Diagram showing viral labeling of corticospinal projections in the lumbar spinal cord.
(I) IHC images showing corticospinal terminals labeled with synaptophysin-tdTomato in the dorsal column, and lamina III in control and AdvillinCre;Pcdhgfcon3/fcon3 mice. White dotted line outlines the shape of the dorsal column with labeling.
(J) Quantification showing the density of Tomato+ corticospinal terminals in lamina III is reduced in the AdvillinCre;Pcdhgfcon3/fcon3 mice (number of puncta per 104 μm2). Each dot represents average number for an animal. Student’s unpaired t test.
(K) Quantification showing the number of excitatory synapses (Homer1+ puncta) surrounding corticospinal terminals in lamina III is reduced in the AdvillinCre;Pcdhgfcon3/fcon3 mice. Student’s unpaired t test.
(L) A schematic summarizing (1) the essential roles of Pcdhg isoform diversity in somatosensory neuron synapse formation (left), peripheral axonal branching (right), and (2) step-wise assembly of central mechanosensory circuitry.
*p < 0.05; **p < 0.01; ***p < 0.001.
Together, these findings show that Pcdhgs function in somatosensory neurons to establish proper numbers, function, and ultrastructural properties of postsynaptic specializations formed between somatosensory neurons and dorsal horn neurons (Figure 3O).
Pcdhgs function in primary sensory neurons for LTMR axonal branching in the skin
Pcdhg proteins are found throughout neurons, including in axons and dendrites37,41,76, and in CNS neurons they regulate many aspects of dendrite development, including dendrite self-avoidance and arborization.36,77-79 To ask whether loss of Pcdhgs in DRG neurons affects LTMR peripheral axon development, the patterns of LTMR endings in the skin of AdvillinCre;Pcdhgfcon3/fcon3 mutants and control littermates were visualized using whole mount immunohistochemistry. In both controls and AdvillinCre;Pcdhgfcon3/fcon3 mutants, all guard hairs were innervated by NFH+ Aβ field-LTMRs and Aβ RA-LTMRs endings (Figures 4A-4C and S4A). However, non-guard hairs, which include awl/auchene and zigzag hairs, exhibited reduced innervation by NFH+ Aβ field-LTMRs and Aβ RA-LTMRs in AdvillinCre;Pcdhgfcon3/fcon3 mutants (Figures 4B and 4C). Of note, the morphology of sensory neuron endings and associated terminal Schwann cells surrounding hair follicles were unchanged (Figures S4B-S4E), and DRG neuron survival is unaffected by the absence of Pcdhgs.39
Figure 4. Pcdhgs function in LTMRs for axonal branching in the skin.
(A) Whole-mount immunostainings of adult back hairy skin sections from control and mutants. Aβ field-LTMRs form circumferential endings around hair follicles and are NFH+ (noted by white arrowheads). Guard hair follicles are noted as “G”. TSCs are labeled using S100 immunostaining (green). Hair follicles without any NFH+ Aβ field-LTMRs are denoted by triangles.
(B-D) Quantification of the percentage of guard and non-guard hair follicles innervated by NFH+ Aβ field-LTMRs circumferential endings and Aβ RA-LTMRs lanceolate endings for AdvillinCre;Pcdhgfcon3/fcon3 mice (n = 958 hair follicles from 3 controls and n = 1357 hair follicles from 3 AdvillinCre;Pcdhgfcon3/fcon3 mice) and DhhCre;Pcdhgfcon3/fcon3 mice (n = 597 hair follicles from 3 controls and n = 863 hair follicles from 4 DhhCre;Pcdhgfcon3/fcon3 mice). Student’s unpaired t test. Each dot represents an imaging field.
(E) Control and AdvillinCre;Pcdhgfcon3/fcon3 mice were injected with AAV-Retro-Flex-PLAP virus to retrogradely label DRG neurons that project to the DCN.
(F) Example whole-mount AP images of the back hairy skin. Reconstructed axons are shown in the right panels (black). Aβ field-LTMR and Aβ RA-LTMR endings associated with hair follicles are marked in the reconstructions using red and blue circles, respectively.
(G and H) Quantification of the number of innervated non-guard hair follicles (G) and the area of innervation (H) by Aβ field-LTMRs (n = 12 neurons from 6 control animals and n = 13 neurons from 5 AdvillinCre;Pcdhgfcon3/fcon3 mice) and Aβ RA-LTMRs (n = 28 neurons from 6 control animals and n = 27 neurons from 6 AdvillinCre;Pcdhgfcon3/fcon3 mice). Student’s unpaired t test. Dots represent individual neurons.
(I and J) Example whole-mount immunostaining images of the Merkel cell complex. TSCs wrapping around major Aβ SAI-LTMRs branches are labeled with S100 (green), and Merkel cells are labeled with Troma-I (blue). Quantifications of the major S100+ branches from AdvillinCre;Pcdhgfcon3/fcon3 mice (n = 3 controls and n = 4 mutants) and DhhCre;Pcdhgfcon3/fcon3 mice (n = 3 control animals and n = 4 mutants) are shown in (J). Each dot represents a touch dome. Student’s unpaired t test.
(K and L) Example immunostaining images of the forepaw glabrous skin. Meissner corpuscles are labeled by S100 (green, for visualizing lamellar cells) and NFH (magenta, for visualizing Aβ RA-LTMRs). Quantifications of the number of Meissner corpuscles normalized by the area of epidermis from AdvillinCre;Pcdhgfcon3/fcon3 (n = 4 animals per genotype) and DhhCre;Pcdhgfcon3/fcon3 mice (n = 3 controls and n = 4 mutant animals) are shown in (L). Dots represent individual skin sections. Student’s unpaired t test.
ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
To directly visualize axonal branching patterns of individual Aβ field-LTMRs and Aβ RA-LTMRs in hairy skin, sparse labeling of LTMRs was done by injecting a Cre-dependent alkaline phosphatase reporter virus into the dorsal column nuclei (DCN) of the brainstem14,80 (Figure 4E). Then, whole mount alkaline phosphatase staining was done to visualize the cell bodies in the DRG and the cutaneous axonal arbors of individual LTMRs (Figures 4F and S4F). This analysis revealed a reduction in both the number of innervated hair follicles per neuron and the area innervated by individual Aβ field-LTMRs and Aβ RA-LTMRs in AdvillinCre;Pcdhgfcon3/fcon3 mutants (Figures 4G and 4H). Moreover, Aβ SAI-LTMRs associated with clusters of Merkel cells (touch domes) that surround guard hairs in hairy skin exhibited a small but significant reduction in the number of the S100+ major branches in the AdvillinCre;Pcdhgfcon3/fcon3 mutants (Figures 4I and 4J), while the total number of Merkel cells in touch domes remained the same (Figure S4G). In glabrous skin, the density of Meissner corpuscles was reduced in the mutants (Figures 4K, 4L, and S4H), suggesting a decrease in axonal branching.
LTMR peripheral axons closely associate with peripheral glial cells.56,81 To address the possibility that Pcdhgs mediate homophilic adhesion between nascent peripheral axons and glial cells to promote axonal branching, DhhCre;Pcdhgfcon3/fcon3 mutants were generated to selectively delete Pcdhgs in developing Schwann cells of the peripheral nervous system, beginning at E1282 (Figure S4I). Remarkably, DhhCre;Pcdhgfcon3/fcon3 mutants phenocopied AdvillinCre;Pcdhgfcon3/fcon3 mutants with respect to the hairy and glabrous skin LTMR innervation deficits (Figures 4A, 4D, and 4I-4L). On the other hand, excitatory synapses in the spinal cord dorsal horn were unaffected in DhhCre;Pcdhgfcon3/fcon3 mutants (Figures S4J-S4M). These findings support a model in which Pcdhgs mediate homophilic interactions between nascent LTMR axonal branches and developing glial cells to promote axonal branching.
We also tested whether Pcdhgs function to maintain central synapses and peripheral axonal branching after they are established, using the tamoxifen-sensitive AdvillinCreERT2 mouse line83 (AdvillinCreERT2; Pcdhgfcon3/fcon3 mice, 1mg per day, P15-P19; Figures S5A and S5B). Late postnatal deletion of Pcdhgs recapitulated the reduction of excitatory synapses and axonal branching observed in AdvillinCre;Pcdhgfcon3/fcon3 mutants (Figures S5C-S5K), suggesting that Pcdhg signaling maintains both LTMR central synapses and peripheral axonal arborization patterns after they are formed.
Differential requirements of Pcdhg isoforms for synapse formation and peripheral axonal branching, with Pcdhgc3 solely required for synapse formation
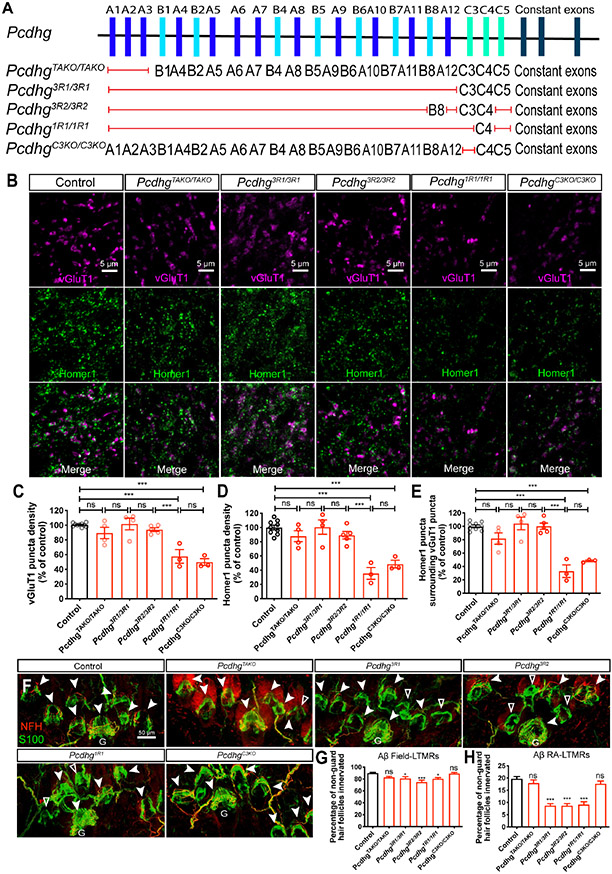
The diversity of Pcdhg isoforms raised the question of whether one or more of the same or different Pcdhg isoforms mediate synapse formation and axonal branching. To address this, we examined a range of Pcdhg mutants that lack one or more of the 22 isoforms (Figure 5A). All mutants tested were viable to adulthood because they have intact Pcdhgc4, which encodes the isoform required for postnatal viability.34
Figure 5. Differential requirements of Pcdhg isoforms for synapse formation and peripheral axonal branching.
(A) Summary of the Pcdhg mutants used. Red lines indicate the corresponding genes are disrupted, while the genes listed are not disrupted.
(B) IHC images of spinal cord dorsal horn lamina III from P21 control and Pcdhg mutants.
(C-E) Normalized densities of vGluT1+ (C) and Homer1+ (D) puncta from P21 control and various Pcdhg mutants. (E) shows the normalized densities of Homer1+ puncta that surrounds vGluT1+ puncta. One-way ANOVA with Tukey’s post hoc test. Each dot represents average value of an animal.
(F) Whole-mount immunostaining of adult back hairy skin sections from control and various Pcdhg mutants. Aβ field-LTMRs innervations are noted by white arrowheads. Hair follicles without any NFH+ Aβ field-LTMRs are denoted by empty arrowheads. Scale bar represents 50 μm.
(G and H) Quantification of the percentage of non-guard hair follicles innervated by NFH+ Aβ field-LTMRs circumferential endings (G) and Aβ RA-LTMRs lanceolate endings (H). n = 649 hair follicles from 5 wildtype animals; n = 336 hair follicles from 2 PcdhgTAKO animals; n = 710 hair follicles from 4 Pcdhg3R1 animals; n = 673 hair follicles from 5 Pcdhg3R2 animals; n = 614 hair follicles from 3 PcdhgC3KO animals. One-way ANOVA with Tukey’s post hoc test.
ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
The triple A-type isoform knockout (TAKO) PcdhgTAKO/TAKO mice showed normal numbers of dorsal spinal cord synapses and Aβ field-LTMR and Aβ RA-LTMR endings in hairy skin (Figures 5B-5H and S5Q).32 Interestingly, while the Pcdhg3R1/3R1 and Pcdhg3R2/3R2 mutants exhibited normal vGluT1 and Homer1 excitatory synapse density in the dorsal horn, they displayed Aβ field-LTMR and Aβ RA-LTMR innervation pattern defects in the hairy skin (Figures 5B-5H and S5Q). These phenotypes suggest that the central and peripheral phenotypes are dissociable and further indicate that the critical isoform(s) for synapse formation is intact in these two mutants (i.e., Pcdhgc3 and Pcdhgc4). Interestingly, the Pcdhg1R1/1R1 mutant (only Pcdhgc4 is intact) exhibited deficits in both synapse formation and peripheral axonal branching (Figures 5B-5H and S5Q), suggesting that Pcdhgc3 is the sole isoform essential for synapse formation.
