Abstract
We analyzed the contribution of structural variants (SVs) to gliomagenesis across 179 pediatric high-grade gliomas (pHGGs). The most recurrent SVs targeted MYC isoforms and receptor tyrosine kinases, including a SV amplifying a MYC enhancer in 12% of diffuse midline gliomas (DMG), revealing an underappreciated role for MYC in pHGG. SV signature analysis revealed that tumors with simple signatures were TP53WT but showed alterations in TP53 pathway members PPM1D and MDM4. Complex signatures were associated with direct aberrations in TP53, CDKN2A, and RB1 early in tumor evolution, and with later occurring extrachromosomal amplicons. All pHGGs exhibited at least one simple SV signature but complex SV signatures were primarily restricted to subsets of H3.3K27M DMGs and hemispheric pHGGs. Importantly, DMGs with complex SV signatures showed shorter overall survival independent of histone mutation and TP53 status. These data inform the impact of SVs in gliomagenesis and mechanisms that shape them.
Introduction
Pediatric high-grade gliomas (pHGGs), encompassing diffuse midline gliomas (DMGs) and hemispheric tumors, represent the most common cause of cancer-related deaths in children age 0-14 years1. Targeted sequencing, including exome sequencing, has revealed recurrent single nucleotide variants (SNVs) in histones including H3.1K27M, H3.3K27M, and H3.3G34R, co-occurring with alterations in the TP53 pathway and receptor tyrosine kinase genes (RTKs)2-9. However, the role of structural variants (SVs) remains underexplored.
SVs represent connections, or rearrangements, between distant genomic loci. They underlie all somatic copy-number alterations (SCNAs) except whole-chromosome gains and losses, thereby altering more of the genome than any other genetic alteration. A single SV can affect dozens to hundreds of genes. In some cases, SVs generate extrachromosomal amplifications (also known as double minutes) that can encode hundreds of copies of an oncogene per cell10-14. The effects of SVs on cellular fitness often result from changes in chromatin structure such as disruption of topologically associated domains (TADs) and gene-enhancer interactions15-20. Therefore, unlike SNVs, the SVs with the largest effects on selection are often outside of the exome and require whole-genome sequencing (WGS) for their characterization. Moreover, these distant effects raise challenges to interpreting the consequences of individual SVs.
Both the frequency with which SVs recur at individual loci, and the mechanisms by which they are generated, can vary widely across cancers. SV signatures can indicate their formation mechanisms21-25, and recent efforts have begun to characterize them in breast22 and other cancer types23,26. However, unlike SNVs, whose signatures have been characterized across tens of thousands of exomes, the relationships between currently described SV signatures across cancer types remain underexplored. For example, high rates of tandem duplications associated with deficiencies in homologous recombination (HR) only in tumors with very high SV burdens24. It remains unclear if these associations translate to other tumor types, including pHGGs, and which other SV signatures or associated variant-generating processes exist.
The differences across lineages indicate the role of epigenetics in shaping the SVs that are observed in cancer21,23. Mutations in core histones in pHGGs8,9 highlight the role of epigenetic dysregulation in these tumors. pHGGs therefore offer a unique perspective on the relationships between patterns of SVs and different alterations in chromatin. Associations between patterns of SVs and other molecular and clinical characteristics of these tumors are also largely unknown.
Historically, the characterization of DMGs lacked pre-treatment tissue due to the risks in biopsies of midline brain structures30-33. A concern with post-treatment samples is that treatment—often involving ionizing radiation—might alter the SV patterns in these tumors. We leveraged samples from the first multi-institutional North American clinical trials to incorporate biopsies of DMGs33 and added published4-7 pre- and post-treatment samples to assemble the largest pHGG WGS cohort to date. We identified recurrent driver events, stratified pHGGs based upon mechanistically informative SV signatures, and detected genetic events and differences in clinical outcome associated with these signatures.
Results
Significantly recurrent SVs
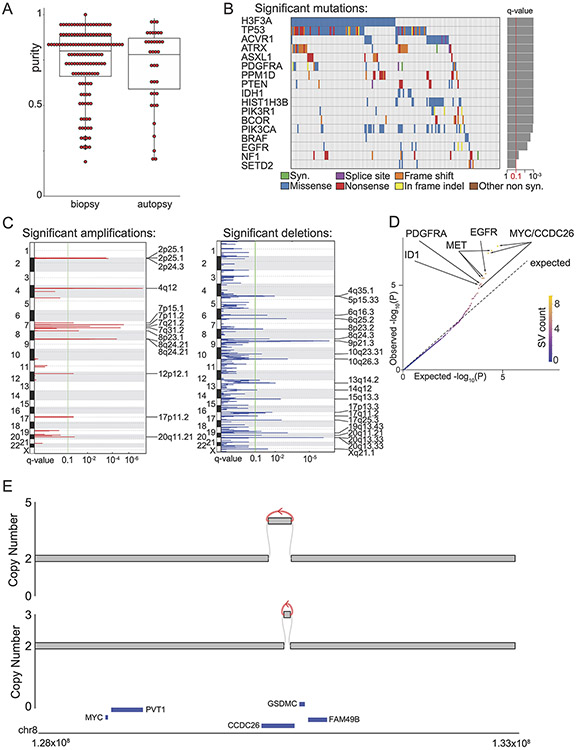
We assembled pHGG WGS from 179 children including 61 hemispheric tumors and 118 DMGs. Of these, 61 were sequenced de-novo for this study. The other 118 samples include 18 from Buczkowicz et al 2014 5, 20 from Taylor et al 2014 7, 30 from Wu et al 2014 6, and 50 from Bender et al 2016 4 (Supp. Table 1). All sequences were subject to a single uniform computational pipeline. Among the DMGs, 84 (71%) were from pre-treatment biopsies including 33 obtained from the first multi-institutional North American clinical trial to incorporate diagnostic biopsies 33. The tumor purity of the pre-treatment biopsies was comparable to autopsy samples (median: 0.8 vs. 0.78, p = 0.5) (Ext. Data Fig. 1A).
We identified significantly recurring somatic copy-number alterations (SCNAs) using GISTIC34, and recurrent SNVs using MutSig2CV. Both analyses largely agreed with prior studies3-7 (Supp. Table 2), including high frequencies of H3.3K27M (50%) or H3.1K27M (12%) mutations reflecting the enrichment for DMGs in this study, and other known drivers of pHGG (Ext. Data Fig. 1B).
The most notable finding was a recurrent amplification in 8q24.21, 2 MB telomeric to MYC. This amplicon, likely not detected in prior array- and exome-based studies because it lies outside of the exome3, was present in 28 tumors (16% of the cohort). All but one of these tumors were DMGs, a significant enrichment (p = 0.0016). Most of these amplicons excluded MYC itself (Ext. Data Fig. 1C). A non-overlapping peak was also detected that did encompass MYC, due to two tumors with extrachromosomal MYC amplicons.
We also found that several regions were recurrently amplified together to high levels, such as 2p25.1 and the MYCN locus at 2p24.3. This pattern of correlated SCNAs in distinct genomic loci suggests underlying recurrent SVs. We therefore comprehensively catalogued SVs using an assembly-based method35 with improved detection of complex and short SVs compared to standard alignment-based methods. We detected 15,485 SVs (Supp. Table 3), averaging 87 per tumor, including 1482 (10%) that were 10-300 bp in span, a “blind spot” in prior analyses35.
To distinguish recurrent SVs, we took two approaches based upon methods that we recently developed21. In the first, we conducted a “1D” analysis that identified genomic loci with more SV breakpoints than expected (“significantly recurrent breakpoints”, or SRBs; Supp. Table 4A). This analysis splits the whole genome into 50kbp bins and compares the observed number of SVs in each bin to a background distribution that considers sequence, epigenetic, and other features of each locus (all results in Supp. Table 4B). In the second, we conducted a “2D” analysis to detect pairs of loci that are recurrently juxtaposed by SVs (“significantly recurrent juxtapositions”, or SRJs). This analysis evaluates the rate at which pairs of bins are connected by SVs to a background distribution that reflects the rates at which each bin suffers breakpoints and the genomic distance between them. The bins and background model in this analysis were determined in a prior pan-cancer study22; the 2D bin median length was 467kbp. In both analyses, bins with q-values < 0.1 were considered significant.
We identified 10 SRB bins across five TADs (Supp. Table 4A-B, Fig. 1A, and Ext. Data Fig. 1D), and two SRJs (Fig. 2A). The most significant SRB was within the MYC TAD, encompassing breakpoints in 28 tumors—more than any other TAD in the genome. This locus was also a component of an SRJ connecting two adjacent bins at the telomeric end of the MYC TAD. The remaining SRBs corresponded to SVs within the TADs of the receptor tyrosine kinase genes (RTKs) MET (q=0.0025), EGFR (q=0.029), and PDGFRA (q=0.032), as well as an SV within the TAD of the transcription factor ID2 (Supp. Table 4A). This latter SRB was also a component of the second SRJ, which connected ID2 and MYCN.
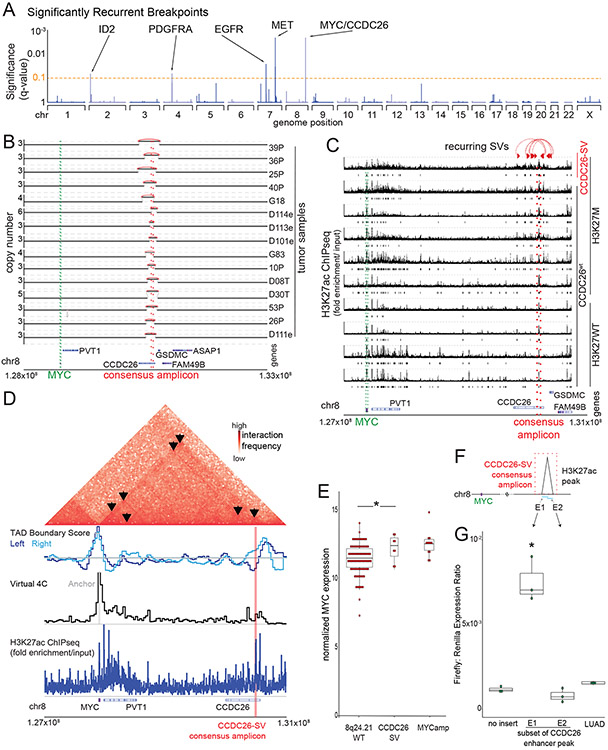
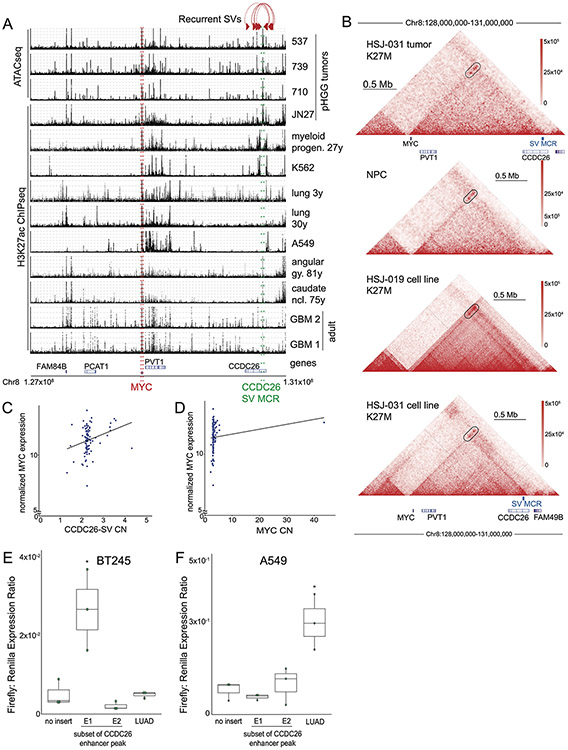
Figure 1. Significantly recurrent breakpoints within CCDC26.
(A) Significance (multiple hypothesis corrected q-values, vertical axis) of recurrent breakpoints (genomic positions on the horizontal axis) across the 179 pHGG genomes. (B) Copy-number profiles across the MYC TAD for the 15 tumors with the recurrent CCDC26 SV. (C) H3K27ac ChIP-seq tracks within the TAD containing MYC (green lines) showing an H3K27ac peak at the location of CCDC26-SV in six H3K27M and four H3K27WT pHGGs tumors. Only the top H3K27ac enrichment track originates from a tumor with a CCDC26-SV. Significantly enriched peaks (q-value < 0.01) are indicated below each H3K27ac ChIP-seq track. The CCDC26 amplicon boundaries for individual samples are indicated by the paired red arrows at the top. The consensus amplicon is indicated by the red dotted lines, and centers on an H3K27ac peak. (D) Hi-C heatmap across the MYC-CCDC26 locus from a midline glioma with the CCDC26-SV. Increasing interaction frequencies are indicated by brighter shades of red. The black arrowheads indicate significant interaction loops. The track beneath the heatmap indicates RobusTAD left and right TAD boundary scores, which represent the likelihood that TAD boundaries are present. The third row contains a virtual 4C track, in which peaks indicate higher interaction frequencies with an anchor sequence in the MYC promoter, which is highlighted in grey. The fourth row shows H3K27ac ChIPseq data from the same sample indicating the location of the enhancer peak within the CCDC26-SV consensus amplicon. (E) Normalized MYC expression in DMG samples with wild-type copy-number profiles at 8q24.21 (n= 92 tumors), CCDC26-SVs (n=8 tumors) or amplifications of the MYC coding sequence (n=12 tumors). *denotes p = 0.04 as determined by two-sided Wilcoxon rank sum test. Center line of the boxplot indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5x inter-quartile range (IQR). (F) Schematic illustrating the luciferase reporter used to validate the enhancer in CCDC26, showing the positions of the E1 and E2 sequences with respect to the enhancer within CCDC26. (G) Luciferase activity in DIPG13 cells following transduction of the E1, E2, and LUAD enhancer reporters or empty vector controls. Values represent the average of four technical replicates in each of three independent experiments. *denotes p = 1.6x10−5 (E1 vs Backbone) and p = 0.89 (LUAD vs Backbone), n = 3 independent experiments, Nested One-Way Anova with Tukey’s post-test, boxplot defined as in E.
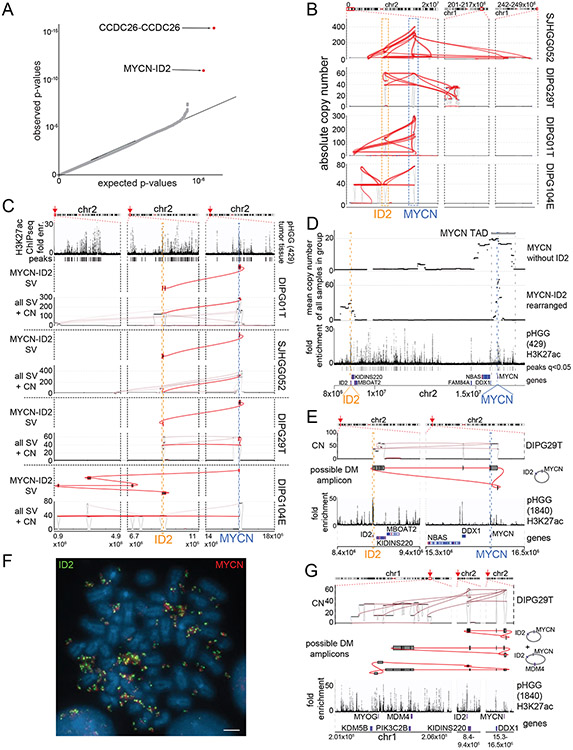
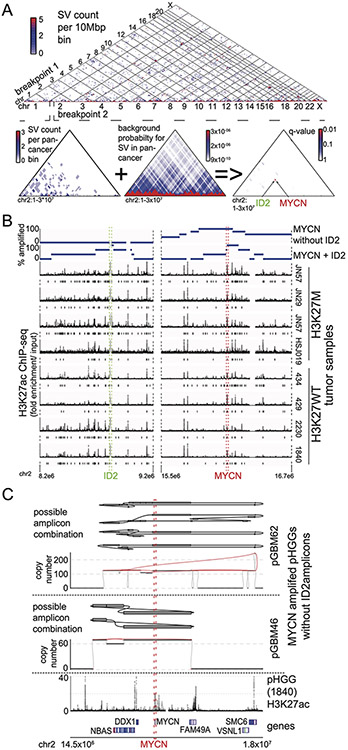
Figure 2. Significantly recurrent juxtapositions: MYCN-ID2.
