Summary
High-throughput screening is a powerful platform that can rapidly provide valuable cytotoxic, immunological, and phenotypical information for thousands of compounds. Human peripheral blood mononuclear cells (PBMCs) cultured in autologous plasma can model the human immune response. Here, we describe a protocol to stimulate PBMCs for 72 h and measure cytokine secretion via AlphaLISA assays and cell surface activation marker expression via flow cytometry. Cryopreserved PBMCs are incubated for 72 h with various small molecule libraries and the supernatants are harvested to rapidly measure secretion levels of key cytokines (tumor necrosis factor alpha, interferon gamma, interleukin 10) via the AlphaLISA assay. Almost simultaneously, the cells can be fixated and stained using antibodies against innate immune activation markers (CD80, CD86, HLA-DR, OX40) for analysis via flow cytometry. This multiplexed readout workflow can directly aid in the phenotypic identification and discovery of novel immunomodulators and potential vaccine adjuvant candidates.
For complete details on the use and execution of this protocol, please refer to Chew et al.1
Subject areas: Flow Cytometry/Mass Cytometry, Cell-based Assays, High-Throughput Screening, Immunology, Biotechnology and bioengineering
Graphical abstract

Highlights
-
•
A rapid, high-throughput workflow for screening small molecule immunomodulation
-
•
Quantification of human peripheral blood mononuclear soluble cytokine expression
-
•
Measurement of cell surface immunological biomarker expression via flow cytometry
-
•
Reproducible high-throughput platform for potential adjuvant discovery
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
High-throughput screening is a powerful platform that can rapidly provide valuable cytotoxic, immunological, and phenotypical information for thousands of compounds. Human peripheral blood mononuclear cells (PBMCs) cultured in autologous plasma can model the human immune response. Here, we describe a protocol to stimulate PBMCs for 72 h and measure cytokine secretion via AlphaLISA assays and cell surface activation marker expression via flow cytometry. Cryopreserved PBMCs are incubated for 72 h with various small molecule libraries and the supernatants are harvested to rapidly measure secretion levels of key cytokines (tumor necrosis factor alpha, interferon gamma, interleukin 10) via the AlphaLISA assay. Almost simultaneously, the cells can be fixated and stained using antibodies against innate immune activation markers (CD80, CD86, HLA-DR, OX40) for analysis via flow cytometry. This multiplexed readout workflow can directly aid in the phenotypic identification and discovery of novel immunomodulators and potential vaccine adjuvant candidates.
Before you begin
Note: The collection of human plasma and PBMCs from human participants will require institutional permissions prior to any studies. Human samples used from this protocol were approved by the Boston Children’s Hospital IRB (protocol number X07-05-0223).
We have successfully developed a reproducible protocol that incorporates human peripheral blood mononuclear cells (PBMCs) in a multiplexed, high-throughput assay system, enabling discovery of immunomodulatory compounds, with a focus on novel adjuvant discovery.
For space limitations, we do not provide the protocols for the isolation and cryopreservation of PBMCs. For details on such methodologies, please refer to ref.2
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD134 (OX40) Antibody, anti-human, APC, REAfinity™ (Clone- REA621) | Miltenyi Biotec | Order Number: 130-122-227 |
| CD80 Antibody, anti-human, PE, REAfinity™ (Clone- REA661). Diluted 1:200 with dPBS by volume. | Miltenyi Biotec | Order Number: 130-123-253 |
| CD19 Antibody, anti-human, VioBlue, REAfinity™ (Clone- REA675). Diluted 1:200 with dPBS by volume. | Miltenyi Biotec | Order Number: 130-120-031 |
| HLA-DR Antibody, anti-human, FITC, REAfinity™ (Clone- REA805). Diluted 1:200 with dPBS by volume. | Miltenyi Biotec | Order Number: 130-111-788 |
| CD86 Antibody, anti-human, PE, REAfinity™ (Clone-REA968). Diluted 1:200 with dPBS by volume. | Miltenyi Biotec | Order Number: 130-116-160 |
