Abstract
The complete nucleotide sequence of the capsular gene cluster (cap8) responsible for the biosynthesis of the capsular polysaccharide of Streptococcus pneumoniae type 8 has been determined. The cap8 gene cluster, located between the genes dexB and aliA, is composed of 12 open reading frames. A 14.7-kb DNA fragment embracing the cap8 genes was sufficient to transform an unencapsulated type 3 S. pneumoniae strain to a strain with the type 8 capsule. A possible scenario for the evolution of pneumococcal types 2 and 8 is outlined.
Streptococcus pneumoniae is an important human pathogen that causes diseases such as pneumonia, bacteremia, meningitis, and otitis media. S. pneumoniae has a polysaccharide capsule that is considered the main virulence factor of this species and is involved in evasion of the host immune system (1). There are 90 distinct capsular serotypes (19), but most cases of pneumococcal pneumonia appear to be caused by a subset of 23 serotypes (11, 44). Efforts to develop a new generation of polysaccharide-protein conjugate vaccines are currently being made (8) because of the limited efficacy of the currently used 23-valent polysaccharide-based vaccine in infants as well as in immunocompromised patients. Nevertheless, technical, immunological, and economic reasons restrict the number of different capsular polysaccharides that can be incorporated into a conjugate vaccine (8). Moreover, it has been predicted that when several serotypes interact epidemiologically, as observed in S. pneumoniae, vaccination against one capsular type may increase carriage of a second type more than it decreases carriage of the first (29). Other possible long-term limitation to the use of conjugate vaccines is the well-known capacity of S. pneumoniae to change its capsular type via in vivo genetic transformation under laboratory conditions (15, 39) as well as in nature (10). Given all of these grounds, it is clear that an understanding of the molecular organization of the genes involved in capsular polysaccharide biosynthesis may facilitate the identification of potential targets and the design of capsule biosynthesis inhibitors. In this context, we have recently proposed that GalU, a pneumococcal UTP:glucose-1-phosphate uridylyltransferase that catalyzes the formation of UDP-glucose (UDP-Glc), represents one of such targets, since UDP-Glc is a key component in the biosynthetic pathway of pneumococcal capsular polysaccharides containing Glc, galactose (Gal), and/or glucuronic acid (GlcA) or galacturonic acid (32).
At present, nucleotide sequence data are available only for a limited number of types, namely, 1, 2, 3, 14, 19F, 19A, 19B, 23F, and 33F (4, 20, 24, 30, 33–36, 38, 40). Type 3 is the only case where the complete biosynthetic pathway for the formation of a capsular polysaccharide has been determined. Only two type 3-specific genes (cap3A and cap3B) are required for capsular biosynthesis (4). Cap3A is a UDP-Glc dehydrogenase (UDP-GlcDH) that converts UDP-Glc into UDP-GlcA (3, 5), whereas cap3B encodes a processive polysaccharide synthase (6). It has long been recognized that type 3 and type 8 polysaccharides cross-react (16–18). Structural analyses of both polysaccharides revealed that they have similar compositions; that is, one half of the type 8 polysaccharide consists of cellobiuronic acid (21), the repeating unit of type 3 capsule (41) (Fig. 1A). Type 8 polysaccharide is included in the 23-valent vaccine (42), and pneumococci of this serotype ranked ninth among sterile-site isolates (44). Furthermore, the relative risk of disease with type 8 pneumococcal strains increases at middle age. We describe here the complete nucleotide sequence of the cap8 locus of a type 8 clinical isolate of S. pneumoniae.
FIG. 1.
(A) Structure of the repeating unit of the type 8 pneumococcal capsular polysaccharide (21). The corresponding structure of the type 3 capsule (41) is shown for comparison. (B) Genetic organization of the S. pneumoniae 6028/95 strain containing the cap8 genes. The capsular gene clusters of types 1 (38), 2 (20), 3 (4), and 33F (30) are compared with type 8 in the text and are also included here. Small arrows correspond to interrupted genes. White and dashed arrows indicate capsular genes and putative insertion sequence elements, respectively. Capsular genes showing more than 90% identical nucleotides in comparisons of sequences of different serotypes are represented by identical shading. The locations of promoters (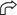 ) and putative transcription terminators (☻) are indicated.