(A) Quantile-Quantile plot indicating the significance of juxtapositions between pairs of genomic loci. SVs that reached statistical significance are depicted in red (based on n=179 tumors). B) SVs (red lines) involving MYCN and ID2 in samples with MYCN-ID2 rearrangements, and the number of copies at each connected locus (vertical axis). The dashed boxes indicate loci encompassing ID2 and MYCN. (C) SV maps as in panel B, with SVs juxtaposing ID2 and MYCN highlighted in red. The top track indicates H3K27ac marks in a pHGG without a known MYCN-ID2 rearrangement, showing a strong enhancer within ID2. In each case, the MYCN-ID2 juxtaposition reduces the somatic distance MYCN and ID2 (each indicated by a dashed line) from 7 Mbp to less than 700kbp. (D) MYCN amplicons in tumors without ID2 amplification (top track) incorporate a larger fraction of the MYCN TAD (mean of 60%, n = 4 tumors) relative to MYCN amplicons in tumors with MYCN-ID2-SV (second track; mean of 23%, n = 4 tumors). As a result, the former tend to include more loci with H3K27ac enrichment (third track) from the MYCN-TAD; significantly enriched H3K27ac peaks (q-value < 0.01) are indicated with black bars between the H3K27ac fold enrichment and the gene track. (E) Example of the simplest possible reconstruction of a circular extrachromosomal amplicon containing MYCN and ID2 from a single DMG. The top track shows copy-number and SVs, the middle track indicates the reconstructed topology, and the bottom track shows H3K27ac binding at the indicated loci in a different pHGG. (F) Metaphase FISH of a pHGG cell line with the MYCN-ID2 rearrangement showing its location on an extrachromosomal amplicon. The ID2 and MYCN locus-targeting probes are colored in green and red, respectively. Scale bar indicates 2μm. Representative image from n=20 metaphases. (G) The chr1 loci connected to the MYCN-ID complex in this DMG. Short-read reconstructions allow for several extrachromosomal amplicons incorporating MYCN-ID2 and MDM4. The difference in the copy number could be explained either by a mix of amplicons containing respectively MYCN-ID2 alone and MYCN-ID2-MDM4 or by more complex amplicons incorporating multiple copies of MYCN-ID2 for each copy of MDM4. Tracks as in (E).
Recurrent simple SVs activate MYC by enhancer amplification
Our 1D and 2D analyses both nominated rearrangements within the MYC TAD. Among the 28 tumors with SVs in this TAD, 6 contained complex rearrangements that amplified MYC, connecting the amplicon to locations outside of the MYC TAD. An additional 15 tumors contained a tandem duplication centering on intron 1 of the lncRNA CCDC26, with a median span of 216 kbp and a minimal common region of amplification (MCR) of only 42 kbp (Fig. 1B). The remaining seven rearrangements exhibited no consistent structure.
The 2 Mb region telomeric to MYC has been shown to contain MYC enhancers in lineage-specific locations in several cancer types36. We therefore hypothesized that the CCDC26 amplicon promotes oncogenesis by amplifying an associated neural-lineage enhancer. We analyzed published H3K27ac enhancer tracks generated from H3K27M and H3wt pHGGs37 and adult glioblastomas38 and observed H3K27ac enhancer peaks within the MCR of the CCDC26 amplicon (Fig. 1C, Ext. Data Fig. 2A). We also confirmed the presence of this enhancer in an independent pHGG ATAC-seq dataset39 and H3K27ac ChIPseq from normal neural tissue40 (Ext. Data Fig. 2A). The H3K27ac peak at chr8:130640182-130641543 was present in all tissues of neural origin but not enhancer maps from non-neural lineages (hematopoietic and lung tissues) (p= 0.0005, Fisher’s exact test). We conclude that the CCDC26 amplicon centers on a neural lineage-specific enhancer.
Although this enhancer is present across neural tissues, histone-mutant DMGs exhibit 31% more H3K27ac binding at this locus than H3WT gliomas—the fourth most differential super-enhancer between these groups (q-value = 0.05; p=0.001)37. Intriguingly, the CCDC26 amplicon also occurred almost exclusively in H3K27M gliomas (14/97 H3K27M vs. 1/82 H3G34R or H3WT tumors; q = 0.0018). We conclude that the CCDC26-SV amplifies an enhancer which is present across neural lineages but is enriched in H3K27M gliomas.
The lineage-specific H3K27ac enhancer at the CCDC26 locus also appears to interact directly with MYC. We evaluated the chromatin topology of an H3.3K27M pHGG that harbored a CCDC26-SV using Hi-C. The CCDC26-SV breakpoints were within the MYC TAD and the H3K27ac peak within the CCDC26-SV formed an interaction peak with the MYC promoter (Fig. 1D). However, this interaction is not restricted to tumors with CCDC26-SVs. Hi-C data generated in a CCDC26wt pHGG, two patient-derived H3.3K27M cell lines, and iPSC-derived neural progenitors (Ext. Data Fig. 2B) also revealed a similar TAD structure and interactions with the MYC promoter. We conclude that the CCDC26-SV amplifies a pre-existing MYC enhancer.
The eight pHGGs with the CCDC26-SV also exhibited increased MYC RNA expression compared to tumors without SVs or amplifications in the MYC TAD (8q24.21-WT, n=92, p=0.04, Figure 1E), and similar levels of MYC as pHGGs with amplifications of the MYC coding sequence (p=0.85). Indeed, the absolute copy number of both MYC and the CCDC26-SV correlated with MYC expression (p = 0.0003 for MYC and p = 0.01 for the CCDC26-SV, Spearman rank correlations, Ext. Data Fig. 2C-D). We conclude that CCDC26-SVs and associated enhancer amplifications activate MYC expression.
We next validated that the H3K27ac peak in CCDC26 represents a functionally relevant, lineage-specific enhancer, leveraging minimal reporter assays. We generated two enhancer reporter systems (E1 and E2), each encompassing slightly more than one half of the enhancer, with a small region of overlap (Figure 1F). We transduced two histone-mutant pHGG cell lines with the reporter constructs and evaluated induction of luciferase expression to mark enhancer activity. In both lines, the E1 enhancer region was sufficient to increase luciferase expression relative to vector control (p-value <0.01 in both cases, n = 3, Nested One-Way Anova: Tukey’s Multiple Comparisons, Fig. 1G, Ext. Data Fig. 2E). We performed similar experiments in the lung cancer cell line A549, and found no increase in luciferase activity, although previously validated lung adenocarcinoma (LUAD) MYC enhancers36 did induce luciferase activity (p-value (E1 vs. control) = 0.96; p-value(LUAD vs. control) = 0.0071, n = 3, Nested One-Way Anova: Tukey’s Multiple Comparisons, Ext. Data Fig. 2F).
MYCN activation through enhancer amplification and hijacking
Somatic enhancer amplification also seems to play a role in the activation of MYCN in pHGG. The SRJ (Ext. Data Fig. 3A) connecting ID2 with MYCN represents a set of complex SVs in tumors with high-level amplifications within this region on chromosome 2 (Fig. 2B). ID2 is a transcription factor regulating neural differentiation41, and the ID2 locus is associated with an H3K27ac enhancer that is present across all analyzed pHGG tumor samples (Ext. Data Fig. 3B). The MCR of the MYCN-ID2 amplification contains both the ID2-associated enhancer and the coding sequence of MYCN (Ext. Data Fig. 3B). The SVs result in juxtaposition of the enhancer in ID2 with MYCN, reducing the distance between the two from the normal 7 Mbp to less than 700 kbp (Fig. 2C). These data suggest these SVs hijack the ID2 enhancer to activate MYCN.
We also identified four pHGGs with MYCN amplifications that did not connect to ID2. However, these latter “localized MYCN” amplicons always encompassed more of the immediate neighborhood to MYCN than the complex MYCN-ID2 amplicons. In contrast to MYCN-ID2 amplicons, which only contained a small fraction of the MYCN TAD (23% on average), localized MYCN amplicons contained most of this TAD (60% on average, p=0.03, T-test), including several enhancers not included in the MYCN-ID2 amplicons (Fig. 2D).
The high-level MYCN amplicons show typical characteristics of extrachromosomal amplicons, reaching copy numbers of 50 – 300 per cell. Other oncogenes with absolute copy numbers greater than 10 have been shown to reside on extrachromosomal amplicons in various cancer types13,42. While projections on the linear reference genome resulted in typical complex patterns14, it was possible to construct circular amplicons containing MYCN and, in the MYCN-ID2 cases, ID2 (Fig. 2E). Indeed, these circular amplicons represent an optimal solution to explain the joint copy number and SV profiles in this region. We therefore sought to validate this using metaphase fluorescence in-situ hybridization (FISH) on a pHGG cell line derived from a tumor with a MYCN-ID2 rearrangement. We found abundant extrachromosomal amplicons containing both the MYCN and ID2 loci (Fig. 2F). These appeared to reflect multiple subclonal amplicons including some containing additional oncogenes such as MDM4 (Fig. 2G). The copy number of MYCN was consistently higher than that of ID2, raising the possibility that ID2 was incorporated into a subset of preexisting MYCN amplicons during tumor development. Alternatively, MYCN could be further amplified within cells that already harbor MYCN-ID2 amplicons.
These data suggest that MYCN-ID2 rearrangements are an example of enhancer hijacking, bringing a strong enhancer in ID2 next to MYCN on amplicons without the endogenous elements of the MYCN TAD, whereas amplifications of MYCN without ID2 amplify enhancers within the MYCN TAD.
Recurrent SV around RTKs suggesting extrachromosomal DNA
The remaining SRBs all involved RTK genes that are known to be amplified and oncogenic in pHGG: PDGFRA, EGFR, and MET3-7,43. These loci, along with MYC and MYCN, were also the only regions with high-level amplifications that recurred in at least three patients (Fig. 3A). The RTK SRBs also comprised both simple SVs that presumably amplify local enhancers (Fig. 3B, Ext. Data Fig. 4C) and SVs that appeared to reflect complex extrachromosomal amplicons that integrate distant sites and reach as many as 200 copies (Fig. 3C and E).
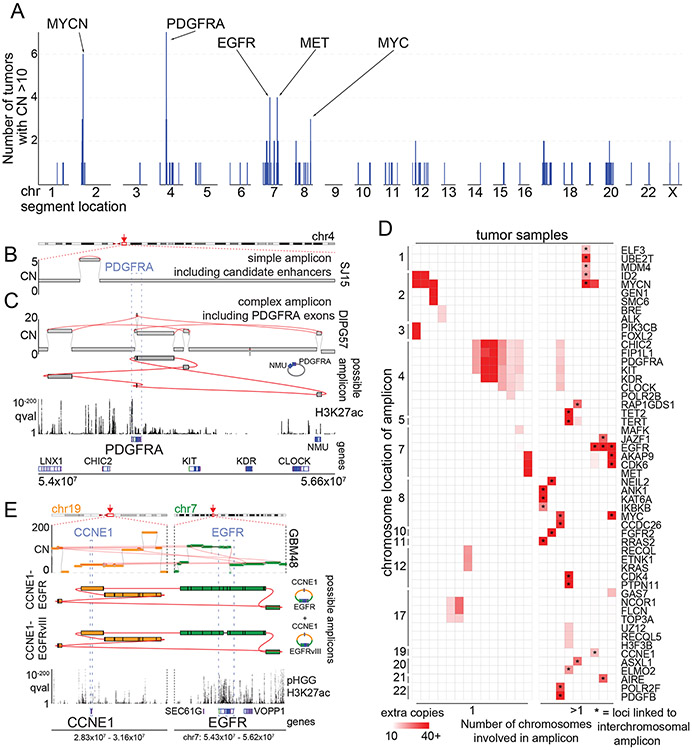
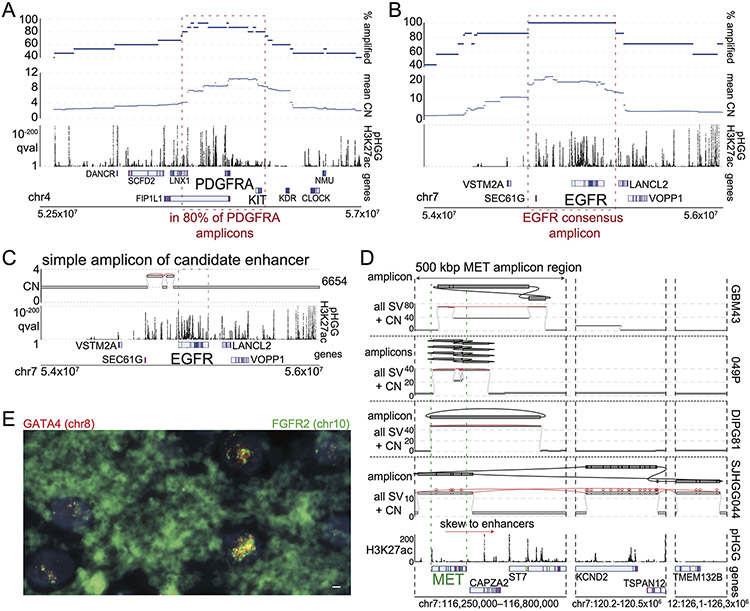
Figure 3. High-level amplicons.
(A) For each genomic locus (horizontal axis), the number of tumors containing a high-level (CN>10) amplicon is indicated (vertical axis). SRBs are highlighted at the top. (B-C) Simple and complex SVs exhibit distinct mechanisms to active PDGFRA. The top and bottom tracks indicate copy-numbers and the significance of H3K27ac enrichment (as calculated from eight pHGGs), respectively. SVs are highlighted in red. Selected gene loci are indicated on the bottom. (B) A simple amplicon of a region with known18,44 PDGFRA enhancers. (C) A complex high-level PDGFRA amplicon, displayed as in panel (B) with the addition of a track (second from top) indicating the topology of the amplicon. The complex-SV cluster around PDGFRA connects several segments on chr4, which are amplified to ten absolute copies. The SV calls support the reconstruction of an extrachromosomal amplicon incorporating PDGFRA exons and these segments. (D) Cancer genes involved in high-level amplicons (>10 copies) within the cohort. 9/23 tumors (grouped on right) contain high-level amplicons encompassing loci from two or more chromosomes. These linked loci are marked by *. The color of each cell represents the number of extra copies due to the amplicon. (E) Example of a tumor with an extrachromosomal amplicon including two oncogenes from different chromosomes. This tumor shows a cluster of SVs connecting the EGFR and CCNE1 loci. The regions of both oncogenes are amplified to different CNs but in both cases reach several dozen absolute copies. (top) The complexity of the SVs allows for the reconstruction of several possible extrachromosomal amplicons. The CN differences in the bulk profile (middle) could be explained by either a mix of different circles or by more complex circles incorporating some segments repeatedly. The SV calls also reveal that a small fraction of the EGFR amplicons in this patient already show the EGFRvIII variant. The bottom two tracks show the genes of interest at the location and q-value H3K27ac track calculated from eight pHGG tumors.