| Chemicals, peptides, and recombinant proteins | ||
| Dimethyl sulfoxide ACS | MP Biomedicals | Catalog Number: 191418 |
| Paraformaldehyde, 16% w/v aq. Soln., methanol free | Alfa Aesar | Stock Number: 43368 |
| Sterile water for irrigation, USP, 100mL | Baxter International | SKU: 2F7114 |
| Reagent alcohol (denatured alcohol), 70% (v/v) | Ricca Chemical Company | Catalog Number: 2546.70-1 |
| UltraPure™ distilled water | Invitrogen | Reference Number: 10977-015 |
| Ficoll-PaqueTM sterile solution | Cytiva | Item Code: 17544203 |
| Dulbecco’s Modified Eagle Medium | Gibco | Reference Number: 10566-106 |
| Trypan blue stain (0.4%) | Gibco | Reference Number: 15250-061 |
| Dulbecco’s phosphate buffered saline | Gibco | Reference Number: 14190-144 |
| UltraPure™ 0.5 M EDTA, pH 8.0 | Invitrogen | Reference Number: 15575-038 |
| R848, 25 μM | Invivogen | TLRL-R848-5 |
| CpG ODN-2395, 1 μM | Invivogen | TLRL-2395-5 |
| Critical commercial assays | ||
| TNF-α AlphaLISA Detection Kit, 5,000 Assay Points | PerkinElmer | Product Number: AL208F |
| IFN-γ AlphaLISA Detection Kit, 5,000 Assay Points | PerkinElmer | Product Number: AL217F |
| IL-10 AlphaLISA Detection Kit, 5,000 Assay Points | PerkinElmer | Product Number: AL218F |
| Customized fluorometric bead-based array multiplex kit | Sigma-Aldrich | Product Code: HCYTA-60K |
| Software and algorithms | ||
| GraphPad Prism v8.3.1 for MacOS | GraphPad | https://www.graphpad.com |
| FlowJo v10 | Becton Dickinson & Company | https://www.flowjo.com |
| Other | ||
| Corning 3656 384-Well Clear Round Bottom Non-Treated Plates | Corning | Product Number: 3656 |
| AlphaPlate-384 Light Gray, Untreated Plates | PerkinElmer | Part Number: 6005350 |
| Thermo Multidrop™ Combi Reagent Dispenser | Thermo Scientific | Catalog Number: 5840300 |
Materials and equipment
| Equipment name | Screening functionality |
|---|---|
| Seiko Compound Transfer Robot | Compound Pinning |
| Agilent (Velocity11) VPrep | Supernatant Extraction |
| Perkin Elmer EnVision Plate Reader | High Throughput Cytokine Quantification |
| Intellicyt iQue Screener PLUS | High Throughput Flow Cytometry |
| Thermo Multidrop™ Combi Reagent Dispenser | Reagent and Cell Dispensation |
| Forma Steri-Cult Humidity Controlled CO2 Incubator | Cell Incubation |
| Biosafety Cabinet Class II (BSCII) | Sterile Handing of Cell Cultures and Buffers |
| Water Bath (37°C) | Thawing of Cell, Heating of Buffers |
| Material name | Screening functionality |
|---|---|
| Cell Media (90% Dulbecco’s Modified Eagle Medium, 10% PPP. Volume differs based on screening throughput) | Medium for Appropriate Cell Growth/Survival |
| EDTA, 250 μM (500 μM Stock Diluted with 50% dPBS by Volume. Volume differs based on screening throughput). | Extraction of Adherent Cells for Flow Cytometry |
| Paraformaldehyde, 1% w/v. (16% Stock Diluted with 93.75% dPBS by Volume. Volume differs based on screening throughput). | Fixation of Cells for Flow Cytometry |
Antibody cocktail and AlphaLISA assay materials
| Reagent | Final concentration | Amount |
|---|---|---|
| Antibody Cocktail. | Each Antibody combined and diluted 1:200 with dPBS by Volume. | ∗ |
| AlphaLISA Acceptor Bead Cocktail. | 5 mg/mL Acceptor Beads and 500 nM Biotinylated Antibody combined 1:100 in 1X AlphaLISA buffer by volume | ∗ |
| AlphaLISA Donor Bead Cocktail. | 5 mg/mL Donor Beads diluted 1:125 in 1X AlphaLISA buffer by volume. | ∗ |
∗Volume can be amended based on screening throughput using the concentration scales indicated.
Step-by-step method details
Thawing cryopreserved cells
Timing: 45 min
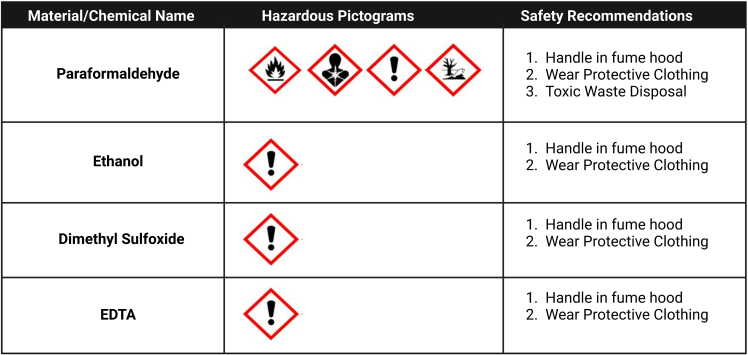
Using cryopreserved rather than fresh blood samples enhances throughput as cryopreserved cells can be kept in long term storage with a relatively small decline in cell viability.3,4 In theory, fresh cells may be alternatively utilized, though this may limit longitudinal validation experiments across study participants. For all steps in this experiment, attention to the potential toxicity of the reagents (Figure 1) is important.
-
1.
Fully thaw autologous platelet poor plasma (PPP) in 15 mL tube in 37°C water bath.
