Abstract
We have previously described the hMYH cDNA and genomic clones (M. M. Slupska et al., J. Bacteriol. 178:3885–3892, 1996). Here, we report that the enzyme expressed from an hMYH cDNA clone in Escherichia coli complements the mutator phenotype in a mutY mutant and can remove A from an A · 8-hydroxydeoxyguanine mismatch and to a lesser extent can remove A from an A · G mismatch in vitro.
Reactive oxygen species generated either as a by-product of cellular respiration or by ionizing radiation and other oxidizing agents can cause damage to DNA. Cells have developed mechanisms to prevent mutations from different types of oxidative damage. Among oxidatively damaged DNA bases, 7,8-dihydro-8-oxodeoxyguanine (also termed 8-hydroxydeoxyguanine [8-oxodG]) is particularly mutagenic. It can easily mispair with adenine, and if it is not repaired before replication, such a mispair can result in a G · C-to-T · A transversion at the next round of replication (19). Cells evolved repair systems to prevent mutation from 8-oxodG. One of the enzymes involved in that process in Escherichia coli is the MutY protein (11), a simple glycosylase that removes A when paired with 8-oxodG or G (1, 10) and to a lesser extent removes A from A · C and A · 7,8-dihydro-8-oxodeoxyadenine (A · 8-oxodA) (21). Recently, it was reported that after prolonged incubation, and with a large excess of the enzyme, E. coli MutY can also remove G from G · 8-oxodG mispairs (24). The 39-kDa E. coli MutY protein belongs to the superfamily of base excision repair glycosylases, in which a helix-hairpin-helix motif is the most conserved structural element (15). The crystal structure of the catalytic core of MutY was recently determined (3). The resolution of crystal structures revealed that, as in other related glycosylases (4), the target adenine is flipped out of the double-stranded DNA helix and accommodated in a positively charged substrate-binding pocket of the MutY protein (3).
We have previously described the cloning and sequencing of hMYH, a human homolog of the E. coli mutY gene (20). Here, we report functional expression and purification of the hMYH enzyme. The cloned hMYH gene, when expressed in E. coli, complements the mutator phenotype of a mutY mutant. hMYH is a 59-kDa protein that has glycosylase activity but no AP-lyase activity. hMYH protein can remove A from an A · 8-oxodG mismatch and to a lesser extent can remove A from an A · G mismatch. After prolonged incubation, it can also remove G from a G · 8-oxodG mispair.
Complementation of the E. coli mutY mutation by hMYH.
The whole cDNA sequence of hMYH was subcloned into the pCR-Script SKII(+) vector (Stratagene, La Jolla, Calif.) vector from different clones described previously (20). For expression, a polyhistidine (His6) affinity tag was attached to the amino terminus of hMYH by subcloning into pQE30 (Qiagen, Inc., Chatsworth, Calif.), and this plasmid was used to check whether hMYH cDNA complements the mutY deficit in E. coli. The expressed hMYH gene had a toxic effect on E. coli cells, and transformation of the pQE30/hMYH plasmid into E. coli cells was difficult. The pQE30 vector is based on the T5 promoter transcription-translation system. It contains the phage T5 promoter and two lac operator sequences (Qiagen, Inc.). Since pQE30 has a high level of background transcription, transformation was improved by using a recipient strain carrying an additional plasmid, pREP4, that constitutively expresses the lac repressor. Transformation of pQE30/hMYH into a strain carrying the pREP4 plasmid was 1,000 times more efficient than its transformation into a strain not carrying pREP4 (data not shown). It was also easier to maintain pQE30/hMYH in strains with pREP4. The addition of 1 to 10 μM IPTG (isopropyl-β-d-thiogalactopyranoside) induced expression of hMYH, but induction with IPTG at concentrations higher than 100 μM resulted in lysis of bacteria.
The E. coli CC104 strain carries the F′ plasmid with the lac gene engineered for detection of G · C-to-T · A transversions (2). Previous work has shown that strains with defects in mutY have increased G · C-to-T · A transversions (16), so we used the CC104 mutY::Tn10 (12) derivative to check whether hMYH complements the mutY mutator phenotype. The complementation experiment was performed as described previously (14). Cells of strain CC104 mutY carrying both the pREP4 and pQE30/hMYH plasmids showed suppression of the mutator phenotype. The suppression was complete when cells were grown in the presence of 1 μM IPTG (Table 1). Use of higher concentrations of IPTG resulted in a very low survival rate for the CC104 mutY strain, loss of the hMYH insert from the plasmid, or both (data not shown).