) and putative transcription terminators (☻) are indicated.
The type 8 clinical strain of S. pneumoniae 6028, isolated in a Spanish hospital in 1995 from the blood of a patient, was kindly provided by A. Fenoll (Pneumococcus Reference Laboratory, Instituto de Salud Carlos III, Majadahonda, Spain). The unencapsulated (S3−) mutant strain M24 (cap3A) (5, 12) was used as the recipient for intertype transformation. All of the experimental procedures used in this work have been recently described (30). These include conditions for growth, transformation, and serotyping of S. pneumoniae, DNA isolation and amplification by PCR, determination of the nucleotide sequence, and computer analysis of DNA and protein sequences. To amplify long fragments of DNA we used the oligonucleotide primers DEX86 (5′-TTGAACATATCGGTCTTCAGTATCAGGAAGGTCAGCC-3′) and D41 (5′-ACTGCCGCGTATTCTTCACC-3′), which are located, respectively, in the 5′ region of dexB (30) and in the complementary strand of the sequence (EMBL database accession no. Z83335) corresponding to the aliA gene in type 1 DNA (38).
According to results obtained previously for other serotypes (13, 14), the gene cluster (cap) responsible for pneumococcal capsular synthesis is located between dexB and aliA, two genes that do not participate in capsule formation. Consequently, we amplified DNA prepared from strain 6028 using oligonucleotide primers DEX86 and D41, and a DNA fragment of about 15 kb was obtained. When this fragment was used to transform the S3− pneumococcal strain M24, encapsulated type 8 transformants were found (data not shown), confirming that we were dealing with the locus responsible for type 8 capsular polysaccharide biosynthesis. Combined PCR and restriction mapping of the DNA prepared from one of these type transformants revealed the integration of the 12 type 8 genes into the type 3 recipient DNA (unpublished data). Sequence analysis of the amplified DNA fragment (14,730 bp) showed the presence of 12 complete and 2 partial open reading frames (ORFs) (Fig. 1B). The two incomplete ORFs correspond to the genes dexB and aliA, as expected. The whole organization of the cap8 locus closely matches that of cap (cps) loci previously studied (13, 14). However, significant differences were found when cap8 and cap3 gene clusters were compared (Fig. 1B), i.e., (i) between dexB and the cap8 promoter (cap8p) there is a 351-bp region that is absent in type 3 DNA; this sequence might correspond to an insertion sequence-like element on the basis of sequence similarities (20); (ii) the cap8p region is intact and presumably functional (30), whereas in type 3 DNA, the −10 box of the promoter is completely missing (4); and (iii) the ORFs cap8A through cap8D are neither deleted nor mutated, in contrast to the situation reported for type 3 DNA.
The fifth ORF of the cluster (cap8E) putatively codes for an undecaprenyl-phosphate Glc-1-phosphate transferase since the product of this gene is 84% similar to Cps14E, one of the few capsular gene products of S. pneumoniae that have been biochemically characterized (23). The highest similarities (>90% identity) were found when Cap8E was compared with Cps2E (20), Cps19aE (34), Cps23fE (33, 40), or Cap33fE (30) (Table 1). The genes cap8G and cap8H appear according to sequence similarities to encode glycosyltransferases (Table 1). Cap8F and Cap8G are similar to Cps14F and Cps14G, respectively. Cps14G is a β-1,4-Gal transferase responsible for the second step in the subunit synthesis of pneumococcal capsular polysaccharide type 14, that is, the transfer of Gal to lipid-linked Glc (25). The function of Cps14F is not clear, although the reduced glycosyltransferase activity detected in Escherichia coli transformants expressing cps14E and cps14G but lacking cps14F suggested that Cps14F has an enhancing role in glycosyltransferase activity (25). Cap8F and Cap8G also have sequence similarity with Orf5 and Orf6, two proteins involved in the synthesis of K40 capsular polysaccharide of E. coli that have been suggested to represent, respectively, the N- and C-terminal domains of a β-GlcA transferase (2). Moreover, it has been proposed that Orf6 encodes the catalytic domain of the enzyme since it contains a conserved amino acid (aa) sequence, termed domain A, and two separated catalytic Asp residues characteristic of β-glycosyltransferase (43). These features are also present in Cap8G, suggesting that this protein might be a β-GlcA transferase.