Note: Fully autologous PPP can be isolated from whole blood by collecting the supernatant from platelet rich plasma spun down at 3000 g for 10 min (high acceleration/medium deceleration) as described.2
-
2.
Centrifuge thawed plasma at 3000 g for 8 min (high acceleration / medium deceleration).
Note: Utilizing lower to medium deceleration in this step ensures proper separation of any solid particulates that may be isolated from the thawed plasma.
-
3.
Transfer supernatant of the centrifuged plasma to new 15 mL tube under sterile conditions.
Note: Avoid collecting any sediments (e.g., white pellet) at the bottom of the tube.
-
4.
Thaw human PBMCs (5 e7/mL) in 37°C water bath.
-
5.
Transfer 1 mL of the PBMC from its 2 mL cryotube to new 50 mL tube under sterile conditions.
-
6.
Use 2 mL of autologous PPP to collect any remaining cells on the side of the cryotube and transfer this PPP to the 50 mL tube.
-
7.Add 27 mL of Dulbecco’s Modified Eagle Medium (DMEM) to the PBMC/plasma mixture in the 50 mL tube.
-
a.Media stocks are stored at 4°C until expiration and diluted media kept at 4°C for day of use.
-
a.
Note: This constitution can be scaled up if more cells are needed for a 10% cells/plasma and 90% DMEM composition. Additionally, a plasma-supplemented media is used because plasma contains age-specific soluble mediators of immunity that can contribute to relevant leukocytic responses.5 While alternative, commercial serums can be utilized, this may affect the accurate capture of these responses.
-
8.
Centrifuge cells at 500 g for 10 min (high acceleration / medium deceleration).
-
9.
While cells are centrifuged, create a freshly prepared media composition of 50 mL with 10% autologous PPP and 90% DMEM.
-
10.
Discard supernatant from centrifuged cells and resuspend cell pellet with 10 mL of media.
-
11.
Dilute samples 1:10 in trypan blue (10 μL cells + 90 μL 0.4% trypan blue) and count cells.
Note: Bear in mind typical cell loss is 40–70% from each freeze-thaw cycle.
-
12.
Dilute cells to 1.67 e6 cells/mL, which will allow distribution of 50,000 cells/well. Keep cells on ice until ready for use.
-
13.
Dilute assay-specific controls (e.g., DMSO for negative control, R848, ODN-2395) in media for manual addition later in step 18. The dilution of DMSO should be calculated to match the concentration in experimental wells.
Figure 1.
Recommended safety procedures
Pictograms represent the Globally Harmonized System of Classification labels that accompany each compound. Recommended safety handling procedures are depicted. A more thorough handling guideline as well as additional hazards may be found through the safety data sheet for each chemical.
Pinning compounds and controls
Timing: 1 h + 72-h incubation
Pinning compounds allow for a rapid plating of experimental conditions, which is optimal for a high-throughput framework.
-
14.
Wash a standard-bore Combi cassette with 50 mL of sterile water for irrigation (St. H2O) followed by a 50 mL ethanol (EtOH) wash, and another wash with 50 mL of St. H2O using the Multidrop™ Combi Reagent Dispenser.
Note: Conduct all steps utilizing reagent or cell dispensing with the Combi reagent under sterile conditions to limit contamination.
-
15.Prime approximately 10–15 mL of the cell suspension through the manifold.
-
a.Prior to dispensing cells to each microplate, lightly agitate the cell suspension to ensure the cells stay in solution and do not settle.
-
a.
Note: Solutions dispensed using the Combi cassette are primed to prevent dilution from adherent fluids in the cassette from previous washes.
-
16.
Dispense 30 μL per well to 384-well Corning 3656 microplates (Corning, Corning, NY) on the low-speed setting to ensure minimal cell loss.
Note: The Combi equipment can be adapted to fill plates partially in the settings of the machine, allowing for adjustments in the case of limited reagent/cell availability.
-
17.
Wash the Combi cassette with 50 mL of St. H2O, followed by a 50 mL EtOH wash, and another wash with 50 mL of St. H2O using the combi reagent dispenser.
-
18.
Transfer 100 nL of compound from source library plates into the destination cell plates using the Seiko Compound Pin Transfer Workstation, allowing for a final concentration of 33 μM for our selected library plate.
Note: This volume transfer can be optimized for different library plates. Each library plate was tested over four biological replicates with two technical replicates per donor.
-
19.
Manually add 5 μL of positive and negative controls to each plate on designated wells that do not have experimental compounds pinned into them, using a multichannel pipette.