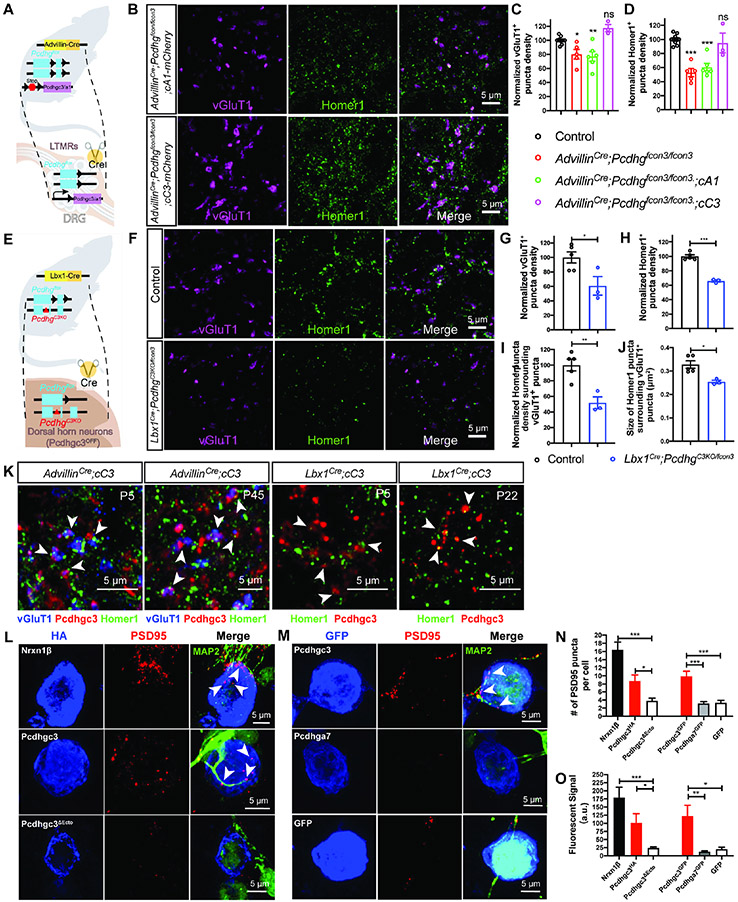
To further test this idea, additional genetic loss-of-function and gain-of-function approaches were taken. First, we found that the PcdhgC3KO/C3KO mutant, which lacks only Pcdhgc3, phenocopied the synapse deficits observed in AdvillinCre;Pcdhgfcon3/fcon3 mice and showed a reduction in excitatory synapse density; however this mutant did not exhibit an alteration of Aβ field-LTMR and Aβ RA-LTMR innervation patterns in hairy skin (Figures 5B-5H and S5Q) or sensory axon targeting in the spinal cord (Figures S5L-S5P). Second, we asked whether expressing Pcdhgc3 in somatosensory neurons of AdvillinCre;Pcdhgfcon3/fcon3 mutants is sufficient to rescue the synapse formation phenotype of the cluster knockout. For this, the cC3 and cA1 mouse lines, in which the Pcdhgc3 or Pcdhga1 isoform fused to a fluorescent protein (mCherry) is expressed in a Cre-dependent manner (ROSA26-CAG::lox-Stop-lox-Pcdhgc3-mCherry and ROSA26-CAG::lox-Stop-lox-Pcdhga1-mCherry),36 were used (Figure 6A). Expression of Pcdhgc3, but not Pcdhga1, in somatosensory neurons of AdvillinCre;Pcdhgfcon3/fcon3 mice rescued the synapse formation deficits observed in the Pcdhg cluster knockout (Figures 6B-6D).
Figure 6. Pcdhgc3 is the only isoform that mediates sensory neuron synapse formation in vivo and can promote postsynaptic specialization in vitro.
(A) Experiment design for expressing single Pcdhg isoforms (Pcdhga1 or Pcdhgc3) in AdvillinCre;Pcdhgfcon3/fcon3 mice.
(B) IHC images of spinal cord dorsal horn lamina III from AdvillinCre;Pcdhgfcon3/fcon3;cA1 and AdvillinCre;Pcdhgfcon3/fcon3;cC3 mice.
(C and D) Normalized densities of vGluT1+ (C) and Homer1+ (D) puncta. Each dot on the plot indicates a normalized value for an animal. One-way ANOVA test.
(E) Experiment design for deleting both copies of the Pcdhgc3 isoforms only in the dorsal horn neurons in the Lbx1Cre;PcdhgC3KO/fcon3 mice.
(F) IHC images of spinal cord dorsal lamina III from control and Lbx1Cre;PcdhgC3KO/fcon3 mice.
(G-J) Quantifications of the synapses in lamina III, showing that deleting Pcdhgc3 in the dorsal horn neurons led to reduced densities of vGluT1+ (G), Homer1+ (H) puncta, as well as the density of Homer1+ puncta surrounding vGluT1+ terminals (I). Similarly, the average size of Homer1+ puncta is reduced in Lbx1Cre;PcdhgC3KO/fcon3 mice (J). Each dot on the plot indicates a normalized value for an animal. Student’s unpaired t test.
(K) IHC showing the localizations of Pcdhgc3-mCherry in the spinal cord lamina III of P5 and P45 AdvillinCre;cC3 mice and P22 Lbx1Cre;cC3 mice (n = 3 animals for each genotype).
(L) Representative images of artificial synapse formation assays in which Nrxn1β, Pcdhgc3, or Pcdhgc3ΔEcto is expressed in HEK293T cells that are cocultured with neonatal spinal cord neurons.
(M) Representative images of in vitro synapse formation assays. Note the MAP2+ neuronal cell body in the upper right corner of the panel on the right (Pcdhga7 condition).
(N and O) Average number (N) and intensity (O) of PSD-95 puncta per HEK 293 cell for each condition. Data are means ± SEM from three independent replicates. One-way ANOVA with Tukey’s post hoc test.
ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
To further test the idea that Pcdhgc3 is required in cutaneous sensory neurons for dorsal horn synapse formation, and that the dorsal horn synapse deficits are not secondary to a proprioceptor deficit,39 AAV9-hSyn-Cre-P2A-dTomato was injected into hindpaw hairy skin of P0 Pcdhgfcon3/C3KO animals to sparsely delete Pcdhgc3 in postnatal LTMRs and other cutaneous sensory neurons but not in proprioceptors (Figures S6A-S6E). Indeed, selective removal of Pcdhgc3 from cutaneous sensory neurons led to synapse formation deficits in laminae III and II (Figures S6F-S6K), suggesting that Pcdhgc3 acts autonomously in LTMRs and other cutaneous neurons to promote synapse formation in the dorsal horn.
Pcdhgc3 functions both presynaptically and postsynaptically to mediate excitatory synapse formation between sensory neurons and spinal cord neurons
Pcdhgs bind strictly homophilically when expressed in cell lines, and can form isoform-specific homophilic trans dimer parallel arrays to generate large two-dimensional structures bridging adjacent membranes.44,84-88 These findings led us to hypothesize that Pcdhgc3 functions in both sensory neurons and spinal cord neurons to promote postsynaptic specializations. To test this idea, Lbx1Cre;Pcdhgfcon3/fcon3 animals were generated to excise all 22 Pcdhgs in dorsal horn neurons.89 However, the size of the dorsal horn in Lbx1Cre;Pcdhgfcon3/fcon3 animals was greatly reduced (Figures S6L-S6O), consistent with the role of Pcdhgs, and Pcdhgc4 in particular34, in survival of spinal cord neurons.42,65 Therefore, we generated Lbx1Cre;PcdhgC3KO/fcon3 compound mutant animals, in which both copies of Pcdhgc3 are absent only in dorsal horn neurons (Figure 6E). Lbx1Cre;PcdhgC3KO/fcon3 animals did not exhibit an alteration in dorsal horn neuron survival or sensory axon targeting (Figures S6P-S6R and S7A-S7C), but they did exhibit a significant decrease in the number of dorsal horn excitatory synapses (Figures 6F-6J). Similarly, Lbx1Cre;PcdhgC3KO/fcon3 animals exhibited reduced density and size of synapses around nonpeptidergic nociceptor terminals in lamina II (Figure S7D-S7H). Thus, Pcdhgc3 functions in both DRG neurons and spinal cord neurons to promote excitatory synapse formation between sensory neurons and spinal cord neurons.
Pcdhgc3 localizes to synapses in the dorsal horn and in a reduced preparation is sufficient to induce PSD-95 clustering in dendrites of spinal cord neurons
Pcdhgs are present in synaptic clefts and concentrated PSD fractions of synaptosomes,40,42,90,91 yet whether Pcdhgs can directly promote postsynaptic specialization is unclear. Therefore, we next visualized the localization of Pcdhg proteins in the central terminals of somatosensory axons by expressing Pcdhgc3 and Pcdhga1 isoforms fused to mCherry in sensory neurons.36 Interestingly, while both isoforms were distributed in a punctate manner in central somatosensory neuron axons, they were distributed evenly in peripheral axons (Figures S7I). Moreover, Pcdhgc3, but not Pcdhga1, was detected in vGluT1+ presynaptic puncta at both P5 and in adults (Figure 6K; Figure S7I). Similarly, Pcdhgc3 was present in a punctate pattern when expressed exclusively in dorsal horn neurons, and approximately 50% of Pcdhgc3+ puncta were found to contain one or more Homer1+ puncta (Figure 6K, n = 3 animals, P20–45). These findings suggest that Pcdhgc3 can localize to both presynaptic and postsynaptic compartments in the dorsal horn.
To directly test whether Pcdhgc3 has synaptogenic activity, we next used an in vitro synapse formation assay51,92,93 to ask whether Pcdhgc3 is capable of promoting clustering of postsynaptic proteins in neonatal spinal cord neurons. As a positive control, HA-tagged neurexin-1β, a known synaptogenic protein, was expressed in HEK 293 cells and found to robustly induce clusters of PSD-95 in the dendrites of spinal cord neurons that contacted the HEK 293 cells (Figures 6L, 6N and 6O).94,95 Pcdhgc3 expressed in HEK 293 cells also promoted a significant increase in the number and intensity of PSD-95 clusters in spinal cord neurons, when compared to cells expressing Pcdhgc3ΔEcto, which lacks all 6 extracellular cadherin repeats,96 or GFP control (Figures 6N and 6O). In contrast, Pcdhga7-GFP did not induce clustering of PSD-95 in contacting dendrites (Figures 6M-6O). These findings suggest that Pcdhgc3 is sufficient to induce excitatory postsynaptic specialization in spinal cord neurons.
Altered spinal cord dorsal horn response to touch in response to reduced mechanosensory inputs
The behavioral alterations and reductions in both LTMR central synapses and peripheral axonal branching in AdvillinCre;Pcdhgfcon3/fcon3 mice raise the question about how changes in mechanosensory synaptic inputs to the dorsal horn affect dorsal horn somatosensory circuit function in the mutants. To address this question, in vivo multielectrode array recordings of the spinal cord dorsal horn were performed to assess dorsal horn neuron responses to a series of mechanical step indentations applied to hindpaw glabrous skin (Figure 7A). Interestingly, the baseline firing rates of dorsal horn units were significantly increased in AdvillinCre;Pcdhgfcon3/fcon3 mice (Figure 7B). When step indentations were applied to the skin, dorsal horn neurons in control animals exhibited responses at both the onset and offset of the step indentations within the low force range, beginning around 5 mN (Figure 7C). In comparison, dorsal horn neurons in AdvillinCre;Pcdhgfcon3/fcon3 mice exhibited reduced responses to both the onset and offset of step indentations, but not during the sustained phase (Figures 7D, 7E and S7J). Using a grid of indentation sites to map functional receptive fields of dorsal horn neurons (Figure 7F), dorsal horn units recorded in AdvillinCre;Pcdhgfcon3/fcon3 mice showed significantly smaller receptive fields (Figure 7G). Taken together, the Pcdhg mutant animals have fewer synaptic inputs to the spinal cord dorsal horn, which results in an increase in tonic firing in dorsal horn neurons, reduced firing in response to innocuous as well as noxious mechanical stimuli acting on the skin, and smaller functional receptive fields.
Reduced corticospinal input to the dorsal horn is secondary to reduced mechanosensory inputs
The altered tonic and evoked firing patterns in the dorsal horn prompted us to ask whether loss of Pcdhgs in sensory neurons affects other aspects of dorsal horn mechanosensory circuit assembly. Indeed, our genetic analyses had suggested that the decrease of vGluT1+ terminals in AdvillinCre;Pcdhgfcon3/fcon3 mice may reflect changes in the corticospinal inputs in lamina III (Figure 3I). To directly test this possibility, AAV8-CAG-Synaptophysin-tdTomato virus was injected into the hindlimb region of primary somatosensory cortex (S1) of control and AdvillinCre;Pcdhgfcon3/fcon3 mice to visualize corticospinal synapses in the dorsal horn (Figure 7H). Viral injections labeled comparable numbers of axons in the corticospinal tract in control (79.7 ± 21.1 axon per dorsal column section) and AdvillinCre;Pcdhgfcon3/fcon3 mice (81.4 ± 17.2 axon per dorsal column section, p = 0.95 , Figure 7I). However, the density of corticospinal terminals in medial lamina III was significantly reduced in AdvillinCre;Pcdhgfcon3/fcon3 mice (Figure 7J). The number of Homer1+ synapses associated with Tomato+ corticospinal terminals was also reduced in the mutants (Figure 7K). Finally, we asked whether the absence of synaptic structures, per se, or the absence of synaptically-evoked activity in the dorsal horn accounts for the deficits in corticospinal inputs to lamina III observed in AdvillinCre;Pcdhgfcon3/fcon3 mice. For this, we used Cdx2Cre; Piezo2f/f mice, in which Piezo2, the principal mechano-sensitive ion channel for LTMRs,97-99 was deleted broadly in neurons below cervical level C2. Cdx2Cre; Piezo2f/f mice lack most if not all low-threshold, mechanically evoked responses in DRG neurons98 and the spinal cord dorsal horn.99 Cdx2Cre;Piezo2flox/flox mutants exhibited a reduction in the density of corticospinal neuron terminals as well as their synapses in dorsal horn lamina III (Figures S7K-S7M). These findings suggest that the deficits in corticospinal synapses in the dorsal horn of AdvillinCre;Pcdhgfcon3/fcon3 mutants is caused by a reduction in mechanically-evoked sensory neuron synaptic drive in the dorsal horn.
Discussion
Here, we report unique functions of Pcdhg isoforms across different subcellular compartments of somatosensory neurons for the assembly of the somatosensory circuitry and behavioral reactivity to touch. Spatially coordinated patterns of expression of three or more clustered protocadherin gamma isoforms underlie the formation of synapses between LTMRs and dorsal horn neurons (Pcdhgc3), peripheral axonal branching (possibly Pcdhga7 and others), dorsal horn neuron survival (Pcdhgc4),42 and corticospinal synapses with dorsal horn neurons (Pcdhgc3, non-cell autonomously), and thus step-wise assembly of dorsal horn somatosensory circuitry (Figure 7L).