TABLE 1.
Complementation of E. coli mutY defect with cloned hMYHa
| Expression plasmid | IPTG | Avg no. of Lac+ revertants/108 cells |
|---|---|---|
| pQE30 | − | 98.3 ± 12.8 |
| pQE30 | + | 127.9 ± 22.7 |
| pQE30/hMYH | − | 8.9 ± 8.8 |
| pQE30/hMYH | + | 1.2 ± 1.1 |
Four or five single colonies of the CC104 mutY mutant carrying pREP and expression plasmids were inoculated into Luria-Bertani medium with 100 μg of ampicillin and 25 μg of kanamycin per ml, with or without 1 μM IPTG, and grown overnight at 37°C. Samples were plated on lactose minimal medium and titers were determined on glucose minimal medium. The number of Lac+ revertants (mean ± standard deviation) of strain CC104 resulting from G · C-to-T · A transversion is shown.
Expression and purification of hMYH.
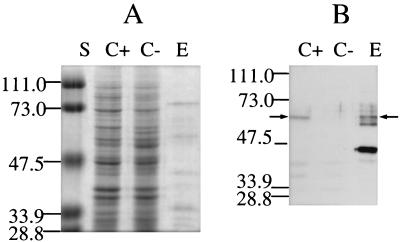
The hexahistidine-tagged hMYH protein was purified by Ni2+ affinity chromatography, followed by single-stranded DNA (ssDNA) affinity chromatography. Conditions for Ni2+-agarose purification were set according to the protocols of the manufacturer (Qiagen, Inc.), except that a pH of 6.5 and no NaCl were used for binding and the column was washed with buffer containing 100 mM NaCl and 30 mM imidazole at pH 6.5. Conditions for ssDNA-cellulose were as described for the E. coli MutY purification (6). The purified protein reacts with antibodies against the histidine tag [anti-RGS(H)4; Qiagen, Inc.] (Fig. 1).
FIG. 1.
(A) Sodium dodecyl sulfate-polyacrylamide gel analysis of hMYH purified by Ni2+-agarose affinity chromatography, followed by ssDNA-cellulose chromatography (E). The proteins were separated on a 10% polyacrylamide gel in the presence of sodium dodecyl sulfate and stained with Coomassie blue. Lane S, molecular mass standards. Molecular masses, in kilodaltons, are marked on the side. Lane C+, lysate of cells of the CC104 mutY mutant containing pQE30/hMYH; lane C−, lysate of the same strain with pQE30. (B) Western blot analysis of hMYH. Proteins were separated on a 10% polyacrylamide gel, transferred onto a Hybond enhanced chemiluminescence nitrocellulose membrane (Amersham Pharmacia Inc., Piscataway, N.J.), and reacted with antibodies against the histidine tag [anti-RGS(H)4; Qiagen, Inc.]. Western blotting was performed by enhanced chemiluminescence analysis (ECL; Amersham). Arrows point to the position of full-length hMYH. Lanes C+, C−, and E are as described for panel A.
The expressed hMYH was rapidly degraded, in both a CC104 mutY mutant (12) (normally used for expression) and a strain that lacked the E. coli outer membrane protease (ompT mutant) (BL 21; Novagen Inc., Madison, Wis.). The degrees of degradation were similar during growth at different temperatures (15 to 37°C). Purified fractions contained other proteins, some of which cross-reacted with anti-RGS(H)4 antibody (Fig. 1).
The cloned hMYH gene has seven AGA and AGG codons in the 5′ terminus of the mRNA. These are rare arginine codons in E. coli and can result in poor expression in E. coli (7), as was the case for hMYH. It has been shown that cloning the argU gene, encoding the arginine tRNA that reads AGA and AGG codons, can improve the expression of some eukaryotic genes with rare arginine codons (18). Therefore, we cloned the argU gene with its natural promoter into the same expression plasmid but did not observe any improvement in hMYH levels. We also cloned and expressed N-terminally truncated clones of hMYH. We expressed a 54-kDa clone (missing the first 41 amino acids), a 52-kDa clone (missing the first 62 amino acids), and a 45-kDa clone (missing the first 120 amino acids). We observed high-level expression only for the 45-kDa clone (data not shown). It turned out that the protein expressed by this clone was inactive (data not shown). Also, this was the only clone that did not have rare arginine codons in the 5′-terminal part of the gene.
hMYH activity.