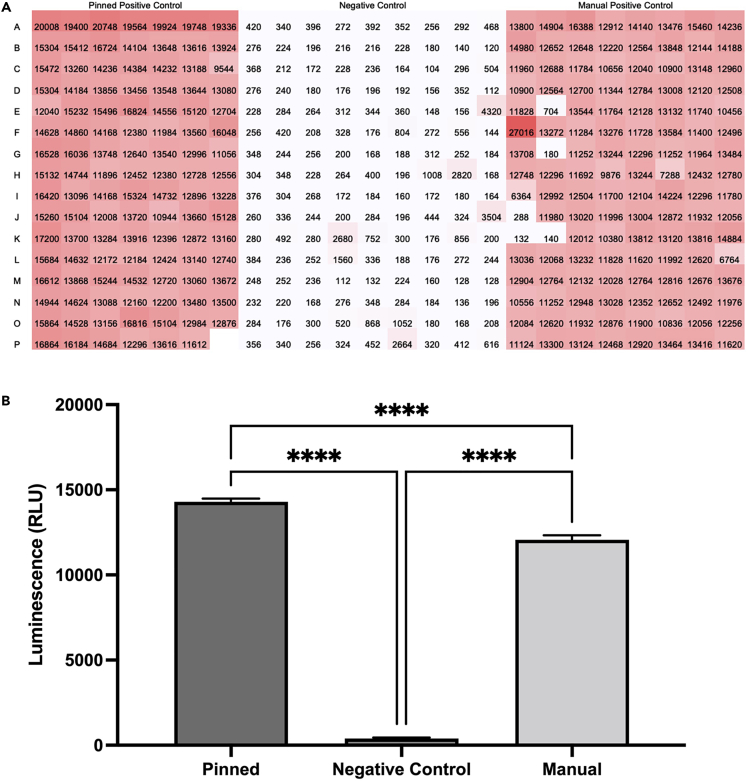
Note: R848 and ODN-2395 serve as strong positive controls while DMSO, which is a typical solvent of choice for chemical libraries, serves as an effective negative control. The controls are manually added in this step because many library plates do not have positive controls on them so they need to be directly added to cell plates. There are minimal differences found between manually added and pinned positive controls (Figure 4). While this manual addition is feasible for positive controls, automated addition of experimental compounds allows for scalability of this protocol to assay 10–30 library plates at a time.
-
20.
Cover and incubate plate for 72 h in 37°C, 5% CO2 humidified incubator.
Note: Chemical screening libraries are often produced from diverse sources. This protocol is optimized for commercially available, small molecule libraries that were dissolved in DMSO. Thus, we used DMSO at 1% as a negative control to match the precise diluent used for screening. This would be used to asses any background activity that may have been caused due to the solvent on the screened cells. For other libraries, the selection and volume of negative control should be optimized to match experimental conditions.
Figure 4.
Representative cytokine induction results
(A) Raw luminescence values (RLU) of a TNF AlphaLISA assay are depicted. Color gradients from white to red indicate lower to higher values. The negative and positive controls for the AlphaLISA assay were DMSO and R848, respectively.
(B) Mean raw luminescence values plotted as bar plots with errors shown as SEM. Each group was compared by one way ANOVA. For statistical analyses: ns p > 0.05, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
Fixation and staining of cells
Timing: 4.5 h
Fixation of cells limits cell degradation and decomposition, enabling preservation of cell protein structures.6 This is particularly important for flow cytometry readouts.7 In this high throughput context, fixation is imperative to prevent cell atrophy and to capture accurate cell activation.
-
21.
Centrifuge plate at 500 g for 10 min (9 g acceleration, 7 g deceleration).
-
22.Aspirate 2 μL of supernatant from each well on the cell plate
-
a.Dispense into a labeled PerkinElmer AlphaPlate-384 (#6005359) using the Agilent VPrep.
-
b.Repeat this 2 μL transfer process into another labeled AlphaPlate.
-
a.
Note: Supernatant is harvested prior to staining and is analyzed for cytokine quantification starting at step 54.
-
23.
Replace the cover the cell plate and store at 4°C, while completing supernatant transfer for any remaining cell plates. This is to minimize additional cell death or degradation for larger throughput experiments.
-
24.
Seal the two AlphaPlates with aluminum seals and store overnight at −80°C.
-
25.
Remove the cell plate from 4°C storage and centrifuge at 700 g for 10 min. Flick the supernatant off.
Note: Flick the supernatant off only once after centrifugation. This is to prevent excessive cell loss. If a considerable amount of supernatant is retained in some wells, re-centrifuge the plate before flicking again.
-
26.
Wash a standard-bore Combi cassette with 50 mL St. H2O, followed by a 50 mL EtOH wash, and 50 mL St. H2O wash using the Combi.
-
27.
Dilute 500 mM EDTA 1:2 with PBS to generate a 250 mM stock of EDTA. Keep the 250 mM EDTA solution on ice.
-
28.
Prime approximately 10–15 mL of the 250 mM EDTA through the manifold, then dispense 20 μL per well.
-
29.
Wash the standard tube cassette with 50 mL distilled water, followed by a 50 mL ethanol wash, and another wash with 50 mL distilled water using the Combi.
-
30.
Shake covered cell plate at 700 rpm for 10 min at room temperature.
-
31.
Centrifuge plate at 750 g for 10 min (9 g acceleration, 7 g deceleration) at room temperature.
