Abstract
Toxic agents are broadly present in the environment, households, and workplaces. Contamination of food and drinking water with these agents results in entry of these materials to the body. The crosstalk between these agents and microRNAs (miRNAs) affects pathoetiology of several disorders. These agents can influence the redox status, release of inflammatory cytokines and mitochondrial function. Altered expression of miRNA is involved in the dysregulation of several pathophysiological conditions and signaling pathways. These molecules are also implicated in the adaption to environmental stimuli. Thus, the interactions between miRNAs and toxic materials might participate in the hazardous effects of these materials in the body. This review describes the effects of the toxic materials on miRNAs and the consequences of these interactions on the human health.
Keywords: miRNA, Hazardous material, Cancer
1. Introduction
Hazardous materials are radioactive substances and/or chemical compounds which can be harmful to individuals or animals and the environment upon exposure [1]. As chemical materials, they fall into a category such as cadmium, lead, mercury, arsenic, chromium, and asbestos [[2], [3], [4], [5]]. The United States Environmental Protection Agency (USEPA) has considered some of them to be probable human carcinogen agents. These materials are found in drinking water [6], air [7], and food [8], and therefore they could be absorbed via dermal contact, ingestion, and/or inhalation, leading to damage to body organs such as the lungs, kidneys, and liver. Different levels of poisoning occur after exposure to such substances. For example, arsenic is a well-known carcinogenic agent and is strongly linked to the development of lung, bladder, liver, and kidney cancers [9,10]. Or, asbestos fibers, including chrysotile, are highly associated with the development of lung cancer, mesothelioma, and pulmonary fibrosis [11,12].
A number of studies have shown that development of several diseases such as hypertension, gastrointestinal disorders, and osteoporosis is resulted from long-term exposure to these materials [[13], [14], [15], [16], [17]]. It has been also reported that some of these hazardous agents such as mercury, lead, and cadmium could pass from the placenta and cause a disruption in the normal process of fetal development [18,19].
In recent years, scientists have focused on a wide range of molecular alterations and mechanisms involved in hazardous material-related diseases [20,21]. Their results have shown that these mentioned materials could affect normal cell function and lead to cell death via a number of mechanisms including DNA methylation, inflammation, oxidative stress, autophagy, and apoptosis [22,23]. However, among these mechanisms, it has been reported that microRNAs (miRNAs) are associated with multiple organ injuries [24,25]. miRNAs are categorized as a form of the molecules of non-coding RNAs with nearly ∼22 nucleotides in length [26,27]. Although they are not involved in protein coding, they could modify target mRNAs via the posttranscriptional mechanism. Both genetic and epigenetic mechanisms could also regulate miRNAs expression [28]. In this regard, for example, in human bronchial epithelial cells (HBECs) exposed to arsenic, elevated promoter methylation has led to suppression of miR-200. Moreover, arsenic has caused malignant transformation via altering epithelial-mesenchymal transition (EMT) signaling pathways [29]. Moreover, in lung cancer asbestos could alter the miRNA expression, where the expression of some miRNAs such as miR-202, miR-605, and miR-939 decreased, while the expression of several miRNAs, such as miR-96, let-7d/e, and/or miR-374a, increased [30]. In this review, we investigated the interaction of miRNAs and some important hazardous materials.
2. Interaction between miRNAs and arsenic compounds
Arsenic compounds have been used for treatment of leukemia. For instance, arsenic trioxide (As [2]O(3), ATO) has been used for treatment of acute promyelocytic leukemia. Gao et al. have assessed possible synergy between miR-15a/16-1 and ATO in K562 cells. They have reported that combination of miR-15a/16-1 and ATO induces growth suppression and apoptosis in these Bcr-Abl positive leukemic cells. Mechanistically, apoptosis is induced through regulation of mitochondrial functions. In fact, this process involves release of cytochrome c and loss of mitochondrial transmembrane potential. Yet, ATO and/or miR-15a/16-1 could not affect expression of Bcr-Abl in these cells. Besides, miR-15a/16-1 and ATO could induce apoptosis in Bcr-Abl negative leukemic cell lines and primary leukemic cells in a synergic manner [31]. Another study has shown that anti-miR-21 oligonucleotide (AMO-miR-21) and ATO inhibit growth of K562 cells and induce apoptosis and G1 arrest in these cells. Mechanistically, AMO-miR-21 induces sensitivity to ATO through induction of apoptosis via up-regulation of PDCD4 levels [32].
miRNAs expression has also been found to be altered after arsenic exposure participating in the arsenic-induced multiorgan damage. Upregulation of miR-155 has been shown to be involved in the arsenic induced skin injury. Moreover, expression levels of miR-21 and miR-145 have been found to be associated with liver damage, while levels of miR-191 have been linked with kidney damage (Fig. 1). Furthermore, miR-155 has can be used as a diagnostic marker for arsenic-induced skin damage. Moreover, miR-21 and miR-145 have been suggested as diagnostic markers for liver damage. Finally, miR-191 can be used as a diagnostic marker for kidney damage [33]. Table 1 summarizes the results of studies that evaluated interactions between miRNAs and arsenic compounds.
Fig. 1.
A schematic representation shows the routes of arsenic exposure in humans and miRNA interaction signaling with arsenic in different types of cancer.
Table 1.
Interaction between miRNAs and arsenic compounds.
| miRNA | Human/animal Study | Dose | Cell line | Dose | Targets | Observations | Ref |
|---|---|---|---|---|---|---|---|
| miR-15a, miR-16-1 (−) | Raji, HL60, RPMI8226, K562, U937 | 1 μM, for 48 h | Caspase-3, Cytochrome-c | In K562 cells, both mentioned miRNAs could sensitize apoptosis induced by ATO. | [31] | ||
| miR-21 (−) | – | – | K562 | 0.5–4.0 μM, 4 h | PDCD4 | The sensitization of K562 cells exposed to ATO increased because of the anti-miR-21 oligonucleotide. | [32] |
| miR-21, miR-200b, miR-191, miR-155, miR-145 (Up) | Human | – | – | – | – | Exposure to arsenic by elevating the expression of mentioned miRNAs could lead to multiorgan (such as skin, liver, and kidney) damage. | [33] |
| miR-21, miR-155, miR-200b (Up) | miR-21 knockout mice, wild-type mice | 20 ppm, 6 months | HBE, MRC-5 | 0–8 μM, 24 h | PTEN, AKT, α-SMA | In the pulmonary fibrosis model induced by NaAsO₂, miR-21 via glycolysis could elevate the differentiation of myofibroblast. | [34] |
| miRNA profile, miR-29a (−) | – | – | HepG-2 | 0–6 μM, 24 h | Wip-1, PPM1D | In HepG-2 cells, miR-29a could mediate ATO induction of cell death. | [35] |
| miR-27a (Down) | – | – | MDA-MB-231, SK-BR-3 | 0–14 μM, for 24 h | Fbw7, Cyclin-E | In breast cancer cells, ATO via suppressing miR-27a could inhibit tumorigenesis. | [36] |
| miR-31 (Down) | – | – | BEAS-2B | 0–10 μM, 48 h, 2 μM, 6 weeks | SATB2 | Arsenic via reducing miR-31 and elevating SATB2 could induce malignant transformation in BEAS-2B cells. | [37] |
| miR-34a, miR-133b, (Down) | Human | – | U251, SH-SY5Y | 5 or 10 μM, 24 h | hERG | miR-133b via sponging the hERG could lead to apoptosis. | [38] |