Pcdhgs are found at synaptic clefts and enriched in postsynaptic density (PSD) fractions of synaptosomes40-43 and their presence is associated with synapse maturation.41,76,100 Moreover, many synapse-associated molecules are found in Pcdhg protein complexes.79,101 However, the roles of Pcdhgs in synapse formation and function have remained enigmatic,102,103 in part due to their requirements in other developmental processes.29,34,37,42,43,77,79,104,105 Indeed, in the spinal cord, removing all Pcdhgs leads to loss of spinal cord neurons.42 Although a reduction in synaptic density was observed when inhibiting apoptosis in the spinal cord of Pcdhg mutants,43 it had remained unclear whether these synapse alterations were a reflection of Pcdhg functions in other developmental processes, such as astrocyte-neuron contacts or dendrite development.77,79,106,107 Here, we took advantage of the dispensability of Pcdhgs for DRG neuron survival39 and identified a direct role of Pcdhgc3 in synapse formation. It remains to be determined whether the observed deficit in proprioceptor synapse formation is caused by changes in proprioceptor targeting39 or a direct role of Pcdhg in promoting synapse formation, or both. Consistent with previous observations that mammals lacking synaptic adhesion molecules rarely exhibit more than ~50% loss of synapses in the brain, presumably because of redundancy,103 many synapses between sensory neurons and spinal cord neurons remain in the Pcdhg mutants. It is likely that other synaptic adhesion molecules act in concert with Pcdhgs to promote formation of the full range of synapses the embryonic or neonatal spinal cord.
We propose that Pcdhgc3 functions as a cell-adhesion molecule that tethers the pre- and postsynaptic membranes and instructs postsynaptic specialization through its unique intracellular domain, encoded by its variable exon, and association with postsynaptic proteins. Based on the loss of synapses observed following postnatal deletion of Pcdhgs using AdvillinCreER;Pcdhgfcon3/fcon3 mice and the finding that Pcdhgc3 is the only isoform required for synapse formation, it is likely that Pcdhgc3 is also required for synapse maintenance. Future work will determine whether the unique juxtamembrane region of the Pcdhgc3 intracellular domain96 engages a specific downstream signaling event that recruits postsynaptic scaffolding proteins, such as PSD-95, and promotes postsynaptic specialization and synaptic maintenance. It is also possible that shared intracellular regions of the Pcdhgs may directly promote synapse formation, allowing other Pcdhg isoforms to directly promote synapse formation. While the present work reveals a synaptogenic role for Pcdhgc3 in somatosensory neurons, other Pcdhg isoforms are widely expressed throughout the developing nervous system with extraordinarily complex patterns. It will be interesting to determine whether Pcdhgc3 and other isoforms promote synapse formation elsewhere in the nervous system, thereby contributing to a synaptic specificity code for nervous system wiring.108
Our findings also highlight a remarkable compartmentalization of Pcdhg function via homophilic interactions in the peripheral and central axonal branches of somatosensory neurons for distinct developmental processes. This compartmentalized signaling could be achieved by local differences in the proteins that interact with Pcdhgs to form unique membrane complexes.96 Future genetic and molecular dissection of Pcdhg isoforms and their intracellular signaling mechanisms will provide insights into how Pcdhgs may function in the same neuron to promote synapse formation and peripheral axonal branching.
We found that the sensory neuron clustered protocadherins, presumably Pcdhgc3, are crucial for corticospinal synaptic inputs of the dorsal horn. We consider this to be a non-cell autonomous function of Pcdhgc3 because corticospinal synapses form onto spinal cord neurons and not onto the axon terminals of the primary sensory neurons that lacked Pcdhgs in the conditional mutants.109,110 Spontaneous and evoked sensory activity play essential roles in wiring the nervous system,111-113 although in the mechanosensory system less is known. Postnatal functional development of A-fiber sensory connectivity in the dorsal horn requires NMDA receptor activation114. In the rat, LTMRs exhibit spontaneous activity only during embryonic development, while evoked responses are detected beginning at late embryonic stages.115,116 Central deficits in somatosensory neuron Pcdhg conditional knockout animals appeared later than P0, suggesting that deficits in evoked synaptic activity may be responsible for the alterations in corticospinal synaptic input observed in the mutants. Consistent with this, corticospinal innervation of the lumbar spinal cord (the area analyzed in the present study) begins ~P4,117 suggesting that postnatal evoked synaptic activity controls the formation of synapses between nascent corticospinal axon terminals and spinal cordinterneurons. Future studies should aim to determine how mechanosensory synaptic transmission in the dorsal horn instructs this assembly.
STAR METHODS
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, David Ginty (David_Ginty@hms.harvard.edu).
Materials availability
This study did not generate new unique reagents or mouse lines.
Data and code availability
Data reported in this paper are available from the lead contact upon reasonable request. RNA sequencing dataset is deposited: https://doi.org/10.7910/DVN/BJGETY. This paper does not report original codes. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experiments were conducted in accordance with Harvard Medical School (Institutional Animal Care and Use Committee) IACUC protocols. Animals used in the study were group housed on a 12-hour light/dark cycle, with control and mutant animals in the same litters and cages. Littermates from the same genetic crosses were used as controls, to control for variability in mouse strains/backgrounds. No differences were observed between wildtype animals and floxed (Pcdhgfcon3/+ and Pcdhgfcon3/fcon3) or Cre littermate control groups for the experiments performed. Wildtype C57BL/6J mice were used for the comparison among various Pcdhg deletion mutants.
Mouse Lines.
Pcdhg lines used in this study include Pcdhgfcon3 (Lefebvre, et al., 2008)37, R26LSL-Pcdhga1-mCherry (cA1-mCherry), and R26LSL-Pcdhgc3-mCherry (cC3-mCherry) (Lefebvre, et al., 2012)36, PcdhgC3KO, Pcdhg1R1, Pcdhg3R1, and Pcdhg3R2 (Garrett, et al., 2019)34, and PcdhgTAKO (Chen et al., 2012)32 were previously described. Pcdhg mutant mice were maintained on a C57/B6J background. Other mouse lines used include R26LSL-synaptophysin-tdTomato (Ai34) (Jax#012570); R26LSL-tdTomato (Ai14) (Jax# 007908)89; AdvillinCre (Hasegawa et al., 2007)52; AdvillinCreERT2 (Lau et al., 2011)83; Lbx1Cre (Sieber et al., 2007)89; DhhCre (Jaegle et al., 2003)82; Cdx2Cre (Coutaud and Pilon, 2013)118; Piezo2f/f (Woo et al., 2014)119; Rpl22HA (Sanz et al., 2009)59; PVCre (Hippenmeyer et al., 2005)120; TrkBCreER (Rutlin et al., 2014)13 and RetCreER (Luo et al., 2009)53. Male and female mice of C57BL/6J and CD1 backgrounds were used for these studies. Expression of GFP in the Pcdhgfcon3 allele was assessed using histological verification. The genotyping primers used were listed in Table S1.
METHOD DETAILS
Tissue Fixation.
Male and female mice of ages younger than 6 days were anesthetized in ice until unconscious, then sacrificed quickly by decapitation. Male and female mice of ages 6 days and older were anesthetized with isofluorane and a transcardial perfusion was performed with 10-15 mL modified Ames Media with heparin, followed by 15-25 mL 4% PFA. Back hairy skin was dissected following application of Nair commercial depilatory cream and consequent hair removal, and the underside of the skin was scrapped clean of fatty tissues and muscles. Back hairy skin and glabrous skin of the paws were dissected and post-fixed in Zamboni’s Fixative overnight at 4°C. Spinal cords and DRGs were post-fixed in 4% PFA overnight at 4°C. Following post-fixation, samples were washed in 1xPBS for 3-5 hours and stored in PBS + 0.01% sodium azide at 4°C for long-term storage.
Tamoxifen Treatment.
Tamoxifen (Sigma, Cat# T5648-1g) was dissolved in 100% ethanol (20 mg/ml) on a rotator for 10 minutes, aliquoted into Eppendorf tubes and mixed with an equal volume of sunflower seed oil (Sigma, Cat# s5007). The mixture was vortexed for 15 minutes and centrifuged in a pressurized vacuum concentrator for 45 minutes to remove the ethanol. When removed from the vacuum concentrator, all tubes were checked carefully to ensure only sunflower seed oil remained, then stored at −20°C for long-term use. To calculate embryonic timepoint, females were checked every morning to monitor when mating occurred. The tamoxifen solution was delivered to pregnant females via oral gavage for embryonic tamoxifen treatment at the timepoints indicated. For all analyses, the morning after coitus was designated as E0.5 and the day of birth as P0. Tamoxifen was administered to control and littermate mice in order to excise the Pcdhgfcon3 alleles using AdvillinCreERT2 in peripheral somatosensory neurons. Intraperitoneal injections of tamoxifen (1 mg per day) were administered to mice for 5 consecutive days, from either P15-P19. All mice in this AdvillinCreERT2 experiments received this tamoxifen regimen, and no changes in health or behaviors were observed in controls or mutant mice.
Immunohistochemistry.
Spinal cord, DRG, and finger pad samples were cryoprotected in 30% sucrose in PBS overnight at 4°C. For synaptic protein analysis, lumbar levels L2 and L3 were dissected (Mohan et al, 2014)121, using the last rib as a landmark to identify thoracic 13 level of the spinal cord. Samples were then rinsed in PBS, embedded in OCT in tissue molds over a 100% ethanol and dry ice bath, and stored at −80°C for at least an hour before cryosectioning. 20-25 μm sections were produced for spinal cord, DRG, and brainstem, and 30 μm transverse sections for fingertip sections. Mutant and littermate control sections were collected on the same slide. Slides were allowed to dry on the benchtop overnight and stored at −20°C.
Slides were defrosted at room temperature for at least 30 minutes, then a hydrophobic barrier was applied with a PAP pen (H-4000, VWR/Vector Lab) around the samples. Slides were then rehydrated with filtered 1xPBS for 3x5 minutes, blocked for 1 hour at room temperature with 1xPBS with 0.1% Triton X-100 and 5% normal goat or donkey serum (Jackson Immuno, 005-000-121), and incubated with primary antibodies in blocking solution (5% serum without detergent) overnight at 4°C. Slides were washed 4x5 minutes with 1xPBS with 0.02% Tween-20, incubated with secondary antibodies in blocking solution (5% serum without detergent) at room temperature for 2 hours, washed 4x5 minutes with 1xPBS with 0.02% Tween-20 detergent, and mounted. Samples were mounted with fluoromount-G (Southern Biotech) or DAPI Fluoromount-G® (Southern Biotech) when visualization of nucleus is needed. All slides were stored at 4°C for up to 1 week until imaged.
The following primary antibodies were used: guinea pig anti-vGluT1 (Millipore Sigma, RRID: AB_2301751; 1:1000), rabbit anti-Homer1 (Synaptic Systems, Cat# 160003, 1:1000), Isolectin B4 647 (Invitrogen, RRID: SCR_014365, 1:500), chicken anti-NFH (Aves, Cat#NFH0211, 1:500); rabbit anti-DsRed (Clontech, RRID: AB_10013483, 1:500), goat anti-mCherry (Sicgen, RRID: AB_2333092, 1:500), goat anti-GFP (Abcam, RRID: AB_305635, 1:500), mouse anti-NeuN (Millipore, RRID: AB_2298772, 1:500), rabbit anti-S100 (Proteintech, Cat# 15146-1-AP, RRID:AB_2254244, 1:500) and rat anti-Troma1 (DSHB, RRID: AB_531826, 1:50). Secondary antibodies include Alexa 405, 488, 546 or 647 conjugated donkey or goat anti-mouse, rabbit, chicken, goat or guinea pig, and used at a 1:500 dilution (Life Technologies or Jackson ImmunoResearch).
Wholemount hairy skin immunostaining.
Back hairy skin was dissected, and fat tissue under the skin was scraped off with a stainless-steel spatula. The skin was then fixed in Zamboni fixative (Fisher/VWR, NC9335034) at 4°C overnight. The tissue was rinsed in PBS three times and then washed with PBS containing 0.3% Triton X-100 (0.3% PBST) every 30 min for 5 to 8 hours. Then, the skin was incubated with primary antibodies in 0.3% PBST containing 5% goat or donkey serum and 20% DMSO at room temperature for 3 to 4 days. Skins were then washed with 0.3% PBST every 30-60 min for 5 to 8 hours. The tissues were transferred to secondary antibodies in 0.3% PBST containing 5% goat/donkey serum and 20% DMSO and incubated at room temperature for 2 to 3 days. The tissues were then washed with 0.3% PBST every 30-60 min for 5 to 8 hours. The skins were dehydrated in 50% methanol for 15 min, 80% methanol for 15 min, and 100% methanol for 15 min, and 100 % methanol overnight. For imaging, skins were cleared in BABB (Benzyl Alcohol, sigma 402834; Benzyl Benzoate, sigma B-6630; 1:2) at room temperature for 10 min. Individual confocal focal planes were examined for hair follicle innervation by an experimenter who was blind to the genotypes.
RNA sequencing and data analysis.