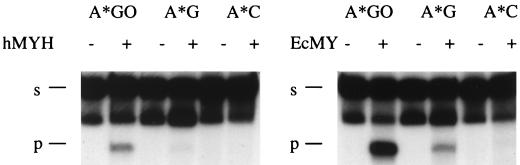
To determine the specificity of hMYH, we investigated its glycosylase activity on various mismatches placed in 96-mers. The sequence of 96-mers containing 8-oxodG or G was 5′AATTGTCCTTCCCTTCCTTTCTCGCC ACGTTCGCCGAATTGGGCTTTCCCCGTCAAGCTCTA AATCGGGGGCTCCCTTTAGGGTTCCGATCCGGCC3′ (the bold G marks the position of 8-oxodG). We tested A · 8-oxodG, A · G, A · C, A · A, A · T, C · G, C · 8-oxodG, C · T, C · C, G · 8-oxodG, G · T, G · G, T · T, and T · 8-oxodG mispairs. We detected activity for the A · 8-oxodG and A · G mispairs but not for the A · C mispair (Fig. 2). As for E. coli MutY, cleavage of the oligomer by hMYH was seen only for the strand containing A (data not shown). After prolonged incubation and strong overexposure of the film, we could also see a small degree of cleavage for the G · 8-oxodG mispair but not for the A · C mispair (data not shown). The same substrates were processed by the mouse MutY homolog (data not shown). The only difference we observed between the hMYH and the E. coli MutY enzymes was the lack of glycosylase activity for the A · C mispair. This activity was also undetectable in HeLa cell extracts (23) and very weak for the Schizosaccharomyces pombe MutY homolog (5), but this activity for a partially purified bovine MutY homolog has been reported (8).
FIG. 2.
Reaction of hMYH and E. coli MutY (EcMY) with different mispair-containing 96-mer templates. Reactions were carried out for 3 h at 37°C; 3 μl of the protein preparation described in the legend to Fig. 1 and 6 ng of E. coli MutY protein were used for all templates. E. coli MutY was purified as described previously (6). The asterisks each indicate an A from a radiolabelled strand. s, substrate; p, product.
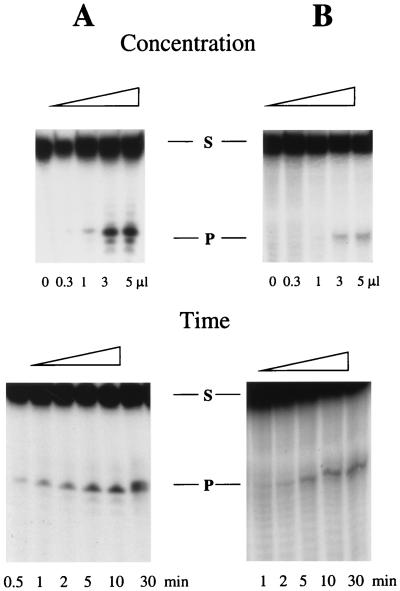
We also tested hMYH activity towards A · 8-oxodG and A · G mispairs placed in shorter oligomers. We could easily detect the activity for the A · 8-oxodG mispair, but surprisingly, we did not see any activity for the A · G mispair placed in a 23-mer. We saw, however, the activity of hMYH with the A · G mispair in 45-mer (Fig. 3) and 72- and 96-mer (data not shown) templates. To detect hMYH glycosylase activity on A · G mismatches we had to use more enzyme and longer reaction times (Fig. 3). The time and concentration courses for the A · 8-oxodG mispair in a 23-mer template and the A · G mispair in a 42-mer template are shown in Fig. 3. The lack of activity of hMYH with an A · G mispair in a 23-mer is an unexpected observation. An S. pombe MutY homolog (5), a partially purified calf MutY homolog (8), and a mouse MutY homolog from our lab (data not shown) all cleave a 20- or 23-bp DNA containing an A · G mismatch. The A · 8-oxodG-containing oligomers are, however, much better substrates for both hMYH and mMYH (mouse MutY homolog), and this difference is particularly obvious when short reaction times and smaller amounts of enzyme are used (data not shown). One explanation for this phenomenon might be that both hMYH and mMYH have low affinity to the A · G mismatch placed in shorter templates but the affinity of the mouse MutY homolog to A · G-containing short oligomers is higher than that of hMYH. The differences between DNA glycosylase homologs of closely related species have already been described for methylpurine-DNA glycosylase from mice and humans (17). It was also observed that human methylpurine-DNA glycosylase has a 10-fold-lower binding capacity for shorter oligomers (<15 bp) than for longer ones (9). We also cannot exclude the possibility that the lack of hMYH glycosylase activity on 23-mer substrates is due to the neighboring sequence context. The investigated mispairs were placed in a different context in the 23-mers than those of the 45-, 72-, and 96-mers used in this work, and the sensitivity to sequence context of the MutY glycosylase activity towards A · G mispairs has been noted for E. coli (22).