| miR-98 (Up) | SD rats | 0.4 mg/kg, 2 weeks | A549 | 0–10 μM, for 48 h | Stat3, α-SMA, E-cadherin, Bax, Bcl-2 | Pulmonary fibrosis induced by bleomycin could be decreased after ATO treatment via increasing miR-98 expression. | [39] |
| miR-126 (Down), miR-155 (−) | Human (Mexican children) | – | – | – | – | Inorganic arsenic could change the expression of miR-126. | [40] |
| miR-148a (Up) | – | – | Multiple drug-resistant (MDR) Bel-7402 | 0–3.5 μM, 24 h | NF-κB | ATO via demethylating miR-148a and suppressing the NF-κB could promote cell sensitivity to chemotherapeutic agents. | [41] |
| miR-155 (Up) | Mice | 2 mg/kg, 2/week, 2 months | PC-3, HUVEC, LNCaP | 0–4 μM, 48 h | VEGF, TGF-β, SMAD | In prostate cancer, ATO could induce anti-angiogenic effects via elevating miR-155 and suppressing the TGF-β/SMAD pathway. | [42] |
| miR-155 (Up) | – | – | A549, A549R | 0–30 μM, 0–150 μM, 72 h | HO-1, Nrf-2, NQO1, Bax, NQO1 | miR-155 via suppressing apoptosis and enhancing Nrf2 could mediate cell resistance to ATO. | [43] |
| miR-184, miR-576-3p, (−) | Human | 59–172 ppb | – | – | – | In skin lesions (west Bengal people) induced by arsenic, the expression of miRNAs could change. | [44] |
| miR-190 (Up) | – | – | BEAS-2B, A549 | 0–20 μM (6 h), | PHLPP, AKT, Talin-2 | Exposed cells to AsCl3 by activating AKT, elevating miR-190, and suppressing PHLPP could lead to carcinogenesis. | [45] |
| miR-191 (Up) | Human | – | – | – | IL-2/6, TGF-β, TNF-α | miR-191 via activating inflammatory response could lead to renal dysfunction induced by coal-burning arsenic. | [46] |
| miR-199a-5p (Down) | Female CrTac: NCrFoxn1nu mice | – | AsT, BEAS-2B | 0–2 μM (24 h), | HIF-1α, COX-2 | Overexpression of miR-199a via targeting COX-2 and HIF-1α could suppress angiogenesis in bronchial epithelial cells. | [47] |
| miR-203 (−) | – | – | K562 | 1.25–20 μg/mL, 48 h | Caspase-3/9, Cytochrome-c | hsa-miR-203 could increase leukemia cell sensitivity to ATO. | [48] |
| miR-222 (Up) | – | – | BEAS-2B | 1 μM, 26 weeks | ARID1A, PTEN, AKT | Administration of anti-miR-222 could inhibit tumor growth induced by arsenic. | [49] |
| miR-301a (Up) | Athymic nude mice | – | BEAS-2B, BEAS-2B–As | 0–10 μM 12 h | SMAD4, IL-6, STAT3 | NaAsO₂ via increasing miR-301a could lead to malignant transformation of BEAS-2B cells. | [50] |
| miR-425-5p (Down) | C57BL/6J mice | (0, 1, or 10 ppm), 3 months | HUVECs, 293T | 1–40 μM, for 24–48 h | CCM3, Notch, VEGF/p38 | NaAsO2 had anti-angiogenesis effects in HUVECs. | [51] |
| miR-539 (Down) | Human, Male athymic nu/nu mice | 5 mg/kg, one injection/3 days, 18 days | Primary human hepatocytes, HepG2, Hep3B, Huh7, PLC/PRF/5, Sk-Hep-1, PLC-ATR, HepG2-ATR | 0–64 μM, for 48 h | Stat3, Bcl-2, Bcl-xL | In hepatocellular carcinoma, miR-539 could decrease cell chemoresistance induced by ATO. | [52] |
| miR-1294 (Up) | female BALB/C athymic nude mice | Huh6/7, HepG2, SMMC7721, Hep3B | 0–12 μM, 48 h | TEAD1, PIM1, caspase-3, Bax, Bcl-2 | In hepatocellular carcinoma, arsenic trioxide via upregulating miR-1294 and sponging PIM1/TEAD1 axis could inhibit tumor growth. | [53] | |
| miR-2909 (Up) | – | – | PBMCs | 0.5–2 μM, 72 h | Cyclin-D1, SP1 KLF4, NF-kB, BCL3 | Arsenic via miR-2909 could mediate the regulation of Cyclin-D1. | [54] |
| miR-4665-3p (Down) | Human (gastric cancer patients) | – | MGC803, AGS, HCT116 | 133.36 (stock solution) μM, 24 h | GSE-1, VEGF, E-cadherin, Vimentin | As4S4 via increasing miR-4665-3p could suppress migration or invasion in gastric cancer cells. | [55] |
2.1. Interaction between miRNAs and cadmium compounds
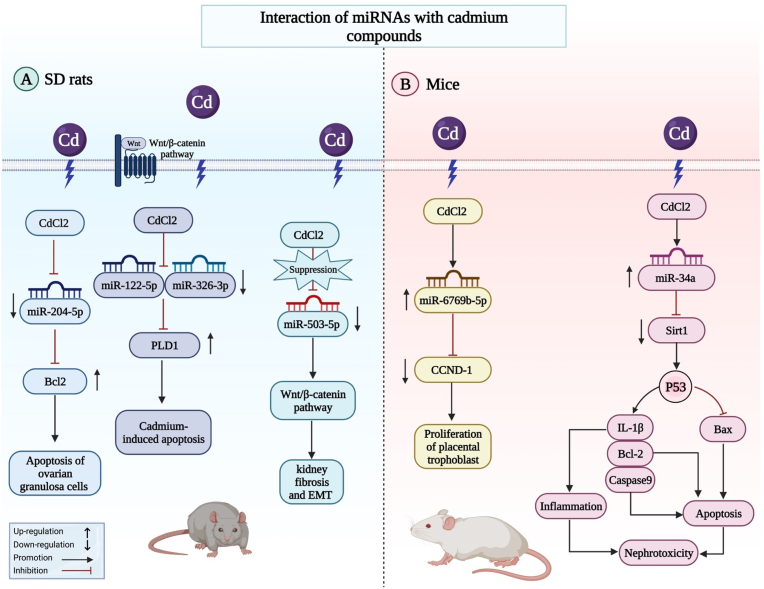
Cadmium (Cd) is an important hazardous agent that has toxic effects on fish and aquatic animals. Expression profiling of miRNAs in cultured common carp (Cyprinus carpio L.) has shown differential expression of a number of miRNAs during Cd exposure. In fact, 7 and 16 miRNAs have been found to be up-regulated and down-regulated, respectively. miR-122, novel-miR6, miR-193a-3p and miR-27a-5p have been among differentially expressed miRNAs (Fig. 2a). Moreover, expressions of BAX, BAD, BAK, CASPASE9 and PIDD have been enhanced, while BCL2 expression has been reduced following Cd exposure. Changes in the expression levels of mentioned miRNAs might be involved in the oxidative stress-induced apoptosis following exposure to Cd [56]. Another study has demonstrated inflammation-related injury in the spleens of common carp following Cd exposure. In fact, 17 miRNAs have been up-regulated, while 6 miRNAs have been down-regulated. Theses miRNAs have been functionally related with NF-κB, Jak-STAT, MAPK, Th1 and Th2 cell differentiation, and Toll-like receptor signaling pathways [57]. Another experiments in rat ovarian granulosa cells has shown that Cd is cytotoxic to these cells affecting expression of a number of miRNAs. In fact, Cd-induced damage to these cells is mediated by mitochondrial apoptosis [57]. In mice animal model, miR-6769b-5p via sponging CCND-1 might involve in the proliferation of placental trophoblast treated with CdCl2. Furthermore, via modulating the miR-34a/Sirt1/p53 signaling pathway, cd can damage the kidneys of mice and can control the apoptosis and inflammation (Fig. 2b).
Fig. 2.
The interaction of miRNAs and cadmium compounds with signaling pathways in Sprague Dawley (SD) rats and mice animal models is shown schematically in the diagram.
Table 2 shows the interaction between miRNAs and Cd.
Table 2.
Interaction between miRNAs and cadmium compounds.
| miRNA | Human/animal Study | Dose | Cell line | Dose | Targets | Observations | Ref |
|---|---|---|---|---|---|---|---|
| novel-miR-6, (Down), miR-27a-5p (Up), miR-122 (Down) | Common carp | 0.275 mg/L, 1 month | – | – | Caspase-9, Bax, Bak, Bcl-2 | In carp exposed to CdCl2, miRNA changes could be a biomarker. | [56] |
| 17 upregulated miRNAs (such as miR-7-1-5p) and 6 downregulated miRNAs (such as miR-9-6-5p) | Juvenile fish | 0.26 mg/L, 96h | – | – | MAPK, Jak/STAT, NF-κB, IL-4/13A, COX-2, PTGES, | CdCl2 in common carp spleens via targeting miRNA-mRNA networks could lead to inflammation. | [57] |
| miR-9a-5p, miR-29a-3p, miR-204-5p (Up) | Female SD rats | – | Rat ovarian granulosa cells, PC-12 | 0–20 μM, h | Bcl-2, Bax, Fas | Exposure to CdCl2 in rat ovarian granulosa cells via miR-204-5p and Bcl-2 could regulate apoptosis. | [57] |
| miR-21, miR-29b, (Up) | Human, SD rats | 2.0 mg/kg, 2 weeks | – | – | – | The expression of miR-21 could be a potential biomarker for the dysfunction of the kidney. | [58] |
| 44 upregulated (such as miR-21-5p, miR-3084c-3p), 54 downregulated (such as miR-455-3p, miR-193b-3p) | Male SD rats | 0.6 mg/kg, 5 days a week,3 months | – | – | – | CdCl2 could induce nephrotoxicity and change the expression of miRNAs. | [59] |
| miR-25-3p (Down) | juvenile common carp | 0.26 mg/L, 45 days | – | – | Hsp70/90, AMPK, PTEN, ULK1, mTOR, Atg-5/12, Beclin-1, LC3-II | Pollutants with CdCl2 in common carp could lead to autophagy and oxidative stress via miR-25-3p. | [60] |
| miR-26a, miR-155, (−) | Human | – | JEG-3 | 1–25 μM, 24–48 h | TGF-β, Smad-2/3 | miRNAs could regulate the TGF-β pathway in trophoblast cells exposed to cadmium. | [61] |
| miR-26a (−) | – | – | JEG-3 | 0–25 μM, 48 h | TGF-β | The migration of placental trophoblast cells could be inhibited by exposure to CdCl2 via the miR-26a/TGF-β axis. | [62] |
| miR-27b-3p (Up), miR-877-5p (Down) | – | – | 16HBE | 5 μmol/L, 14 weeks | CCM2 | Mentioned miRNAs could act as malignant transformation. | [24] |
| miR-30 family (a, b, d, c, e) (Down) | – | – | BEAS-2B, BEP2D | 0–10 μM, 72 h | SNAIL, ZEB1, Vimentin, E-cadherin | In human lung epithelial cells, exposure to CdCl2 via suppressing miR-30 could promote SNAIL and fibrosis. | [63] |