-
32.While plate is centrifuged, dilute antibody cocktail 1:200 in dPBS by volume. Prepare 2 mL of cocktail per plate with 10–15 mL excess for priming and deadweight loss.
-
a.Antibody cocktail is stored at 4°C until expiration.
-
a.
-
33.
Attach and wash a small-bore Combi cassette with 30 mL St. H2O, followed by a 30 mL EtOH wash, and 30 mL St. H2O wash using the Combi.
-
34.
Prime approximately 5–10 mL of the antibody solution through the cassette, then dispense 5 μL per well.
-
35.
Wash the small-bore Combi cassette with 30 mL St. H2O, followed by a 30 mL EtOH wash, and 30 mL St. H2O wash using the Combi.
-
36.
Incubate cell plate at 4°C for 30 min during antibody incubation.
-
37.
Attach and wash a standard-bore Combi cassette with 50 mL St. H2O, followed by a 50 mL EtOH wash, and another wash with 50 mL St. H2O using the Combi.
-
38.
Prime approximately 10–15 mL of PBS through the cassette, then dispense 40 μL per well.
-
39.
Centrifuge plate at 750 g for 10 min (9/7 g acceleration and deceleration).
-
40.
Wash the standard-bore Combi cassette with 50 mL St. H2O, followed by a 50 mL EtOH wash, and another wash with 50 mL St. H2O using the Combi.
-
41.
While plate is centrifuged, dilute 16% PFA to 1% PFA in dPBS, calculating for 10 mL per plate and an excess 10–15 mL for priming and deadweight loss.
-
42.
When plate finishes centrifugation, flick the supernatant off.
-
43.
Prime approximately 10mL of 1% PFA through the cassette, then dispense 25 μL per well.
-
44.
Wash the standard-bore Combi cassette with 50 mL St. H2O, followed by a 50 mL EtOH wash, and another wash with 50 mL St. H2O using the Combi.
-
45.
Incubate the cell plate at 4°C during cell fixation.
Pause point: The cell plates can be kept covered with plate lids in this fixative solution up to a week. We avoid keeping the cells in this condition longer than 7 days and recommend shorter storages when using tandem dye conjugated antibodies. This fixation step can be alternatively skipped if the flow cytometry can be run immediately. However, at large enough throughputs, we recommend fixating your cells to prevent undesirable cell death or degradation.
Flow cytometry
Timing: 30 min to 1 h
High throughput flow cytometry is now compatible with multiplexed cell-based screening.8 This is particularly useful for immunological screening with diverse cell types such as PBMCs. We measured the presence of cell-surface activation markers for each of our profiled cell types (CD80/86 and HLA-DR for Monocytes and B cells, OX40 for T Cells) to detect any general innate immune responses that the screened compounds induced (Figure 2). Alternatively, the panel of antibodies can be tailored to the focus of different screening efforts. However, the REAffinity antibodies used in this protocol prevent their binding to Fc receptors. Thus, if other antibodies are used, blocking steps may be necessary to prevent non-specific binding. Similarly, antibodies should be carefully selected and assessed for potential spectral overlap of fluorophores, in which case compensation should be performed.
-
46.
Retrieve cell plates from 4°C storage and centrifuge at 750 g for 10 min (9/7 g acceleration and deceleration).
-
47.
When plate finishes centrifugation, flick the supernatant off.
Note: This step can be alternatively skipped to reduce cell loss in aspiration steps if proper disposal of PFA can be arranged for the waste content from the flow cytometry.
-
48.
Wash a standard-bore Combi cassette with 50 mL St. H2O, followed by a 50 mL EtOH wash, and 50 mL St. H2O wash using the Combi.
-
49.
Prime approximately 10 mL of PBS through the cassette, then dispense 25 μL per well.
-
50.
Centrifuge plate at 750 g for 10 min (9/7 g acceleration and deceleration).
-
51.
When plate finishes centrifugation, flick the supernatant off.
-
52.
Using the Combi, dispense 20 μL of PBS per well.
-
53.
Wash the standard-bore Combi cassette with 50 mL St. H2O, followed by a 50 mL EtOH wash, and 50 mL St. H2O wash using the Combi.
-
54.
Assess plates on the iQue to collect flow cytometry results.
Note: One of the added benefits of using this set-up is the throughput of the IQue Flow Cytometer, as each cell plate can be ideally run in less than an hour. Alternatively, other flow cytometers may be used, though with elongated flow collection periods that may limit throughput.
Figure 2.
Representative schematic of the high throughput flow cytometry system
A total of 5 μL of antibodies diluted 1:200 in PBS were added to each well containing 30 μL of cells (50,00 cells/well). This flow cytometry system captures the total events or cells per well in addition to the antibody activity, which measures markers including the surface receptors Ox40, CD80/86, and HLA-DR. The flow cytometry approach also identifies B cells, T cells, and monocytes.