TABLE 1.
Type 8 capsular locus-encoded proteins: properties and similarities to proteins in the databases
| Type 8 gene | Location in nucleotide sequence | Predicted protein (no. of aa/no. of kDa) | GRAVYa score | Proposed function or product | Similar polypeptide(s) (no. of aa) | Database accession no. | Degree of identity (%)/degree of similarity (%) | Organism |
|---|---|---|---|---|---|---|---|---|
| cap8E | 5782–7149 | 455/52.0 | 0.159 | Undecaprenyl-phosphate glucose-1-phosphate transferase | Cap33fE (455) | O86888 | 96/97 | S. pneumoniae type 33F |
| Cps2E (455) | AAD10174 | 95/96 | S. pneumoniae type 2 | |||||
| Cps23fE (455) | AAC69528 | 94/95 | S. pneumoniae type 23F | |||||
| Cps19aE (453) | AAC78667 | 92/95 | S. pneumoniae type 19A | |||||
| cap8F | 7129–7633 | 165/19.5 | 0.002 | Unknown | Orf5 (161) | O30630 | 37/58 | E. coli |
| Cps14F (149) | P72514 | 35/56 | S. pneumoniae type 14 | |||||
| cap8G | 7634–8113 | 159/18.2 | −0.053 | β-GlcA transferase | Cps14G (167) | P72515 | 34/60 | S. pneumoniae type 14 |
| Orf6 (153) | O30631 | 33/52 | E. coli | |||||
| cap8H | 8126–9190 | 354/40.0 | −0.114 | Galactosyltransferase | Cap33fG (346) | O86890 | 27/51 | S. pneumoniae type 33F |
| PH1844 (381) | O59512 | 30/52 | Pyrococcus horikoshii | |||||
| cap8I | 9190–10650 | 486/55.3 | 0.818 | Oligosaccharide repeat unit transporter | Cps19bJ (481) | O07870 | 33/56 | S. pneumoniae type 19B |
| Cps14L (487) | O07342 | 26/43 | S. pneumoniae type 14 | |||||
| Cap33fL (471) | O86895 | 25/52 | S. pneumoniae type 33F | |||||
| cap8J | 10663–11391 | 242/28.4 | −0.381 | Unknown | C17G8.11C (356) | Q10323 | 35/49b | Schizosaccharomyces pombe |
| Cps14K (282) | O07341 | 32/50b | S. pneumoniae type 14 | |||||
| cap8K | 11375–12538 | 387/46.0 | 0.822 | Polysaccharide polymerase | Cps23fG (397) | AAC69530 | 21/41 | S. pneumoniae type 23F |
| cap8L | 12607–13845 | 412/46.5 | −0.250 | UDP-GlcDH | Cps2K (412) | AAD10180 | 96/98 | S. pneumoniae type 2 |
| Cap1K (410) | P96482 | 78/90 | S. pneumoniae type 1 | |||||
| Cap3A (394) | Q57346 | 57/74 | S. pneumoniae type 3 |
Grand average of hydropathy (26).
Sequence similarity is restricted to the N-terminal part of the protein (aa positions 1 to 100).
Cap8I is predicted to be a 55.3-kDa protein having 12 membrane-spanning regions. This finding together with sequence similarities (Table 1) strongly suggests that this protein is involved in transport of the type 8 repeating unit. Cap8K also appears to be an integral membrane protein because nine transmembrane segments can be predicted. Moreover, sequence comparison (Table 1) suggests that Cap8K may be the type 8 polysaccharide polymerase. The cap8J gene putatively codes for a 28.4-kDa protein of unknown function. A similar putative protein has been found in the capsular gene cluster of type 14 pneumococci (25).