| miR-33-5p (Down) | Hy-Line Brown laying hens | 150 mg/kg, 3 months | – | – | BNIP3, LC3-I/II, Beclin-1, AMPK, AKT/mTOR, NF-kB/JNK, | In the chicken spleen, CdCl2 via regulating AMPK and miR-33 could induce autophagy, dependent on BNIP3. | [64] |
| miR-34a (Up) | Male Kunming mice | 1.5 mg/kg, 1 month | – | – | Sirt1, p53, Bax, Bcl-2, IL-1β Caspase-9, | miR-34a via targeting p53 or Sirt1 could induce nephrotoxicity. | [65] |
| miR-34a (Up) | Male Wistar rats | 10 mg/L | – | – | SIRT1, p53, IL-6, TNF-α, SREBP1/2, | The non-alcoholic fatty liver disease could be induced in an animal model by CdCl2. | [66] |
| miR-92a-2-5p (Down), miR-181b-5, (Up) | SD rats | 0.5–8 mg/kg | granulosa cells | 0–20 μM, 12 h | Bcl-2, Bax | miRNA profile could be changed in ovarian granulosa cells of rats exposed to CdCl2 during the prenatal period. | [67] |
| miR-92a-2-5p (Up) | Female SD rats | 8 mg/kg, postnatal day [56] | granulosa cells, COV434 | 0–20 μM, 24 h | c-Myc, Bcl-2, DNMT3B, DNMT1, DNMT3A | After cadmium exposure, in rat ovarian granulosa cells, c-Myc could promote the transcription of miR-92a-2-5p. | [68] |
| miR-101 (Down) | – | – | HUVECs | 0–80 μM, 0–36 h | COX-2, VEGF, eIF2α | miR-101 by sponging COX-2 could suppress angiogenesis induced by CdCl2 in HUVECs. | [69] |
| miR-122 (Down) | farmed tilapia | 12 mg/L, 24 h | – | – | MT 3′UTR | miR-122 via sponging metallothionein gene could act against hepatic oxidants induced by CdCl2. | [70] |
| miR-122-5p, miR-326-3p, (Up) | Human, Male SD rats | 0.6 mg/kg, 3 months | HK-2, NRK-52E | 9.18 and 10 μM, 48 h | – | Both mentioned miRNAs could be an early detective biomarker for CdCl2 exposure. | [71] |
| miR-122-5p, miR-326-3p, (Up) | SD rats | 0.6 mg/kg, 1.5 months | NRK-52E | 10 μM, 48 h | PLD1 | Both mentioned microRNAs via decreasing PLD1 could increase apoptosis in NRK-52E cells treated with CdCl2. | [72] |
| miR-143-3p (Up) | Human | – | hBMSCs | 0–30 μM, 7–24 h | Wnt/β-catenin, ARL6, ALP, RUNX2, LEF1, TCF1 | In hBMSCs exposed to CdCl2, miR-143-3p via targeting ARL6 could inhibit osteogenic differentiation. | [73] |
| miR-155, miR-181a, (Up) | Common carp | 0.005–0.5 mg/L, 1 month | – | – | HO-1, NF-κB, TLR-4, IL-1β, IL-8/10 | Both mentioned miRNAs via targeting HO-1 could lead to immunotoxicity in the carp's kidneys. | [74] |
| miR-155 (−), miR-221 (Down) | Human | – | – | – | IL-17, TNF-α | In workers exposed to CdCl2, there is an association between miRNAs and immune markers. | [75] |
| miR-217 (−) | Common carp | 0.005–0.5 mg/L, 1 month | – | – | SIRT1, TLR-4, NF-kB, TRAF6 | In common carp exposed to CdCl2, the miR-217/SIRT1 axis could lead to immunotoxicity. | [76] |
| miR-363-3p (Up) | Human (occupational chronic Cd poisoning) | – | HK-2, NRK-52E | 0–64 μM, 48 h | PI3K, PARP, Caspase-3 | miR-363-3p via suppressing PI3K could enhance cell death in the kidney. | [77] |
| miR-381 (Down) | – | – | HBEC | 1 μM | EZH2, H3K27me3 | In epithelial cells exposed to CdCl2, the miR-381/EZH2 axis could regulate the expression of the chloride channel. | [78] |
| miR-503-5p (Down) | SD rats | 0.6 mg/kg, 6 or 12 weeks | NRK-52E | 6–10 μM, 24 h | Wnt/β-catenin, α-SMA, Vimentin, Collagen1 | CdCl2 could induce kidney fibrosis and EMT via suppressing miR-503-5p and promoting the Wnt/β-catenin pathway | [79] |
| miR-6769b-5p (Up) | Human, male and female CD-1 mice | 2.5 mg/kg on the 15th gestational day | HTR-8/SVneo, | 0–40 μM, for 24 h | CCND1, PCNA | miR-6769b-5p via sponging CCND-1 could be involved in the proliferation of placental trophoblasts treated with CdCl2. | [80] |
2.2. Interaction between miRNAs and lead compounds
miR-106b-5p has been shown to be up-regulated by lead (Pb2+)-induced stress. miR-106b-5p has been shown to bind with the 3′-UTR of XIAP to down-regulate expression of XIAP. Inhibition of miR-106b-5p has been shown to reverse the decrease in IAP levels and cell viability in Pb2+-treated HT-22 and PC12 cells. Cumulatively, regulation of XIAP by miR-106b-5p might be associated with Pb neurotoxicity [81]. Another study has detected high levels of miR-155 and low levels of miR-126 in Pb exposed women. Moreover, authors have reported a significant simple positive relationship between blood lead levels and serum levels of miR-155. On the other hand, blood lead levels have been inversely correlated with serum miR-126 levels. Taken together, epigenetic changes might be linked with Pb exposure and its effects on health [82]. Besides, the interaction between miR-137 and EZH2 has been shown to contribute to the genome-wide redistribution of H3K27me3 which is responsible for Pb-associated memory impairment [83]. Table 3. Interaction between miRNAs and Pb in different contexts.
Table 3.
Interaction between miRNAs and lead compounds.
| miRNA | Human/animal Study | Dose | Cell line | Dose | Targets | Observations | Ref |
|---|---|---|---|---|---|---|---|
| miR-106b-5p (Up) | – | – | HT-22, PC12 | 0–100 μM, 48 h | XIAP | miR-106b-5p via targeting XIAP could inhibit cell viability. | [81] |
| miR-126 (Down), miR-155 (Up) | Human (Mexican women) | – | – | – | – | The expression mentioned miRNAs changed in Mexican women exposed to Pb. | [82] |
| miR-137 (Up) | Female SD rats | 250 p.p.m. | PC12, primary hippocampal neurons | 5 μM, 24 h | H3K27me3, EZH2, Wnt9b | Pb could lead to memory impairment via the miR-137/EZH2 axis. | [83] |
| miR-143-5p (Down) | C57BL/6 mice | 10 mg/kg, 1 month | Renal interstitial fibroblasts | 0–2 μM, 24 h | CCL20, Smad2/3, AKT, TGF-β1 | Administration of miR-143-5p via sponging CCL20 could decrease renal fibrosis induced by Pb. | [84] |
| miR-146a (−) | C57BL/6 mice | 200 mg/L, 15 days | BMECs | – | IRAK1, IL-2/8, PTX3 | In the mammary gland, Pb could increase inflammation levels. | [85] |
| miR-148a (−) | Human | – | – | – | Exposure to Pb could make an association between the methylation of DNA and miR-148a. | [86] | |
| miR-155 (Up) | Asian Carp | 1–2 mg/L | – | – | ERK, p38, IL-6, TNF-α, IL-1β | In the carp's head kidney, miR-155 could induce inflammation in the MAPK-dependent pathway. | [87] |
| miR-155, miR-221, (Up) | Human (North-Western India) | – | – | – | – | Both mentioned miRNAs increased in workers exposed to Pb. | [88] |
| miR-378a-3p (Up) | male C57 mice | 250–1000 mg/L, 12 weeks | HT22 | 10–40 μM, for 24 h | SLC7A11, GPX4 | In a model of nerve injury induced by Pb, miR-378a-3p via sponging SLC7A11 could be involved in the induction of ferroptosis. | [89] |
2.3. Interaction between miRNAs and asbestos
Comparison of miRNA signature between malignant pleural mesothelioma and benign asbestos-associated pleural effusion has led to identification of several up-regulated miRNAs in the former condition, among them being hsa-miR-484, hsa-miR-320, hsa-let-7a, and hsa-miR-125a-5p. These miRNAs have the potential to discriminate these two conditions [90]. Another study has reported down-regulation of miR-30d is in the pleural malignant mesothelioma cell line NCI–H2452, in the plasma samples of asbestos-exposed persons, and in mesothelial cells exposed to asbestos. Up-regulation of miR-30d could inhibit proliferation, migration, and invasion pleural malignant mesothelioma cells and enhance their apoptosis without affecting cell cycle. Moreover, it could decrease vimentin and TWIST1 levels, and increase CDH1 levels in NCI–H2452 cells. Thus, miR-30d is related to asbestos exposure and suppresses migration and invasion of NCI–H2452 cells through regulation of epithelial-mesenchymal transition [91]. Moreover, extracellular vesicle-levels of miR-103a-3p and miR-30e-3p have been shown to discriminate malignant pleural mesothelioma from past asbestos exposure [92]. Table 4 shows interactions between miRNAs and asbestos.
Table 4.
Interaction between miRNAs and asbestos.