Overall, 35/179 tumors showed at least one >50 kbp amplicon with an absolute copy number greater than ten. Among 34 of these 35 tumors, the high-level amplicons contained at least one well-known oncogene, and apart from the coding sequence of the oncogene, they recurrently incorporated the same genomic loci around the oncogene (Ext. Data Fig. 4A-B for PDGFR and EGFR). In several pHGGs, we detected SVs that allowed for the reconstruction of circular extrachromosomal amplicons containing multiple oncogenes from different chromosomes (Fig. 3D-E). We again validated this by performing FISH on a tissue slide of a tumor with a high-level amplification and SVs connecting segments on chr8 (including GATA4) and on chr10 (including FGFR2). This showed massively increased numbers of foci for both the GATA4 and FGFR2 probes. In many cases the signal of both probes overlapped, indicating colocalization (Ext. Data Fig. 4E). The number of copies per cell was highly variable and the amplicons were distributed in heterogeneous clusters throughout the nucleus. All these features have been associated with extrachromosomal amplicons13. These data suggest that only a subset of pHGGs develop high-level amplicons, which recurrently contain the same (presumably regulatory) sequences in addition to the target oncogene and can contain segments originating from different chromosomes.
To further understand the structure of these amplicons, we first focused on high-level amplicons containing PDGFRA. These amplicons span more than 2.5 Mbp and are superimposed upon low-level amplicons of the surrounding region, often starting from the centromere. The amplicons included KIT in 80% of cases and reached KDR in 60%. All but one (14/15, 93%) of amplicons in the PDGFRA TAD amplified PDGFRA itself, often to the highest copy numbers reached in the region (Ext. Data Fig. 4A). The sole exception amplified of a short sequence centromeric to PDGFRA containing H3K27ac peaks (Fig. 3B) that have been shown to interact with the PDGFRA promoter in pHGGs18 and adult GBMs44, suggesting use of enhancer amplification to activate PDGFRA. Indeed, this region was included in the PDGFRA amplicon in nearly all tumors (14/15, 93%) (Ext. Data Fig. 4A). These data suggest that SVs in pHGG recurrently incorporate an upstream enhancer-rich region into high-level PDGFRA amplicons.
The high-level EGFR and MET amplicons also extended beyond the RTK coding sequence to recurrently involve associated enhancers (Ext. Data Fig. 4B-D). The EGFR amplicons showed a skew towards the enhancers in SEC61G, which drive EGFR expression in extrachromosomal amplicons in adult GBM20. Both EGFR and MET amplicons showed subclonal SVs within the coding sequence, potentially allowing for expression of alternate transcripts. In two tumors, these resulted in the EGFRvIII variant. EGFR amplicons also showed complex subclonal structures with incorporation of distant oncogenes (Fig. 3E). MET amplicons skewed towards a region including enhancers in CAPZA2 (Ext. Data Fig. 4D). We conclude that the RTK gene amplicons are shaped by the epigenetic machinery necessary to drive their expression.
SV signatures relate to genetic and epigenetic tumor states
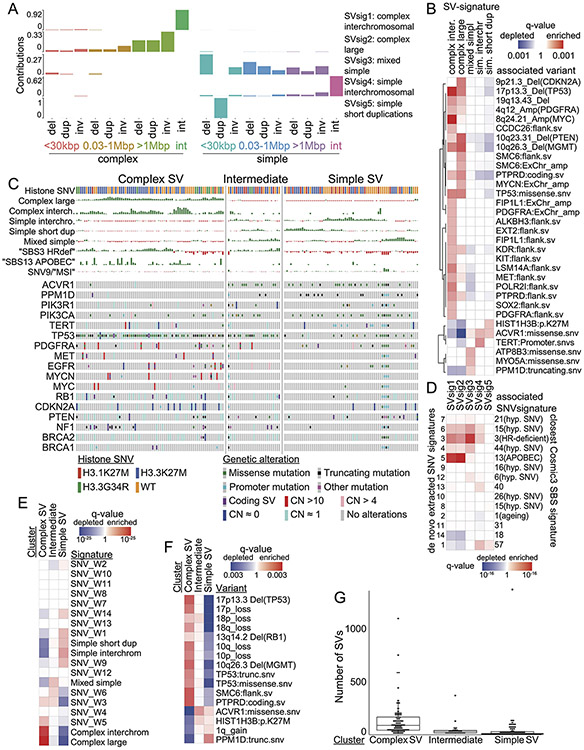
The discovery of these two distinct classes of recurrent SVs, the simple enhancer amplifications and the complex ecDNA-based amplicons, raises the question whether these alterations are part of distinct variant patterns in different pHGGs. Unsupervised identification of SV signatures can reveal tumor subgroups with distinct SV generating mechanisms26. Recent work has identified SV signatures that are present across cancer types, albeit in tissue-specific conformations and with tissue-specific variant associations26. We performed manual review of individual SVs to assess their likely mechanisms of formation above, but for genome-wide analyses automated classifiers have recently been developed45. Using methods developed by the Pan-Cancer Analysis of Whole Genomes (PCAWG) group23, we detected 10,385 complex/clustered SVs. Among these, automated methods further classified 21% by their likely formation mechanism. We therefore combined both approaches to identify SV signatures, using the more precise formation mechanisms when available and noting the rest of the complex events as Complex-NOS.
We observed nine resulting SV signatures (Fig 4A). Six signatures were complex (BFB, Chromothripsis, Chromoplexy, Complex-NOS, DM, Tyfonas) and three were simple (Deletion, Duplication, Translocation/Inversion). The DM signature comprised only DMs, of which 6% were Complex DMs according to the Hadi et al45 categorization. Templated Insertion Chains (TICs) contributed 6% to the Translocation/Inversion, 1% to the deletion, and 1% to the Complex-NOS SV signature. Inversions contributed 35% to the Translocation/Inversion signature and 13% to the duplication signature. Four signatures comprised entirely a single type of feature: Chromothripsis, BFB, Chromoplexy, and Tyfonas, respectively (Fig. 4B).
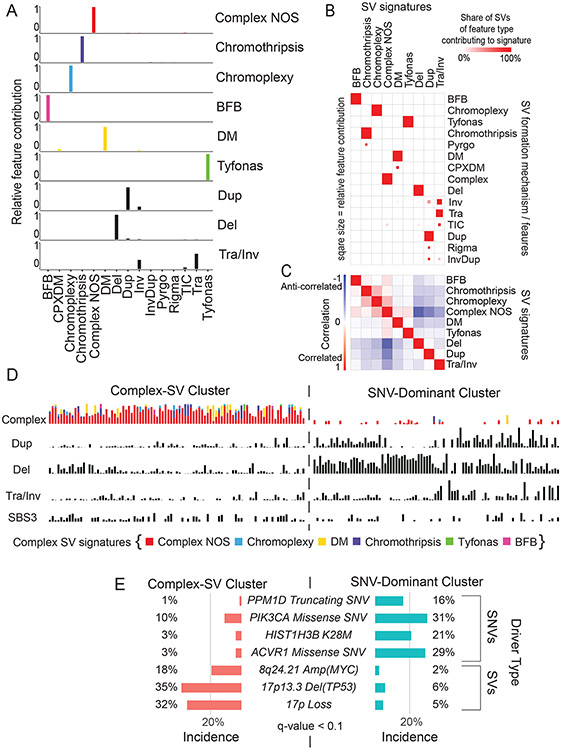
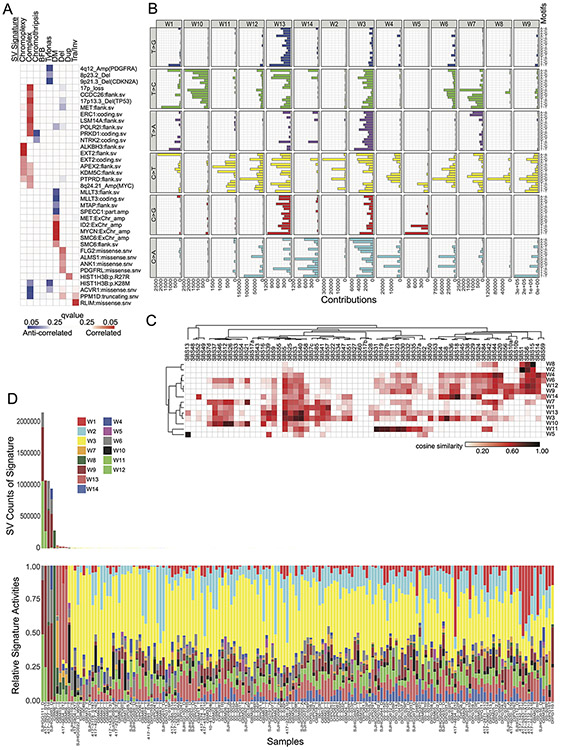
Figure 4. SV signatures in pHGG.
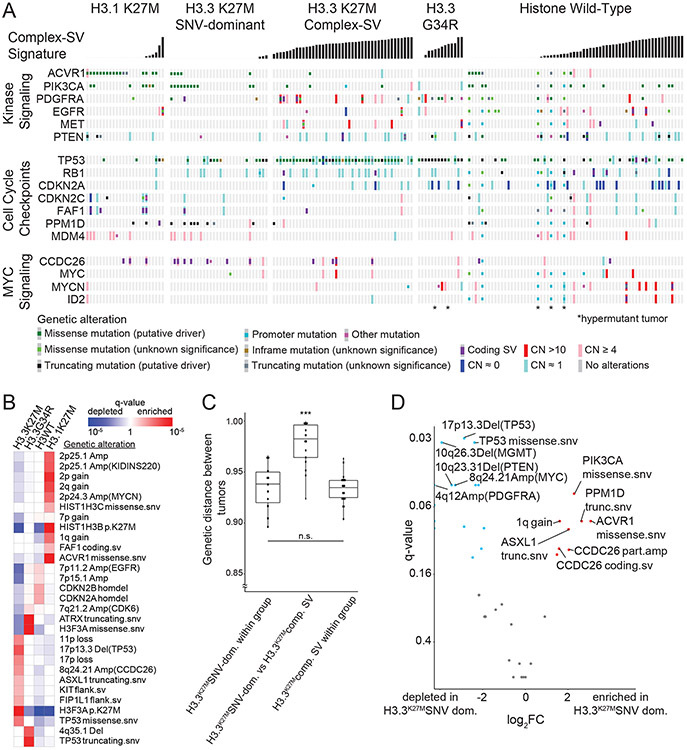
(A) Contribution of different SV features to each of the identified SV signatures. The horizontal axis indicates SV features - Deletions (Del), duplication (Dup), inversion (Inv), breakage fusion bridge cycle (BFB), complex double minute (cpxdm), double minute (dm), template insertion chains (tic), and translocation (tra). The vertical axis indicates the fraction of SVs with each of these features within the nine identified SV signatures. (B) Heatmap indicating the contribution of each SV type to each signature. Deeper red color indicates a larger fraction of all SVs with each type contributing to each signature. The size of the squares indicates the fraction of all SVs contributing to a signature that belong to this SV type. (C) Heatmap indicating the correlation between the SV-signatures as determined by Pearson. Shading in red indicates positive correlation coefficient and blue indicates anti-correlation. (D) Normalized SNV and SV signature activities within the Complex-SV and SNV-Dominant clusters. Each column represents an individual tumor and rows indicate signature activities in the individual samples. Contribution of complex SV signatures within the Complex SV group are represented by the colors shown (n=179 tumors). (E) Genetic variants (SV and SNVs) significantly enriched in the Complex-SV and SNV-Dominant clusters (n=179 tumors), with correlations reaching q < 0.1. q-values were calculated using Fisher’s exact tests.
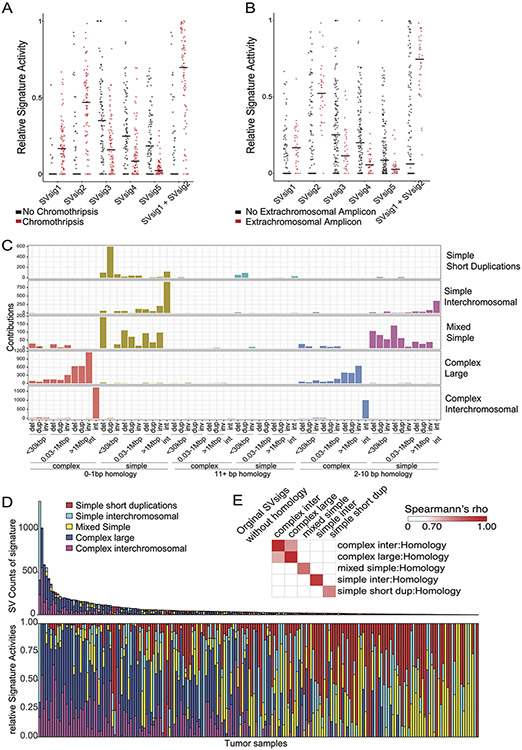
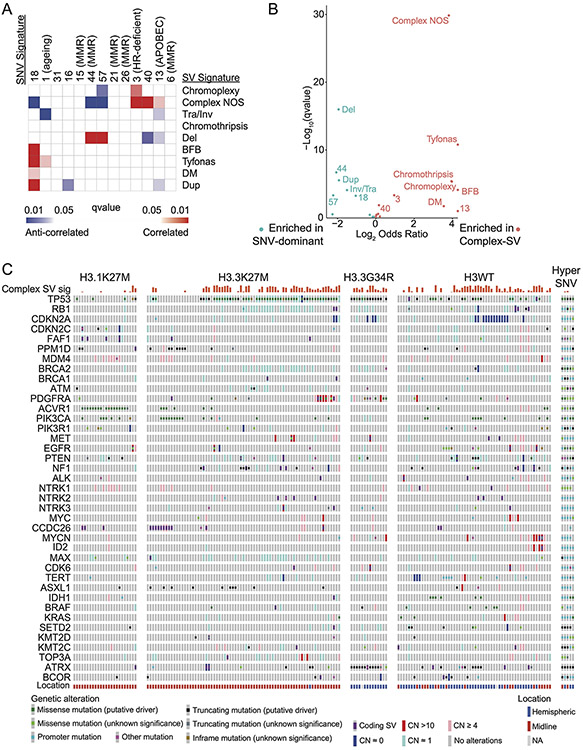
We next looked for possible causes and consequences of the pHGG SV signatures by testing for associations between the activity of each signature and presence of recurrent and known oncogenic46 variants (Supp. Table 5). As expected, the DM-signature associated with the ecDNA amplifications of MYCN and ID2. The complex-SV-NOS signature closely associated with focal TP53 disruption and loss of 17p (encompassing TP53) and anticorrelated with oncogenic mutations in PPM1D, ACVR1 and HIST1H3B (Ext. Data Fig. 5A). Notably, we observed complex-SV signatures in pHGGs with high SV counts (rho = 0.68, p < 2.2e-16, Spearman's). Unlike several high-SV-count adult cancers with disrupted DNA-damage responses (DDR)22,23,28,29 and homologous recombination (HR)/BRCA23, where tandem duplications signature were dominant, pHGGs with the simple tandem duplication signature tended to have few SVs. None of the genes previously implicated in tandem duplication signatures or loss of HR in adults23 reached a significant level of association with any signature in pHGG.
We next asked whether pHGGs separate into subsets with different DNA damage and damage response characteristics based on patterns of both SVs and SNVs. We detected an anticorrelation between the complex SV signatures and the three simple SV signatures (Fig. 4C). Evaluation of SNV mutation patterns revealed 14 SNV signatures, including signatures similar to known aging, APOBEC (COSMIC signature SBS1347), HR-deficiency (SBS325) and hypermutation SNV signatures25 (Ext. Data Fig. 5B-D). We also performed signature analyses using alternative methods26 which generated similar results (Ext. Data Fig. 6-7).