Cap8L is a 412-aa UDP-GlcDH. Interestingly, the cap8L gene is 92% identical to the cps2K gene, which encodes the type 2 UDP-GlcDH. This was unexpected since, to date, the pneumococcal genes encoding this type of enzyme, namely, cap3A (5), cap1K (38), and cps2K (20), were noticeably divergent. Actually, the evolutionary distance between cap1K and cap3A is similar to that found between any of these pneumococcal genes and the hasB gene, which encodes a UDP-GlcDH of Streptococcus pyogenes (37).
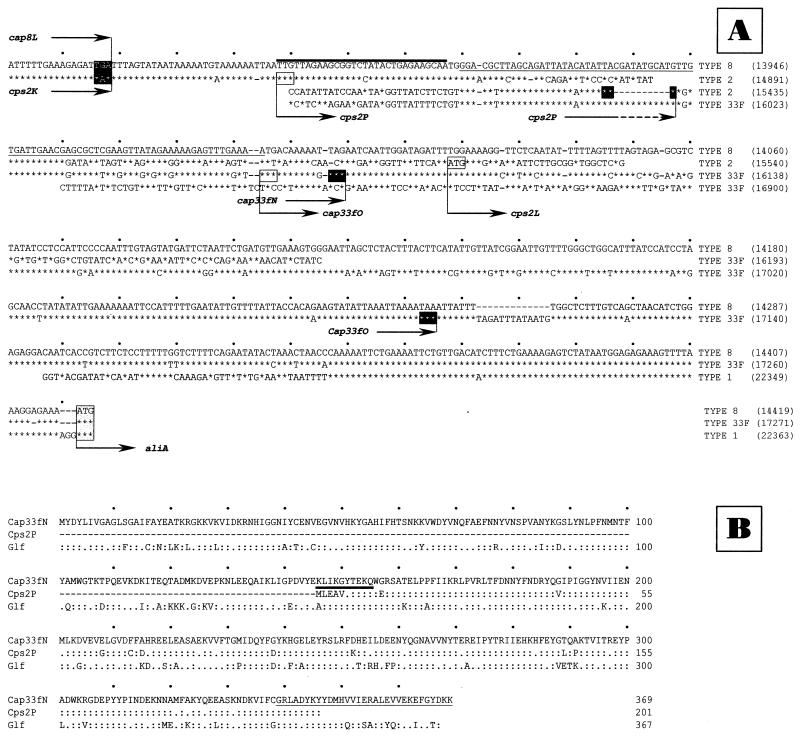
The 573-bp-long cap8L-aliA intergenic region is one of the shortest segments found so far in this flanking region of the capsular gene clusters of S. pneumoniae (Fig. 2A). It is noteworthy that the similarity between cap8L and cps2K extends beyond the corresponding termination codons as far as the proposed 5′ end of the following gene of the cps2 gene cluster (cps2P) (20). The proposal that Cps2P, suggested to be a UDP-galactopyranose (UDP-Galp) mutase, participates in the biosynthesis of type 2 capsule is controversial since Galf is not a structural component of this polysaccharide (45). In fact, a close examination of the nucleotide sequences of types 2, 33F, and 8 capsular gene clusters showed that cps2P is most probably a deleted version of an ancestral gene. This was particularly evident when Cps2P (201 aa residues), Cap33fN (369 aa residues) (30), and the E. coli UDP-Galp mutase Glf (367 aa residues) (27) were aligned (Fig. 2B). Csp2P apparently lacks the 151 N-terminal as well as the 23 C-terminal aa residues found in the other two proteins examined. Thus, the concept that cps2P starts with a TTG codon (20) cannot be supported. The corresponding region in type 8 pneumococci contains remnants of a gene coding for the 10 N-terminal aa residues of Cps2P (Fig. 2A and B) and the 32 C-terminal residues of Cap33fN (Fig. 2A and B). Figure 2A also shows that cps2P has undergone a 10-bp deletion that caused the appearance of a premature TAG termination codon and that the nucleotide sequence between positions 13906 and 14050 is very similar to the region containing the 3′ and 5′ ends of genes cap33fN and cap33fO, respectively (Fig. 1B). Further downstream (from position 14060 to 14255) the sequence was very similar to that of the 3′ end of the gene cap33fO, whereas from position 14344 to the ATG initiation codon of aliA, the type 8 sequence is almost identical to that of pneumococcal types 1, 2, 19F, 19A, and 23F, although only the sequence corresponding to type 1 is shown in Fig. 2A.