| miRNA | Human/animal Study | Dose | Cell line | Dose | Targets | Observations | Ref |
|---|---|---|---|---|---|---|---|
| hsa-let-7a, miR-125a-5p, miR-320, miR-484, (Up) | Human (Malignant pleural mesothelioma patients) | – | – | – | WNT3, FGF9, TGFB2 | The expression of miRNAs could be changed in pleural effusion induced by asbestos. | [90] |
| miR-30d (Down) | Human (pleural malignant mesothelioma patients) | – | NCI–H2452 | Vimentin, CDH1, TWIST1 | Exposure of NCI–H2452 cells to asbestos could suppress invasion or migration via miR-30d. | [91] | |
| miR-30e-3p, miR-103a-3p, (Down) | Human (malignant pleural mesothelioma patients) | – | – | – | – | Extracellular vesicle miRNAs could be considered a biomarker of mentioned disease. | [92] |
| hsa-miR-98 (Down) | Human (Malignant pleural mesothelioma patients), GSE92636 database | – | – | – | – | Higher expression of miR-98 was associated with poor overall survival in patients with mesothelioma. | [93] |
| miR-126, miR-222, (Up) | Human (samples from malignant mesothelioma patients) | – | HUVECs, BEAS-2B, IMR-90, Met-5A | 5 μg/cm2 | EGFR, AKT, ERK, p38, PARP1 | Exposed cells to asbestos via activating the EGFR pathway could increase the expression of mentioned miRs | [94] |
| miR-126 (Up), miR-520g (−), miR-222 (Up), miR-205 (−) | Human (non–small cell lung cancer patients) | – | – | – | – | Mentioned-miRNAs could be changed in lung malignancies caused by asbestos. | [20] |
| miR-197-3p (dysregulated) | Human | – | – | – | Serum levels of miR-197-3p could be dysregulated in workers exposed to asbestos. | [95] | |
| miRNA profile, miR-197-3p, miR-1281, (Up) | Human (malignant pleural mesothelioma patients) | – | – | – | – | In workers who are ex-exposed to asbestos, the level of mentioned miRNAs increased. | [96] |
| miR-199/214 (Up) | specific pathogen-free F1 hybrid rats | – | MeT5A | – | Twist1, Akt, ERK, PTEN | In an animal model of sarcomatoid mesothelioma induced by asbestos, higher expression of miR-199/214 via targeting Twist1 could increase tumorigenesis. | [97] |
2.4. Interaction between miRNAs and mercury
Exposure to mercury is regarded as a public health problem in the world. Hsa-miR-92a and hsa-miR-486 have been suggested as novel diagnostic markers for detection of occupational mercury poisoning. These two miRNAs have been found to be over-expressed in individuals exposed to occupational mercury. Over-expression of these miRNAs contributes to mercury toxicity through activation of NF-κB signaling via influencing expressions of KLF4 and Cezanne, respectively [98]. Another study has shown significant differences in the plasma levels of miR-124-3p, miR-125-5p, and miR-127-3p between patients with amalgam filling, dentists, and control group. Serum mercury concentration and plasma miR-125-5p and miR-127-3p levels have been positively correlated. Serum mercury has also been correlated with plasma miR-125-5p levels among dentists. This study shows the impact of amalgam filling in enhancement of serum mercury and plasma miRNA levels [99]. Besides, two distinct miRNA signatures have been reported to be activated upon neuronal differentiation and following MeHgCl-induced toxicity. Principally, exposure to MeHgCl could induce down-regulation of six out of the ten most up-regulated neuronal pathways in neural models. In fact, miRNAs expression profiling has been suggested as a possible way for evaluation of developmental neurotoxicity pathway [100]. Table 5 shows the interaction between miRNAs and mercury.
Table 5.
Interaction between miRNAs and mercury.
| miRNA | Human/animal Study | Dose | Cell line | Dose | Targets | Observations | Ref |
|---|---|---|---|---|---|---|---|
| hsa-miR-92a, hsa-miR-486, (Up) | Human (workers exposed to mercury) | – | 293T, HUVECs | 0–10 μM, 24 h | NF-κB, KLF4, COX-2 | Both mentioned miRNAs via afecting NF-κB activity could lead to mercury toxicity. | [98] |
| miR-124-3p (−), miR-125-5p (Up), miR-127-3p (Up) | Human (amalgam filling patients and Dentists) | – | – | – | – | There is a relationship between serum mercury levels of miRNAs. | [99] |
| 9 upregulated (such as miR-141, miR-196b), 5 downregulated (such as miR-217, miR-296) | – | – | H9, hESCs | 0–500 nM | SOX2, FGF4, DNMT3B, COL2A1 | Methyl Mercury could Induce neuronal toxicity and change the expression of miRNAs. | [100] |
2.5. Interaction between miRNAs and chromium compounds
Hexavalent chromium [Cr(VI)] has been shown to induce various kind of cancer including lung cancer. Cr(VI) treatment can also increase expression of Nrf2, a redox sensitive transcription factor with protective effcets on normal cells. Mechanistically, expression of redox sensitive miRNAs miR-27a and miR-27b is ecreased after 1 month exposure to Cr(VI), leading to alteration sin levels of their target Nrf2. Taken together, suppression of miR-27a/b leads to up-regulation of Nrf2 at early and late stages of exposure to Cr(VI) [101]. Cr(VI) has also been found to induce malignant transformation in lung bronchial epithelium through ROS-dependent induction of miR-21-PDCD4 signals [102]. Table 6 shows the interaction between miRNAs and chromium compounds.
Table 6.
Interaction between miRNAs and chromium compounds.
| miRNA | Human/animal Study | Dose | Cell line | Dose | Targets | Observations | Ref |
|---|---|---|---|---|---|---|---|
| miR-21 (Up) | Human, female BALB/c mice, Athymic nude mice | 1.2 mg/ml, once/week, 3 months | BEAS-2B, NL20, A549, H23, H2030, H460 | 0–5 μM, 24 h | PDCD4, β-catenin, c-Myc, TCF4, E-cadherin | Cr(VI) via upregulating miR-21 could lead to malignant transformation. | [102] |
| miR-27a/b (Down) | nude mice, BALB/cJ mice | zincchromate (1.0 mg/mL), 3 months | BEAS-2B | 1 μM, 6 months | Nrf-2, KEAP1, HO-1 | Overexpression of miR-27a/b via sponging Nrf-2 could act against Cr(VI) and lead to tumor suppression. | [101] |
| miR-223-3p, miR-327, miR-466f-3p, (Up) | C57BL mice offspring | 0.14 and 1.19 mg Cr/kg | – | – | Akt1, Pik3ca | In adult mice offspring, maternal chromium restriction via targeting miRNAs could lead to insulin resistance. | [103] |
| 9 downregulated miRNAs (such as miR-451, miR-301) | Human | – | – | – | – | Urinary chromium levels could change the expression of miRNAs in patients with cardiovascular diseases and metabolic diseases. | [104] |
| miR-494 (Down) | nude mouse | – | BEAS-2B | K2Cr2O7 (0.25 μM), 5 months | c-Myc | Cr(VI) via promoting c-Myc expression could lead to tumorigenesis. | [105] |
| miR-3940-5p (Down) | Human | – | – | – | XRCC2, BRCC3 | In workers exposed to Cr(VI), miR-3940-5p could lead to genetic damage | [106] |
2.6. Interactions between miRNAs and beryllium sulfate
The carcinogenic material beryllium sulfate (BeSO4) can affect expression of a number of non-coding RNAs in human bronchial epithelial cells. This substance has been found to up-regulate expression of 36 circRNAs and down-regulate other 35 circRNAs in these cells. Hsa_circ_0004214 and hsa_circ_0003586 have been among up-regulated circRNAs; and hsa_circ_0047958, hsa_circ_0001944, and hsa_circ_0008982 have been among down-regulated ones. These circRNAs can affect expression of a number of miRNAs that regulate cellular senescence, as well as TNF, NF-κB, HIF-1, and Hippo signaling pathways. The toxic effects of this substance is mainly mediated through sponging miR-663b and regulating JAK/STAT signaling [107]. Another study has shown that BeSO4 increases expression of some inflammatory molecules, including IL-10, TNF-α, IFN-γ, iNOS, and COX-2. Most notably, expression of 179 miRNAs has also been found to be changed by this substance. A number of these miRNAs have been shown to contribute to the transcription regulation, or modulation of MAPK, and VEGF signaling pathways [108].
2.7. Interaction between miRNAs and fluoride
Expression of several miRNAs has been shown to be changed in rat renal cortex following subchronic exposure to fluoride. These miRNAs have been mainly associated with extracellular matrix-receptor interactions, Mucin type O-glycan synthesis and Gap junctions. Moreover, expressions of miRNAs involved in cancer and proliferation have been changed after exposure to fluoride [109]. Combination of fluoride and aluminum (FA) has been shown to trigger apoptosis of rat hippocampal neurons and NG108-15 cells, enhance expression of miR-34b-5p, and decrease levels of Gnai2, PKA, ERK and CREB. Notably, suppression of miR-34b-5p expression could ameliorate FA-associated apoptosis and changes in the expressions of mentioned genes. Besides, miR-34b-5p has been found to modulate expression Gnai2 through targeting its 3′-UTR, indicating that miR-34b-5p participate in FA-associated neuron apoptosis through negatively targeting Gnai2 and suppressing activity of PKA/ERK/CREB cascade [110].
Furthermore, fluoride has been demonstrated to affect expression of 35 miRNAs, particularly those associated with glycolipid metabolism in the liver. In fact, these miRNAs could mediate fluoride-induced disturbance in the glycolipid metabolism, possibly through affecting activity of insulin, PPAR, and FOXO pathways [111].
2.8. Interaction between miRNAs and a combination of hazardous compounds
A number of studies have compared the effects of different hazardous materials in cell lines or animal models. For instance, experiments in C57BL/6J WT mice have shown that arsenic has more potent effects in disruption of the INS-1 beta cell miRNA landscape thansignature compared with cadmium or manganese [112]. Meanwhile, mixture of As–Cd–Pb has been shown to induce cellular transformation through affecting expression of miR-222 and post-transcriptional regulation of Rad51c levels [113]. Another study has assessed the association between miRNA profile in the cervix during pregnancy and levels of lead and mercury. This study has reported negative associations between levels of 17 miRNAs and toenail mercury levels. Moreover, tibial bone lead levels have been associated with down-regulation of miR-575 and miR-4286. Taken together, miRNAs levels in the human cervix has been suggested as novel markers for maternal exposures during pregnancy [114]. Table 7 shows the interaction between miRNAs and a combination of hazardous compounds.
Table 7.