DRGs from P3 pups were dissected and RiboTag immunoprecipitation was performed as previously described. DRGs from the thoracic levels of five pups were combined and subject to immunoprecipitation to obtain mRNA for each sequencing replicate, and three to five biological replicates were sequenced for each group of neurons. Briefly, pups were sacrificed and DRGs were dissected on ice in DMEM Complete Medium with 1% PenStrep (Cat# 15140122, Thermo Fisher Scientific). DRGs were snap-frozen and stored at −80°C. Samples were homogenized (homogenization buffer with 5mg/ml cycloheximide, cOmplete™ protease inhibitor cocktails, 100mg/ml heparin, RNAsin and 1M Dithiothreitol) and centrifuged at 10,000 rpm at 4 °C for 10min. Lysis buffer (RLT buffer from RNeasy kit, Cat# 74004, Qiangen) was added to each sample, and 800 μl were collected from each spun-down lysate. 5 μl of Mouse anti-HA antibody (Cat#H9658, Sigma-Aldrich), was added to each sample and all samples were incubated for 4 hours at 4 °C on a gentle spinner. 200μl of Dynabeads™ Protein G (Cat#10004D, Thermo Fisher) were equilibrated with 1000 μl of homogenization buffer (RNase-free water with 10% NP-40, 1M KCl, 1M MgCl2, 1.5M Tris, pH 7.4) for 10 min at 4 °C. Homogenization buffer was then suctioned off and antibody-tissue homogenate sample was added to the beads and incubated overnight at 4 °C on a gentle spinner/rotator. Next day, beads were washed with 800 μl of high salt buffer for 3 X 10 min at 4C in cold room on rotator. 350 μl of lysis buffer (RLT lysis buffer with βME) were added to each sample after removing final high salt wash buffer and standard RNA purification was performed using the manufacturer’s protocol (RNeasy kit, Cat# 74004, Qiangen).
Purified RNA samples with a concentration of 1-10 ng/μL for 8-10 μL and RNA integrity number (RIN) above 8 (except for one sample for Aβ SAI-LTMRs that had a RIN ~7) were used for library preparation and RNA-sequencing at the Biopolymers Facility at Harvard Medical School. Libraries were prepared using Illumina NexteraXT using the manufacturer’s protocol. The libraries were pooled together and then sequenced using the Illumina NextSeq 500 sequencer with 150 bp paired-end reads at an average depth of ~13,000 genes per sample. Reads were aligned and counted using STAR (Dobin et al., 2013).122 Both read count matrix and normalized data using transcript per million reads (TPM) were obtained. The matrix of read counts was transformed using a regularized logarithm (rlog) implemented in DESeq2 (Love et al., 2014).123 The heatmaps and dotplots were plotted using the ggplot2 package in R124. Subtype uniquely enriched genes and differentially expressed genes analyses were performed as previously described (Zheng et al., 2019).62 Briefly, genes were then grouped based on the neuronal groups their expression is enriched in and ordered based on the enrichment. Genes with rlog values in the top 75% and displayed expression differences over 1 in all replicates were used for displaying the expression pattern. Differentially expressed genes (DEGs) were defined using the criteria of a 1.5-fold change with a false discovery rate (FDR) <0.01. Subtype uniquely enriched genes (SUEGs) were selected for genes that are enriched in each group based on pairwise comparison, thus the FDR for SUEGs after correction for multiple comparisons is ~0.04. Rlog difference from the average rlog value was plotted in the main heatmaps using a scale of −2 to 2 and average rlog values were plotted in another heatmap to the right of the main heatmap.
Quantification of Meissner corpuscle density.
Images of serial sections of glabrous pads from forepaw fingertips were sectioned at 30 μm. Images of sections were analyzed using ImageJ125. In each section, the number of Meissner corpuscles, visualized by using S100 antibody (Meissner corpuscles form ovoid masses in the dermal papillae and Schwann cells surrounding axons are S100+) and NFH antibody. Then, the area of the epidermis was traced and measured in ImageJ. Density was calculated by dividing the number of corpuscles by the area of epidermis.
Dorsal column injections.
Mice (8-12 weeks old) were anesthetized via continuous inhalation of isoflurane (2%) from a precision vaporizer for the 20-40 minutes duration of the surgery. Protective eye ointment (Puralube) was applied to the eyes. The animal’s breathing rate was monitored throughout the procedure and the isoflurane dose was adjusted as necessary. Hair from the back of the neck was treated by a commercial hair remover (Nair, Church and Dwight Co.; Princeton, NJ) for 1-3 min, and swabbed with water, Betadine, and 70% ethanol. A 7 mm incision was made in the midline of the skin at the cervical level. Muscles were separated from the midline until the spinal cord cervical vertebrae were exposed. A small incision was made on the dura and arachnoid membranes above the C1 cervical spinal vertebrae to expose the dorsal column. Next, 100 nl of AAV-Retro-Flex-PLAP (obtained from Dr. Sara Prescott from Harvard Medical School) was injected into the dorsal column above the C1 level using a glass pipette under visual guidance. Afterwards, muscles and skin were stitched together with sutures, and SR Buprenorphine (1.5 mg/kg) was applied for analgesia. Mice recovered from anesthesia on a warm pad for ~1 hour and then returned to their home cage. The condition of the mice, including locomotor activity, the healing of wounds, body weight, and grooming, were monitored daily. 3-4 weeks after surgery, mice were sacrificed by CO2 asphyxiation followed by perfusion.
Alkaline phosphatase staining.
AAV-Retro-Flex-PLAP virus was injected into the dorsal column on littermate control and mutant animals. 3-5 weeks following the injection, animal was perfused and back hairy skins were treated with commercial hair remover and rinsed with PBS. Skins were further dissected and carefully scrapped free of remaining fat tissue under the skin. Spinal cords and DRGs were dissected. Samples in PBS were placed in a 65-68°C incubator for 2 hours, washed 3x5 minutes in B3 buffer (0.1M Tris pH 9.5; 0.1M NaCl; 50mM MgCl2; 0.1% Tween-20), and incubated at room temperature for 24 hours in B3 buffer with 3.4 μL/mL each of NBT (Nitro Blue Tetrazolium, Roche 11-383-213-001) and BCIP (5-Bromo-4-chloro-3-indolyl-phosphate, 4-toluidine salt, Roche 11-383-221-001). Samples were then incubated with 4% PFA for 1 hour at room temperature, washed with 1xPBS, and dehydrated for ~30 minutes with 50%, 75%, and 100% ethanol overnight. Samples were stored in 100% ethanol at −20°C until imaging. For imaging, benzyl alcohol and benzyl benzoate mixed 1:2 (BABB) was used to clear tissue for imaging. Samples were cleared with BABB on a rotator for 10 minutes before imaging.
Skin injections.
The hindpaw hairy skin of newborn pups was swabbed with ethanol. A small amount of fast green (Sigma F7252-5G) in 0.9% saline was added to the virus sample. Forceps and tape were used to stabilize the hindpaw while the needle was inserted into the dermis, injecting ~50 nL of AAV9-hSyn-P2A-dTomato virus (Addgene, #107738-AAV9). Pups were then returned to the litter. 24-28 days after injections, mice were sacrificed by transcardial perfusion under isoflurane anesthesia.
Corticospinal neuron labeling.
To label corticospinal neurons, mice underwent unilateral stereotactic injection of AAV8-CAG-synaptophysin-tdTomato virus (Penn viral core) into the hindlimb region of primary somatosensory cortex (S1). Mice were injected with SR Buprenorphine (0.1 mg/kg), anesthetized with isoflurane and placed in a small animal stereotaxic frame (David Kopf Instruments). The skull was exposed under aseptic conditions and one craniotomy was made centered at −0.6 posterior and 1.75 lateral from bregma. A borosilicate glass pipette was lowered to a depth of 0.7 mm from the surface of the brain. 150 nL of virus was injected at a rate of 50 nL·min−1 using a Microinject system (World Precision Instruments). 4 weeks following the injection, animals were perfused and spinal cords were collected for immunohistochemistry.
RNAScope.
P3 animals were anesthetized in ice for 2-4 minutes or until unconscious, then sacrificed quickly by decapitation. P21 mice were euthanized by CO2 followed by decapitation. DRGs and spinal cords from mice were rapidly dissected and the axial level was identified using the T13 DRG as a landmark. DRG were quickly frozen in dry-ice-cooled 2-metylbutane and stored at −80 °C. DRG were sectioned at a thickness of 15 μm and RNAs were detected by RNAscope (Advanced Cell Diagnostics) using the manufacturer’s protocol. The following probes were used: mm-Pcdhga2 (Cat# 2835811), mm-Pcdhga7-C2 (Cat# 835781-C2), Mm-Pcdhgb1-O1 (Cat# 837621), Mm-Pcdhgc3(Cat# 802841), Mm-Pcdhgc4-C3 (Cat# 835791-C3), Mm-Slc17a6-C3 (Cat# 319171-C3), Mm-Nefh (Cat# 443671), and Mm-Plp1 (Cat# 428181).
Electron microscopy and image analysis.
Electron microscopy of the spinal cord was conducted as previously described (Zhang et al., 2019).75 Briefly, animals were transcardially perfused and fixed with 2% PFA (Electron Microscopy Sciences) and 2.5% glutaraldehyde (Electron Microscopy Sciences), and postfixed in the same fixative overnight. Spinal cords were dissected out and sectioned at 200 μm with a Leica VT1000 S vibratome. These sections were then osmicated with reduced osmium (1% osmium tetroxide (Electron Microscopy Sciences)/1.5% potassium ferrocyanide (MilliporeSigma), stained with 1% uranyl acetate (Electron Microscopy Sciences), dehydrated with an ethanol series followed by propylene oxide, and infiltrated and embedded with an epoxy resin mix (LX-112, Ladd Research). Samples were then cured in the oven at 60 °C for 48-72 hours. Samples were sectioned using a Leica EM UC7 ultramicrotome with Diatome diamond knives, and ultrathin sections (40-60 nm) were picked up on glow discharged formvar/carbon films on slot grids (Ted Pella). Grids were post stained with 2% uranyl acetate in 50% acetone and 0.2% lead citrate (Electron Microscopy Sciences) before imaging. Ultrathin sections were imaged using a JEOL 1200EX transmission electron microscope at 80 kV accelerating voltage and 10,000× nominal magnification with an AMT XR-611 CCD camera at a final pixel size of 1.84 nm.
Micrographs of the glomeruli were taken in lamina III of the spinal cord, based on morphological characteristics of glomeruli previously described (Ribeiro-da-Silva et al., 1985).74 Experimenter was blind to the genotypes. For quantification of the micrographs, images were adjusted with normalization using Fiji/ImageJ to enhance contrast. Morphometric classification of synapses and analysis of ultrastructural PSD parameters were performed as previously described (Robbins et al., 2010).126 Quantifications of the length and thickness of PSDs were performed with experimenter blind to the genotypes.
Synaptic puncta analysis.
Z stack images for puncta analysis were obtained on the Zeiss LSM700 confocal microscope with the 63x objective. Images were taken in lamina III, within 150 μm of lamina Iliv marked by IB4. Imaging parameters were held constant for littermate control and mutants, which were immunostained on the same glass slide to reduce variation. At least 3 animals of each genotype or age were used, with at least ~2000 vGluT1 and ~8000 Homer1 puncta per animal. For each animal used in analysis, 4-6 sets of images, each image set comprising (4 to 5) 1 μm z stacks from a minimum of 3 separate spinal cord sections, was used for analysis. Homer and vGluT1 puncta analyses were performed using a custom script in ImageJ, creating a mask of vGluT1 terminals between 0.5 μm and 5 μm in diameter, thresholding Homer1, and counting puncta of 0.1 μm to 5 μm in diameter contained within that mask.
Whole-cell patch-clamp recordings using acute spinal cord slices.
Mice (P13-P16) were anesthetized via continuous inhalation of isoflurane (2%) and then perfused with ice-cold choline solution. Lumbar enlargements (L1-L5) were dissected out from vertebral columns in ice-cold choline solution (in mM: 110 Choline Chloride, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 25 Glucose, 5 Sodium Ascorbate, 7 MgCl2, 2 Ethylpyruvate, 0.5 CaCl2) and mounted in 4% LMP agarose. The lumbar spinal cords were sliced in a sagittal plane (200 μm, VT1200S, Leica), and the spinal cord slices were recovered at room temperature for 30 minutes in oxygenated (95% O2 and 5% CO2) ACSF solution (in mM: 2.5 CaCl2 2H2O, 1.0 NaH2PO4 H2O, 119 NaCl, 2.5 KCl, 1.3 MgSO4 7H2O, 26 NaHCO3, 25 Glucose, 1.3 Sodium L-ascorbate). Spinal cord slices were then transferred to a recording and cells were visualized by infrared differential interference contrast microscopy for patching (SliceScope Pro 6000, Scientifica). Whole-cell voltage-clamp recordings of random spinal cord neurons right below substantia gelatinosa were obtained at room temperature. The pipette resistance ranged from 3 to 6 MΩ, and the electrodes were filled with an intracellular solution (in mM: 130 Cs-gluconate, 5 TEA, 0.1 CaCl2, 1 EGTA, 10 HEPES, 1 QX-314, 0.2 D-600). Signals were acquired using a Multiclamp 700 B (Molecular Devices). Spontaneous miniature excitatory postsynaptic currents (mEPSCs) were monitored in the presence of tetrodotoxin (0.5 μM, Tocris Cat#1069) and 4-Aminopyridine (1 mM)., at −60 mV holding potential. Series resistance was left uncompensated. Synaptic currents were sampled at 10 kHz using a Digidata 1440A (Molecular Devices) and analyzed offline using Clampfit 9 (Molecular Devices) software. Events were analyzed using template matching search and each event was visually inspected for inclusion or rejection by an experimenter blind to genotype.
Behavioral Analysis.
Both male and female mice of the C57BL6J background with a combination of AdvillinCre, Pcdhgfcon3/fcon3, and Pcdhgfcon3/+ transgenes were used for the behavioral tests. Pregnant females were separated from breeding males before giving birth, and the progeny were allowed to reach weaning age when ear notching and genotyping was performed. Both male and female mutant animals were group-housed with their control littermates. For these assays, AdvillinCre; Pcdhgfcon3/+, AdvillinCre, Pcdhgfcon3/+ and Pcdhgfcon3/fcon3 were used as littermate control animals and were used to account for variabilities in the backgrounds of the animals tested. No differences were observed among these control genotypes. Testing was initiated at 6 to 8 weeks of age, and the animals completed Open Field, EPM, PPI, and Von Frey testing within 2 weeks of the start of testing and in that order. Balance beam and rough floor aversion were completed after this and held consistent within behavior cohorts. Prior to testing dates, animals were habituated to each new testing environment or room for approximately one hour to help eliminate anxiety during testing. All testing and analysis were performed by experimenters blind to genotypes.