FIG. 2.
(A) Multiple sequence alignment of the region located between the 3′ end of the cap8 gene cluster and the aliA gene. Asterisks indicate identical nucleotides, and hyphens indicate gaps introduced to maximize similarity. Gray and black boxes indicate initiation and termination codons, respectively. Numbers at the right of each line correspond to the nucleotide positions of the sequences included in the data banks. (B) Multiple sequence alignment of the cap33fN, cps2P, and glf gene products. Colons and dots indicate identical and conserved amino acid residues, respectively. The amino acid sequences highlighted with either a thick line or underlining correspond to the translation of the corresponding sequence shown in panel A. Numbers on the right indicate amino acid positions.
Pneumococcal polysaccharides of types 2, 19F, 19A, and 23F contain rhamnose (Rha) as a structural component of their capsular polysaccharides (45). On the other hand, it is noticeable that the four genes involved in the synthesis of dTDP-Rha were always found to be located at the right end of the capsular gene cluster (13). Interestingly, these four genes were also found to be located downstream of the cap1 gene cluster in up to 19 different type 1 pneumococcal isolates examined (38) (indicated as orf1 through orf4 in Fig. 1B), although Rha is not a component of the type 1 polysaccharide. It has been suggested that the ancestor of the type 1 strains of S. pneumoniae might have been a strain of a Rha-containing serotype which, upon natural transformation with DNA from an unknown origin, retained the recipient dTDP-Rha-encoding genes, possibly as a consequence of an abnormal transformation event (38). In recent years, transformation of pneumococcal types has been documented as the underlying mechanism of capsular shifting in various clinical isolates (7, 9, 22, 31). Recently, the multiresistant Spanish clone 23F (MMSp23F) was shown to be transformed, on at least four separate occasions, to type 19F through recombination events that usually took place upstream of dexB and downstream of aliA. In two cases, however, the recombinational crossover point was identified within the genes involved in the synthesis of dTDP-Rha (10). Such transformation events can also be recognized by noting the presence of remnants, as in type 1 pneumococci, where an IS1167-like element is located between the 3′ end of the cap1 cluster and the four genes originally involved in dTDP-Rha biosynthesis (38) (Fig. 1B). It is conceivable that the cps2P gene of type 2 pneumococci and part of the region located downstream of cap8L might also represent, respectively, remnants of the ancestors of type 2 and type 8 S. pneumoniae isolates. In a putative scenario, a type 33F pneumococcus might have been the parent of the contemporary type 8 strains whereas a strain having both Galf and Rha in its capsule might have generated type 2 pneumococcal isolates. Both of these sugars are in fact components of the capsular polysaccharides of several pneumococcal types, i.e., 17A, 22F, and 31 (45). It is interesting that these capsules also contain GlcA in addition to Galf and Rha.
The flexibility of pneumococci in exchanging capsular types is important since serious pneumococcal diseases can be caused by isolates expressing a considerable number of the 90 different capsular polysaccharides (44). The conserved organization of the cap8 locus with respect to the cap loci previously reported undoubtedly facilitates switching of cap8 to other capsular gene clusters. In this sense, the results reported here add most-needed information on genetic variability of the genes governing capsule formation in types that are potential candidates for the conjugate vaccine formulations currently under experimentation. Frequent recombinational events occurring at the capsular polysaccharide biosynthesis loci facilitate clonal variability and should represent a noticeable limitation for a worldwide use of these newly developed vaccines. In trials of pneumococcal vaccines type replacements which may increase the incidence of disease from serotypes not included in the vaccine have already occurred (28). A detailed molecular knowledge of the different pneumococcal types might provide insights in future approaches to controlling this variability by selecting the most appropriate strains to diminish serotype replacement.