The entire pHGG cohort separated into two groups reflecting different amplitudes of the 9 SV and 14 SNV signatures (Fig. 4D). One cluster (“Complex-SV”) was dominated by Complex-SV signatures (q-values ranging from q= 0.02 for DM to q=1.4x10−30 for Complex-NOS, Ext. Data Fig 8A). SBS3 and SBS13 were also enriched in this cluster, following their close correlation with the Complex-NOS SV signature (Ext. Data Fig. 8B). The Complex-SV cluster was enriched for TP53 inactivation (q<0.1, Fig 4E), SVs surrounding/amplification of PDGFRA, EGFR and MET (q<0.1). In contrast, the other cluster (“SNV-dominant”) was dominated by the simple SV signatures (q<8x10−5), lacked TP53 disruption (q<0.1), and was enriched for PPM1D mutations. This cluster seemed to be driven instead by a combination of SNVs including ACVR1, PPM1D, H3.1K27M, and PIK3CA mutations (all q<0.1). Both clusters included hemispheric and midline gliomas. H3.3K27M showed no enrichment in either cluster (q= 0.46). These data suggest that pHGG genomes are shaped by at least two distinct variant generating processes, which associate with distinct driver combinations.
Signatures indicate two groups: Complex-SV and SNV-Dominant
We next evaluated whether SV signatures could inform pHGG subtypes. Currently, pHGGs are classified according to their location and histone mutation; different histone mutations are known associate with distinct recurrent SNVs and SCNAs. We confirmed these known relationships3,48 and detected two additional associations with SVs (Fig. 5A-B and Ext. Data Fig. 8C). The SV in CCDC26 resulting in MYC enhancer amplification was enriched in H3.3K27M gliomas (q = 0.008) and H3.1K27M pHGGs were enriched for a focal deletion of CDKN2C with breakpoints in the adjacent gene FAF1 (q = 0.04).
Figure 5. Overview of somatic variants and associated features within subgroups defined by histone mutations.
(A) Co-mut plot of the 176/179 (98.3%) tumors with somatic variants in at least one well-known cancer gene. Columns represent tumors, ordered within histone mutation-defined subgroups by increasing activity of the complex SV signatures. Rows represent cancer genes that harbor somatic variants, grouped by pathways. Type of genetic alterations are depicted by the colors shown in the key below the comut plot. *indicates hypermutant tumors. (B) Heatmap indicating significance of histone subgroup associations for all variants with correlations reaching q < 0.05 (based on Fisher’s exact tests) within any subgroup. Red indicates enrichment and blue indicates depletion. Shading reflects q values as shown in the legend. (C) Jaccard distances based upon the variants in each tumor (vertical axis), for pairs of tumors within the H3.3K27M SNV-dominant group (left column), within the H3.3K27M Complex-SV group (right column) or paired between these groups (middle column). H3.3K27M tumors were considered to be in the Complex-SV group if complex SV signatures comprised more than 20% of its SV signature activity. *** denotes q=5.8x10−8 for within-group H3.3K27M SNV-dominant vs. between groups and q=1.3x10−10 for within-group H3.3K27M Complex-SV vs. between groups by a two-sided Wilcoxon test, n = 102 DMGs. Center line of the boxplot indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5xIQR. (D) Volcano plot indicating the significance (vertical axis; FDR-corrected Fisher’s exact tests) of associations in H3.3K27MSNV-dominant [n=30 DMGs] relative to H3.3K27MComplex-SV [n=43 DMGs], for all genetic variants observed in at least 10% of tumors in one subgroup. Variants enriched or depleted to q<0.15 are highlighted in red on the right or in blue on the left, respectively.
We also found that inclusion of SV signatures identified two pHGG subtypes, both deriving from H3.3K27M pHGGs. Most H3.3K27M pHGGs exhibited high Complex-SV signature activity, but 42% did not. The combinations of genetic alterations in known cancer-related genes differed significantly between these H3.3K27M Complex-SV and H3.3K27M SNV-Dominant groups (q < 6x10−8; Fig. 5C). Indeed, the H3.3K27M SNV-Dominant pHGGs were as different from the H3.3K27M Complex-SV pHGGs as the H3.1K27M pHGGs were (Ext. Data Fig. 9A). The H3.3K27M SNV-Dominant pHGGs showed a variant pattern resembling H3.1K27M DMGs; (Fig. 5D) mutations in PPM1D, ACVR1, and PIK3CA, gains of 1q encompassing MDM4, and amplifications in CCDC26, encompassing MYC enhancers, were enriched in these tumors relative to H3.3K27M Complex-SV pHGGs (all q < 0.09 except for CCDC26 amplifications, where q=0.11). In contrast, loss of TP53 (by SNV or SCNA) and amplifications of PDGFRA and MYC were depleted (all q <0.05) in H3.3K27M SNV-Dominant. These data suggest that the propensity of pHGGs to develop complex SVs influences the combination of driver alterations they accrue, even within groups defined by their histone mutations.
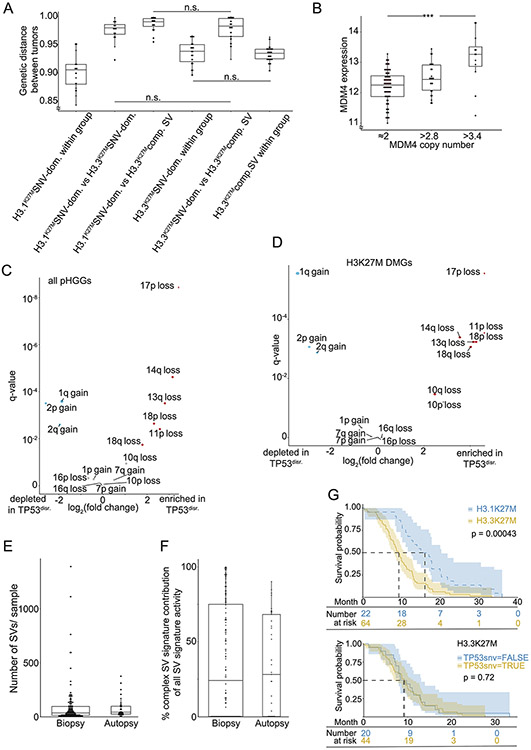
Across the DMGs with more than 20% Complex-SV signature activity (“H3K27M Complex-SV”, including H3.3K27M [n=43] and H3.1K27M [n=3] DMGs), the TP53 pathway was inactivated almost universally through direct disruption of TP53 (44/46 cases, 96%; Fig. 5A). In contrast, the majority of DMGs with less complex signature activity (“H3K27M SNV-Dominant”, H3.3K27M [n=30] and H3.1K27M [n=21]) lacked direct TP53 disruption (37/51, 73% TP53WT; q = 2.3x10−5) but appeared to suppress the TP53 pathway through other mechanisms. Mutations in PPM1D were more prevalent in this group, though still in a minority (7/30 H3.3K27M, 2/21 H3.1K27M, 20% in total; vs 1/46 H3K27M-complex tumors; q=0.008). It is possible that gains of 1q, encompassing MDM4, also served to suppress the TP53 pathway in these tumors. Although 1q spans approximately 2580 genes, we observed two sources of evidence that their prevalence in SNV-Dominant DMGs related to MDM4 and TP53 pathway suppression. First, MDM4 was significantly overexpressed in 1q-amplified pHGGs of all types in our cohort (q=0.004; Ext. Data Fig. 9B). Second, 1q gains were the only arm-level SCNAs that anti-correlated with disruption of TP53 (q=2.8x10−6 in H3K27M-DMGs; q=0.0003 across all pHGGs, Ext. Data Fig. 9C-D) apart from gains of chromosome 2 (q < 0.0025). In contrast, seven of the other thirteen significantly recurrent arm-level SCNAs were positively correlated with TP53 disruption in H3K27M DMGs (all q < 0.018), presumably due to the role of TP53 in generating aneuploidies49. Indeed, across TP53WT H3K27M DMGs, gains of 1q were among the most common genetic events, observed in 85% of tumors (16/20 H3.3K27M, 17/19 H3.1K27M), as opposed to 31% of TP53-disrupted H3K27M DMGs (p= 1.4x10−6). No pHGGs in our cohort exhibited focal, high-level amplifications of MDM2. These data suggest that direct disruption of TP53 contributes to a different pattern of SVs compared to other mechanisms of TP53 pathway inactivation, including primarily alterations of PPM1D and MDM4.
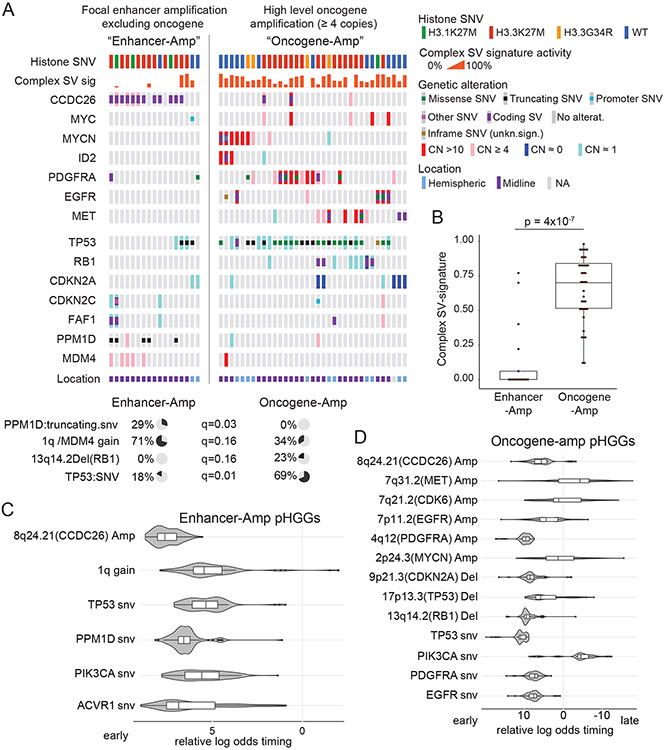
Focusing on the tumors that harbored significantly recurrent SVs, we observed two groups. One group contained the tumors with high-level amplicons of PDGFRA, EGFR, MET, MYC, and MYCN (“Oncogene-Amp”). In contrast, the second group amplified presumed enhancer elements within the TADs of these oncogenes without amplifying their coding sequences (“Enhancer-Amp”). The Oncogene-Amp pHGGs exhibited significantly higher activity of the complex SV signatures (p = 4x10−7; Fig. 6A-B). The two groups also harbored inactivating alterations in different DDR genes (Fig. 6A). Oncogene-Amp pHGGs were enriched for TP53 SNVs (69% of Oncogene-Amp vs. 18% of Enhancer-Amp pHGGs, q=0.01) and RB1 deletions (23% of Oncogene-Amp vs. 0% of Enhancer-Amp pHGGs, q=0.16). In contrast, Enhancer-Amp pHGGs were enriched with PPM1D SNVs (29% of Enhancer-Amp vs. 0% of Oncogene-Amp pHGGs, q=0.03) and gains of 1q encompassing MDM4 (71% of Enhancer-Amp vs. 34% of Oncogene-Amp pHGGs, q=0.16). In sum, alterations in TP53 and RB1 associate with complex SV signatures and high-level amplifications of oncogenes, while PPM1D SNVs and 1q gains more frequently occur with simple SV signatures and amplifications of enhancer elements near oncogenes. These data raise the possibility that alterations in the DDR not only shape the processes that generate SVs but also the types of driver alterations they exhibit in MYC, MYCN and RTKs genes.
Figure 6. Context of the significantly recurrent SVs.
(A) Co-mut plot for Enhancer-Amp and Oncogene-Amp significantly recurrent SVs. Enhancer-Amp significantly recurrent SVs generate focal amplifications in the TAD of an oncogene without amplifying the protein coding sequence; Oncogene-Amp significantly recurrent SVs generate high-level (CN >3.4) amplifications or fusions of the coding sequence. The top two rows show associated metadata. The next seven rows indicate the genes affected by the significantly recurrent SVs. The bottom seven rows show genes in DNA damage response pathways. Significant associations with the two groups are illustrated with pie charts below the plot, based on Fisher’s exact tests. (B) Enhancer-Amp pHGGs show significantly lower combined Complex-SV signature activity than Oncogene-Amp pHGGs (p = 4x10−7, two-sided Wilcoxon, n = 52 tumors, center line of the boxplot indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5xIQR). (C-D) Timing analysis of somatic variant acquisition in Enhancer-Amp (C) and Oncogene-Amp (D) pHGGs based on a Bradley-Terry model. The horizontal axis shows the log odds of the variant being an early event. The distributions indicate the results of 100 random subsamples of the data (C: n = 17 pHGGs, D: n = 35 pHGGs). Center line of the boxplot indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5xIQR. Only variants in the SRSV-affected genes and pathways (growth factor and MYC signaling) and in DNA damage response genes altered in more than two samples are shown.
Temporal evolution of genetic variants
The correlation between the presence of a variant and the activity of a signature by itself cannot tell us anything about the direction of the link between the two. This is most obvious for the inactivation of tumor suppressors through copy number loss or the amplification of oncogenes and their association with the complex SV signatures. These events could be direct consequences of the activity of this signature. On the other hand, these genetic variants could drive survival after catastrophic SVs, increase genomic instability and thereby drive the activity of the signature after their initial random occurrence in tumor evolution.
Specifically, we considered two hypotheses regarding tumor evolution. First, disruption of DDR could be an early event that activates the Complex-SV generating process and culminates in the development of a specific class of genetic events including the high-level amplicons described earlier. Alternatively, both disruption of the DDR and the high-level oncogene amplification could happen later in tumor development as a consequence of complex SVs involving these genes.
Notably, we observed no effects of therapy on SV patterns, suggesting that the SVs occurred during gliomagenesis. Although radiation treatment has been shown to induce DNA breaks50, we found no differences in the number of SVs per sample (median 35 vs 42; q= 0.6) or in the activity of the Complex-SV signatures (median 24% vs 28%; q=0.7) between pre-treatment biopsy and autopsy samples (Ext. Data Fig. 9E-F).
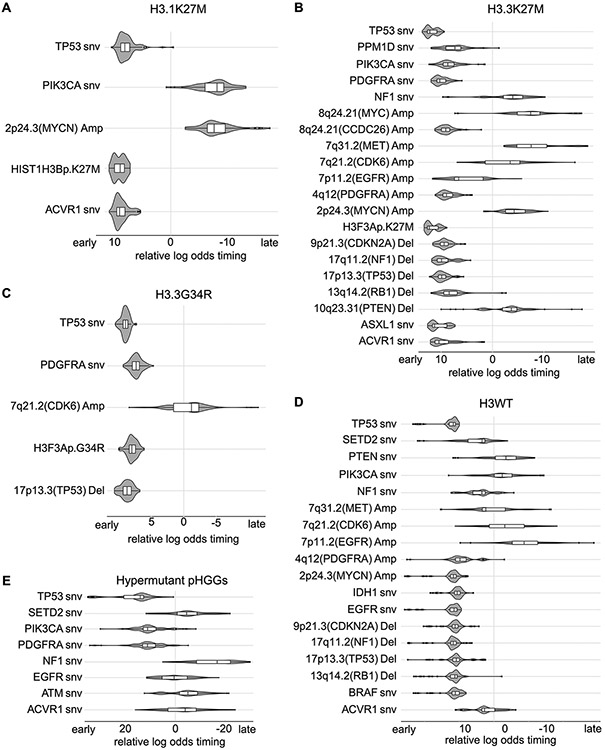
We performed a timing analysis reflecting the relative ordering of mutations and SCNAs during gliomagenesis51. Focusing on the subset of pHGG with simple enhancer amplifications (Enhancer-Amp pHGGs) we found that the focal amplification of the MYC enhancer in CCDC26 is one of the earliest variants in these samples (Fig. 6C), occurring earlier than alterations in PPM1D and 1q/MDM4 gain. In contrast, the amplification of the MYC isoform and RTK genes in the Oncogene-Amp samples happened after the loss of the tumor suppressors TP53, RB1 and CDKN2A/B (Fig. 6D). These data suggest that simple tandem duplications can arise in tumors without major disruptions of DDR, potentially contributing to tumor initiation, whereas the creation of the complex high-level oncogene amplicons requires prior direct genetic disruption of TP53, RB1, or CDKN2A/B.