Open Field Testing.
Animals were habituated to the testing room in their cages with dim lights for 1 hour prior to testing. The lighting in the room was dimmed to limit anxiety as much as possible and to ensure the assays were as accurate to the natural behavior of the animals as possible. For this test, two 40 cm3 plexiglass arenas (40 cm x 40 cm x 40 cm) were used to test 2 animals concurrently and labeled for easy identification of the animals in each chamber during video analysis. The chambers utilized were opaque black and were cleaned thoroughly with 70% ethanol and water with paper towels before and after each trial. For the test, an animal was placed in the chamber and allowed to explore the chamber for 10 minutes. Following testing, videos of the animals’ position within the chamber were analyzed with custom MATLAB scripts. Total distance traveled and time spent away from walls (time in center) were analyzed for each video.
Elevated Plus Maze (EPM) Test.
The EPM test was used to measure anxiety-like behavior. The custom-made maze utilized was constructed in the shape of a plus sign. The arms were each 30 cm long x 5 cm wide and two of the opposing arms have 15-cm high walls. The maze itself was raised on stilts to measure 40 cm above ground. Testing occurred in a dimly lit and quiet room, and the maze was positioned under a camera for video analysis following testing. After the animals were habituated to the testing room for at least 1 hour, they were placed on the center of the EPM construct and allowed to freely explore the open and closed arms of the maze for 10 minutes. MATLAB scripts were used to quantify the amount of the time each animal spent in the open and closed arms, as well as the center of the maze.
Prepulse Inhibition (PPI).
A San Diego Instruments startle reflex system (SR-LAB Startle Response System) was used to quantify the startle response of animals subjected to tactile and acoustic stimuli and measure the ability of a tactile or acoustic stimulus to inhibit startle response to a loud acoustic stimulus. Tactile PPI assay utilized a 50 ms 0.9 PSI air puff pre-stimulus delivered to the back of the animal, followed by a 20 ms 125 dB startle tone stimulus, to determine the responsiveness to tactile sensitivity in the back hairy skin.
For the tactile PPI test, ventilated cylindrical holders with removable and adjustable doors were used to contain the animals during testing. These holders also had a small opening on the top of the chamber to apply air puff to the back hairy skin. Both before and after testing, these cylindrical holders and the testing chambers were thoroughly cleaned with 70% ethanol and paper towels. The animals were placed in the holder, and the length of the holder were adjusted to match the length of the animal and eliminate extraneous movement during testing. The cylindrical holders were placed in the sanitized soundproof testing chambers. The testing consisted of an acclimation phase, and block I, block II, block III and block IV trials. The acclimation phase lasted 5 minutes, allowing the animal to become adjusted to the chambers and background noise of 75 dB. Block I of testing consisted of 5 startle stimuli alone and utilized 120 or 125 dB white noise to determine the initial startle reflex of the animal for measuring the initial startle reflex. Block II consisted of 5 prepulse stimuli alone (0.9 PSI air puff) to measure response to the prepulse stimulus. Block III utilized pseudorandomized prepulse/pulse, pulse alone and no stimulation trials. Finally, Block IV tested the animals’ habituation to the startle stimuli over the approximate total PPI session by delivering startle stimuli alone. Each trial lasted an average of 30 seconds. Postnatal day 5 mice were tested for reactivity to air puff stimuli applied to the back. This behavioral assay will be described in detail elsewhere.
For the acoustic PPI test, the acclimation tone was 65 dB during the acclimation phase. For Block I, the broadband white noise was 120 dB for determination of the initial startle reflex. For Block II, the prepulse stimuli consisted of 5 80 dB broadband white noise. Block III also utilized prepulse and pulse, pulse alone, and no stimulation trials which were used in a pseudo-randomized fashion. Block IV tested the animal’s habituation to the acoustic startle stimuli with the delivery of 5 startle stimuli.
Von Frey.
Materials for this assay included transparent 4.5 in x 2 in x 2 in chambers with numerous 1cm wide ventilation holes, a wire grid stabilized between an acrylic frame and elevated to 14.5 in by metal columns, and an assortment of filaments with various forces. Prior to testing, animals were habituated to the testing setup by being placed on the wire grid inside the plastic chambers for 2 consecutive days for an hour. On the third day, the animals were habituated for 45 min before testing. Following habituation, the Von Frey filaments were applied to hairless plantar surface of the right hind paw until the filament buckled. Filaments were used beginning with the lightest force to the greatest, completing all trials with one filament before beginning the next. Each filament was used for 10 trials, with number of responses recorded. A withdrawing, sharp movement of the paw, or shaking of the paw behavior was defined as a response.
Balance beam assay.
One hour prior to testing animals were habituated to testing room and investigator handling by undergoing tail inking. To ink the tail, each animal was gently lifted and placed on the wire food hopper facing away from investigator. Firmly holding the tail midway from the tail base, a blue soy ink marker was rolled across the tail forming parallel lines to indicate identifying ear notch number. Once inked animals were gently transferred back into the home cage.
The balance beam rod was constructed of 1 meter matte white acrylic with a flat surface measuring 12 mm or 6 mm in width. The beam rods were mounted on two poles 50 cm above the bench top. A black matte acrylic box was placed at each end of the beam to provide a start and end enclosure for the animal. To encourage the animal to traverse the beam home cage nesting material was placed in the end box to draw the animal to the end box. A hammock was stretched below the beam 10cm above the bench top to serve as a soft landing surface to cushion falls and contain the animal should a fall occur. Digital USB 2.0 CMOS video cameras were mounted above and to side of the balance beam to record crossings.
Balance Beam Training: Day 1 and Day 2. Animals are transported to the testing room at least 30 minutes prior to testing to acclimate to room lighting, sound, odor, and temperature. To train the animals to cross the beams unassisted, each animal was placed on the 12 mm rod and encouraged to cross at least 3 times before returning to the home cage. Once the cohort has rested for 30 minutes, they are then placed on the 6 mm rod and encouraged to complete 3 crossings. If animals refuse to cross, the investigator may gently touch the hindquarters to encourage forward movement. The beam and chambers were cleaned with dilute soapy water and 70% ethanol between animals.
Balance Beam Test: Day 3. First, the 12 mm beam was mounted onto the support poles with the start and end chambers connected to the beam ends. Each beam habituated animal was place into the start chamber with home cage bedding added to the end chamber. At least 2-4 successful crossings were video recorded for each animal. Each animal was allowed to rest for 10 seconds before each crossing. The animals were returned to their home cage once 2-4 successful trials were completed for each beam width. Time to cross, number of slips, number of falls and number of stalls were analyzed.
Texture aversion assay.
One hour prior to testing animals were habituated to testing room and investigator handling by undergoing tail inking. The two test chambers constructed of black matte (inside) acrylic, 12 in x 11.8 in x 6 in, 0.25 in thick, centered side by side on a white matte acrylic floor under diffuse warm white light, 2700 K. A digital USB 2.0 CMOS video camera was mounted directly above the two test chambers. Testing for each texture was conducted over three days and consisted of two chamber habituation days and one test day. On day 1 and day 2 each animal was removed from the home cage placed in an empty test chamber resting on the white matte acrylic floor. Animals were allowed to freely explore and acclimate to the test environment for 10 minutes, then returned to their home cages. On day 3, the texture aversion assay consisted of two phases: a 10 minute explore phase and a 10 minute test phase. In the explore phase, the white matte acrylic floor was covered with a fresh sheet of construction paper to act as the control texture. Animals were placed in the test chamber and allowed to explore for 10 minutes, then returned to their home cage. The control paper was then discarded, and the test chambers were cleaned with 70% ethanol and unscented soapy water, then reset for the test phase. In the aversive texture test phase, two distinct sandpaper textures, extra fine grit SP400 (smooth) and medium coarse grit SP150 (rough) or SP400 (smooth) and averse coarse grit SP60 were taped together using 3M 1-inch scotch tape on the back of the paper with a ½ inch overlap. The test chambers were then placed onto the sandpaper sheets with the seams of the joined sandpaper centered in the middle of the test chamber, ensuring each texture covered half of the testing chamber. Animals were once again transferred from their home cage to the test chamber and allowed to explore the two sandpaper textures SP400 vs SP150 or SP400 vs SP60 for 10 minutes. At the completion of the 10 minute averse texture test animals were removed from the test chambers and returned to their home cage. Both exploration and test phases were video-recorded and custom Matlab tracking codes were used to track animal position in the chamber, time spent on each texture and preference for smooth vs rough textures were analyzed.
Artificial synapse formation assay.
Primary spinal cord cultures were prepared from dorsal spinal cords of P0 mice and maintained in neurobasal medium with B27 supplement. P0 C57BL/6 mouse pups were anesthetized on ice and then decapitated using scissors. Spinal columns were dissected and washed with filter-sterilized HABG medium (Hibernate A with 2% B-27 and 0.5 mM GlutaMAX) to remove excess blood and tissue, as previously described (Eldeiry et al., 2017).93 22 G needle and syringe filled with filter-sterilized HABG was inserted into the caudal end of the spinal column and flushed cranially, allowing the cord to exit into a Petri dish. Dorsal spinal cord is cut into small pieces about 0.5 mm in size. Spinal cord tissues were placed in HABG medium in a 30 °C incubator for 30 min and then digested with Papain (Sigma, Cat#10108014001) for 30 min at 30 °C (HA-Ca 10 mL, Papain, 25 mg, and 0.5 mM GlutaMAX). Papain solution was aspirated and spinal cord tissues were washed with 10 ml neurobasal media (Neurobasal A, 2% B-27, 0.5 mM GlutaMAX, and 1% Pen/Strep). Tissues were gently dissociated by pipetting in neurobasal media with a flame-polished Pasteur pipette. Cells were plated on laminin-covered cover glasses in tissue culture plates and placed in the 37 °C cell culture incubator.
Artificial synapse formation assays were performed as previously described (Biederer and Scheiffele, 2007). HEK 293 cells were maintained in DMEM (Thermo Fisher Scientific, Cat# 10564-011) with 10% HyClone™ Cosmic Calf™ Serum (VWR, Cat# SH3008703), 1% Sodium Pyruvate (Gibco, Cat# 11360-070), 1% MEM Non-Essential Amino Acids Solution (Gibco, Cat# 11140-050) and 1% Pennicillin-Streptomycin (Simga, Cat# P4333). HEK 293 cells were cultured in six-well plates and transfected at 50-70% confluency by using polyethylenimine according to standard protocol (4 ug of DNA mixed with 8 ug of polyethylenimine). One day after transfection, HEK 293 cells were resuspended and plated into dorsal spinal cord neurons at day 8 in vitro. Cells were fixed with 4% PFA for 10 min at room temperature and then were washed three times for 5 min with 1 ml of 0.3% PBST. The fixed cells were blocked with blocking buffer (5% normal goat serum in 0.3% PBST) for 2 h at RT. Then, primary antibody diluted in the blocking buffer was added to the wells at 4°C overnight. Cells were washed 3 × 10 min with 1 ml of 0.1% PBST, followed by secondary antibodies incubation for 1 hour at room temperature. The coverslips were washed for 3X 10 min before mounting on the Superfrost slides.
For fluorescent image analysis, cells were chosen randomly and imaged from four independent replicates. Fluorescent images were acquired with a Zeiss 700 confocal microscope using a Zeiss Achroplan 63/0.95 at 1,024 by 1,024-pixel resolutions. No detectable bleed-through occurred between different channels. The pixel intensities for the brightest samples were below saturation, with the exception that when contours of the HA or GFP signal from transfected HEK 293 cells need to be clearly defined. MAP2 channel was used to confirm dendrites contacting HEK 293 cells. vGlut2 channel was used to exclude extensive neuronal synapses formed near or within HEK293 cells. Images were using identical confocal settings. For analysis, images were thresholded by intensity, and PSD-95 puncta were quantified by counting the number of thresholded puncta. The puncta number and total integrated intensity of PSD-95 punta within the HEK 293 cells were calculated using ImageJ software. Both image acquisition and quantification were performed by experimenters who were blind to the experimental conditions.
In vivo spinal cord multielectrode array electrophysiology.
Adult (>6 weeks) animals were administered dexamethasone (2 mg/kg) 1-2 hours prior to recording to prevent tissue swelling and were then anesthetized with urethane (1.5 g/kg, Sigma). Isoflurane (1%) was administered during surgery and removed prior to multielectrode array recordings. Surgical plane of anesthesia and the temperature of the animal were monitored and maintained throughout the recording session. Briefly, the hair surrounding the dorsal hump was shaved and the spinal column of T13 to L6 was exposed. The vertebrae above the recording site were stabilized using clamps. The vertebrae between L4 and L5 were retracted to expose the dorsal surface of the spinal cord and the dura was removed. The surface of the cord was bathed in mineral oil/submerged in saline. A 32-channel silicon probe (Neuronexus A1x32-Poly3-5mm-25s-177-A32 or Cambridge Neurotech ASSY-37 H4 optrode) was inserted into the dorsal horn and advanced up to ~700 μm below the dorsal surface. The probe was then kept in place for 20 minutes to ensure a stable recording. Signals were amplified, filtered (0.1 - 7.5 kHz bandpass), and digitized (20 kHz) using a headstage amplifier and recording controller (Intan Technologies RHD2132 and Recording Controller). Data acquisition was controlled with Intan Technologies Recording Controller (version 2.07). A 150-200 μm diameter Teflon-tipped indenter controlled by a dual-mode force controller (Aurora Scientific 300C-I) was used to stimulate the glabrous hindpaw. For mapping receptive field (RF) areas, the position of the indenter was controlled with two linear translation piezo stages and a stage controller (Physik Instrumente U-521.24 and C-867.2U2). The position, force, and displacement of the indenter were commanded with custom-written Matlab scripts.