Nucleotide sequence accession number.
The nucleotide sequence of the cap8 locus has been deposited in the EMBL, GenBank, and DDBJ databases under nucleotide sequence accession no. AJ239004.
Acknowledgments
This work was supported by grant PB96-0809 from the Dirección General de Investigación Cientifica y Técnica and the Programa de Cooperación Cientifica con Iberoamérica from the Subdirección General de Cooperación Internacional. R. Muñoz was the recipient of a Contrato Temporal de Investigadores from the CSIC.
REFERENCES
- 1.AlonsoDeVelasco E, Verheul A F M, Verhoef J, Snippe H. Streptococcus pneumoniae: virulence factors, pathogenesis, and vaccines. Microbiol Rev. 1995;59:591–603. doi: 10.1128/mr.59.4.591-603.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amor P A, Whitfield C. Molecular and functional analysis of genes required for expression of group IB K antigens in Escherichia coli: characterization of the his-region containing gene clusters for multiple cell-surface polysaccharides. Mol Microbiol. 1997;26:145–161. doi: 10.1046/j.1365-2958.1997.5631930.x. [DOI] [PubMed] [Google Scholar]
- 3.Arrecubieta C, García E, López R. Demonstration of UDP-glucose dehydrogenase activity in cell extracts of Escherichia coli expressing the pneumococcal cap3A gene required for the synthesis of type 3 capsular polysaccharide. J Bacteriol. 1996;178:2971–2974. doi: 10.1128/jb.178.10.2971-2974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrecubieta C, García E, López R. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniae type 3. Gene. 1995;167:1–7. doi: 10.1016/0378-1119(95)00657-5. [DOI] [PubMed] [Google Scholar]
- 5.Arrecubieta C, López R, García E. Molecular characterization of cap3A, a gene from the operon required for the synthesis of the capsule of Streptococcus pneumoniae type 3: sequencing of mutations responsible for the unencapsulated phenotype and localization of the capsular cluster on the pneumococcal chromosome. J Bacteriol. 1994;176:6375–6383. doi: 10.1128/jb.176.20.6375-6383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arrecubieta C, López R, García E. Type 3-specific synthase of Streptococcus pneumoniae (Cap3B) directs type 3 polysaccharide biosynthesis in Escherichia coli and in pneumococcal strains of different serotypes. J Exp Med. 1996;184:449–455. doi: 10.1084/jem.184.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnes D M, Whittier S, Gilligan P H, Soares S, Tomasz A, Henderson F W. Transmission of multidrug-resistant serotype 23F Streptococcus pneumoniae in group day care: evidence suggesting capsular transformation of the resistant strain in vivo. J Infect Dis. 1995;171:890–896. doi: 10.1093/infdis/171.4.890. [DOI] [PubMed] [Google Scholar]
- 8.Briles D E, Tart R C, Swiatlo E, Dillard J P, Smith P, Benton K A, Ralph B A, Brooks-Walter A, Crain M J, Hollingshead S K, McDaniel L S. Pneumococcal diversity: considerations for new vaccine strategies with emphasis on pneumococcal surface protein A (PspA) Clin Microbiol Rev. 1998;11:645–657. doi: 10.1128/cmr.11.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 10.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 11.Feldman C, Klugman K P. Pneumococcal infections. Curr Opin Infect Dis. 1997;10:109–115. [Google Scholar]
- 12.García E, García P, López R. Cloning and sequencing of a gene involved in the synthesis of the capsular polysaccharide of Streptococcus pneumoniae type 3. Mol Gen Genet. 1993;239:188–195. doi: 10.1007/BF00281617. [DOI] [PubMed] [Google Scholar]
- 13.García E, Llull D, López R. Functional organization of the gene cluster involved in the synthesis of the pneumococcal capsule. Int Microbiol. 1999;2:169–176. [PubMed] [Google Scholar]