Prior studies have shown histone mutations to be the initiating event in the pHGGs in which they occur8,9,37,43. However, the studies investigating pHGG evolution in human tumor tissues were limited to exomic alterations in fewer than 15 patients52-55. Using the power of the WGS data from 179 tumors, we confirmed the findings of these previous studies52-55 including that H3K27M mutations are the earliest mutations, followed by SNVs in ACVR1 and TP53 in H3.1K27M and H3.3K27M gliomas, respectively. (Ext. Data Fig. 10). This large WGS cohort also allowed us to time focal SCNAs based on the ratio of SNVs acquired before and after each change in each copy number51. We found that losses of TP53, CDKN2A/B, and RB1 precede RTK gene amplifications across the H3.3K27M, H3.3G34R and H3WT subgroups of pHGG. In H3.3K27M DMGs, the simple amplifications of the MYC enhancer in CCDC26 were early events while the complex amplifications of MYC itself occurred later in tumor development.
Complex-SV DMGs are associated with shorter survival
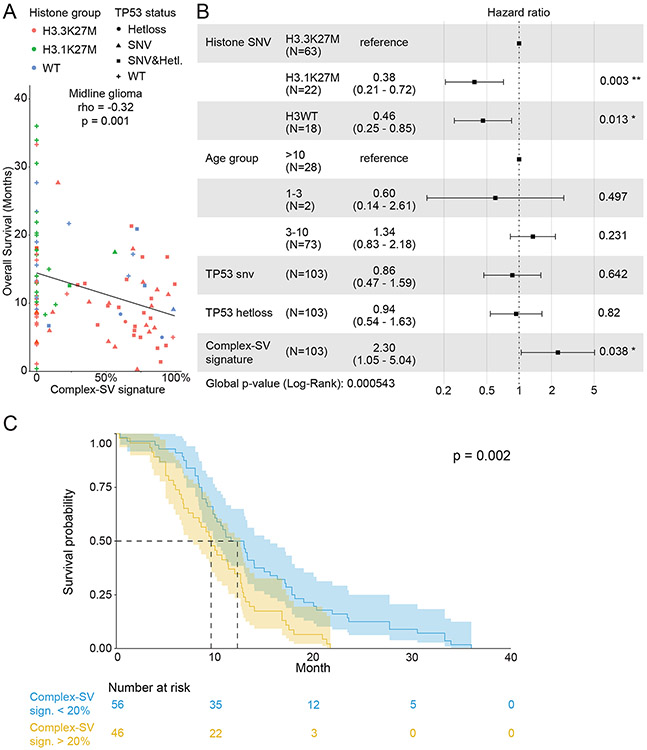
We suspected that the differences in SV-generating processes across DMGs could associate with clinical phenotypes including survival. First, we confirmed the known association48,56 between H3.1K27M and longer overall survival (OS) compared to H3.3K27M (9.3 vs 16.1 months; p=0.0004; Ext. Data Fig. 9E top), and the lack of association between TP53 mutation and OS within H3.3K27M pHGGs (p=0.72; Ext. Data Fig. 9E bottom). To address SV signatures specifically, we also investigated the correlations between the numeric values of the combined Complex-SV signature and OS. Across all DMGs, this Complex-SV signature was significantly anti-correlated with OS (Fig. 7A; p=0.001).
Figure 7. Association between the complex SV signatures and overall survival in midline gliomas.
(A) Associations between the fraction of SV signature activity attributed to complex SV signatures and overall survival for all midline gliomas (n= 103; rho = −0.32 and p=0.001, two-sided Spearman correlation test). (B) Cox proportional hazards analysis incorporating histone group, age, TP53 status and the combined complex SV signatures. Error bars show the 95% confidence interval of the hazard ratio. (C) Kaplan-Maier plot of a univariate analysis of the association between the complex SV signatures and overall survival, in which tumors were classified as Complex-SV if complex SV signatures contributed more than 20% of all SV signature activity. The number of patients in each group is indicated below the plot. The p-value represents a log-rank test. Error bands show the 95% confidence interval.
The combined Complex-SV signature also significantly associated with shorter survival in a multivariate Cox regression analysis of DMGs that controlled for the known predictors of survival3,48 (Histone SNV and age) and for TP53 status (Fig. 7B; p=0.038). This analysis confirmed a significantly increased hazard ratio for H3.3K27M compared to both H3.1K27M and H3WT DMGs, and a lack of significant associations between TP53 disruptions and OS in multivariate analyses as previously described48. However, associations with age did not reach significance, probably due to our low representation of the under-three and over-ten age groups. While all patients with DMGs in our study died from their disease, the combined effects of these factors caused survival differences of several months. For example, children with DMGs with at least 20% Complex-SV activity survived a median of 9.6 months, about 3 months less than the 12.3-month survival of children with less than 20% Complex-SV activity (Fig. 7C).
Discussion
These analyses found recurrent SVs, including a tandem duplication in 12% of all DMGs encompassing a MYC enhancer; revealed distinct SV signatures; and indicated two classes of DMG, whose driver alterations are either largely complex SVs or dominated by SNVs.
The MYC enhancer amplifications highlight an underrecognized role for MYC in pHGGs. MYC is the most frequently amplified gene in all of cancer, with focal amplifications observed in 15% of tumors49. In contrast, MYC amplifications only occur in 5% of pHGGs3. The MYC enhancer amplification in pHGGs, without amplification of MYC itself, start to address this discrepancy. While tissue-specific amplifications of MYC enhancers occur in other cancers36, the CCDC26 duplication is a pHGG-specific location apparently driven by differences in enhancers across cell types. Altogether, when including high-level MYCN amplifications, 14% of pHGGs harbored SVs predicted to activate MYC pathways. Given this high rate, the role of MYC in pHGG formation requires further study.
Although both SNV-Dominant and Complex-SV pHGGs activate MYC signaling pathways, they do so in strikingly different ways. While SNV-Dominant pHGGs amplify only the MYC enhancer in CCDC26, pHGGs with Complex-SV signatures contain high-level amplicons of both the MYC coding sequence and segments of CCDC26, PVT1 or other distant regions. Amplifications of MYC-PVT1 were reported in DMGs and other cancers57,58. These additional segments could contain independent oncogenic activity as has been proposed for PVT1 or represent regulatory elements that got hijacked to drive MYC expression. The complex MYC amplicons are often extrachromosomal, as indicated by their circular topology and high copy number. In this respect, MYC serves as an example for other oncogenes, including MYCN, PDGFRA, EGFR and MET. Extrachromosomal amplicons (also known as double minutes, or DMs) containing recurrent oncogene-enhancers-combinations occur in several cancers12,20,59. However, their regulatory elements differ from pHGG’s and appear to reflect the tissue specificity of regulatory loci19.
DMs have been shown to originate as byproducts of chromothripsis10. Our data suggest that, in pHGG, they often contain multiple oncogenes from different chromosomes. These DMs would therefore either require simultaneous chromothripsis of two chromosomes or need to develop sequentially by a less-clear mechanism. Our data also suggest multiple variants of DMs within individual pHGGs. This could correlate to different descendants of the initial DM as suggested by recent mechanistic and long-read sequencing studies10,14. In cases where two oncogenes are integrated into a DM that is subsequently amplified, the number of copies of each oncogene should be identical. However, pHGGs often exhibit different amplification levels of these oncogenes, suggesting sequential incorporation into the amplicon. The exact mechanism for this remains elusive and could range from sequential chromothripsis events10 to reversible DM integration in proximity to oncogenes11, or deletions within the DMs. It is tempting to speculate that the evolution and optimization of DMs13 could contribute to the rapid, lethal growth of pHGGs and their poor response to available therapies. RTK inhibition is still a promising goal in pHGG4, but our study highlights that understanding how these DMs evolve might inform about resistance mechanisms.
We also observed an association between H3.3K27M, Complex-SV signatures, and TP53 loss. While TP53 disruption is known to associate with higher SV burden49, the reason for its association with H3.3K27M over H3.1K27M is unclear. H3.1K27M and H3wt pHGGs also comprised both Complex-SV and SNV-Dominant tumors, although H3.1K27M DMGs were enriched with the SNV-Dominant subgroup. The split into Complex-SV and SNV-Dominant observed in H3.3K27M pHGGs could exist in H3.1K27M and H3wt tumors—and indeed these distinctions may exist in other tumor types—but our cohort is insufficient to address this possibility.
We found TP53 disruption to be an early event in tumors with complex SVs. TP53 disruption also precedes and might facilitate survival after chromothripsis in medulloblastoma60. Notably, while almost all DMGs with Complex-SV signatures were TP53 disrupted, not all TP53-disrupted DMGs showed Complex-SV signatures. Additionally, hemispheric pHGGs with Complex-SV signatures were frequently TP53WT but often harbored early loss of CDKN2A/B. This indicates that while TP53 loss and H3.3K27M are correlated with Complex-SV signatures they are neither necessary nor sufficient, either alone or in combination, for the generation of the Complex-SV signatures in pHGG.
Finally, we found variants in known cancer genes in 98.3% (176/179) of pHGGs, significantly expanding the share of patients with identified potential drivers compared to prior exome-sequencing based studies. Many of our observed alterations were in non-coding regions of the genome, targeting regulatory elements such as enhancers. WGS also allowed us to determine which patients had Complex-SV or SNV-Dominant signatures, which associated with survival, controlling for histone and TP53 status. The association between the complex SV signatures and survival might be causative and indicate potential therapeutic targets or it could be quantifiable biomarker for underlying factors like genome instability. In any case, these findings indicate that both research and clinical sequencing of these tumors should encompass the whole genome.
Methods
Sample acquisition
This study includes published4-7 data available under EGAS00001000575, EGAS00001001139, EGAS00001000572 and EGAS00001000192. Novel data was generated from samples obtained from the DIPG-BATs clinical trial (NCT01182350)33, the Dana-Farber Tissue Bank or collaborating institutions, under protocols approved by the institutional review board of the Dana-Farber/Harvard Cancer Center with informed consent (DFCI protocols 10417, 10201 and DFCI 19293), without participant compensation. DNA and RNA were extracted from single DMG cores, pHGG biopsies, and autopsy samples using Qiagen AllPrep DNA/RNA extraction kits. Previously published pHGG whole genome sequencing (WGS) data4-7 and, paired RNA-seq were acquired from public repositories.
Whole-genome sequencing
Library preparation for paired end whole genome sequencing (WGS) was performed16. Genomic DNA was fragmented and prepped for sequencing (60X depth for tumors and 30X depth for normal samples) on an Illumina HiSeq 2000. Reads from both novel and published data were aligned to hg19/GRCh37 with BWA, duplicate-marked, and indexed using SAMtools and Picard. Base quality score was bias adjusted for flowcell, lane, dinucleotide context, and machine cycle and recalibrated, and local realignment around insertions or deletions (indels) was achieved using the Genome Analysis Toolkit. All paired samples underwent quality control.
SNV and SCNA analyses
SNVs were detected using Mutect2 and filtered for common sequencing artifacts, gnomad SNPs, and SNVs present in a panel of whole genome-sequenced normal samples. Significance of recurrent SNVs in non-hypermutant samples (SNV counts < 100,000/sample) was determined with MutSigCV61. SCNAs were called using the GATK4 somatic CNV pipeline with normalization against a panel of blood normals from 184 samples (174 from this cohort, 10 from TCGA). Purity and ploidy were determined using ABSOLUTE62. All somatic copy number alteration (SCNA) calls were purity- and ploidy-adjusted. Significantly recurrent SCNAs were identified using GISTIC2.034 with the following parameters: amp_thresh=0.5; del_thresh=0.7; arm_peel=0; broad_length_cutoff=0.5; cap=3.6; conf_level=0.99; max_sample_segs=3500; and qv_thresh=0.1.
Structural variant detection and significance analysis
SvABA35 was used to call SV in paired tumor normal mode with default parameters. In addition to filtering germline variants against the paired normal, telomers and centromeres were blacklisted. The significance of recurrence analysis was performed separately for breakpoints (1D) and juxtapositions (2D) adapted from pan-cancer.21
The analysis of recurrent breakpoints (1D)21, binned the genome into 50kbp bins with 500bp overlap. The bins were annotated by the overlapping TAD with TAD names derived from Cosmic Cancer Gene Census46 gene presence. Germinal zone TAD boundaries from GSE7756563 were used as the closest available normal neural progenitor. Eligible territory was defined by masking low-complexity genomic loci based on https://github.com/lh3/sgdp-fermi/releases/download/v1/um35-hs37d5.bed.gz. Only one SV per sample per bin was counted. Fishhook64 calculated a background model for the likelihood of a breakpoint in each bin, based on six covariates: replication timing (from http://mskilab.com/fishHook/hg19/RT_NHEK_Keratinocytes_Int92817591_hg19.rds), GC content (from http://hgdownload.cse.ucsc.edu/goldenPath/hg19/gc5Base/hg19.gc5Base.txt.gz), presence of SINE elements (http://www.repeatmasker.org), fragility (from https://data.broadinstitute.org/pcawg6sv/1D_covariates/fragile_genes_smith.hg19fp.txt (adapted from 65, mappability (from http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeMapability/), and heterochromatin status. ChromHMM for the caudate nucleus was chosen for the heterochromatin track based epigenetic similarity to pHGG (Ext. Data Fig. 2A). These data were downloaded from https://egg2.wustl.edu/roadmap/data/byFileType/chromhmmSegmentations/ChmmModels/coreMarks/jointModel/final/E068_15_coreMarks_mnemonics.bed.gz and subset to ['8_ZNF/Rpts', '9_Het', '15_Quies', '14_ReprPCWk'] to define heterochromatin. These covariates predict the probability of a SV occurring21,23,45,64. p-values for the influence of each individual covariate on the model ranged from 2x10−16 to 0.21. p-values of breakpoint enrichment were calculated and corrected using Benjamini–Hochberg procedure. If several bins within one TAD were significant, they were considered as one significant locus and linked to any COSMIC Cancer Gene46 gene in that TAD.
Recurrent juxtapositions (the 2D analysis; Ext. Data Fig. 3A) were detected using a background model determined from 2658 cancers across several types21 and a binning scheme (5583 bins, median span: 467kbp, interquartile range: 347kbp). One SV from each sample was allowed to contribute to connections between any two bins (a “tile”). SVs with at least four supporting reads and a span of >1kbp were included in this analysis. p-values reflecting the significance of enrichment of SVs within each tile were corrected using the Benjamini–Hochberg procedure. Only significantly recurrent juxtapositions which did not occur at the same nucleotide position, had a mean SvABA-assigned quality score of larger than 20, included at least one SV detected with post-assembly (ASDIS or ASSMB) evidence, occurred in more than two samples, and had a q-value smaller than 0.1 were considered for further analysis.
SV signature analysis
SV signature analysis followed published approaches22,23. SVs were stratified according to the span between the two breakpoints (0-30kbp, 0.03-1Mbp, >1Mbp, interchromosomal); read orientation (deletion, duplication, inversion and interchromosomal); and whether they were clustered, as determined by clusterSV23. This was analyzed with Bayesian NMF using SignatureAnalyzer24,25 .