FIG. 3.
Concentration dependence and time course for the enzymatic reaction of hMYH with A · 8-oxodG placed in a 23-mer (A) and A · G placed in a 45-mer (B). Reaction conditions were as described previously (13). The sequence of the A-containing 23-mer was 5′AGAGGAAAGGAGAGAAGGGAGAG3′. The sequence of the 45-mer for the A-containing strand was 5′TTAGAGCTTGACGGGGAAAGCCAAATTCGGCGAACGTGGCGAGAA3′ (bold A marks the position of the mispaired A for both templates). In the variable-concentration experiment, reaction with A · 8-oxodG was carried out for 10 min, whereas reaction with A · G was carried out for 60 min. In the time course experiment for the A · 8-oxodG substrate, 0.3 μl of the enzyme solution was used, and for the A · G substrate, 1 μl of the enzyme was used. The A-containing oligonucleotide was radiolabelled. S and P, substrate and product, respectively.
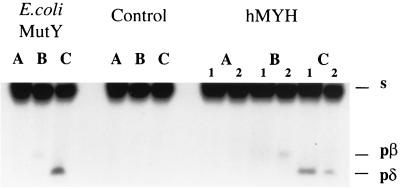
To determine whether hMYH has both glycosylase and AP-lyase activities, we repeated conditions for a similar experiment with E. coli MutY, described previously (25). In this experiment, the product of the same reaction was resolved on acrylamide gels under different, carefully controlled heating conditions. As can be seen in Fig. 4, for hMYH as well as E. coli MutY, under the “no-heat” conditions the product of DNA nicking is barely visible, whereas when the same reaction mixture was heated at 95°C for 5 min, the β-elimination product is obvious. Additional treatment with piperidine (known to cleave AP sites) resulted in a shift of the β-elimination product into a δ-elimination product (Fig. 4). These results indicate that, similar to the E. coli MutY protein, hMYH is a simple glycosylase with only residual, if any, AP-lyase activity.
FIG. 4.
Products generated by hMYH under different conditions of heating. Reactions were stopped by freezing in a dry ice bath (lanes A), loading buffer was added and reaction mixtures were heated for 5 min at 95°C (lanes B), and panels an equal volume of 20% piperidine was added and the reaction mixture was heated for 30 min at 95°C, dried under vacuum, and dissolved in formamide loading buffer (lanes C). Products were resolved on a 15% polyacrylamide–8 M urea gel under conditions described previously (25). Numbers 1 and 2 indicate the enzyme from two different preparations of hMYH. s, migration of substrate; pβ, product of β-elimination; pδ, product of δ-elimination. Samples without enzyme were used for the control. Reactions were carried out with an A · 8-oxodG mispair placed in a 23-mer, with the A-containing strand radiolabelled, for 10 min at 37°C. The sequence of the 23-mer is in the legend to Fig. 3.
In summary, the data presented in this paper show that hMYH, cloned in our laboratory on the basis of DNA sequence homology to the E. coli mutY gene, is the true homolog of the bacterial MutY glycosylase.
Acknowledgments
This work was supported by a grant to J.H.M. from the National Institutes of Health (GM 32184).