Interaction between miRNAs and a combination of hazardous compounds.
| Compounds | miRNA | Human/animal Study | Dose | Cell line | Dose | Targets | Observations | Ref |
|---|---|---|---|---|---|---|---|---|
| Inorganic arsenic (iAs), manganese (Mn), CdCl2 | miR-146a, (Up in iAS), (Down in CdCl2) | C57BL/6J WT mice | 0.1 mg As/L | INS-1832/13, | iAs (1 μM), CdCl2 (5 μM), Mn (25 μM), 24 h | NF-κB, Camk2a | In INS-1832/13 cells (i.e. insulinoma cells), arsenic had more powerful damaging effects compared to others. | [112] |
| Pb, As, and CdCl2 | miR-222 (Up) | – | – | Balb/c 3T3 | NaAsO2 (2 μM), Pb (5 μM), and CdCl2 (2 μM) | Rad51c | A mixture of all hazardous materials could lead to cellular transformation via the miR-222/Rad51c axis. | [113] |
| Pb and Mercury | miR-575, miR-4286, (Down) | Human (pregnant women) | – | – | – | – | In the human cervix of pregnant women exposed to mercury and Pb, the miRNA expression was altered. | [114] |
| CdCl2 and NaAsO2 | miR-let7a, miR-146a, (Down) | male Wistar rats | NaAsO2 (5 mg/kg), CdCl2 (1 mg/kg), one month | – | – | – | Arsenite and CdCl2 could lead to organ toxicity. | [115] |
2.9. The effects of antioxidants on expression of miRNAs during exposure with hazardous compounds
Treatment with antioxidants can ameliorate the effects of hazardous materials on body organs through modulation of expression of miRNAs (Table 8). For instance, treatment with quercetin via inhibiting miR-21 could attenuate liver fibrosis and steatosis induced by cadmium [116]. Moreover, Se–Y via increasing the expression of miR-26a-5p could act against necroptosis induced by CdCl2 in the kidney of the chicken [117] (Fig. 3). Other examples are shown in Table 8.
Table 8.
Interaction between miRNAs, hazardous compounds, as well as antioxidants.
| Type | miRNA | Human/Animal Study | Dose | Other treatments (Source of antioxidants) | Cell line | Dose of hazardous materials | Targets or Pathways | Results | Ref |
|---|---|---|---|---|---|---|---|---|---|
| CdCl2 | miR-21a (Up) | Male Wistar rats | 10 moml/L, 5 months | Quercetin; 50 mg/kg, 5 months | – | – | NF-kB P65, Nrf2, Smad3, SREBP1, TGF-β1 | Treatment with quercetin via inhibiting miR-21 could attenuate liver fibrosis and steatosis induced by cadmium. | [116] |
| CdCl2 | miR-26a-5p (Down) | Hy-Line Brown strain | 150 mg/kg, 42 days | Selenium yeast (Se–Y); 0.5 mg/kg, 42 days | – | – | HSP60/80/90, PTEN, PI3K/AKT, RIP1/3 | Se–Y via increasing the expression of miR-26a-5p could act against necroptosis induced by CdCl2 in the kidney of the chicken. | [117] |
| CdCl2 | miR-30a (Down) | Hy-Line Brown chickens | 150 mg/kg, 3 months | Se; (0.2 mg/kg of Na2SeO3), 3 months | – | – | GRP78, JNK, IRE-1, ATG5, LC-3I/II, Beclin-1 | In the chicken kidneys, CdCl2 via mediating GRP78 and miR-30a could cause JNK-dependent autophagy. | [118] |
| CdCl2 | miR-125a, miR-125b, (Down) | – | – | Selenium; 5–20 μM, 0.5 h before the Cd administration | LLC-PK1 | 20 μM, 12 h, | Bax, Bak, Caspase-3 | Treatment with selenium via targeting miR-125a/b could inhibit apoptosis induced by CdCl2. | [119] |
| CdCl2 | miR-146a (Up) | Male albino rats | 3 mg/kg, daily, 2 months | N-acetylcysteine (NAC); 100 mg/kg, daily, 2 months | – | – | NF-κB p65, TNF-α, IL-1β, TRAF6 | NAC could attenuate Cd-induced hepatotoxicity by decreasing the expression of miR-146a and inflammation. | [120] |
| CdCl2 | miR-182-5p (Down) | male Kunming mice | 1.5 mg/kg, | CAPE; 10 μmol/kg body weight |
– | – | TLR4, IL-1β, IL-6, TNF-α, PI3K/AKT, mTOR, Caspase-3 | CAPE could downregulate hepatotoxicity induced by CdCl2. | [121] |
| CdCl2 | miR-216a (Up) | common carps | – | Se; (10−6 mol/L of Na2SeO3), 6 h | Lymphocyte | 4 × 10−5 mol/L, 6 h | PI3K/AKT, Bax, Bcl-2, Caspase-3/9, RIP3, MLKL | Se could act against the promotion of the miR-216a, necrosis, and apoptosis induced by CdCl2 in the lymphocytes of common carp. | [122] |
| CdCl2 | miR-661 (Down) | – | – | Caffeic acid phenethyl ester (CAPE); 10 μM | HepG2 | 0–30 μM, 24 h | Caspase-9 | CAPE could downregulate apoptosis induced by CdCl2. | [123] |
| Pb | miR-16-5p (Up) | Hy-Line Brown chickens | – | Se; (1 μM of Na2SeO3) | Neutrophil | 12.5 μM | IGF1R, PiK3R1, p53, Bcl-2, Bax, Caspase-3/8/9 | In chicken neutrophil cells, Se via targeting miR-16-5p had an antagonistic impact against lead-induced apoptosis. | [73] |
| Pb | miR-224 (Up) | Male Wistar rats | 30 mg/kg, once every 2 days, less than 4 months | Selenium nanoparticles (Se-NPs); 0.5 mg/kg, less than 4 months | – | – | ID1 | Se-NPs via inhibiting miR-224 could attenuate adverse effects of Pb on thyroid tissues. | [124] |
| ATO | miR-182-5p (down) | – | – | NAC; 10 mM, 4h | U87MG, S1 GBM primary cells, A549, H1299 | 0–5 μM | SESN2, HO-1 | ATO via inhibiting miR-182-5p and increasing SESN2 could impede oxidative stress. | [125] |
Fig. 3.
The illustration shows the effects of antioxidants on miRNA expression during exposure to hazardous compounds. (A) CAPE could downregulate hepatotoxicity induced by CdCl2 through upregulation of miR-182-5p. (B) Selenium in common carp lymphocytes may inhibit the promotion of miR-216a, necrosis, and apoptosis caused by CdCl2. (C) Quercetin, via inhibiting miR-21, could attenuate liver fibrosis and steatosis induced by cadmium. (D) Selenium yeast in the kidney of the chicken could protect against necroptosis caused by CdCl2 by increasing the expression of miR-26a-5p. (E) N-acetylcysteine in male albino rats could attenuate Cd-induced hepatotoxicity by decreasing the expression of miR-146a and inflammation.
3. Conclusions
Several compounds have been shown to affect expressions of miRNAs, thus disturbing activity of several signaling pathways in different tissues and contributing to diverse disorders. The impacts of the environmental exposure to hazardous materials on the epigenome have attracted a substantial interest in the recent years. miRNAs as important regulators of gene expression are of considerable importance in this regard. Several miRNAs have been shown to be dysregulated during exposure to these toxic agents being responsible for alterations in the physiological processes after exposure to toxins. Therefore, expression profiling of miRNAs represents a possible route for determination of the effects of hazardous materials on the body organs. Since miRNAs are stable in the circulation and are protected from endogenous RNase, miRNAs are regarded as suitable blood-based biomarkers not only for detection of human diseases but also for estimation of the amount of exposure to hazardous materials. However, the underlying mechanisms of contribution of miRNAs in toxic effects of these materials have not been elucidated yet.
More research is needed to establish a reliable profile of miRNA alterations after exposure to each hazardous material. These putative well-defined miRNA signatures can be used for early detection of disorders being associated with these compounds. Examples of these disorders include cancers, neurodegenerative disorders and pulmonary disorders. In addition, identification of the altered miRNAs during exposure to toxins can help in design of novel therapeutic modalities for complex disorders that are associated with environmental exposure. Finally, certain antioxidants have been found to ameliorate the effects of hazardous materials, particularly CdCl2 and Pb on miRNAs profile, thus amending the organ impairment/dysfunction associated with hazardous materials. Future high throughput studies are needed to find the suitable antioxidant for amelioration of each condition. These antioxidants are expected to reverse the effects of these materials on body organs; thus, they can be prescribed for persons that environmental or occupational exposure to hazardous materials. The off-target effects of antioxidants should be assessed in future studies.
Ethics approval and consent to participant
Not applicable.
Consent of publication
Not applicable.
Funding
Not applicable.
Authors’ contributions
SGF wrote the draft and revised it. MT and EJ designed and supervised the study. SRA, BMH, SDO and HS collected the data and designed the figures and tables. All the authors read the submitted version and approved it.
Declaration of competing interest
The authors declare they have no conflict of interest.
Acknowledgement
The authors would like to thank the clinical Research Development Unit (CRDU) of Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran for their support, cooperation and assistance throughout the period of study.
Contributor Information
Mohammad Taheri, Email: mohammad.taheri@uni-jena.de.
Elena Jamali, Email: elena.jamali@yahoo.com.