AlphaLISA assay
Timing: 2.5 h
AlphaLISA assays demonstrate a unique rapidity and ease of use.9 This makes it uniquely compatible in a high throughput setting for cytokine induction measurements. We used detection of TNF-α, IFN-λ, and IL-10 secretion as a broad panel to capture induction of innate immune responses (Figure 3). Alternatively, this panel of cytokines may be adapted to fit the focus of other screening efforts.
Pause point: The AlphaPlates can be kept at −80°C up to two weeks. We typically conduct the assays on these plates the next day to increase throughput and enable real-time monitoring of assay performance.
-
55.Prepare AlphaLISA buffers and the AlphaLISA Acceptor beads + Biotinylated Antibody mixtures for each cytokine of interest.
-
a.Prepare 15 mL of extra reagent for priming on the Combi as well as potential deadweight volume loss.
-
b.AlphaLISA assay systems are stored at 4°C until expiration and any dilutions are prepared immediately prior to conducting the assay.
-
a.
-
56.
Reconstitute AlphaLISA standards in 100 μL of MilliQ water and prepare standard dilutions as described in the AlphaLISA protocols.
-
57.
Manually add 2 μL of standards on a new AlphaPlate using a multichannel pipette.
Note: The volume of standard added per well was scaled down to 2 μL to match the volume of supernatant being tested. This enables consistent reagent dispensing.
-
58.
Retrieve AlphaPlates with supernatant from −80°C storage and remove aluminum seal.
-
59.
Wash a small-bore Combi cassette with 30 mL St. H2O, followed by a 30 mL EtOH wash, and 30 mL St. H2O wash using the Combi.
-
60.
Prime approximately 10 mL of the Acceptor beads+ Biotinylated Antibody mixture through the cassette, then add 8 μL per well.
-
61.
Repeat steps 5–6 for each cytokine/AlphaLISA kit of interest.
-
62.
Allow the AlphaPlates to incubate at room temperature for an hour.
Note: AlphaLISA kits are light sensitive so incubate in the dark without sealing the plates.
-
63.
Wash a different small-bore Combi cassette with 30 mL St. H2O, followed by a 30 mL EtOH wash, and 30 mL St. H2O wash using the Combi.
-
64.Prepare Streptavidin Donor Beads for each cytokine of interest in the dark, as described in their published protocols.
-
a.Prepare 10 mL for priming on the Combi as well as potential deadweight volume loss.
-
a.
-
65.
Prime approximately 5–10 mL of donor beads through the cassette, then add 10 μL per well.
-
66.
Allow the AlphaPlates to incubate at room temperature in the dark for 30 min.
-
67.
Quantitate fluorescent signal in each well using the EnVision to determine cytokine levels in the supernatant.
Note: The following apertures must be used: Mirror Module = Barcode 444, EMS filter = Barcode 244 (570 nm).
Figure 3.
Representative schematic of the AlphaLISA assay system
A total of 8 μL of acceptor bead solution, containing biotinylated antibodies as well as acceptor beads, were added to each well containing 2 μL of supernatant. After a 1 h incubation, a 10 μL solution of Streptavidin donor beads were added to this solution and incubated for 30 min. Both beads are manufactured to be specific to a binding site for the cytokine of interest. When read on the EnVision Plate Reader, bound donor and acceptor beads, due to their proximity, will emit a 615 nm luminescence when excited by a 680 nm wavelength. The intensity of this luminescence reflects the concentration of bound beads or the concentration of the cytokine.
Expected outcomes
For cytokine quantification, increased luminescence values indicate higher cytokine induction. With a multi-cytokine readout system, broad and multifaceted immunological profiles of different compounds can be constructed. The luminescence across different cytokines measured for each compound screened will typically be highly variable (Figure 4). This variability is to be expected and serves as an indication of the utility of the assay system in identifying the immunological profiles of different compounds. This can be useful in the context of phenotypic screens for immunomodulators or drug-repurposing screens, as new immunological functions of established biologically active compounds can be discovered.
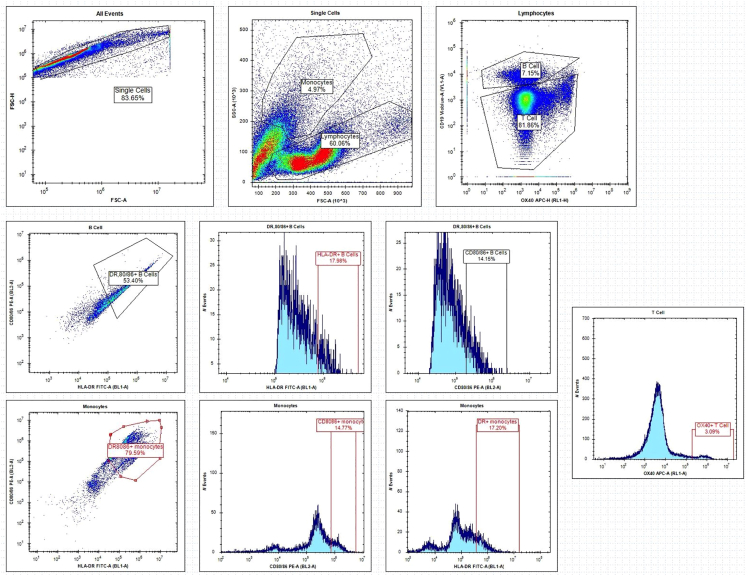
For flow cytometry, the extensive cell-identifying and cell-surface activity markers can be highly informative of the immunological mechanism of action (Figure 5). CD14 is used as a marker for monocytes, and CD19 for B cells. T cells can be further distinguished by size and granularity (light scatter) and absence of CD19. The degree of activation of each cell type is evaluated by increases in the surface expression of CD80, CD86, HLA-DR, or OX40. These markers can determine which compounds activate a proportionally higher percentage of a given cell type. This can be useful for identifying compounds that demonstrate an activity phenotype of interest, such as T cell or monocytic activators.