- 14.García E, López R. Molecular biology of the capsular genes of Streptococcus pneumoniae. FEMS Microbiol Lett. 1997;149:1–10. doi: 10.1111/j.1574-6968.1997.tb10300.x. [DOI] [PubMed] [Google Scholar]
- 15.Griffith F. The significance of pneumococcal types. J Hyg. 1928;27:113–159. doi: 10.1017/s0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heidelberger M. Precipitating cross-reactions among pneumococcal types. Infect Immun. 1983;41:1234–1244. doi: 10.1128/iai.41.3.1234-1244.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heidelberger M, Kabat E A, Mayer M M. Further study of the cross-reaction between the specific polysaccharides of types III and VIII pneumococci in horse antisera. J Exp Med. 1942;75:35–47. doi: 10.1084/jem.75.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heidelberger M, Kabat E A, Shrivastava D L. Quantitative study of the cross reaction of type III and VIII pneumococci in horse and rabbit antisera. J Exp Med. 1937;65:487–496. doi: 10.1084/jem.65.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henrichsen J. Six newly recognized types of Streptococcus pneumoniae. J Clin Microbiol. 1995;33:2759–2762. doi: 10.1128/jcm.33.10.2759-2762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iannelli F, Pearce B J, Pozzi G. The type 2 capsule locus of Streptococcus pneumoniae. J Bacteriol. 1999;181:2652–2654. doi: 10.1128/jb.181.8.2652-2654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones J K N, Perry M B. The structure of the type VIII pneumococcus specific polysaccharide. J Am Chem Soc. 1957;79:2787–2793. [Google Scholar]
- 22.Kell C M, Jordens J Z, Daniels M, Coffey T J, Bates J, Paul J, Gilks C, Spratt B G. Molecular epidemiology of penicillin-resistant pneumococci isolated in Nairobi, Kenya. Infect Immun. 1993;61:4382–4391. doi: 10.1128/iai.61.10.4382-4391.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolkman M A B, Morrison D A, van der Zeijst B A, Nuijten P J M. The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: identification of the glycosyl transferase gene cps14E. J Bacteriol. 1996;178:3736–3741. doi: 10.1128/jb.178.13.3736-3741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolkman M A, Wakarchuk W, Nuijten P J, van der Zeijst B A. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 25.Kolkman M A B, van der Zeijst B, Nuitjen P J M. Functional analysis of glycosyltransferases encoded by the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae serotype 14. J Biol Chem. 1997;272:19502–19508. doi: 10.1074/jbc.272.31.19502. [DOI] [PubMed] [Google Scholar]
- 26.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 27.Lee R, Monsey D, Weston A, Duncan K, Rithner C, McNeil M. Enzymatic synthesis of UDP-galactofuranose and assay for UDP-galactopyranose mutase based on high-performance liquid chromatography. Anal Biochem. 1996;242:1–7. doi: 10.1006/abio.1996.0419. [DOI] [PubMed] [Google Scholar]
- 28.Lipsitch M. Bacterial vaccines and serotype replacement: lessons from Haemophilus influenzae and prospects for Streptococcus pneumoniae. Emerg Infect Dis. 1999;5:336–345. doi: 10.3201/eid0503.990304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipsitch M. Vaccination against colonizing bacteria with multiple serotypes. Proc Natl Acad Sci USA. 1997;94:6571–6576. doi: 10.1073/pnas.94.12.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Llull D, López R, García E, Muñoz R. Molecular structure of the gene cluster responsible for the synthesis of the polysaccharide capsule of Streptococcus pneumoniae type 33F. Biochim Biophys Acta. 1998;1443:217–224. doi: 10.1016/s0167-4781(98)00213-9. [DOI] [PubMed] [Google Scholar]