Abbreviations
- TEAD1
TEA domain transcription factor 1
- PIM1
Pim-1 proto-oncogene
- NQO1
NAD(P)H quinone oxidoreductase 1
- hERG
human ether-a-go-go-related gene
- SATB2
special AT-rich sequence-binding protein 2
- SP1
Specificity Protein 1
- PHLPP
PH domain leucine-rich repeat protein phosphatase
- LEF1
lymphoid enhancer factor
- TCF1
T-cell factor 1
- XIAP
X-linked inhibitor of apoptosis protein
- CCL20
CC chemokine ligand 20
- CCM3
cerebral cavernous malformation 3
- PLD1)
Phospholipase D1
- Camk2a
Ca2+/calmodulin-dependent protein kinase IIα
- KLF4
Krüppel-like factor 4
- COX-2
cyclooxygenase-2
- PDCD4
Programmed cell death 4
- IGF1R
insulin-like growth factor 1 receptor
- ID1
Iodothyronine deiodinase type 1
- SESN2
Sestrin-2
- HBEC
human bronchial epithelial cells
- hBMSCs
human bone marrow mesenchymal stem cells
- BMECs
bovine mammary epithelial cells
- HUVECs
Human Umbilical Vein Endothelial Cells
- ATO
Arsenic trioxide
- NaAsO₂
Sodium arsenite
- AsCl3
Arsenic chloride
- As4S4
Arsenic sulfide
- CdCl2
Cadmium
- Pb
Lead
- Cr
chromium
- Cr(VI)
Hexavalent chromium
References
- 1.Henretig F.M., Kirk M.A., McKay C.A., Jr. Hazardous chemical emergencies and poisonings. N. Engl. J. Med. 2019;380(17):1638–1655. doi: 10.1056/NEJMra1504690. [DOI] [PubMed] [Google Scholar]
- 2.Genchi G., Sinicropi M.S., Lauria G., Carocci A., Catalano A. The effects of cadmium toxicity. Int. J. Environ. Res. Publ. Health. 2020;17(11):3782. doi: 10.3390/ijerph17113782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DesMarias T.L., Costa M. Mechanisms of chromium-induced toxicity. Curr. Opin.Toxicol. 2019;14:1–7. doi: 10.1016/j.cotox.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdul K.S.M., Jayasinghe S.S., Chandana E.P., Jayasumana C., De Silva P.M.C. Arsenic and human health effects: a review. Environ. Toxicol. Pharmacol. 2015;40(3):828–846. doi: 10.1016/j.etap.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Hanna-Attisha M., Lanphear B., Landrigan P. American Public Health Association; 2018. Lead Poisoning in the 21st Century: the Silent Epidemic Continues; p. 1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feenstra L., Erkel J., Vasak L. International Groundwater Resources Assessment Centre. 2007. Arsenic in groundwater: overview and evaluation of removal. [Google Scholar]
- 7.Avataneo C., Petriglieri J.R., Capella S., Tomatis M., Luiso M., Marangoni G., et al. Chrysotile asbestos migration in air from contaminated water: an experimental simulation. J. Hazard Mater. 2022;424 doi: 10.1016/j.jhazmat.2021.127528. [DOI] [PubMed] [Google Scholar]
- 8.Oymak T., Tokalıoğlu Ş., Yılmaz V., Kartal Ş., Aydın D. Determination of lead and cadmium in food samples by the coprecipitation method. Food Chem. 2009;113(4):1314–1317. [Google Scholar]
- 9.Steinmaus C., Moore L., Hopenhayn-Rich C., Biggs M.L., Smith A.H. Arsenic in drinking water and bladder cancer: environmental carcinogenesis. Cancer Invest. 2000;18(2):174–182. doi: 10.3109/07357900009038249. [DOI] [PubMed] [Google Scholar]
- 10.Wei S., Zhang H., Tao S. A review of arsenic exposure and lung cancer. Toxicol. Res. 2019;8(3):319–327. doi: 10.1039/c8tx00298c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kameda T., Takahashi K., Kim R., Jiang Y., Movahed M., Park E.-K., et al. Asbestos: use, bans and disease burden in Europe. Bull. World Health Organ. 2014;92:790–797. doi: 10.2471/BLT.13.132118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carbone M., Kratzke R.A., Testa J.R., editors. Seminars in Oncology. Elsevier; 2002. The pathogenesis of mesothelioma. [Google Scholar]
- 13.Larsson S.C., Wolk A. Urinary cadmium and mortality from all causes, cancer and cardiovascular disease in the general population: systematic review and meta-analysis of cohort studies. Int. J. Epidemiol. 2016;45(3):782–791. doi: 10.1093/ije/dyv086. [DOI] [PubMed] [Google Scholar]
- 14.da Cunha Martins A., Jr., Carneiro M.F.H., Grotto D., Adeyemi J.A., Barbosa F., Jr. Arsenic, cadmium, and mercury-induced hypertension: mechanisms and epidemiological findings. J. Toxicol. Environ. Health, Part A B. 2018;21(2):61–82. doi: 10.1080/10937404.2018.1432025. [DOI] [PubMed] [Google Scholar]
- 15.Bishak Y.K., Payahoo L., Osatdrahimi A., Nourazarian A. Mechanisms of cadmium carcinogenicity in the gastrointestinal tract. Asian Pac. J. Cancer Prev. APJCP. 2015;16(1):9–21. doi: 10.7314/apjcp.2015.16.1.9. [DOI] [PubMed] [Google Scholar]
- 16.Ghafouri-Fard S., Shoorei H., Anamag F.T., Taheri M. The role of non-coding RNAs in controlling cell cycle related proteins in cancer cells. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.608975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghafouri-Fard S., Shirvani-Farsani Z., Taheri M. The role of microRNAs in the pathogenesis of thyroid cancer. Non-coding RNA Res. 2020;5(3):88–98. doi: 10.1016/j.ncrna.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z., Myers R., Wei T., Bind E., Kassim P., Wang G., et al. Placental transfer and concentrations of cadmium, mercury, lead, and selenium in mothers, newborns, and young children. J. Expo. Sci. Environ. Epidemiol. 2014;24(5):537–544. doi: 10.1038/jes.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobo-Estrada T., Santoyo-Sánchez M., Thévenod F., Barbier O. Cadmium handling, toxicity and molecular targets involved during pregnancy: lessons from experimental models. Int. J. Mol. Sci. 2017;18(7):1590. doi: 10.3390/ijms18071590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santarelli L., Gaetani S., Monaco F., Bracci M., Valentino M., Amati M., et al. Four-miRNA signature to identify asbestos-related lung malignanciesmiRNAs in asbestos-related malignancies. Cancer Epidemiol. Biomarkers Prev. 2019;28(1):119–126. doi: 10.1158/1055-9965.EPI-18-0453. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y., Guo S., Wang S., Li X., Hou D., Li H., et al. LncRNA OIP5-AS1 inhibits ferroptosis in prostate cancer with long-term cadmium exposure through miR-128-3p/SLC7A11 signaling. Ecotoxicol. Environ. Saf. 2021;220 doi: 10.1016/j.ecoenv.2021.112376. [DOI] [PubMed] [Google Scholar]
- 22.Shafiq S., Zeb Q., Ali A., Sajjad Y., Nazir R., Widemann E., et al. Lead, cadmium and zinc phytotoxicity alter DNA methylation levels to confer heavy metal tolerance in wheat. Int. J. Mol. Sci. 2019;20(19):4676. doi: 10.3390/ijms20194676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y., Li Z.-F., Zhang D., Wang Z.-Y., Wang L. Quercetin alleviates Cadmium-induced autophagy inhibition via TFEB-dependent lysosomal restoration in primary proximal tubular cells. Ecotoxicol. Environ. Saf. 2021;208 doi: 10.1016/j.ecoenv.2020.111743. [DOI] [PubMed] [Google Scholar]
- 24.Liu Q., Zheng C., Shen H., Zhou Z., Lei Y. MicroRNAs-mRNAs expression profile and their potential role in malignant transformation of human bronchial epithelial cells induced by cadmium. BioMed Res. Int. 2015;2015 doi: 10.1155/2015/902025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassan F., Nuovo G.J., Crawford M., Boyaka P.N., Kirkby S., Nana-Sinkam S.P., et al. MiR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0050837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghafouri-Fard S., Abak A., Talebi S.F., Shoorei H., Branicki W., Taheri M., et al. Role of miRNA and lncRNAs in organ fibrosis and aging. Biomed. Pharmacother. 2021;143 doi: 10.1016/j.biopha.2021.112132. [DOI] [PubMed] [Google Scholar]
- 27.Dexheimer P.J., Cochella L. MicroRNAs: from mechanism to organism. Front. Cell Dev. Biol. 2020;8:409. doi: 10.3389/fcell.2020.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vinci S., Gelmini S., Mancini I., Malentacchi F., Pazzagli M., Beltrami C., et al. Genetic and epigenetic factors in regulation of microRNA in colorectal cancers. Methods. 2013;59(1):138–146. doi: 10.1016/j.ymeth.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z., Zhao Y., Smith E., Goodall G.J., Drew P.A., Brabletz T., et al. Reversal and prevention of arsenic-induced human bronchial epithelial cell malignant transformation by microRNA-200b. Toxicol. Sci. 2011;121(1):110–122. doi: 10.1093/toxsci/kfr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nymark P., Guled M., Borze I., Faisal A., Lahti L., Salmenkivi K., et al. Integrative analysis of microRNA, mRNA and aCGH data reveals asbestos‐and histology‐related changes in lung cancer. Gene Chromosome Cancer. 2011;50(8):585–597. doi: 10.1002/gcc.20880. [DOI] [PubMed] [Google Scholar]