JabBa generated genome graphs45. SV events were called on the gGraph output from JabBa using the gGnome::events function. Events were mapped to individual SVs with gGnome designations when available. SVs without annotations from JabBa/gGnome were classified as ‘Complex NOS’ if they showed >2 cluster size using the clusterSV method23 or as inversion, translocation, deletion, and duplication based on the orientation of their supporting reads. The count matrix was analyzed with Bayesian NMF using SignatureAnalyzer24,25.
RNAseq analysis
RNAseq data were available for 112/179 tumors (57 sequenced de novo and 55 previously published4,6). For de novo samples, cDNA libraries were prepped16 using the Tru-Seq Strand Specific Large-Insert kit, and sequenced to a depth of 50 million paired ends using Illumina Hi-Seq. All reads were aligned to the hg19 reference genome using STAR and quantified with RNA-SeQC following the GTEX analysis pipeline66. Counts were normalized using the VST transform as implemented in DESeq267 and batch-corrected with COMBAT68 as implemented in sva69.
Chromatin Immunoprecipitation
Active Motif (Carlsbad, CA) performed ChIP-Seq. Cells were fixed with 1% formaldehyde (15min) and quenched with 0.125 M glycine. Chromatin was isolated by adding lysis buffer, then disrupted with a Dounce homogenizer. Lysates were sonicated and DNA sheared to an average length of 300-500 bp with Active Motif’s EpiShear probe sonicator (cat# 53051). Genomic DNA was prepared by treating with RNase, proteinase K and heat for de-crosslinking, followed by SPRI beads clean up (Beckman Coulter) and quantitation by Clariostar (BMG Labtech).
An aliquot of chromatin (30 ug) was precleared with protein A agarose beads (Invitrogen). Genomic DNA regions of interest were isolated using 4 ug of antibody against H3K27ac. Complexes were washed, eluted from the beads with SDS buffer, and subjected to RNase and proteinase K treatment. Crosslinks were reversed by incubation overnight at 65 C, and ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation.
ChIP Sequencing
Illumina sequencing libraries were prepared from the ChIP and Input DNAs by the standard consecutive enzymatic steps. Steps were performed on an automated system (Apollo 342, Wafergen Biosystems/Takara). After PCR amplification, the DNA libraries were sequenced on Illumina’s NextSeq 500 (75 nt reads, single end). Reads were aligned to hg19 using the BWA algorithm (default settings). Duplicate reads were removed, and only mapping quality >= 25 were used for further analysis. Alignments were extended in silico at their 3’-ends to a length of 200 bp and assigned to 32-nt bins along the genome. Published H3K27ac ChIP-seq sequencing data from primary DMGs were downloaded from GSE12874537. Peaks were called using MACS270 callpeak with -B –SPMR to save the fragment pileup per million reads track. The bdg-files were used to calculate fold enrichment and q-value tracks with MACS2 bdgcmp, transformed into bigwig files with rtracklayer, and visualized with copy number and SV calls in gtrack. Additional bigwig files for adult GBM H3K27ac, pHGG ATAC seq and non-cancerous or non-brain tissues were downloaded from GSE5479238, GSE12631939 and the Encode project40 respectively.
Hi-C
Library generation and sequencing
In-situ Hi-C libraries were generated from 5 million cultured H3.3K27M glioblastoma cell lines HSJ-019 and HSJ-031 as well as from H3.3K27M primary tumors HSJ-031 and 039 following published protocols71 with minor modifications. Briefly, (1) crosslinking cells with formaldehyde, (2) digesting the DNA using a 4-cutter restriction enzyme (e.g., DpnII) within intact permeabilized nuclei, (3) filling in, biotinylating the resulting 5′-overhangs and ligating the blunt ends, (4) shearing the DNA, (5) pulling down the biotinylated ligation junctions with streptavidin beads, (6) library amplification and (7) analyzing these fragments using paired end sequencing. Quality control for efficient sonication was performed through the combination agarose DNA gel electrophoresis and for appropriate size selection using the Agilent Bioanalyzer on final amplified libraries, followed by low-pass sequencing on the Illumina HiSeq 2500 (~30M reads/sample) to assess quality of the libraries using percent of reads passing filter, percent of chimeric reads, and percent of forward-reverse pairs.
Data processing
Additional Hi-C files for NPCs (neural progenitor cells) were downloaded from www.synapse.org/#!Synapse:syn12979101 (registration required; Data Download - Study “iPSC-HiC”)72. Analysis of Hi-C generated and downloaded fastq files was performed using Juicer73. Contact maps were generated using Juicer with the following parameters: -s DpnII -g hg19. Map resolution was determined by using Juicer’s “calculate_map_resolution.sh” script. Hi-C contact maps and associated annotations were visualized using Juicebox. The HIFI algorithm was used to process 5-kb resolution Hi-C data to obtain higher accuracy estimates of interaction frequencies, using the following parameters: bandSize=1000, outputNormalized, boundaryKS=1000. TAD boundaries were determined using RobusTAD74. Composite figure panels including HiC and other genomic data was created using plotgardener (https://github.com/PhanstielLab/plotgardener)
Luciferase reporter
Cell lines
pHGG cell line DIPG1375 was a gift from the Michelle Monje lab. BT245 was a gift from the Keith Ligon lab. Cell lines were grown in ULA flasks in a 1:1 ratio of Neurobasal A (Gibco) and DMEM/F-12 (Gibco) and 1% of each HEPES Buffer Solution 1M, Sodium Pyruvate Solution 100nM, MEM-NEAA Solution 10mM, Glutamax-I Supplement, and Penicillin/Streptomycin solution. The culture medium was supplemented with epidermal and fibroblast growth factor (H-EGF & H-FGF; StemCell Tech, Inc.) at 20 ng/mL, platelet derived growth factors (H-PDGF-AA & H-PDGF-BB StemCell Tech, Inc.) at 10 ng/mL, Heparin Solution (0.2%; StemCell Tech, Inc.) at 2 ug/mL, and 50X B-27 Minus Vitamin A (Invitrogen). Cells were passaged every 2-4 days and were dissociated into single cells at the time of passage using Accutase (StemCell Tech, Inc.).
Cell line authenticity and mycoplasma surveillance
Cell line authenticity was confirmed using STR profiling. All cell lines were monitored and negative for mycoplasma infection using the MycoAlert Mycoplasma Detection Kit (Lonza), following the manufacturer’s protocol.
Luciferase reporter construction
A lentiviral firefly luciferase reporter system was constructed from pGL4.26 (Promega) and the pLKO.1 backbone via Gibson Assembly. The pLKO.1 backbone was digested with FastDigest® KflI and EcoRI. The minimal promoter firefly reporter cassette was PCR amplified from pGL4.26 using the lucminP primer set (Supp. Table 6) using NEB Q5 polymerase. These two fragments were assembled into the lentiviral firefly luciferase reporter using the NEBuilder HiFi DNA Assembly Cloning Kit according to manufacturer’s instructions. The DNA sequence in the H3K27ac peak in the consensus CCDC26-SV amplicon was split into two fragments (E1/E2) and PCR amplified from DIPG13 genomic DNA with the primers listed in Supp. Table 6 using NEB Q5 polymerase. The resulting E1 and E2 fragments were cloned into the vector using KPN1 and NHE1 restriction sites. The lentiviral constitutively active pLX313-Renilla construct was obtained from Addgene (Plasmid # 118016) to serve as intrinsic control.
Viral Production
HEK-293T (ATCC CRL-3216) cells were cultured in T75 tissue culture treated flasks in DMEM (Gibco) supplemented with 10% FBS (Gemini Bio). Lipofectamine-3000 (Invitrogen) was used to transfect with plasmid of interest in addition to packaging plasmids VSV-G and psPAX2, according to manufacturer’s protocol. Media was replaced with DMEM supplemented with 20% FBS six hours after transfection. Media was harvested 24-48 hours post-transfection and virus concentrated (20x) using Lenti-X Concentrator (Takara Bio) per manufacturer’s protocol.
Lentiviral infection of BT245 and DIPG13
Cells were dissociated and plated in a 12-well tissue culture plate at a density of 1.5 million cells/mL. Concentrated virus was added to the media and the cells were centrifuged for 120 minutes at 850g and 30C. Cells were placed into T-75 ULA flask with selection (1μg/ml puromycin for the firefly reporter, 300μg/ml hygromycin for pLX313-Renilla) was added the following day to achieve survival of 40-80% in the infected conditions.
A549 Transduction
A549 (ATCC CCL-185) cells were cultured in a T75 tissue culture flask in RPMI (Gibco) supplemented with 10% FBS. Lipofectamine, following manufacturer’s protocol, was used to transduce the enhancer reporter plasmids.
Luciferase Reporter Readout
The Dual-Glo Luciferase Assay System (Promega) was used following manufacturers protocols for all measurements four days post spinfection (two days post puro selection).
Visualization and reconstruction of complex MYCN and RTK amplicons
JaBbA45 was used to generate cancer genome graphs using SvABA SV, GATK CNV and absolute purity and ploidy as inputs. Tracks were visualized in gGnome/gTrack which also calculated distances between loci in the cancer genome. Extrachromosomal amplicons were inferred by sub-setting to only circular path segments with CN > 20 and reconstructed with the gGnome walks() function.
Genomic loci recurrently incorporated into the amplicons were determined by the distribution of amplicons around the oncogene19,20 . TADs adjacent to the amplified oncogene were divided into 10kbp windows. Average copy number per 10kbp window was calculated in all tumors with an amplicon of CN >5 anywhere in the TAD of the oncogene (using the germinal zone TAD boundaries from GSE7756563). Among tumors with an amplification of CN>5 anywhere within the TAD of the oncogene, the fraction of tumors with an amplification in each 10kbp window was determined. The location of likely enhancer elements, necessary to drive expression of the amplified oncogene, was inferred from the direction of the skew of the observed distribution compared to the expected symmetric normal distribution.
GATA4/3’FGFR2 Interphase Enumeration FISH
Probe Specifics
GATA4/3’FGFR2 enumeration was analyzed with FISH. Human bacterial artificial chromosomes (BACs) covering the GATA4 gene region were identified using the University of California Santa Cruz (UCSC) August 2021 Assembly hg38. The GATA4 clones (RP11-241B23, RP11-235I5, and RP11-737E8) were labeled were labeled by nick translation with Spectrum Orange (Abbott Molecular) and the 3’FGFR2 clones (RP11-878D1, CTD-2542P10, RP11-984I17, CTD-2291K12, and CTD-3237E5) were labeled by nick translation with Spectrum Green. Labeled clones were combined to create an enumeration probe set.
Slide processing for Paraffin Embedded Tissue Samples
Slides were placed in a 90°C oven for 15 minutes. Slides were then deparaffinized with xylene (2 times, 15 minutes each) at room temperature (RT), dehydrated in 100% ethanol for 5 minutes at RT, and placed in 10mM Citric Acid (pH 6.0) and microwaved for 10 minutes. Following this, the slides were immersed in 2x standard saline citrate (SSC) for 5 minutes at 37°C followed by digestion in 0.2 % pepsin working solution (1.2 grams pepsin/600 mL 0.9% NaCl pH 1.5) at 37°C for 12 minutes. Immediately after digestion, the slides were dehydrated using an ethanol series (70, 85, 100%) 2 minutes each at RT. Working solution of GATA4/3’FGFR2 (Mayo Clinic laboratory developed probe) was made by mixing 2 °L of concentrated 3’FGFR2 probe and 1 μL of concentrated GATA4 probe with 7 μL of LSI/WCP® hybridization buffer (Abbott Laboratories). The working solution was applied to the target areas, coverslipped, co-denatured with a ThermoBrite® at 83°C for 5 minutes, and hybridized overnight in a 37°C humidified oven. Following hybridization, slides were soaked in RT 2xSSC/0.1% NP-40 to remove coverslips, placed in 2xSSC/0.1% NP-40 at 74°C for 2 minutes and then placed into RT 2xSSC/0.1% NP-40 for 2 min. The slides were stained with 4’−6,-diamidino-2-phenylindole (DAPI) (Vector Laboratories) and coverslipped.
ID2/MYCN Metaphase FISH
Probe Specifics
ID2/MYCN enumeration was analyzed with FISH. Human bacterial artificial chromosomes (BACs) covering the ID2 gene region were identified using the University of California Santa Cruz (UCSC) August 2021 Assembly hg38. The ID2 clone (CTD-2131H8) was labeled by nick translation with Spectrum Green (Abbott Molecular) and the MYCN probe was commercially available from Abbott Molecular. ID2 probe and MYCN probe were combined to create an enumeration probe set.
Slide processing for Metaphase Samples
Slides were air dried at RT overnight. Following this, the slides were immersed in 2x standard saline citrate (SSC) for 30 minutes at 37°C. The slides were dehydrated using an ethanol series (70, 85, 100%) 2 minutes each at RT. Working solution of ID2/MYCN was made by mixing 2 μL of concentrated ID2 probe and 1 μL of concentrated MYCN probe with 7 μL of LSI/WCP® hybridization buffer (Abbott Laboratories). The working solution was applied to the target areas, coverslipped, co-denatured with a ThermoBrite™ at 73°C for 5 minutes and hybridized overnight in a 37°C humidified oven. Following hybridization, slides were soaked in RT 2xSSC/0.1% NP-40 to remove coverslips, placed in 2xSSC/0.1% NP-40 at 74°C for 2 minutes and then placed into RT 2xSSC/0.1% NP-40 for 2 min. The slides were stained with 10% 4’−6,-diamidino-2-phenylindole (DAPI) (Vector Laboratories) and coverslipped.
SNV signature analysis
De novo SNV signature extraction was performed using Bayesian NMF in SignatureAnalyzer24,25. The resulting SNV signatures were compared to the COSMICv3 SBS signatures using cosine similarity to annotate known etiologies and signature names. DeconstructSig76 extracted SBS signatures with the highest degree of similarity to the de novo signatures, including a designation of “unknown”.
Signature integration and definition of signature clusters with similar variant generating processes
To better understand the information contained in each of the 9 SV and 14 SNV signatures, consensus clustering was applied to the tumor x signature proportion matrix, comprising the 23 values representing the proportion of each of the SV and SNV signatures out of all SV and SNV signatures in each tumor. The proportions for each signature were median-centered across all tumors before consensus clustering with the ConsensusClusterPlus R package using the parameters: reps=1000, pItem=0.9, pFeature=0.9, clusterAlg="hc",distance="spearman". The resulting most stable and informative clusters were named “Complex-SV” and “SNV-dominant”, after the signatures with the highest enrichment in the cluster.
Chromothripsis and Extrachromosomal amplicons
To define regions of chromothripsis, we used Shatterseek77. Regions in the genome with CN > 10 extending for more than 50kbp were defined as likely-extrachromosomal or derived from an extrachromosomal stage.
Comut plots and variant combination matrix
SNVs were annotated using Oncotator. SVs were annotated and linked to a gene based on if the SV breakpoints were in exons of the gene (named ‘coding SV’), intronic (‘intron SV’) or in the TAD of the gene (‘flank SV’). Absolute purity- and ploidy-adjusted copy number (CN) was determined for every gene using the width-weighted mean CN from all segments overlapping the gene.