The detailed data analyses will be described elsewhere. Briefly, JRCLUST (version 3.2.2) was used to automatically sort action potentials into clusters, manually refine clusters, and classify clusters as single or multi units. Action potentials were detected with a threshold of 4.5 times the standard deviation of the noise. Action potentials with similar times across sites were merged and then sorted into clusters. Outlier spikes were removed from each cluster. To isolate putative single units, manual cluster curation was performed with JRCLUST split and merge tools. Clusters were classified as single units if (1) the waveforms were large with respect to baseline; (2) there was a clear refractory period in the cross-correlogram (interspike intervals > 1 ms); (3) waveforms were clearly distinct from nearby clusters. Spike event times for clusters classified as single units were exported and processed in Python.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data are presented as the mean ± the standard error of the mean (SEM), unless otherwise noted in the figure legend. Student’s t test is used to compare the morphological/physiological comparisons between littermate control and mutants. One-way ANOVA tests are used to compare three or more groups in one condition and are expressed as an F-statistic and P value within brackets, and post hoc comparisons were performed using the post hoc test noted in the figure legend. Two-way ANOVA tests are used to compare at least two groups with multiple conditions. The number of cells or animals per group used in each experiment is described in the figure legends. Unless otherwise noted, *, p < 0.05, **, p < 0.01, and ***, p < 0.001. All statistics were performed using GraphPad Prism.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Guinea pig polyclonal anti-VGlut1 | Millipore Sigma | Cat# AB5905; RRID: AB_2301751 |
| Rabbit polyclonal anti-Homer1 | Synaptic Systems | Cat# 160003 |
| Monoclonal Anti-HA antibody produced in mouse | Sigma-Aldrich | Cat# H3663; RRID:AB_262051 |
| Isolectin B4 647 | Invitrogen | Cat# I32450; RRID: SCR_014365 |
| Chicken polyclonal anti Neurofilament (NFH) | Aves | Cat# NFH0211 |
| Rabbit polyclonal anti DsRed | Clontech | Cat# 632496; RRID: AB_10013483 |
| Goat polyclonal anti mCherry | Sicgen | Cat# Ab0040-200; RRID: AB_2333092 |
| Goat polyclonal anti GFP-FITC | Abcam | Cat# ab6662; RRID: AB_305635 |
| Mouse monoclonal anti-Neuronal Nuclei (NeuN) | Millipore | Cat# MAB377; RRID: AB_2298772 |
| Rat polyclonal anti-Troma1 (cytokeratin endo A) supernatant | U of Iowa (DSHB) | Cat# TROMA-I; RRID: AB_531826 |
| Rabbit anti GFP | Thermo Fisher Scientific | Cat# A-11122; RRID:AB_221569 |
| Rabbit anti S100 | Proteintech | Cat# 15146-1-AP; RRID:AB_2254244 |
| Mouse anti PSD-95 | Thermo Fisher Scientific | Cat# MA1-045; RRID:AB_325399 |
| Rabbit anti Human HA | Sigma-Aldrich | Cat# H6908; RRID:AB_260070 |
| Chicken anti MAP2 | Millipore | Cat# AB5543; RRID:AB_571049 |
| Guinea pig anti VGLUT2 | Millipore | Cat# AB2251; RRID:AB_1587626 |
| Rabbit anti CGRP | ImmunoStar | Cat# 24112; RRID:AB_572217 |
| Mouse anti Kv4.3 | Antibodies Incorporated | Cat# 75-017; RRID:AB_2131966 |
| Rabbit anti Parvalbumin | Swant | Cat# PV27; RRID:AB_2631173 |
| Rabbit anti NeuN | Millipore | Cat# ABN78; RRID:AB_10807945 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11008; RRID:AB_143165 |
| Goat anti-Guinea Pig IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11073; RRID:AB_2534117 |
| Goat anti-Guinea Pig IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-21435; RRID:AB_2535856 |
| Alexa Fluor 647-AffiniPure Donkey Anti-Chicken | Jackson ImmunoResearch Labs | Cat# 703-605-155; RRID:AB_2340379 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 546 | Thermo Fisher Scientific | Cat# A-11056; RRID:AB_2534103 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31573; RRID:AB_2536183 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody | Thermo Fisher Scientific | Cat# A-21206; RRID:AB_2535792 |
| Alexa Fluor 647-AffiniPure Donkey Anti-Guinea Pig IgG (H+L) | Jackson ImmunoResearch Labs | Cat# 706-605-148; RRID:AB_2340476 |
| Bacterial and Virus Strains | ||
| AAV8-CAG-Synaptophysin-tdTomato | Penn Viral Core | |
| AAV-Retro-Flex-PLAP | Sara L. Prescott | Prescott et al., 2020 |
| AAV9-hSyn-Cre-P2A-dTomato | Addgene | Cat# 107738-AAV9; RRID:Addgene_107738 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Zamboni’s Fixative | Fisher Scientific | Cat# NC9335034 |
| Paraformaldehyde, reagent grade, crystalline | Millipore Sigma | Cat# P6148-500G |
| 4-nitro blue tetrazolium chloride | Sigma | Cat# 11383213001 |
| 5-bromo-4-chloro-3-indolyl-phosphate, 4-toluidine salt | Sigma | Cat# 11383221001 |
| Benzyl Benzoate | Sigma-Aldrich | Cat# B-6630 |
| Benzyl Alcohol | Sigma-Aldrich | Cat# 402834 |
| Tamoxifen | Sigma | Cat# T5648-1g |
| NAIR, commercial hair remover | Church and Dwight Co., Princeton NJ | ASIN# B001G7PTWU |
| Isofluorane | Henry Schein, supplied by Covetrus | Cat# 029405; |
| Sunflower seed oil | Sigma | Cat# S5007 |
| Tetrodotoxin citrate | Tocris | Cat# 1069 |
| Critical Commercial Assays | ||
| RNAscope Fluorescent Multiplex Assay | ACD Bio | Cat# 320850 |
| RNeasy Mini Kit | Qiagen | Cat# 74104 |
| Nextera XT DNA Library Prep Kit | illumina | Cat# FC-131 |
| SsoAdvanced Universal SYBR Green Supermix | Bio-Rad | Cat# 1725271 |
| Deposited Data | ||
| RNA-sequencing Data | This manuscript | GEO (https://www.ncbi.nlm.nih.gov/geo/) |
| Experimental Models: Cell Lines | ||
| HEK 293T cells | DSMZ | Cat# CRL-3216, RRID: CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| Mouse: AdvillinCre | Hasegawa et al., 2007 | RRID: IMSR_JAX:032536; MGI:1333798 |
| Mouse: AdvillinCreER | Lau et al., 2011 | Strain #:026516 RRID:IMSR_JAX:0265 16 |
| Mouse: Lbx1Cre | Sieber et al., 2007 | N/A |
| Mouse: DhhCre | Jaegle et al., 2003 | Strain #:0129 29 RRID:IMSR_JAX:012929 |
| Mouse: Cdx2Cre | Coutaud and Pilon, 2013 | N/A |
| Mouse: Piezo2fl | JAX | 027720 |
| Mouse: Pcdhgfcon3 | Prasad et al., 2008; Lefebvre et al., 2008 | Strain #:0126 44; RRID:IMSR_JAX:012644 |
| Mouse: Rosa26LSL-synaptophysin-tdTomato (Ai34) | Jackson Laboratory stock | Jax:012570 |
| Mouse: B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato) Hze/J (Ai14) | Jackson Laboratory stock | Jax:007908 |
| Mouse: Pcdhg3R1 | Garrett et al., 2019 | N/A |
| Mouse: Pcdhg3R2 | Garrett et al., 2019 | N/A |
| Mouse: Pcdhg1R1 | Garrett et al., 2019 | N/A |
| Mouse: Pcdhgc3KO | Garrett et al., 2019 | N/A |
| Mouse: PcdhgTAKO | Chen et al., 2012 | N/A |
| Mouse: TrkBCreER | Rutlin et al., 2014 | MGI:5616440 |
| Mouse: RetCreER | Luo et al., 2009 | N/A |
| Mouse: PVires-Cre | Hippenmeyer et al., 2005 | MGI:3590684 |
| Mouse: Rpl22HA | Sanz et al., 2009 | Strain #:0110 29 RRID:IMSR_JAX:011029 |
| Mouse: R26LSL-Pcdhga1-mCherry (cA1-mCherry) | Lefebvre, et al., 2012 | |
| Mouse: R26LSL-Pcdhgc3-mCherry (cC3-mCherry) | Lefebvre, et al., 2012 | |
| Oligonucleotides | ||
| Primers for mouse genotyping (see Table S1) | This paper | N/A |
| Mm-Pcdhga2 | ACD Bio | Cat# 2835811 |
| Mm-Pcdhga7-C2 | ACD Bio | Cat# 835781-C2 |
| Mm-Pcdhgb1-O1 | ACD Bio | Cat# 837621 |
| Mm-Pcdhgc3 | ACD Bio | Cat# 802841 |
| Mm-Pcdhgc4-C3 | ACD Bio | Cat# 835791 |
| Mm-Slc17a6-C3 | ACD Bio | Cat# 319171 |
| Mm-Nefh | ACD Bio | Cat# 443671 |
| Mm-Plp1 | ACD Bio | Cat# 428181 |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | imagej.nih.gov/ij |
| Clampex 10 | Molecular Devices | https://www.moleculardevices.com; RRID: SCR_011323 |
| ZEN | Zeiss | https://www.zeiss.com/microscopy/us/downloads.html; RRID: SCR_013672 |
| R and RStudio | R and RStudio |
https://www.r-project.org/ https://www.rstudio.com/; RRID: SCR_000432 |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html; RRID: SCR_015687 |
| ComplexHeatmap | Gu et al., 2016 | https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html |
| MATLAB | Mathworks | https://www.mathworks.com/products/matlab.html; RRID: SCR_001622 |
| JRCLUST | Jun et al., 2017 | https://www.biorxiv.org/content/10.1101/101030v1 |
Highlights.
Pcdhg isoforms have compartmentalized roles in touch sensory neuron development.
Pcdhgc3 promotes synapse formation between sensory neurons and spinal cord neurons.
Pcdhgs promote peripheral axonal branching through neuron-glia interactions.
Sensory inputs to the spinal cord regulate the assembly of spinal cord circuitry.
Acknowledgements
We thank members of the Ginty lab for feedback on the manuscript. We thank Josef Turecek for help with electrophysiology, the Harvard Medical School Neurobiology Imaging Facility and Electron Microscopy Facility. We thank Annie Chen for mouse breeding. This work was supported by a Hanna Gray Fellowship (S.M.), NSF GRFP fellowship (E.H.), NIH 1R01 AT011447-01 (D.D.G.), NIH R35 5R35NS097344-05 (D.D.G.), the Edward R. and Anne G. Lefler Center for Neurodegenerative Disorders (D.D.G.), NIH R01 NS055272 (J.A.W.), NIH R01 EY031690 (A.M.G.), NIMH R01 5R01MH108579 (T.M.), and Canada Research Chair and CIHR PJT-148961 (J.L.L.). D.D.G. is an investigator of the Howard Hughes Medical Institute. This article is subject to HHMI’s Open Access to Publications policy. HHMI lab heads have previously granted a nonexclusive CC BY 4.0 license to the public and a sublicensable license to HHMI in their research articles. Pursuant to those licenses, the author-accepted manuscript of this article can be made freely available under a CC BY 4.0 license immediately upon publication.