Figure 5.
Representative flow cytometry results and gating strategy
Representative gating strategy of the multiplexed flow cytometry readout system. Black lines indicate the traditional gating system, which may have to be qualitatively adjusted depending on screening environments.
For both assay systems, considering the high levels of biological variability, positive and negative control data (Figure 4) serve as quantitative markers of the assay system’s effectiveness. There should be a broad identifiable difference between the positive and negative controls, which should clarify the relative spectrum of activity that screened compounds possess. This invites further evaluation of hit compounds using, for example, a Z-score based hit-calling algorithm. This z-score metric formulaically assesses the relative performance of compounds relative to the median or mean performance of other experimental conditions, allowing a pre-determined threshold to be set in order to aid in down-selecting lead compounds. This allows for a consistent and quantitative framework to evaluate relative compound performance while controlling for technical and day-to-day variability.
Limitations
The limitation of a primary cell-based screen is that improved biological relevance comes with increased biological variability. Immune responses of PBMCs can vary between and within donors across time. Further approaches to data analysis standpoint to account for this level of variation via donor-specific metrics for compound performance may prove important in future applications of this high throughput platform.
An additional limitation, due to the cell culture being used, is that the media to supplement the cells is aqueous in nature. With a small fraction of dispensed compound being prepared in DMSO, this assay system may not be optimized to test certain compounds with very low aqueous solubilities due to potential crashing out of compounds in solution. This could potentially lead to overlooking potential immunomodulatory yet insoluble compounds but is a natural limitation of culturing and using human primary immune cells.
Additionally, the choice for a 72-h incubation was made for optimal detection of IFN-γ, whilst still being able to detect IL-10 and TNF, the peak secretion of which is around the 24-h time point. This longer incubation period may bias towards the activity of more robust leukocytes that can survive the longer incubation without supplemental media components at least in the cell-based readouts measured through flow cytometry. Further, with the additional cell-loss from staining, we measured ∼10,000 viable cells per well via flow cytometry, which indicates a notable loss in viable cells in this workflow. Thus, if more powerfully measuring broader immunological activity phenotypes captured by flow cytometry is preferred to detecting IFN production for discovery-based strategies, a 24-h incubation may be more suitable.
Troubleshooting
Problem 1
Low monocyte counts found in flow cytometry results, following step 53.
Potential solution
Activated monocytes are highly adherent,10 leading to adhesion to the bottom of cell plates. Washing with cold EDTA and shaking plates at 700 rpm can help loosen the monocytes into solution. If low monocyte counts are a recurrent problem, incorporation of multiple cold EDTA washes will likely improve the retention of activated monocytes. Additionally, inter-well shaking at 3000 rpm during flow cytometry readouts on the iQue should be incorporated.
Problem 2
High background noise on the EnVision plate reader for AlphaLISA results, following step 64.
Potential solution
High background on the EnVision plate reader can sometimes be traced down to the aluminum seals that are used during the freezing of the AlphaPlate with supernatant. Typically, the seals are highly adherent and during the thaw process, a significant portion of the adherents can remain on the plates, which affects luminescence values. We found that quickly removing the seals right after taking the plates out of the freezer has helped avoid this issue. Additionally, the choice of solvent is important. AlphaLISA assays rely on a biotinylated bead system that is sensitive to trace amounts of biotin in common cell culture media like RPMI. This sensitivity was noted (Figure 6) and highlights the choice media of DMEM in our screening platform.
Figure 6.
RPMI and DMEM as cell culture media options for screening platform
Heat map and density distribution of luminescence readouts for a representative TNF AlphaLISA readout using DMEM (left) and RPMI (right). The trace amounts of biotin in RPMI causes high levels of interference and induces artificially high luminescence readouts in many wells.
Problem 3
Limited number of human PBMCs to complete a study, discovered after step 11.
Potential solution
Depending on the number of plates to be screened, and the limits on peripheral blood sampling collections/donations allowed at a time, it is important to plan ahead regarding potential need for repeated blood collections from a given study participant. This is particularly important when also considering the deadweight volume of cells necessary for priming the dispenser system. We found an effective strategy to address this was through increasing the daily throughput of our experimental design, which would naturally decrease the proportion of cells and materials that were sunk costs for priming and volume losses.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, David Dowling, David.Dowling@childrens.harvard.edu.