To create the variant combination matrix, we subset to only Cancer Gene Census46 genes and genes which showed significantly recurrent variants in this cohort. For SNVs, the variant classification was simplified to truncating_snvs = ("Nonsense_Mutation", "Frame_Shift_Del", "Frame_Shift_Ins", "Splice_Site", 'Start_Codon_SNP', 'START_CODON_SNP', 'Translation_Start_Site') and missense.snvs = ("Missense_Mutation", "In_Frame_Del", "Stop_Codon_Del", 'DE_NOVO_START_IN_FRAME', 'DE_NOVO_START_OUT_FRAME', 'Nonstop_Mutation', 'In_Frame_Ins', 'START_CODON_INS'). SCNAs of genes with a ploidy- and purity-adjusted copy number of <0.4 were annotated as ‘homdel’, CN > 5.4 as amp, CN > 10 as ExChr_amp and amplifications covering only parts of a gene with a CN > 3.1 as ‘part.amp’ based on the CN histogram across all tumors defining recurrent CN states. A genetic variant had to recur in at least three samples (excluding the hypermutant samples for SNVs) to be kept in the matrix. For SV in the TAD of a gene (‘flank SV’) this threshold was increased to at least ten. GISTIC peaks in each sample was used to incorporate the SCNAs of lower amplitude.
cbioportal oncoprinter was used to visualize variants. Column order represents the samples within each subgroup determined by hierarchical clustering (HC). HC with one minus spearman rank correlation metric was applied in Ext. Data Fig. 6C and HC with one minus cosine similarity metric on the respective subsets of variant combination matrix for Fig. 6A and Ext. Data Fig. 8C with average linkage in all cases. Genes of interest were manually selected based on the variants with the highest enrichment in the subgroups.
Distances between sample groups in variant space
We calculated Jaccard distances between genetic profiles across 369 variants in genes from the Cancer Gene Census46 for every pair of tumors. Tumor groups were determined by mutations in H3.1 or H3.3 and complex SV signature contribution more or less than 20% to all SV signature activity.
Variant timing analysis
palimpsest78 R package determined single patient timing of SNVs79. SNVs were classified into clonal vs. subclonal based on their cancer cell fraction (CCF, variant allele fraction adjusted by local copy number and purity/ploidy). SNVs overlapping with SCNA could further be timed into early or late depending on if they happened before or after the SCNA51. MutationTimeR51 was used to determine the timing of SCNAs in individual patients, including clone clusters as input. Mobster80 was used to define the clone clusters based on the distribution of the absolute62 CCFs. The resulting molecular time for the SCNA segments was assigned to the GISTIC peaks present in the respective sample with a width-weighted mean and categorized based on the timing quarters. For each subgroup, in addition to the timed GISTIC peaks, single patient timed SNVs in consensus cancer genes were tallied into winning tables reflecting the frequency of this variant being an early event using published code79. The BradleyTerryScalable R package was applied to estimate the winning probability across the subgroup for each variant, which in this setting is a measure for the probability of this variant being an early event in the tested subgroup. The Bayesian maximum a posteriori probability (MAP) estimate was used to fit the model as previously described79. To control for outlier samples the analysis was performed on 100 random samples of 70% of each subgroup. The resulting distributions for the strength parameters (on log scale) were plotted for variants recurrent at least three times in the tested subgroup.
Survival analysis
Univariate correlations for differences in survival were analyzed using the Kaplan-Maier method and significance was determined with a log-rank test. Spearman rank correlation tests were used to determine correlations between overall survival and the complex-SV signature activity. This was possible because all children with DMGs died within the observed period, resulting in an absence of censored data. Variables included in the multivariate analysis (Cox-model) were Histone-SNV, age, TP53 status combined Complex-SV signature activity.
Statistics and Reproducibility
No statistical method was used to predetermine sample size. Three previously published samples were excluded because their fastq files could not be successfully realigned using our pipelines. Exclusion criteria were pre-established. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment. All statistical analyses were performed in R 3.6.3. Unless otherwise indicated, statistical comparisons were performed using Fisher's exact tests or Wilcoxon tests, as appropriate. The data met the assumptions of the statistical tests used. Unless otherwise specified data was assumed to be not normal but this was not formally tested. p-values < 0.05 were considered significant. Multiple testing was accounted for by using false discovery rate q-values unless otherwise indicated. In all box plots the boxes represent the range between the 25th and 75th percentiles and the central line indicates the median. Statistical comparisons for the luciferase reporter were performed in Prism 9 using a Nested One-Way Anova and Tukey’s Multiple Comparison Test.
Extended Data
Extended Data Fig. 1. Sample characteristics and significantly recurrent variants.
(A) Purity of pre-treatment biopsy and autopsy samples were not significantly different (p = 0.5, two-sided Wilcoxon, n= 174 tumors, center line of the boxplot indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5xIQR). (B) Significantly recurrent SNVs in non-hypermutant tumors (n= 179 tumors). (C) Significantly recurrent SCNAs (n= 179 tumors). All of these SCNAs have been noted3 except for a non-protein-coding locus in 8q.24.21, near MYC—which is also within a separate recurrently amplified locus. (D) Q-Q plot for the analysis of significantly recurrent SV breakpoints. The most significantly recurrent breakpoints are within the long non-coding RNA CCDC26, within the TAD encompassing MYC (based on n= 179 tumors). (E) Representative examples of the enhancer amplification through simple tandem-duplications within the long non-coding RNA encoding CCDC26.
Extended Data Fig. 2. Lineage specificity of the enhancer peak in CCDC26.
(A) ATAC-seq (top) and H3K27ac ChIP-seq (bottom) enrichment (vertical axis) of samples from different lineages (indicated on right; “27y” indicates the sample was obtained from a 27-year-old person) across the TAD encompassing MYC (horizontal axis). The location of the MYC coding sequence is highlighted in red. The CCDC26 amplicon boundaries for the 15 samples with the amplicon are indicated by the paired red arrows at the top. The consensus amplicon is indicated by the green dotted lines and centers on an H3K27ac peak present only in glial samples. (B) Hi-C heatmaps depicting DNA interaction profiles (5 kb resolution) from a midline glioma (top), iPSC-derived neural progenitor cells (2nd from top) and two cell lines harboring H3.3K27M mutations (bottom). Red and white indicate high and low interaction frequencies, respectively. MYC interacts more frequently with the H3K27ac peak within CCDC26 (black oval) relative to neighboring loci. The minimal common region of the CCDC26 amplicon is indicated at the bottom of the heatmaps (SV MCR; blue rectangle). (C-D) Correlation between MYC expression and genomic copy number of (C) its enhancer amplified in the CCDC26-SV (p = 0.01, two-sided Spearman rank correlation test, samples with MYC CN>2.5 excluded, n=94 tumors), or (D) the MYC coding sequence (p = 0.0003, two-sided Spearman rank correlation test, n=114 tumors). (E-F) Lineage specificity of E1 enhancer activity in (E) neural lineage/BT245 p-value(E1vs.Backbone) = 0.0056; p-value(LUAD vs. Backbone) = 0.99 and (F) lung epithelial lineage/A549, p-value(E1 vs. Backbone) = 0.96 ; p-value(LUAD vs. Backbone) = 0.0071, n = 3 independent experiments, Nested One-Way Anova: Tukey’s Multiple Comparisons. Analogous to Fig. 1G; center line of the boxplot indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5xIQR.
Extended Data Fig. 3. Significantly recurrent juxtaposition between MYCN and ID2.
(A) (top) Count matrix showing all possible juxtapositions between pairs of genomic loci. (bottom) Illustration of the principle behind the analysis of recurrent juxtapositions, as exemplified by the MYCN-ID2 loci. First, we count the number of SVs connecting each pair of genomic loci. Using a background model for the probability of juxtapositions generated from an analysis of 2658 cancers21, we then determine the probability of observing this number of SVs due to chance alone, corrected for multiple hypothesis testing. This analysis revealed the MYCN-ID2 juxtaposition as the only significantly recurrent juxtaposition in the window shown. (B) Overlay of amplification frequencies on ChIP-seq data in the ID2 and MYCN loci. The top two tracks show, among tumors with MYCN-ID2 rearrangements (top track, n=4 tumors or MYCN amplifications without ID2 involvement (second track, n=4 tumors), the percentage of tumors with amplifications (y-axis) at each genomic locus (x-axis). The bottom eight tracks indicate H3K27ac ChIP-seq profiles across these loci for four H3K27M and four H3WT pHGGs tumors. Coding sequences of ID2 and MYCN are highlighted with yellow and red lines respectively. Significantly enriched H3K27ac peaks (q-value < 0.01) are indicated below each ChIP-seq track. The small region at the ID2 locus that is amplified in all MYCN-ID2 pHGGs shows an H3K27ac peak in the ChIP tracks from all six pHGG tumor samples. Tumors that amplify MYCN without ID2 take in a much larger region of the MYCN TAD into the amplicon. (C) G-track plots indicating copy-number profiles and genome topology after consideration of local SVs, for two examples of pHGGs with focal MYCN amplicons without incorporation of ID2. For both tumors the copy number and SV profiles support several possible reconstructions of extrachromosomal circular amplicons. All are limited to the neighborhood of MYCN, presumably incorporating endogenous enhancers from the MYCN TAD.
Extended Data Fig. 4. Structures of recurrent RTK amplicons.
(A) Average amplicon profile for all pHGGs with amplifications in the PDGFRA TAD reaching a copy number (CN) of at least four (n = 15 tumors). The top track shows the percentage of those tumors with amplifications (vertical axis) at each location (horizontal axis). The track below shows the average CN across all tumors with amplifications in the PDGFRA TAD. The segments included in the PDGFRA amplicon in 80% of tumors are highlighted in the red box. Most amplicons range over several Mbp, often including KIT. (B) Average amplicon profile for all pHGGs with amplifications in the EGFR TAD reaching at least four copies (n = 7 tumors) displayed as in (A). The segments included in all the EGFR amplicons are highlighted in the red box. The pHGG EGFR amplicons always include upstream enhancers elements around SEC61G. (C) Structure of a simple EGFR-TAD amplicon that encompasses enhancers that are also amplified in all tumors with EGFR amplifications. (D) SVs, CN tracks and reconstructions for all pHGGs with high-level MET amplifications. The observed high-level MET amplicons are a few 100kbp in size. Three out of four MET-amplified pHGGs incorporate a downstream region including an enhancer (see bottom H3K27ac track) into the amplicon. For all four MET-amplified pHGGs possible reconstructions of the extrachromosomal amplicon are shown above the CN and SV track. (A-D) From bottom to top the tracks show: the genes of interest at the location, a q-value H3K27ac track calculated from eight pHGG tumor samples, the CN and SV for the indicated tumor at the location and reconstructions of possible extrachromosomal amplicons if applicable. (E) FISH with probes for the GATA4 locus on chr8 (red) and the FGFR2 locus on chr10 (green) in tissue from a pHGG with SVs within high-level amplicons that connect these two loci. Scale bar indicates 2μm. Representative image from n=200 nuclei.
Extended Data Fig. 5. Associations between SV signatures, genetic variants and SNV signatures.
(A) The statistical significance (as determined by Wilcoxon Tests) of positive (enriched, shown in red) and negative (depleted, shown in blue) associations between each SV signature and of all recurrently altered somatic genetic alterations that are documented in the Cancer Gene Census46. Shading within each box indicates level of significance as determined by the q value. (B) De-novo extracted SNV signatures (based on n=179 tumors). (C) Cosine similarity between de novo extracted SNV signatures and the COSMICv3 SBS-signatures. (D) SNV signature activity in every tumor. The hypermutant tumors on the left show signatures associated with hypermutation in COSMICv3. Signature 3, which is similar to the SBS3 homologous recombination deficiency signature, features prominently in many non-hypermutant tumors.
Extended Data Fig. 6. SV signatures in pHGG based on size, SV type and complexity.
(A) The horizontal axis indicates the size and type of SVs. Del stands for deletion, dup for duplication, inv for inversion, and int for interchromosomal rearrangement. The vertical axis indicates the fraction of SVs within each signature that are contributed by each SV type (based on n=179 tumors). (B) The statistical significance of positive (enriched) and negative (depleted) associations (Wilcoxon Tests) between each SV signature and of all recurrently altered somatic genetic alterations that are documented in the Cancer Gene Census46. (C) Consensus clustering of the normalized SNV and SV signature activities in each tumor sample (columns). Rows indicate signature activities (top) and potentially oncogenic variants (bottom). (D) Correlations between SV and SNV signatures. Signature labels from this analysis are indicated on the left; the nearest COSMICv3 signatures are indicated on the right, with their proposed mechanisms in parentheses. Complex-SV signatures show a close correlation with APOBEC and homologous recombination deficiency SNV signatures (SBS3). q-values are based on Spearman rank correlations. (E) Enrichment analysis for signature activities in each cluster from panel B. FDR q-values are based on Wilcoxon tests. (F) Significance of signature cluster associations for all variants with correlations reaching q < 0.1; q-values are based on Fisher’s exact tests. Tumors in the complex-SV clusters are enriched for copy-number changes in cancer genes and SNVs in TP53, whereas simple-SV pHGGs tend to exhibit SNVs in different cancer genes. (G) Number of SVs per tumor in each cluster (n=179 tumors). All differences are significant to q<0.003 by Wilcoxon tests, center line of the boxplot indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5xIQR.
Extended Data Fig. 7. Chromothripsis and homology length at the breakpoints in size/SV-type/complexity SV signatures.
(A-B) SV signature activities in samples that contain or lack (A) chromothriptic regions or (B) extrachromosomal amplifications (n=179 tumors, center line indicates the median). (C) SV signature analysis including homology length channels reveals five signatures. The horizontal axis indicates the size and type of SVs. Del stands for deletion, dup for duplication, inv for inversion, and int for interchromosomal rearrangement. The vertical axis indicates the amount of SVs within each signature that are contributed by each SV type. (D) SV signature activity in every tumor of the homology SVsigs. Tumors with higher SV counts on the left show complex-SV signature activity whereas tumors with lower SV counts on the right show a mix of simple-SV signatures mimicking the SVsig distribution without homology information (n=179 tumors). (E) By sample correlation between SVsigs with and without homology
Extended Data Fig. 8. Associations between SV-defined groups and extended co-mut plot split by histone groups.
(A) Correlations between SVs and SNV signatures in included in the COSMICv3 signatures database. SNV signatures with well-established links to mechanisms are indicated in parentheses. These include mismatch repair (MMR), homologous recombination (HR) deficiency, APOBEC and aging. q-values were calculated using Spearman rank correlations. (B) Enrichment analysis for signature activities present in Complex-SV and SNV-dominant clusters. q-values were calculated using Wilcoxon tests. (C) Co-mut plot of the 176/179 (98.3%) tumors with somatic variants in at least one well-known oncogene. Columns represent tumors, ordered within histone mutation-defined subgroups by hierarchical clustering of all potential driver variants. The top two rows show signature metadata.
Extended Data Fig. 9. Associations between SV + histone defined groups.
(A) Jaccard distances between tumor pairs (vertical axis), calculated from the combination of variants in each tumor, across subgroups of H3K27M DMGs. Tumor groups were determined by H3.1 or H3.3 mutations and the combined complex SV signatures exceeding 20% of all SV signature activity. Tumors were paired within or between these groups, as indicated on the horizontal axis. All differences were significant with q < 0.005 (FDR-corrected two-sided Wilcoxon test) unless indicated otherwise. n = 165 DMGs) (B) Association between MDM4 expression (vertical axis) and copy number (horizontal axis). MDM4 gains universally represent arm-level gains of 1q. *** indicates adjusted p = 0.004, ANOVA with two-sided Tuckey post-test, n = 114 tumors. (C-D) Volcano plot indicating the significance (vertical axis; FDR-corrected Fisher’s exact tests) of associations between genetic variants and pHGG subgroups (horizontal axis). (C) Arm-level SCNAs in TP53-disrupted (n=97 tumors) vs TP53WT pHGGs (n = 77 tumors). TP53 disruption represented SNVs (n=88 tumors, often with copy loss) or copy loss alone (n=9 tumors). (D) Arm-level SCNAs in TP53-disrupted (n=56 tumors) vs TP53WT H3K27M mutant DMGs (n = 39 tumors). TP53 disruption represented SNVs (n=53 tumors often with copy loss) or copy loss alone in (n=3 tumors). Only significantly recurrent arm-level SCNAs are shown. (E-F) Comparison between pre-treatment biopsy and autopsy samples. These groups exhibit no significant differences in (E) the number of SVs per sample (q= 0.6, Wilcoxon, n= 174 tumors) or (F) the activity of the combined complex-SV signatures (q= 0.7, Wilcoxon, n= 174 tumors). Center line of the boxplots indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5xIQR. (G) Kaplan-Maier plot indicating overall survival for (top) H3.1K27M and H3.3K27M DMGs and (bottom) H3.3K27M DMG with and without TP53 SNVs. p-values are from log-rank tests. Error bands show the 95% confidence interval.