Footnotes
Declaration of Interests
D.D.G. is a member of the Neuron Advisory Board.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hertenstein MJ, Verkamp JM, Kerestes AM, and Holmes RM (2006). The communicative functions of touch in humans, nonhuman primates, and rats: a review and synthesis of the empirical research. Genetic, social, and general psychology monographs 132, 5–94. 10.3200/mono.132.1.5-94. [DOI] [PubMed] [Google Scholar]
- 2.Main M, and Stadtman J (1981). Infant response to rejection of physical contact by the mother: aggression, avoidance, and conflict. Journal of the American Academy of Child Psychiatry 20, 292–307. 10.1016/s0002-7138(09)60990-0. [DOI] [PubMed] [Google Scholar]
- 3.Orefice LL (2020). Peripheral Somatosensory Neuron Dysfunction: Emerging Roles in Autism Spectrum Disorders. Neuroscience 445, 120–129. 10.1016/j.neuroscience.2020.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Handler A, and Ginty DD (2021). The mechanosensory neurons of touch and their mechanisms of activation. Nat Rev Neurosci 22, 521–537. 10.1038/s41583-021-00489-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abraira VE, and Ginty DD (2013). The Sensory Neurons of Touch. Neuron 79, 618–639. 10.1016/j.neuron.2013.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abraira VE, Kuehn ED, Chirila AM, Springel MW, Toliver AA, Zimmerman AL, Orefice LL, Boyle KA, Bai L, Song BJ, et al. (2017). The Cellular and Synaptic Architecture of the Mechanosensory Dorsal Horn. Cell 168, 295–310.e19. 10.1016/j.cell.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olausson H, Lamarre Y, Backlund H, Morin C, Wallin BG, Starck G, Ekholm S, Strigo I, Worsley K, Vallbo AB, et al. (2002). Unmyelinated tactile afferents signal touch and project to insular cortex. Nature Neuroscience 5, 900–904. 10.1038/nn896. [DOI] [PubMed] [Google Scholar]
- 8.Jenkins BA, and Lumpkin EA (2017). Developing a sense of touch. Dev Camb Engl 144, 4078–4090. 10.1242/dev.120402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meltzer S, Santiago C, Sharma N, and Ginty DD (2021). The cellular and molecular basis of somatosensory neuron development. Neuron 109, 3736–3757. 10.1016/j.neuron.2021.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olson W, Dong P, Fleming M, and Luo W (2016). The specification and wiring of mammalian cutaneous low-threshold mechanoreceptors. Wiley Interdiscip Rev Dev Biology 5, 389–404. 10.1002/wdev.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horch KW, Tuckett RP, and Burgess PR (1977). A key to the classification of cutaneous mechanoreceptors. The Journal of investigative dermatology 69, 75–82. 10.1111/1523-1747.ep12497887. [DOI] [PubMed] [Google Scholar]
- 12.Burgess PR, Petit D, and Warren RM (1968). Receptor types in cat hairy skin supplied by myelinated fibers. Journal of neurophysiology 31, 833–848. 10.1152/jn.1968.31.6.833. [DOI] [PubMed] [Google Scholar]
- 13.Rutlin M, Ho C-Y, Abraira VE, Cassidy C, Bai L, Woodbury CJ, and Ginty DD (2014). The Cellular and Molecular Basis of Direction Selectivity of Aδ-LTMRs. Cell 159, 1640–1651. 10.1016/j.cell.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai L, Lehnert BP, Liu J, Neubarth NL, Dickendesher TL, Nwe PH, Cassidy C, Woodbury CJ, and Ginty DD (2015). Genetic Identification of an Expansive Mechanoreceptor Sensitive to Skin Stroking. Cell 163, 1783–1795. 10.1016/j.cell.2015.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Rutlin M, Abraira VE, Cassidy C, Kus L, Gong S, Jankowski MP, Luo W, Heintz N, Koerber HR, et al. (2011). The functional organization of cutaneous low-threshold mechanosensory neurons. Cell 147, 1615–1627. 10.1016/j.cell.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson KO (1983). Neural mechanisms of tactual form and texture discrimination. Federation proceedings 42, 2542–2547. [PubMed] [Google Scholar]
- 17.Zimmerman AL, Bai L, and Ginty DD (2014). The gentle touch receptors of mammalian skin. Science 346, 950–954. 10.1126/science.1254229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neubarth NL, Emanuel AJ, Liu Y, Springel MW, Handler A, Zhang Q, Lehnert BP, Guo C, Orefice LL, Abdelaziz A, et al. (2020). Meissner corpuscles and their spatially intermingled afferents underlie gentle touch perception. Science 368. 10.1126/science.abb2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuehn ED, Meltzer S, Abraira VE, Ho C-Y, and Ginty DD (2019). Tiling and somatotopic alignment of mammalian low-threshold mechanoreceptors. Proc National Acad Sci 116, 9168–9177. 10.1073/pnas.1901378116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirnics K, and Koerber HR (1995). Prenatal development of rat primary afferent fibers: II. Central projections. Journal of Comparative Neurology 355, 601–614. 10.1002/cne.903550409. [DOI] [PubMed] [Google Scholar]
- 21.Ozaki S, and Snider WD (1997). Initial trajectories of sensory axons toward laminar targets in the developing mouse spinal cord. Journal of Comparative Neurology 380, 215–229. . [DOI] [PubMed] [Google Scholar]
- 22.Andrews K, and Fitzgerald M (1994). The cutaneous withdrawal reflex in human neonates: sensitization, receptive fields, and the effects of contralateral stimulation. Pain 56, 95–101. 10.1016/0304-3959(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 23.Fitzgerald M (1985). The post-natal development of cutaneous afferent fibre input and receptive field organization in the rat dorsal horn. The Journal of physiology 364, 1–18. 10.1113/jphysiol.1985.sp015725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Q, and Maniatis T (1999). A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 97, 779–790. 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 25.Obata S, Sago H, Mori N, Davidson M, John TS, and Suzuki ST (1998). A common protocadherin tail: multiple protocadherins share the same sequence in their cytoplasmic domains and are expressed in different regions of brain. Cell adhesion and communication 6, 323–333. 10.3109/15419069809010791. [DOI] [PubMed] [Google Scholar]
- 26.Sugino H, Hamada S, Yasuda R, Tuji A, Matsuda Y, Fujita M, and Yagi T (2000). Genomic organization of the family of CNR cadherin genes in mice and humans. Genomics 63, 75–87. 10.1006/geno.1999.6066. [DOI] [PubMed] [Google Scholar]
- 27.Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, and Maniatis T (2002). Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Molecular Cell 10, 21–33. 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Su H, and Bradley A (2002). Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes & Development 16, 1890–1905. 10.1101/gad.1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flaherty E, and Maniatis T (2020). The role of clustered protocadherins in neurodevelopment and neuropsychiatric diseases. Curr Opin Genet Dev 65, 144–150. 10.1016/j.gde.2020.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schreiner D, and Weiner JA (2010). Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proceedings of the National Academy of Sciences of the United States of America 107, 14893–14898. 10.1073/pnas.1004526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thu CA, Chen WV, Rubinstein R, Chevee M, Wolcott HN, Felsovalyi KO, Tapia JC, Shapiro L, Honig B, and Maniatis T (2014). Single-cell identity generated by combinatorial homophilic interactions between α, β, and γ protocadherins. Cell 158, 1045–1059. 10.1016/j.cell.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen WV, Alvarez FJ, Lefebvre JL, Friedman B, Nwakeze C, Geiman E, Smith C, Thu CA, Tapia JC, Tasic B, et al. (2012). Functional Significance of Isoform Diversification in the Protocadherin Gamma Gene Cluster. Neuron 75, 402–409. 10.1016/j.neuron.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen WV, Nwakeze CL, Denny CA, O’Keeffe S, Rieger MA, Mountoufaris G, Kirner A, Dougherty JD, Hen R, Wu Q, et al. (2017). Pcdhαc2 is required for axonal tiling and assembly of serotonergic circuitries in mice. Science 356, 406–411. 10.1126/science.aa13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrett AM, Bosch PJ, Steffen DM, Fuller LC, Marcucci CG, Koch AA, Bais P, Weiner JA, and Burgess RW (2019). CRISPR/Cas9 interrogation of the mouse Pcdhg gene cluster reveals a crucial isoform-specific role for Pcdhgc4. PLOS Genetics 15, e1008554. 10.1371/journal.pgen.1008554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasegawa S, Kumagai M, Hagihara M, Nishimaru H, Hirano K, Kaneko R, Okayama A, Hirayama T, Sanbo M, Hirabayashi M, et al. (2016). Distinct and Cooperative Functions for the Protocadherin-α, -β and -γ Clusters in Neuronal Survival and Axon Targeting. Frontiers in Molecular Neuroscience 9, 155. 10.3389/fnmol.2016.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, and Sanes JR (2012). Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 488, 517–521. 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lefebvre JL, Zhang Y, Meister M, Wang X, and Sanes JR (2008). gamma-Protocadherins regulate neuronal survival but are dispensable for circuit formation in retina. Development 135, 4141–4151. 10.1242/dev.027912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mountoufaris G, Canzio D, Nwakeze CL, Chen WV, and Maniatis T (2018). Writing, Reading, and Translating the Clustered Protocadherin Cell Surface Recognition Code for Neural Circuit Assembly. Annual Review of Cell and Developmental Biology 34, 471–493. 10.1146/annurev-cellbio-100616-060701. [DOI] [PubMed] [Google Scholar]
- 39.Prasad T, and Weiner JA (2011). Direct and Indirect Regulation of Spinal Cord Ia Afferent Terminal Formation by the γ-Protocadherins. Front Mol Neurosci 4, 54. 10.3389/fnmol.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loh KH, Stawski PS, Draycott AS, Udeshi ND, Lehrman EK, Wilton DK, Svinkina T, Deerinck TJ, Ellisman MH, Stevens B, et al. (2016). Proteomic Analysis of Unbounded Cellular Compartments: Synaptic Clefts. Cell 166, 1295–1307.e21. 10.1016/j.cell.2016.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phillips GR, Tanaka H, Frank M, Elste A, Fidler L, Benson DL, and Colman DR (2003). Gamma-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 23, 5096–5104. 10.1523/jneurosci.23-12-05096.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Weiner JA, Levi S, Craig AM, Bradley A, and Sanes JR (2002). Gamma protocadherins are required for survival of spinal interneurons. Neuron 36, 843–854. 10.1016/s0896-6273(02)01090-5. [DOI] [PubMed] [Google Scholar]
- 43.Weiner JA, Wang X, Tapia JC, and Sanes JR (2005). Gamma protocadherins are required for synaptic development in the spinal cord. Proceedings of the National Academy of Sciences 102, 8–14. 10.1073/pnas.0407931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brasch J, Goodman KM, Noble AJ, Rapp M, Mannepalli S, Bahna F, Dandey VP, Bepler T, Berger B, Maniatis T, et al. (2019). Visualization of clustered protocadherin neuronal self-recognition complexes. Nature 569, 280–283. 10.1038/s41586-019-1089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodman KM, Rubinstein R, Dan H, Bahna F, Mannepalli S, Ahlsen G, Thu CA, Sampogna RV, Maniatis T, Honig B, et al. (2017). Protocadherin cis-dimer architecture and recognition unit diversity. Proceedings of the National Academy of Sciences of the United States of America 114, E9829–E9837. 10.1073/pnas.1713449114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodman KM, Rubinstein R, Thu CA, Bahna F, Mannepalli S, Ahlsen G, Rittenhouse C, Maniatis T, Honig B, and Shapiro L (2016). Structural Basis of Diverse Homophilic Recognition by Clustered α- and β-Protocadherins. Neuron 90, 709–723. 10.1016/j.neuron.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodman KM, Rubinstein R, Thu CA, Mannepalli S, Bahna F, Ahlsen G, Rittenhouse C, Maniatis T, Honig B, and Shapiro L (2016). γ-Protocadherin structural diversity and functional implications. eLife 5. 10.7554/elife.20930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicoludis JM, Vogt BE, Green AG, Schärfe CP, Marks DS, and Gaudet R (2016). Antiparallel protocadherin homodimers use distinct affinity- and specificity-mediating regions in cadherin repeats 1-4. eLife 5. 10.7554/elife.18449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubinstein R, Thu CA, Goodman KM, Wolcott HN, Bahna F, Mannepalli S, Ahlsen G, Chevee M, Halim A, Clausen H, et al. (2015). Molecular logic of neuronal self-recognition through protocadherin domain interactions. Cell 163, 629–642. 10.1016/j.cell.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peek SL, Mah KM, and Weiner JA (2017). Regulation of neural circuit formation by protocadherins. Cellular and molecular life sciences : CMLS 74, 4133–4157. 10.1007/s00018-017-2572-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Südhof TC (2018). Towards an Understanding of Synapse Formation. Neuron 100, 276–293. 10.1016/j.neuron.2018.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasegawa H, Abbott S, Han B-X, Qi Y, and Wang F (2007). Analyzing Somatosensory Axon Projections with the Sensory Neuron-Specific Advillin Gene. J Neurosci 27, 14404–14414. 10.1523/jneurosci.4908-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo W, Enomoto H, Rice FL, Milbrandt J, and Ginty DD (2009). Molecular Identification of Rapidly Adapting Mechanoreceptors and Their Developmental Dependence on Ret Signaling. Neuron 64, 841–856. 10.1016/j.neuron.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gutierrez-Mecinas M, Kuehn ED, Abraira VE, Polgár E, Watanabe M, and Todd AJ (2016). Immunostaining for Homer reveals the majority of excitatory synapses in laminae I–III of the mouse spinal dorsal horn. Neuroscience 329, 171–181. 10.1016/j.neuroscience.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dani A, Huang B, Bergan J, Dulac C, and Zhuang X (2010). Superresolution imaging of chemical synapses in the brain. Neuron 68, 843–856. 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meltzer S, Boulanger KC, Osei-Asante E, Handler A, Zhang Q, Sano C, Itohara S, and Ginty DD (2022). A role for axon–glial interactions and Netrin-G1 signaling in the formation of low-threshold mechanoreceptor end organs. Proc. Natl. Acad. Sci. U.S.A 119, e2210421119. 10.1073/pnas.2210421119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meltzer S, Santiago C, Sharma N, and Ginty DD (2021). The cellular and molecular basis of somatosensory neuron development. Neuron 109, 3736–3757. 10.1016/j.neuron.2021.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jenkins BA, Fontecilla NM, Lu CP, Fuchs E, and Lumpkin EA (2019). The cellular basis of mechanosensory Merkel-cell innervation during development. Elife 8, e42633. 10.7554/elife.42633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanz E, Yang L, Su T, Morris DR, McKnight GS, and Amieux PS (2009). Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America 106, 13939–13944. 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma N, Flaherty K, Lezgiyeva K, Wagner DE, Klein AM, and Ginty DD (2020). The emergence of transcriptional identity in somatosensory neurons. Nature 577, 392–398. 10.1038/s41586-019-1900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggstr̈om J, Kharchenko O, Kharchenko PV, et al. (2015). Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nature Neuroscience 18, 145–153. 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 62.Zheng Y, Liu P, Bai L, Trimmer JS, Bean BP, and Ginty DD (2019). Deep Sequencing of Somatosensory Neurons Reveals Molecular Determinants of Intrinsic Physiological Properties. Neuron 103, 598–616.e7. 10.1016/j.neuron.2019.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Südhof TC (2012). The presynaptic active zone. Neuron 75, 11–25. 10.1016/j.neuron.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Togashi H, Sakisaka T, and Takai Y (2009). Cell adhesion molecules in the central nervous system. Cell adhesion & migration 3, 29–35. 10.4161/cam.3.1.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prasad T, Wang X, Gray PA, and Weiner JA (2008). A differential developmental pattern of spinal interneuron apoptosis during synaptogenesis: insights from genetic analyses of the protocadherin-gamma gene cluster. Development 135, 4153–4164. 10.1242/dev.026807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Orefice LL, Zimmerman AL, Chirila AM, Sleboda SJ, Head JP, and Ginty DD (2016). Peripheral Mechanosensory Neuron Dysfunction Underlies Tactile and Behavioral Deficits in Mouse Models of ASDs. Cell 166, 299–313. 10.1016/j.cell.2016.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Orefice LL, Mosko JR, Morency DT, Wells MF, Tasnim A, Mozeika SM, Ye M, Chirila AM, Emanuel AJ, Rankin G, et al. (2019). Targeting Peripheral Somatosensory Neurons to Improve Tactile-Related Phenotypes in ASD Models. Cell 178, 867–886.e24. 10.1016/j.cell.2019.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.von Frey M (1896). Untersuchunger über die Sinnesfunctionen der menschlichen Haut. bei S. Hirzel 23, 175–266. [Google Scholar]