- 31.Gao S-m, Chen C., Wu J., Tan Y., Yu K., Xing C.-Y., et al. Synergistic apoptosis induction in leukemic cells by miR-15a/16-1 and arsenic trioxide. Biochem. Biophys. Res. Commun. 2010;403(2):203–208. doi: 10.1016/j.bbrc.2010.10.137. [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Zhu X., Gu J., Dong D., Yao J., Lin C., et al. Anti‐miR‐21 oligonucleotide sensitizes leukemic K562 cells to arsenic trioxide by inducing apoptosis. Cancer Sci. 2010;101(4):948–954. doi: 10.1111/j.1349-7006.2010.01489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng Q., Zou Z., Wang Q., Sun B., Liu Y., Liang B., et al. Association and risk of five miRNAs with arsenic-induced multiorgan damage. Sci. Total Environ. 2019;680:1–9. doi: 10.1016/j.scitotenv.2019.05.042. [DOI] [PubMed] [Google Scholar]
- 34.Wang P., Xiao T., Li J., Wang D., Sun J., Cheng C., et al. miR-21 in EVs from pulmonary epithelial cells promotes myofibroblast differentiation via glycolysis in arsenic-induced pulmonary fibrosis. Environ. Pollut. 2021;286 doi: 10.1016/j.envpol.2021.117259. [DOI] [PubMed] [Google Scholar]
- 35.Meng X.Z., Zheng T.S., Chen X., Wang J.B., Zhang W.H., Pan S.H., et al. microRNA expression alteration after arsenic trioxide treatment in HepG‐2 cells. J. Gastroenterol. Hepatol. 2011;26(1):186–193. doi: 10.1111/j.1440-1746.2010.06317.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang S., Ma C., Pang H., Zeng F., Cheng L., Fang B., et al. Arsenic trioxide suppresses cell growth and migration via inhibition of miR-27a in breast cancer cells. Biochem. Biophys. Res. Commun. 2016;469(1):55–61. doi: 10.1016/j.bbrc.2015.11.071. [DOI] [PubMed] [Google Scholar]
- 37.Chen Q.Y., Li J., Sun H., Wu F., Zhu Y., Kluz T., et al. Role of miR‐31 and SATB2 in arsenic‐induced malignant BEAS‐2B cell transformation. Mol. Carcinog. 2018;57(8):968–977. doi: 10.1002/mc.22817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J., Li Y., Jiang C. MiR-133b contributes to arsenic-induced apoptosis in U251 glioma cells by targeting the hERG channel. J. Mol. Neurosci. 2015;55(4):985–994. doi: 10.1007/s12031-014-0455-8. [DOI] [PubMed] [Google Scholar]
- 39.Gao S.-Y., Zhou X., Li Y.-J., Liu W.-L., Wang P.-Y., Pang M., et al. Arsenic trioxide prevents rat pulmonary fibrosis via miR-98 overexpression. Life Sci. 2014;114(1):20–28. doi: 10.1016/j.lfs.2014.07.037. [DOI] [PubMed] [Google Scholar]
- 40.Pérez-Vázquez M.S., Ochoa-Martínez Á.C., RuÍz-Vera T., Araiza-Gamboa Y., Pérez-Maldonado I.N. Evaluation of epigenetic alterations (mir-126 and mir-155 expression levels) in Mexican children exposed to inorganic arsenic via drinking water. Environ. Sci. Pollut. Res. 2017;24(36):28036–28045. doi: 10.1007/s11356-017-0367-6. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y., Jiang F., Jiao K., Ju L., Liu Q., Li Y., et al. De-methylation of miR-148a by arsenic trioxide enhances sensitivity to chemotherapy via inhibiting the NF-κB pathway and CSC like properties. Exp. Cell Res. 2020;386(2) doi: 10.1016/j.yexcr.2019.111739. [DOI] [PubMed] [Google Scholar]
- 42.Ji H., Li Y., Jiang F., Wang X., Zhang J., Shen J., et al. Inhibition of transforming growth factor beta/SMAD signal by MiR‐155 is involved in arsenic trioxide‐induced anti‐angiogenesis in prostate cancer. Cancer Sci. 2014;105(12):1541–1549. doi: 10.1111/cas.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu S., Lai Y., Chen H., Liu Y., Zhang Z. miR-155 mediates arsenic trioxide resistance by activating Nrf2 and suppressing apoptosis in lung cancer cells. Sci. Rep. 2017;7(1):1–13. doi: 10.1038/s41598-017-06061-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Eryani L., Jenkins S.F., States V.A., Pan J., Malone J.C., Rai S.N., et al. miRNA expression profiles of premalignant and malignant arsenic-induced skin lesions. PLoS One. 2018;13(8) doi: 10.1371/journal.pone.0202579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beezhold K., Liu J., Kan H., Meighan T., Castranova V., Shi X., et al. miR-190-mediated downregulation of PHLPP contributes to arsenic-induced Akt activation and carcinogenesis. Toxicol. Sci. 2011;123(2):411–420. doi: 10.1093/toxsci/kfr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han Y., Cai Y., Lai X., Wang Z., Wei S., Tan K., et al. lncRNA RMRP prevents mitochondrial dysfunction and cardiomyocyte apoptosis via the miR-1-5p/hsp70 Axis in LPS-induced sepsis mice. Inflammation. 2020:1–14. doi: 10.1007/s10753-019-01141-8. [DOI] [PubMed] [Google Scholar]
- 47.He J., Wang M., Jiang Y., Chen Q., Xu S., Xu Q., et al. Chronic arsenic exposure and angiogenesis in human bronchial epithelial cells via the ROS/miR-199a-5p/HIF-1 α/COX-2 pathway. Environ. Health Perspect. 2014;122(3):255–261. doi: 10.1289/ehp.1307545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He J.-H., Li Y.-M., Li Y.-G., Xie X.-Y., Wang L., Chun S.-Y., et al. hsa-miR-203 enhances the sensitivity of leukemia cells to arsenic trioxide. Exp. Ther. Med. 2013;5(5):1315–1321. doi: 10.3892/etm.2013.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang M., Ge X., Zheng J., Li D., Liu X., Wang L., et al. Role and mechanism of miR-222 in arsenic-transformed cells for inducing tumor growth. Oncotarget. 2016;7(14) doi: 10.18632/oncotarget.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong M., Huang Z., Wang L., Lin Z., Cao Z., Li X., et al. Malignant transformation of human bronchial epithelial cells induced by arsenic through STAT3/miR-301a/SMAD4 loop. Sci. Rep. 2018;8(1):1–12. doi: 10.1038/s41598-018-31516-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao Y., Yin Y., Xing X., Zhao Z., Lu Y., Sun Y., et al. Arsenic-induced anti-angiogenesis via miR-425-5p-regulated CCM3. Toxicol. Lett. 2016;254:22–31. doi: 10.1016/j.toxlet.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 52.Zhu C., Zhou R., Zhou Q., Chang Y., Jiang M. microRNA-539 suppresses tumor growth and tumorigenesis and overcomes arsenic trioxide resistance in hepatocellular carcinoma. Life Sci. 2016;166:34–40. doi: 10.1016/j.lfs.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Cai X., Yu L., Chen Z., Ye F., Ren Z., Jin P. Arsenic trioxide-induced upregulation of miR-1294 suppresses tumor growth in hepatocellular carcinoma by targeting TEAD1 and PIM1. Cancer Biomarkers. 2020;28(2):221–230. doi: 10.3233/CBM-190490. [DOI] [PubMed] [Google Scholar]
- 54.Sharma M., Sharma S., Arora M., Kaul D. Regulation of cellular Cyclin D1 gene by arsenic is mediated through miR-2909. Gene. 2013;522(1):60–64. doi: 10.1016/j.gene.2013.03.058. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X., Tan Z., Kang T., Zhu C., Chen S. Arsenic sulfide induces miR-4665-3p to inhibit gastric cancer cell invasion and migration. Drug Des. Dev. Ther. 2019;13:3037. doi: 10.2147/DDDT.S209219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Q., Yang J., Gong Y., Cai J., Zheng Y., Zhang Y., et al. MicroRNA profiling identifies biomarkers in head kidneys of common carp exposed to cadmium. Chemosphere. 2020;247 doi: 10.1016/j.chemosphere.2020.125901. [DOI] [PubMed] [Google Scholar]
- 57.Chen J., Zhang S., Tong J., Teng X., Zhang Z., Li S., et al. Whole transcriptome-based miRNA-mRNA network analysis revealed the mechanism of inflammation-immunosuppressive damage caused by cadmium in common carp spleens. Sci. Total Environ. 2020;717 doi: 10.1016/j.scitotenv.2020.137081. [DOI] [PubMed] [Google Scholar]
- 58.Lei L.-J., Zhang Z., Guo J.-Y., Shi X.-J., Zhang G.-Y., Kang H., et al. MiR-21 as a potential biomarker for renal dysfunction induced by cadmium exposure. Int. J. Clin. Exp. Med. 2019;12(2):1631–1639. [Google Scholar]
- 59.Fay M.J., Alt L.A., Ryba D., Salamah R., Peach R., Papaeliou A., et al. Cadmium nephrotoxicity is associated with altered microRNA expression in the rat renal cortex. Toxics. 2018;6(1):16. doi: 10.3390/toxics6010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z., Shah S.W.A., Zhou Q., Yin X., Teng X. The contributions of miR-25-3p, oxidative stress, and heat shock protein in a complex mechanism of autophagy caused by pollutant cadmium in common carp (Cyprinus carpio L.) hepatopancreas. Environ. Pollut. 2021;287 doi: 10.1016/j.envpol.2021.117554. [DOI] [PubMed] [Google Scholar]
- 61.Brooks S.A., Martin E., Smeester L., Grace M.R., Boggess K., Fry R.C. miRNAs as common regulators of the transforming growth factor (TGF)-β pathway in the preeclamptic placenta and cadmium-treated trophoblasts: links between the environment, the epigenome and preeclampsia. Food Chem. Toxicol. 2016;98:50–57. doi: 10.1016/j.fct.2016.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brooks S.A., Fry R.C. Cadmium inhibits placental trophoblast cell migration via miRNA regulation of the transforming growth factor beta (TGF-β) pathway. Food Chem. Toxicol. 2017;109:721–726. doi: 10.1016/j.fct.2017.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanwar V.S., Zhang X., Jagannathan L., Jose C.C., Cuddapah S. Cadmium exposure upregulates SNAIL through miR-30 repression in human lung epithelial cells. Toxicol. Appl. Pharmacol. 2019;373:1–9. doi: 10.1016/j.taap.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen M., Li X., Fan R., Yang J., Jin X., Hamid S., et al. Cadmium induces BNIP3-dependent autophagy in chicken spleen by modulating miR-33-AMPK axis. Chemosphere. 2018;194:396–402. doi: 10.1016/j.chemosphere.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 65.Hao R., Song X., Sun-Waterhouse D., Tan X., Li F., Li D. Mir-34a/sirt1/p53 signaling pathway contributes to cadmium-induced nephrotoxicity: a preclinical study in mice. Environ. Pollut. 2021;282 doi: 10.1016/j.envpol.2021.117029. [DOI] [PubMed] [Google Scholar]