Extended Data Fig. 10. Timing analysis of somatic variant acquisition in histone mutation-defined pHGG subgroups.
For each subgroup the individual (per-sample) timing of recurrent variants is fed into a Bradley-Terry model. This results in a strength parameter for each variant which is indicated on the x-axis in log scale and can be interpreted as the relative log odds of the variant being an early event in this subgroup. Each distribution indicates the results of 100 random subsamples of the respective subgroup. Only potential driver variants recurrent in more than two samples are shown. Subgroups: (A) H3.1K27M, n = 24 tumors (B) H3.3K27M, n = 73 tumors (C) H3.3G34R, n = 14 tumors (D) H3WT, n = 63 tumors (E) hypermutant pHGGs, n = 5 tumors. Center line of the boxplot indicates the median, bounds of the box the 25th and 75th percentiles and whiskers extend from the box to the largest or smallest value no further than 1.5xIQR
Supplementary Material
Acknowledgements
We thank and acknowledge the children and their families who made this research possible through their participation in pHGG clinical trials and tissue banking protocols. We also thank the many clinicians and researchers who led the DIPG BATS trial. We would also like to thank the members of the Ligon, Beroukhim and Bandopadhayay Laboratories, in addition to Ruolin Liu, Justin Rhoades, Matthew Leventhal, Justin Hwang, Jonathan So, David Mayhew, Jonathan Rennhack, Mehdi Touat, Andrew Cherniack, Beatrice Rondelli, Sophie Polo for their support and helpful discussions, Active Motif®, Inc for performing the H3K27ac ChIP seq and HiC of the CCDC26-SV tumors and the Mayo Clinic’s Cytogenetics Core, specifically Director Patricia T. Greipp, D.O., Carolyn Brandt, Troy Gliem, and Ryan Knudson for FISH analyses.
The authors would like to thank and acknowledge the following funding sources:
The NIH: NIH R37 5R37CA255245-02 (P.B.), NIH R01 CA188228 (R.B., K.L.L.), R01 CA215489 (R.B., K.L.L.), R01 CA219943 (R.B., K.L.L.); The German Research Foundation (DFG) (research fellowship supporting FD), Fund for Innovative Cancer Informatics (RB), the Gray Matters Brain Cancer Foundation (RB), The Bridge Project of MIT and Dana- Farber/Harvard Cancer Center (RB, KLL), The Sontag Foundation (RB), The V Foundation for Cancer Research (PB), Michael Mosier Defeat DIPG Foundation (PB), The ChadTough Foundation (PB), The St. Baldrick’s Foundation (RB, PB, JWT), Prayers from Maria Foundation (PB, MWK), Pediatric Brain Tumor Foundation (PB, RB), Friends of DFCI (PB, RB), The Giving for Gabi Fund (PB), Alex’s Lemonade Stand Foundation (PB, RB, JWT), The Broad Institute Escape Velocity Award (PB), The Jared Branfman Sunflowers for Life Fund (PB, RB), McKennaClaire Foundation (PB, MWK), We Love You Connie Foundation (PB, RB, MWK, NJ), Imagine for Margo and Lemos Family (JG), Frederic Lemos and the Gustave Roussy Foundation (JG), The Pussycat Foundation Helen Gurley Brown Presidential Initiative (JWT, PB), Canadian Institutes of Health Research (NJ, CLK), Genome Canada (CLK), Compute Canada and Calcul Quebec (CLK), Zach Carson Fund (MWK, RB, PB), Ellie Kavalieros DIPG Fund (MWK, RB, PB), Ryan Harvey Fund (MWK, RB, PB), Mikey Czech DIPG Foundation (MWK, RB, PB), Caroline Cronk Fund (MWK, RB, PB), Markoff Art in Giving Foundation (MWK, RB, PB), Brock Fleming Fund (MWK, RB, PB), Stop and Shop Pediatric Brain Tumor Program (MWK, RB, PB, KDW, SNC), the DIPG Collaborative (MWK, RB, PB), The Cure Starts Now (RB). The Mayo Clinic Cytogenetics Core Laboratory is supported by the Mayo Clinic Comprehensive Cancer Center Grant, funded by National Cancer Institute. (P30CA15083)
Footnotes
Conflict of Interest
RB and PB receive grant funding from the Novartis Institute of Biomedical Research for an unrelated project, RB receives grant funding from Merck and PB has received grant funding from Deerfield Therapeutics for an unrelated project, P.B consults for QED Therapeutics, R.B. consults for and owns equity in Scorpion Therapeutics, reports consulting or advisory role for Novartis, Merck (I), Gilead Sciences (I), ViiV Healthcare (I); research funding from Novartis; patents, royalties, other intellectual property—Prognostic Marker for Endometrial Carcinoma (US patent application 13/911456, filed June 6, 2013), SF3B1 Suppression as a Therapy for Tumors Harboring SF3B1 Copy Loss (international application No. WO/2017/177191, PCT/US2017/026693, filed July 4, 2017), Compositions and Methods for Screening Pediatric Gliomas and Methods of Treatment Thereof (international application No. WO/2017/132574, PCT/US2017/015448, filed 1/27/2017). KLL reports grants and personal fees from BMS, grants from Amgen, personal fees and other from Travera LLC, personal fees from Rarecyte, grants from Tragara, grants from Lilly, grants from Deciphera, grants from X4, grants from Charles River Labs, grants from Specicare, outside the submitted work; has a patent US20160032359A1 pending. MWK is now an employee of Day One Biopharmaceuticals. LCG is the Co-Founder of Thromboprotea Inc. All the remaining authors declare no competing interests.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
De-novo generated sequencing data from this study are accessible under dbGaP Accession Number phs002380.v1.p1
Previously published sequencing data4-7 that were re-analysed here are available under accession code EGAS00001000575, EGAS00001001139, EGAS00001000572, EGAS00001000192, GSE12874537, GSE5479238, GSE12631939 and the Encode project40. Cosmic signatures and cancer genes are available at: https://cancer.sanger.ac.uk/cosmic/download. TAD boundaries are from GSE7756563.
Source data have been provided as Source Data files. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code Availability
Publicly available software was used as indicated in the methods. Main custom analysis code is available at: https://github.com/FrankDubois/pHGG_SVs. All custom code to connect and reformat the outputs of the publicly available software as well as code to generate the figures is available upon request.
References
- 1.Ostrom QT et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro-Oncology 20, iv1–iv86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puget S et al. Mesenchymal Transition and PDGFRA Amplification/Mutation Are Key Distinct Oncogenic Events in Pediatric Diffuse Intrinsic Pontine Gliomas. PLOS ONE 7, e30313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackay A et al. Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell 32, 520–537.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender S et al. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nature Medicine 22, 1314 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Buczkowicz P et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nature Genetics 46, 451–456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu G et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nature Genetics 46, 444–450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor KR et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nature Genetics 46, 457–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu G et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nature Genetics 44, 251 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartzentruber J et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Shoshani O et al. Chromothripsis drives the evolution of gene amplification in cancer. Nature (2020) doi: 10.1038/s41586-020-03064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koche RP et al. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nature Genetics 52, 29–34 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nature Genetics 52, 891–897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner KM et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543, 122–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu K et al. Structure and evolution of double minutes in diagnosis and relapse brain tumors. Acta Neuropathologica 137, 123–137 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Northcott PA et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandopadhayay P et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nature Genetics 48, 273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beroukhim R, Zhang X & Meyerson M Copy number alterations unmasked as enhancer hijackers. Nature Genetics 49, 5–6 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Chen CCL et al. Histone H3.3G34-Mutant Interneuron Progenitors Co-opt PDGFRA for Gliomagenesis. Cell (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helmsauer K et al. Enhancer hijacking determines extrachromosomal circular MYCN amplicon architecture in neuroblastoma. Nature Communications 11, 5823 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morton AR et al. Functional Enhancers Shape Extrachromosomal Oncogene Amplifications. Cell 179, 1330–1341.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rheinbay E et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature (2020) doi: 10.1038/s41586-020-1965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nik-Zainal S et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y et al. Patterns of somatic structural variation in human cancer genomes. Nature 578, 112–121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nature Genetics 48, 600–606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandrov LB et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Degasperi A et al. A practical framework and online tool for mutational signature analyses show intertissue variation and driver dependencies. Nature Cancer 1, 249–263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angus L et al. The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nature Genetics 51, 1450–1458 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morganella S et al. The topography of mutational processes in breast cancer genomes. Nature Communications 7, 11383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bayard Q et al. Cyclin A2/E1 activation defines a hepatocellular carcinoma subclass with a rearrangement signature of replication stress. Nature Communications 9, 5235 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puget S et al. Biopsy in a series of 130 pediatric diffuse intrinsic Pontine gliomas. Child’s Nervous System 31, 1773–1780 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Roujeau T et al. Stereotactic biopsy of diffuse pontine lesions in children. Journal of Neurosurgery: Pediatrics PED 107, 1–4. [DOI] [PubMed] [Google Scholar]
- 32.Cage TA et al. Feasibility, safety, and indications for surgical biopsy of intrinsic brainstem tumors in children. Child’s Nervous System 29, 1313–1319 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Gupta N et al. Prospective feasibility and safety assessment of surgical biopsy for patients with newly diagnosed diffuse intrinsic pontine glioma. Neuro-Oncology 20, 1547–1555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mermel CH et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biology 12, R41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wala JA et al. SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Research 28, 581–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nature Genetics 48, 176–182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krug B et al. Pervasive H3K27 Acetylation Leads to ERV Expression and a Therapeutic Vulnerability in H3K27M Gliomas. Cancer Cell 35, 782–797.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suvà ML et al. Reconstructing and Reprogramming the Tumor-Propagating Potential of Glioblastoma Stem-like Cells. Cell 157, 580–594 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagaraja S et al. Histone Variant and Cell Context Determine H3K27M Reprogramming of the Enhancer Landscape and Oncogenic State. Molecular Cell 76, 965–980.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunham I et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paolella BR et al. p53 Directly Represses Id2 to Inhibit the Proliferation of Neural Progenitor Cells. STEM CELLS 29, 1090–1101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frankell AM et al. The landscape of selection in 551 esophageal adenocarcinomas defines genomic biomarkers for the clinic. Nature Genetics 51, 506–516 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pathania M et al. H3.3K27M Cooperates with Trp53 Loss and PDGFRA Gain in Mouse Embryonic Neural Progenitor Cells to Induce Invasive High-Grade Gliomas. Cancer Cell 32, 684–700.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flavahan WA et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hadi K et al. Distinct Classes of Complex Structural Variation Uncovered across Thousands of Cancer Genome Graphs. Cell 183, 197–210.e32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sondka Z et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nature Reviews Cancer 18, 696–705 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoopes JI et al. APOBEC3A and APOBEC3B Preferentially Deaminate the Lagging Strand Template during DNA Replication. Cell Reports 14, 1273–1282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffman LM et al. Clinical, Radiologic, Pathologic, and Molecular Characteristics of Long-Term Survivors of Diffuse Intrinsic Pontine Glioma (DIPG): A Collaborative Report From the International and European Society for Pediatric Oncology DIPG Registries. Journal of Clinical Oncology 36, 1963–1972 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zack TI et al. Pan-cancer patterns of somatic copy number alteration. Nature Genetics 45, 1134 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LETT JT, CALDWELL I, DEAN CJ & ALEXANDER P Rejoining of X-ray Induced Breaks in the DNA of Leukaemia Cells. Nature 214, 790–792 (1967). [DOI] [PubMed] [Google Scholar]
- 51.Gerstung M et al. The evolutionary history of 2,658 cancers. Nature 578, 122–128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffman LM et al. Spatial genomic heterogeneity in diffuse intrinsic pontine and midline high-grade glioma: implications for diagnostic biopsy and targeted therapeutics. Acta Neuropathologica Communications 4, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nikbakht H et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nature Communications 7, 11185 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salloum R et al. Characterizing temporal genomic heterogeneity in pediatric high-grade gliomas. Acta Neuropathologica Communications 5, 78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vinci M et al. Functional diversity and cooperativity between subclonal populations of pediatric glioblastoma and diffuse intrinsic pontine glioma cells. Nature Medicine 24, 1204–1215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castel D et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathologica 130, 815–827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khuong-Quang D-A et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathologica 124, 439–447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cho SW et al. Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element. Cell 173, 1398–1412.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu S et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 575, 699–703 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rausch T et al. Genome Sequencing of Pediatric Medulloblastoma Links Catastrophic DNA Rearrangements with TP53 Mutations. Cell 148, 59–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 61.Lawrence MS et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carter SL et al. Absolute quantification of somatic DNA alterations in human cancer. Nature Biotechnology 30, 413 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Won H et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Imielinski M, Guo G & Meyerson M Insertions and Deletions Target Lineage-Defining Genes in Human Cancers. Cell 168, 460–472.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith DI, Zhu Y, McAvoy S & Kuhn R Common fragile sites, extremely large genes, neural development and cancer. Cancer Letters 232, 48–57 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Aguet F et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson WE, Li C & Rabinovic A Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Leek JT, Johnson WE, Parker HS, Jaffe AE & Storey JD The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biology 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rao SSP et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rajarajan P et al. Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science (1979) 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Durand NC et al. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Systems 3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dali R, Bourque G & Blanchette M RobusTAD: A Tool for Robust Annotation of Topologically Associating Domain Boundaries. bioRxiv 293175 (2018) doi: 10.1101/293175. [DOI] [Google Scholar]
- 75.Grasso CS et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nature Medicine 21, 555–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosenthal R, McGranahan N, Herrero J, Taylor BS & Swanton C deconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biology 17, 31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cortés-Ciriano I et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nature Genetics 52, 331–341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shinde J et al. Palimpsest: an R package for studying mutational and structural variant signatures along clonal evolution in cancer. Bioinformatics 34, 3380–3381 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Amin SB et al. Comparative Molecular Life History of Spontaneous Canine and Human Gliomas. Cancer Cell 37, 243–257.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caravagna G et al. Subclonal reconstruction of tumors by using machine learning and population genetics. Nature Genetics 52, 898–907 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
De-novo generated sequencing data from this study are accessible under dbGaP Accession Number phs002380.v1.p1
Previously published sequencing data4-7 that were re-analysed here are available under accession code EGAS00001000575, EGAS00001001139, EGAS00001000572, EGAS00001000192, GSE12874537, GSE5479238, GSE12631939 and the Encode project40. Cosmic signatures and cancer genes are available at: https://cancer.sanger.ac.uk/cosmic/download. TAD boundaries are from GSE7756563.
Source data have been provided as Source Data files. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Publicly available software was used as indicated in the methods. Main custom analysis code is available at: https://github.com/FrankDubois/pHGG_SVs. All custom code to connect and reformat the outputs of the publicly available software as well as code to generate the figures is available upon request.