- 69.Choi S, Hachisuka J, Brett MA, Magee A, Omori Y, Iqbal N-A, Zhang D, DeLisle MM, Wolfson RL, Bai L, et al. (2020). Parallel ascending spinal pathways for affective touch and pain. Nature 587, 258–263. 10.1038/s41586-020-2860-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paixão S, Loschek L, Gaitanos L, Morales PA, Goulding M, and Klein R (2019). Identification of Spinal Neurons Contributing to the Dorsal Column Projection Mediating Fine Touch and Corrective Motor Movements. Neuron 104, 749–764.e6. 10.1016/j.neuron.2019.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rossignol S, Dubuc R, and Gossard J-P (2006). Dynamic Sensorimotor Interactions in Locomotion. Physiol Rev 86, 89–154. 10.1152/physrev.00028.2005. [DOI] [PubMed] [Google Scholar]
- 72.Panek I, Bui T, Wright ATB, and Brownstone RM (2014). Cutaneous afferent regulation of motor function. Acta Neurobiol Exp 74, 158–171. [DOI] [PubMed] [Google Scholar]
- 73.McGill BE, Bundle SF, Yaylaoglu MB, Carson JP, Thaller C, and Zoghbi HY (2006). Enhanced anxiety and stress-induced corticosterone release are associated with increased Crh expression in a mouse model of Rett syndrome. Proc National Acad Sci 103, 18267–18272. 10.1073/pnas.0608702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ribeiro-Da-Silva A, Pignatelli D, and Coimbra A (1985). Synaptic architecture of glomeruli in superficial dorsal horn of rat spinal cord, as shown in serial reconstructions. J Neurocytol 14, 203–220. 10.1007/bf01258448. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Q, Lee W-CA, Paul DL, and Ginty DD (2019). Multiplexed peroxidase-based electron microscopy labeling enables simultaneous visualization of multiple cell types. Nat Neurosci 22, 828–839. 10.1038/s41593-019-0358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Y, Serwanski DR, Miralles CP, Fiondella CG, Loturco JJ, Rubio ME, and Blas ALD (2010). Synaptic and nonsynaptic localization of protocadherin-gammaC5 in the rat brain. Journal of Comparative Neurology 518, 3439–3463. 10.1002/cne.22390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garrett AM, Schreiner D, Lobas MA, and Weiner JA (2012). γ-protocadherins control cortical dendrite arborization by regulating the activity of a FAK/PKC/MARCKS signaling pathway. Neuron 74, 269–276. 10.1016/j.neuron.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keeler AB, Molumby MJ, and Weiner JA (2015). Protocadherins branch out: Multiple roles in dendrite development. Cell adhesion & migration 9, 214–226. 10.1080/19336918.2014.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Molumby MJ, Keeler AB, and Weiner JA (2016). Homophilic Protocadherin Cell-Cell Interactions Promote Dendrite Complexity. CellReports 15, 1037–1050. 10.1016/j.celrep.2016.03.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Prescott SL, Umans BD, Williams EK, Brust RD, and Liberles SD (2020). An Airway Protection Program Revealed by Sweeping Genetic Control of Vagal Afferents. Cell 181, 574–589.e14. 10.1016/j.cell.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li L, and Ginty DD (2014). The structure and organization of lanceolate mechanosensory complexes at mouse hair follicles. Elife 3, e01901. 10.7554/elife.01901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jaegle M, Ghazvini M, Mandemakers W, Piirsoo M, Driegen S, Levavasseur F, Raghoenath S, Grosveld F, and Meijer D (2003). The POU proteins Brn-2 and Oct-6 share important functions in Schwann cell development. Genes & Development 17, 1380–1391. 10.1101/gad.258203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lau J, Minett MS, Zhao J, Dennehy U, Wang F, Wood JN, and Bogdanov YD (2011). Temporal control of gene deletion in sensory ganglia using a tamoxifen-inducible Advillin-Cre-ERT2 recombinase mouse. Molecular pain 7, 100. 10.1186/1744-8069-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fernandez-Monreal M, Kang S, and Phillips GR (2009). Gamma-protocadherin homophilic interaction and intracellular trafficking is controlled by the cytoplasmic domain in neurons. Molecular and cellular neurosciences 40, 344–353. 10.1016/j.mcn.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frank M, Ebert M, Shan W, Phillips GR, Arndt K, Colman DR, and Kemler R (2005). Differential expression of individual gamma-protocadherins during mouse brain development. Molecular and Cellular Neuroscience 29, 603–616. 10.1016/j.mcn.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 86.Goodman KM, Katsamba PS, Rubinstein R, Ahlsen G, Bahna F, Mannepalli S, Dan H, Sampogna RV, Shapiro L, and Honig B (2022). How clustered protocadherin binding specificity is tuned for neuronal self-/nonself-recognition. eLife 11. 10.7554/elife.72416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Obata S, Sago H, Mori N, Rochelle JM, Seldin MF, Davidson M, John TS, Taketani S, and Suzuki ST (1995). Protocadherin Pcdh2 shows properties similar to, but distinct from, those of classical cadherins. Journal of Cell Science 108 (Pt 12), 3765–3773. 10.1242/jcs.108.12.3765. [DOI] [PubMed] [Google Scholar]
- 88.Sano K, Tanihara H, Heimark RL, Obata S, Davidson M, John TS, Taketani S, and Suzuki S (1993). Protocadherins: a large family of cadherin-related molecules in central nervous system. The EMBO Journal 12, 2249–2256. 10.1002/j.1460-2075.1993.tb05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sieber MA, Storm R, Martinez-de-la-Torre M, Müller T, Wende H, Reuter K, Vasyutina E, and Birchmeier C (2007). Lbx1 acts as a selector gene in the fate determination of somatosensory and viscerosensory relay neurons in the hindbrain. The Journal of neuroscience : the official journal of the Society for Neuroscience 27, 4902–4909. 10.1523/jneurosci.0717-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fernandez-Monreal M, Kang S, and Phillips GR (2009). Gamma-protocadherin homophilic interaction and intracellular trafficking is controlled by the cytoplasmic domain in neurons. Molecular and cellular neurosciences 40, 344–353. 10.1016/j.mcn.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Phillips GR, Tanaka H, Frank M, Elste A, Fidler L, Benson DL, and Colman DR (2003). Gamma-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 23, 5096–5104. 10.1523/jneurosci.23-12-05096.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Biederer T, and Scheiffele P (2007). Mixed-culture assays for analyzing neuronal synapse formation. Nat Protoc 2, 670–676. 10.1038/nprot.2007.92. [DOI] [PubMed] [Google Scholar]
- 93.Eldeiry M, Yamanaka K, Reece TB, and Aftab M (2017). Spinal Cord Neurons Isolation and Culture from Neonatal Mice. J Vis Exp. 10.3791/55856-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Graf ER, Zhang X, Jin S-X, Linhoff MW, and Craig AM (2004). Neurexins Induce Differentiation of GABA and Glutamate Postsynaptic Specializations via Neuroligins. Cell 119, 1013–1026. 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nam CI, and Chen L (2005). Postsynaptic assembly induced by neurexin-neuroligin interaction and neurotransmitter. Proc National Acad Sci 102, 6137–6142. 10.1073/pnas.0502038102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mah KM, Houston DW, and Weiner JA (2016). The γ-Protocadherin-C3 isoform inhibits canonical Wnt signalling by binding to and stabilizing Axin1 at the membrane. Scientific Reports 6, 31665–17. 10.1038/srep31665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ranade SS, Woo S-H, Dubin AE, Moshourab RA, Wetzel C, Petrus M, Mathur J, Bégay V, Coste B, Mainquist J, et al. (2014). Piezo2 is the major transducer of mechanical forces for touch sensation in mice. Nature 516, 121–125. 10.1038/nature13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lehnert BP, Santiago C, Huey EL, Emanuel AJ, Renauld S, Africawala N, Alkislar I, Zheng Y, Bai L, Koutsioumpa C, et al. (2021). Mechanoreceptor synapses in the brainstem shape the central representation of touch. Cell 184, 5608–5621.e18. 10.1016/j.cell.2021.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chirila AM, Rankin G, Tseng S-Y, Emanuel AJ, Chavez-Martinez CL, Zhang D, Harvey CD, and Ginty DD (2022). Mechanoreceptor signal convergence and transformation in the dorsal horn flexibly shape a diversity of outputs to the brain. Cell 185, 4541–4559.e23. 10.1016/j.cell.2022.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.LaMassa N, Sverdlov H, Mambetalieva A, Shapiro S, Bucaro M, Fernandez-Monreal M, and Phillips GR (2021). Gamma-protocadherin localization at the synapse is associated with parameters of synaptic maturation. Journal of Comparative Neurology 529, 2407–2417. 10.1002/cne.25102. [DOI] [PubMed] [Google Scholar]
- 101.Han M-H, Lin C, Meng S, and Wang X (2010). Proteomics analysis reveals overlapping functions of clustered protocadherins. Molecular & Cellular Proteomics 9, 71–83. 10.1074/mcp.m900343-mcp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Missler M, Südhof TC, and Biederer T (2012). Synaptic Cell Adhesion. Csh Perspect Biol 4, a005694. 10.1101/cshperspect.a005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Südhof TC (2017). Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell 171, 745–769. 10.1016/j.cell.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kostadinov D, and Sanes JR (2015). Protocadherin-dependent dendritic self-avoidance regulates neural connectivity and circuit function. Elife 4, e08964. 10.7554/elife.08964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Weiner JA, and Jontes JD (2013). Protocadherins, not prototypical: a complex tale of their interactions, expression, and functions. Frontiers in Molecular Neuroscience 6, 4. 10.3389/fnmol.2013.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garrett AM, and Weiner JA (2009). Control of CNS synapse development by γ-protocadherin-mediated astrocyte-neuron contact. The Journal of neuroscience 29, 11723–11731. 10.1523/jneurosci.2818-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Molumby MJ, Anderson RM, Newbold DJ, Koblesky NK, Garrett AM, Schreiner D, Radley JJ, and Weiner JA (2017). γ-Protocadherins Interact with Neuroligin-1 and Negatively Regulate Dendritic Spine Morphogenesis. Cell Reports 18, 2702–2714. 10.1016/j.celrep.2017.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Canzio D, and Maniatis T (2019). The generation of a protocadherin cell-surface recognition code for neural circuit assembly. Current Opinion in Neurobiology 59, 213–220. 10.1016/j.conb.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hanaway J, and Smith JM (1979). Synaptic fine structure and the termination of corticospinal fibers in the lateral basal region of the cat spinal cord. Journal of Comparative Neurology 183, 471–486. 10.1002/cne.901830303. [DOI] [PubMed] [Google Scholar]
- 110.Valtschanoff JG, Weinberg RJ, and Rustioni A (1993). Amino acid immunoreactivity in corticospinal terminals. Experimental brain research 93, 95–103. 10.1007/bf00227784. [DOI] [PubMed] [Google Scholar]
- 111.Ebert DH, and Greenberg ME (2013). Activity-dependent neuronal signalling and autism spectrum disorder. Nature 493, 327–337. 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kirkby LA, Sack GS, Firl A, and Feller MB (2013). A role for correlated spontaneous activity in the assembly of neural circuits. Neuron 80, 1129–1144. 10.1016/j.neuron.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leighton AH, and Lohmann C (2016). The Wiring of Developing Sensory Circuits-From Patterned Spontaneous Activity to Synaptic Plasticity Mechanisms. Frontiers in neural circuits 10, 71. 10.3389/fncir.2016.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Beggs S, Torsney C, Drew LJ, and Fitzgerald M (2002). The postnatal reorganization of primary afferent input and dorsal horn cell receptive fields in the rat spinal cord is an activity-dependent process. The European journal of neuroscience 16, 1249–1258. 10.1046/j.1460-9568.2002.02185.x. [DOI] [PubMed] [Google Scholar]
- 115.Fitzgerald M (1987). Cutaneous primary afferent properties in the hind limb of the neonatal rat. J Physiology 383, 79–92. 10.1113/jphysiol.1987.sp016397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fitzgerald M (1987). Spontaneous and evoked activity of fetal primary afferents in vivo. Nature 326, 603–605. 10.1038/326603a0. [DOI] [PubMed] [Google Scholar]
- 117.Hsu J-YC, Stein SA, and Xu X-M (2006). Development of the corticospinal tract in the mouse spinal cord: a quantitative ultrastructural analysis. Brain research 1084, 16–27. 10.1016/j.brainres.2006.02.036. [DOI] [PubMed] [Google Scholar]
- 118.Coutaud B, and Pilon N (2013). Characterization of a novel transgenic mouse line expressing Cre recombinase under the control of the Cdx2 neural specific enhancer. Genesis 51, 777–784. 10.1002/dvg.22421. [DOI] [PubMed] [Google Scholar]
- 119.Woo S-H, Ranade S, Weyer AD, Dubin AE, Baba Y, Qiu Z, Petrus M, Miyamoto T, Reddy K, Lumpkin EA, et al. (2014). Piezo2 is required for Merkel-cell mechanotransduction. Nature 509, 622–626. 10.1038/nature13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, and Arber S (2005). A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biology 3, e159. 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mohan R, Tosolini AP, and Morris R (2014). Targeting the motor end plates in the mouse hindlimb gives access to a greater number of spinal cord motor neurons: An approach to maximize retrograde transport. Neuroscience 274, 318–330. 10.1016/j.neuroscience.2014.05.045. [DOI] [PubMed] [Google Scholar]
- 122.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550–21. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gu Z, Eils R, and Schlesner M (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics (Oxford, England) 32, 2847–2849. 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 125.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Robbins EM, Krupp AJ, Arce K.P. de, Ghosh AK, Fogel AI, Boucard A, Südhof TC, Stein V, and Biederer T (2010). SynCAM 1 Adhesion Dynamically Regulates Synapse Number and Impacts Plasticity and Learning. Neuron 68, 894–906. 10.1016/j.neuron.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper are available from the lead contact upon reasonable request. RNA sequencing dataset is deposited: https://doi.org/10.7910/DVN/BJGETY. This paper does not report original codes. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.