REFERENCES
- 1.Au K G, Clark S, Miller J H, Modrich P. Escherichia coli mutY gene encodes an adenine glycosylase active on G-A mispairs. Proc Natl Acad Sci USA. 1989;86:8877–8881. doi: 10.1073/pnas.86.22.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cupples C G, Miller J H. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc Natl Acad Sci USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan Y, Manuel R, Arvai A, Parikh S, Mol C, Miller J, Lloyd S, Tainer J. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat Struct Biol. 1998;5:1058–1064. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- 4.Krokan H, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem J. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu A-L, Fawcett W P. Characterization of the recombinant MutY homolog, an adenine DNA glycosylase, from yeast Schizosaccharomyces pombe. J Biol Chem. 1998;273:25098–25105. doi: 10.1074/jbc.273.39.25098. [DOI] [PubMed] [Google Scholar]
- 6.Manuel R C, Czerwinski E W, Lloyd R S. Identification of the structural and functional domains of MutY, an Escherichia coli DNA mismatch repair enzyme. J Biol Chem. 1996;271:16218–16226. doi: 10.1074/jbc.271.27.16218. [DOI] [PubMed] [Google Scholar]
- 7.Mattes R. Principles of gene expression. In: Rehm H-J, Reed G, editors. Biotechnology. II. Weinheim, Germany: VCH; 1993. pp. 233–256. [Google Scholar]
- 8.McGoldrick J P, Yeh Y-C, Solomon M, Essigmann J M, Lu A-L. Characterization of a mammalian homolog of the Escherichia coli MutY mismatch repair protein. Mol Cell Biol. 1995;15:989–996. doi: 10.1128/mcb.15.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miao F, Bouziane M, O’Connor T R. Interaction of the recombinant human methylpurine-DNA glycosylase (MPG protein) with oligodeoxyribonucleotides containing either hypoxanthine or abasic sites. Nucleic Acids Res. 1998;26:4034–4041. doi: 10.1093/nar/26.17.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michaels M L, Cruz C, Grollman A P, Miller J H. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michaels M L, Miller J H. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michaels M L, Pham L, Nghiem Y, Cruz C, Miller J H. MutY, an adenine glycosylase active on G-A mispairs, has homology to endonuclease III. Nucleic Acids Res. 1990;18:3841–3845. doi: 10.1093/nar/18.13.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michaels M L, Tchou J, Grollman A P, Miller J H. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 14.Miller J H. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 15.Nash H, Bruner S, Schärer O, Kawate T, Addona T, Spooner E, Lane W, Verdine G. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 16.Nghiem Y, Cabrera M, Cupples C G, Miller J H. The mutY gene: a mutator locus in Escherichia coli that generates GC→TA transversions. Proc Natl Acad Sci USA. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy R, Kumar A, Lee J C, Mitra S. The domains of mammalian base excision repair enzyme N-methylpurine-DNA glycosylase. J Biol Chem. 1996;271:23690–23697. doi: 10.1074/jbc.271.39.23690. [DOI] [PubMed] [Google Scholar]
- 18.Schenk P M, Baumann S, Mattes R, Steinbiss H-H. Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. BioTechniques. 1995;19:196–200. [PubMed] [Google Scholar]
- 19.Shibutani S, Takeshita M, Grollman A P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature (London) 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 20.Slupska M M, Baikalov C, Luther W M, Chiang J-H, Wei Y-F, Miller J H. Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J Bacteriol. 1996;178:3885–3892. doi: 10.1128/jb.178.13.3885-3892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai-Wu J-J, Liu H-F, Lu A-L. Escherichia coli MutY protein has both N-glycosylase and apurinic/apyrimidinic endonuclease activities on A-C and A-G mispairs. Proc Natl Acad Sci USA. 1992;89:8779–8783. doi: 10.1073/pnas.89.18.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams S D, David S S. Evidence that MutY is a monofunctional glycosylase capable of forming a covalent Schiff base intermediate with substrate DNA. Nucleic Acids Res. 1998;26:5123–5133. doi: 10.1093/nar/26.22.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh Y-C, Chang D-Y, Masin J, Lu A-L. Two nicking enzyme systems specific for mismatch-containing DNA in nuclear extracts from human cells. J Biol Chem. 1991;266:6480–6484. [PubMed] [Google Scholar]
- 24.Zhang Q-M, Ishikawa N, Nakahara T, Yonei S. Escherichia coli MutY protein has a guanine-DNA glycosylase that acts on 7,8-dihydro-8-oxoguanine:guanine mispair to prevent spontaneous G:C→C:G transversions. Nucleic Acids Res. 1998;26:4669–4675. doi: 10.1093/nar/26.20.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zharkov D O, Grollman A P. MutY DNA glycosylase: base release and intermediate complex formation. Biochemistry. 1998;37:12384–12394. doi: 10.1021/bi981066y. [DOI] [PubMed] [Google Scholar]