Materials availability
This study did not generate new unique reagents.
Acknowledgments
The current study was supported in part by US National Institutes of Health/National Institutes of Allergy and Infectious Diseases Vaccine Adjuvant Discovery Program Contracts (75N93019C00044) to O.L. and D.J.D. The high-throughput screen was conducted at the ICCB-Longwood Screening Facility at Harvard Medical School. Figures 1, 2, and 3 and the graphical abstract were created with BioRender.com.
Author contributions
B.L. and K.C. designed, performed, and analyzed the experiments and wrote the manuscript. A.O., D.J.D., J.A.S., O.L., and S.V.H. conceived the project, designed the experiments, and wrote the manuscript.
Declaration of interests
S.V.H., O.L., and D.J.D. are named inventors on vaccine adjuvant patents assigned to Boston Children’s Hospital. O.L. served as a paid consultant to Moody’s analytics. D.J.D. is on the scientific advisory board of EdJen BioTech and serves as a consultant with Merck Research Laboratories/Merck Sharp & Dohme Corp. (a subsidiary of Merck & Co., Inc.). These commercial or financial relationships are unrelated to the current study.
Contributor Information
David J. Dowling, Email: david.dowling@childrens.harvard.edu.
Simon Van Haren, Email: simon.vanharen@childrens.harvard.edu.
Data and code availability
All data reported in this paper will be shared by the lead contact upon reasonable request.
References
- 1.Chew K., Lee B., van Haren S.D., Nanishi E., O’Meara T., Splaine J.B., DeLeon M., Soni D., Seo H.S., Dhe-Paganon S., et al. Adjuvant discovery via a high throughput screen using human primary mononuclear cells. bioRxiv. 2022 doi: 10.1101/2022.06.17.496630. Preprint at: [DOI] [Google Scholar]
- 2.van Haren S.D., Dowling D.J., Foppen W., Christensen D., Andersen P., Reed S.G., Hershberg R.M., Baden L.R., Levy O. Age-specific adjuvant synergy: dual TLR7/8 and mincle activation of human newborn dendritic cells enables Th1 polarization. J. Immunol. 2016;197:4413–4424. doi: 10.4049/jimmunol.1600282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nazarpour R., Zabihi E., Alijanpour E., Abedian Z., Mehdizadeh H., Rahimi F. Optimization of human peripheral blood mononuclear cells (PBMCs) cryopreservation. Int. J. Mol. Cell. Med. 2012;1:88–93. [PMC free article] [PubMed] [Google Scholar]
- 4.Ticha O., Moos L., Bekeredjian-Ding I. Effects of long-term cryopreservation of PBMC on recovery of B cell subpopulations. J. Immunol. Methods. 2021;495:113081. doi: 10.1016/j.jim.2021.113081. [DOI] [PubMed] [Google Scholar]
- 5.England R., Pak J., Liu M., Rao S., Ozonoff A., Levy O., van Haren S.D. Human blood plasma shapes distinct neonatal TLR-mediated dendritic cell activation via expression of the MicroRNA Let-7g. Immunohorizons. 2021;5:246–256. doi: 10.4049/immunohorizons.2000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang B.Q., Yeung E.C. In: Plant Microtechniques and Protocols. Yeung E.C.T., Stasolla C., Sumner M.J., Huang B.Q., editors. Springer International Publishing; 2015. Chemical and physical fixation of cells and tissues: an overview; pp. 23–43. [DOI] [Google Scholar]
- 7.O'Brien M.C., Bolton W.E. Comparison of cell viability probes compatible with fixation and permeabilization for combined surface and intracellular staining in flow cytometry. Cytometry. 1995;19:243–255. doi: 10.1002/cyto.990190308. [DOI] [PubMed] [Google Scholar]
- 8.Black C.B., Duensing T.D., Trinkle L.S., Dunlay R.T. Cell-based screening using high-throughput flow cytometry. Assay Drug Dev. Technol. 2011;9:13–20. doi: 10.1089/adt.2010.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaudet L., Rodriguez-Suarez R., Venne M.-H., Caron M., Bédard J., Brechler V., Parent S., Bielefeld-Sévigny M. AlphaLISA immunoassays: the no-wash alternative to ELISAs for research and drug discovery. Nat. Methods. 2008;5:an8–an9. doi: 10.1038/nmeth.f.230. [DOI] [Google Scholar]
- 10.Meisel S.R., Xu X.P., Edgington T.S., Dimayuga P., Kaul S., Lee S., Fishbein M.C., Cercek B., Shah P.K. Differentiation of adherent human monocytes into macrophages markedly enhances tissue factor protein expression and procoagulant activity. Atherosclerosis. 2002;161:35–43. doi: 10.1016/s0021-9150(01)00616-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon reasonable request.