- 31.Marchese A, Ramirez M, Schito G C, Tomasz A. Molecular epidemiology of penicillin-resistant Streptococcus pneumoniae isolates recovered in Italy from 1993 to 1996. J Clin Microbiol. 1998;36:2944–2949. doi: 10.1128/jcm.36.10.2944-2949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mollerach M, López R, García E. Characterization of the galU gene of Streptococcus pneumoniae encoding a uridine diphosphoglucose pyrophosphorylase: a gene essential for capsular polysaccharide biosynthesis. J Exp Med. 1998;188:2047–2056. doi: 10.1084/jem.188.11.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morona J K, Miller D C, Coffey T J, Vindurampulle C J, Spratt B G, Morona R, Paton J C. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniae type 23F. Microbiology. 1999;145:781–789. doi: 10.1099/13500872-145-4-781. [DOI] [PubMed] [Google Scholar]
- 34.Morona J K, Morona R, Paton J C. Analysis of the 5′ portion of the type 19A capsule locus identifies two classes of cpsC, cpsD, and cpsE genes in Streptococcus pneumoniae. J Bacteriol. 1999;181:3599–3605. doi: 10.1128/jb.181.11.3599-3605.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morona J K, Morona R, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 36.Morona J K, Morona R, Paton J C. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniae type 19B. J Bacteriol. 1997;179:4953–4958. doi: 10.1128/jb.179.15.4953-4958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mũnoz R, García E, López R. Evidence for horizontal transfer from Streptococcus to Escherichia coli of the kfiD gene encoding the K5-specific UDP-glucose dehydrogenase. J Mol Evol. 1998;46:324–326. doi: 10.1007/pl00006322. [DOI] [PubMed] [Google Scholar]
- 38.Mũnoz R, Mollerach M, López R, García E. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol Microbiol. 1997;25:79–92. doi: 10.1046/j.1365-2958.1997.4341801.x. [DOI] [PubMed] [Google Scholar]
- 39.Ottolenghi-Nightingale E. Competence of pneumococcal isolates and bacterial transformation in man. Infect Immun. 1972;6:785–792. doi: 10.1128/iai.6.5.785-792.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez M, Tomasz A. Molecular characterization of the complete 23F capsular polysaccharide locus of Streptococcus pneumoniae. J Bacteriol. 1998;180:5273–5278. doi: 10.1128/jb.180.19.5273-5278.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reeves R E, Goebel W F. Chemoimmunological studies on the soluble specific substance of pneumococcus. V. The structure of the type III polysaccharide. J Biol Chem. 1941;139:511–519. [Google Scholar]
- 42.Robbins J B, Austrian R, Lee C J, Rastogi S C, Schiffman G, Henrichsen J, Mäkelä P H, Broome C V, Facklam R R, Tiesjema R H, Parke J C J. Considerations for formulating the second-generation pneumococcal capsular polysaccharide vaccine with emphasis on the cross-reactive types within groups. J Infect Dis. 1983;148:1136–1159. doi: 10.1093/infdis/148.6.1136. [DOI] [PubMed] [Google Scholar]
- 43.Saxena I M, Brown R M, Fevre M, Geremia R A, Henrissat N. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott J A, Hall A J, Dagan R, Dixon J M, Eykyn S J, Fenoll A, Hortal M, Jetté L P, Jorgensen J H, Lamothe F, Latorre C, Macfarlane J T, Shlaes D M, Smart L E, Taunay A. Serogroup-specific epidemiology of Streptococcus pneumoniae: associations with age, sex, and geography in 7,000 episodes of invasive disease. Clin Infect Dis. 1996;22:973–981. doi: 10.1093/clinids/22.6.973. [DOI] [PubMed] [Google Scholar]
- 45.van Dam J E, Fleer A, Snippe H. Immunogenicity and immunochemistry of Streptococcus pneumoniae capsular polysaccharides. Antonie Leeuwenhoek. 1990;58:1–47. doi: 10.1007/BF02388078. [DOI] [PubMed] [Google Scholar]