- 66.Alshehri A.S., El-Kott A.F., El-Kenawy A.E., Khalifa H.S., AlRamlawy A.M. Cadmium chloride induces non-alcoholic fatty liver disease in rats by stimulating miR-34a/SIRT1/FXR/p53 axis. Sci. Total Environ. 2021;784 doi: 10.1016/j.scitotenv.2021.147182. [DOI] [PubMed] [Google Scholar]
- 67.Liu J., Liao J., Zhang C., Zeng L., Zong C., Lv Y., et al. The role of miRNAs in regulating the effect of prenatal cadmium exposure on ovarian granulosa cells in a transgenerational manner in female rats. Food Chem. Toxicol. 2021;150 doi: 10.1016/j.fct.2021.112062. [DOI] [PubMed] [Google Scholar]
- 68.Sun Y., Zong C., Liu J., Zeng L., Li Q., Liu Z., et al. C-myc promotes miR-92a-2-5p transcription in rat ovarian granulosa cells after cadmium exposure. Toxicol. Appl. Pharmacol. 2021;421 doi: 10.1016/j.taap.2021.115536. [DOI] [PubMed] [Google Scholar]
- 69.Che L., Wu Z.-L., Huang L.-Y., Wu J.-S., Du Z.-B., Lin J.-X., et al. MicroRNA-101 inhibits cadmium-induced angiogenesis by targeting cyclooxygenase-2 in primary human umbilical vein endothelial cells. Biochem. Pharmacol. 2021;189 doi: 10.1016/j.bcp.2020.114192. [DOI] [PubMed] [Google Scholar]
- 70.Qiang J., Tao Y.-F., He J., Xu P., Bao J.-W., Sun Y.-L. miR-122 promotes hepatic antioxidant defense of genetically improved farmed tilapia (GIFT, Oreochromis niloticus) exposed to cadmium by directly targeting a metallothionein gene. Aquat. Toxicol. 2017;182:39–48. doi: 10.1016/j.aquatox.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 71.Yuan W., Liu L., Liang L., Huang K., Deng Y., Dong M., et al. MiR-122-5p and miR-326-3p: potential novel biomarkers for early detection of cadmium exposure. Gene. 2020;724 doi: 10.1016/j.gene.2019.144156. [DOI] [PubMed] [Google Scholar]
- 72.Yuan W., Liang L., Huang K., Deng Y., Dong M., Wang G., et al. MiR‐122‐5p and miR‐326‐3p promote cadmium‐induced NRK‐52E cell apoptosis by downregulating PLD1. Environ. Toxicol. 2020;35(12):1334–1342. doi: 10.1002/tox.22998. [DOI] [PubMed] [Google Scholar]
- 73.Wu L., Song J., Xue J., Xiao T., Wei Q., Zhang Z., et al. MircoRNA-143-3p regulating ARL6 is involved in the cadmium-induced inhibition of osteogenic differentiation in human bone marrow mesenchymal stem cells. Toxicol. Lett. 2020;331:159–166. doi: 10.1016/j.toxlet.2020.06.001. [DOI] [PubMed] [Google Scholar]
- 74.Li H., Di G., Zhang Y., Xue R., Zhang J., Liang J. MicroRNA-155 and microRNA-181a, via HO-1, participate in regulating the immunotoxicity of cadmium in the kidneys of exposed Cyprinus carpio. Fish Shellfish Immunol. 2019;95:473–480. doi: 10.1016/j.fsi.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 75.Goyal T., Mitra P., Singh P., Ghosh R., Sharma S., Sharma P. Association of microRNA expression with changes in immune markers in workers with cadmium exposure. Chemosphere. 2021;274 doi: 10.1016/j.chemosphere.2021.129615. [DOI] [PubMed] [Google Scholar]
- 76.Li H., Di G., Zhang Y., Liang J., Wang X., Xu Z., et al. miR-217 through SIRT1 regulates the immunotoxicity of cadmium in Cyprinus carpio. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2021;248 doi: 10.1016/j.cbpc.2021.109086. [DOI] [PubMed] [Google Scholar]
- 77.Chen J., Lai W., Deng Y., Liu M., Dong M., Liu Z., et al. MicroRNA-363-3p promotes apoptosis in response to cadmium-induced renal injury by down-regulating phosphoinositide 3-kinase expression. Toxicol. Lett. 2021;345:12–23. doi: 10.1016/j.toxlet.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 78.Singh P., Surolia R., Li F., Wang Z., Dsouza K., Stephens C., et al. American Thoracic Society; 2021. Cadmium Induced EZH2 Regulates Chloride Channel Expression in Epithelial Cells through Transcriptional Regulation of miRNAs. TP64 TP064 TOBACCO PRODUCT EXPOSURES and RESPIRATORY HEALTH; p. A3113. (A) [Google Scholar]
- 79.Liang L., Huang K., Yuan W., Liu L., Zou F., Wang G. Dysregulations of miR-503-5p and Wnt/β-catenin pathway coordinate in mediating cadmium-induced kidney fibrosis. Ecotoxicol. Environ. Saf. 2021;224 doi: 10.1016/j.ecoenv.2021.112667. [DOI] [PubMed] [Google Scholar]
- 80.Xiong Y.-W., Feng Y.-J., Wei T., Zhang X., Tan L.-L., Zhang J., et al. miR-6769b-5p targets CCND-1 to regulate proliferation in cadmium-treated placental trophoblasts: association with the impairment of fetal growth. Ecotoxicol. Environ. Saf. 2022;230 doi: 10.1016/j.ecoenv.2021.113109. [DOI] [PubMed] [Google Scholar]
- 81.Xue C., Kang B., Su P., Wang D., Zhao F., Zhang J., et al. MicroRNA-106b-5p participates in lead (Pb2+)-induced cell viability inhibition by targeting XIAP in HT-22 and PC12 cells. Toxicol. Vitro. 2020;66 doi: 10.1016/j.tiv.2020.104876. [DOI] [PubMed] [Google Scholar]
- 82.Ochoa-Martínez Á.C., Varela-Silva J.A., Orta-García S.T., Carrizales-Yáñez L., Pérez-Maldonado I.N. Lead (Pb) exposure is associated with changes in the expression levels of circulating miRNAS (miR-155, miR-126) in Mexican women. Environ. Toxicol. Pharmacol. 2021;83 doi: 10.1016/j.etap.2021.103598. [DOI] [PubMed] [Google Scholar]
- 83.Gu X., Xu Y., Xue W.-Z., Wu Y., Ye Z., Xiao G., et al. Interplay of miR-137 and EZH2 contributes to the genome-wide redistribution of H3K27me3 underlying the Pb-induced memory impairment. Cell Death Dis. 2019;10(9):1–16. doi: 10.1038/s41419-019-1912-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Han L., Zou Y., Yu C. Targeting CC chemokine ligand (CCL) 20 by miR-143-5p alleviate lead poisoning-induced renal fibrosis by regulating interstitial fibroblasts excessive proliferation and dysfunction. Bioengineered. 2022;13(4):11156–11168. doi: 10.1080/21655979.2022.2062106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Z., Lu Q., Cao X., Wang K., Wang Y., Wu Y., et al. Lead exposure promotes the inflammation via the circRNA-05280/miR-146a/IRAK1 axis in mammary gland. Ecotoxicol. Environ. Saf. 2022;247 doi: 10.1016/j.ecoenv.2022.114204. [DOI] [PubMed] [Google Scholar]
- 86.Araujo MLd, Gomes B.C., Devóz P.P., Duarte NdAA., Ribeiro D.L., Araujo ALd, et al. Association between miR-148a and DNA methylation profile in individuals exposed to lead (Pb) Front. Genet. 2021;12 doi: 10.3389/fgene.2021.620744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jing H., Zhang Q., Li S., Gao X-j. Pb exposure triggers MAPK-dependent inflammation by activating oxidative stress and miRNA-155 expression in carp head kidney. Fish Shellfish Immunol. 2020;106:219–227. doi: 10.1016/j.fsi.2020.08.015. [DOI] [PubMed] [Google Scholar]
- 88.Mitra P., Goyal T., Singh P., Sharma S., Sharma P. Assessment of circulating miR-20b, miR-221, and miR-155 in occupationally lead-exposed workers of North-Western India. Environ. Sci. Pollut. Control Ser. 2021;28(3):3172–3181. doi: 10.1007/s11356-020-10676-5. [DOI] [PubMed] [Google Scholar]
- 89.Wang W., Shi F., Cui J., Pang S., Zheng G., Zhang Y. MiR-378a-3p/SLC7A11 regulate ferroptosis in nerve injury induced by lead exposure. Ecotoxicol. Environ. Saf. 2022;239 doi: 10.1016/j.ecoenv.2022.113639. [DOI] [PubMed] [Google Scholar]
- 90.Ak G., Tomaszek S.C., Kosari F., Metintas M., Jett J.R., Metintas S., et al. vol. 2015. BioMed Research International; 2015. (MicroRNA and mRNA Features of Malignant Pleural Mesothelioma and Benign Asbestos-Related Pleural Effusion). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ju L., Wu W., Yin X., Xiao Y., Jia Z., Lou J., et al. miR‐30d is related to asbestos exposure and inhibits migration and invasion in NCI‐H2452 cells. FEBS Open bio. 2017;7(10):1469–1479. doi: 10.1002/2211-5463.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cavalleri T., Angelici L., Favero C., Dioni L., Mensi C., Bareggi C., et al. Plasmatic extracellular vesicle microRNAs in malignant pleural mesothelioma and asbestos-exposed subjects suggest a 2-miRNA signature as potential biomarker of disease. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0176680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim K., Ko Y., Oh H., Ha M., Kang J., Kwon E.J., et al. MicroRNA-98 is a prognostic factor for asbestos-induced mesothelioma. J. Toxicol. Environ. Health, Part A. 2020;83(3):126–134. doi: 10.1080/15287394.2020.1734891. [DOI] [PubMed] [Google Scholar]
- 94.Gaetani S., Monaco F., Alessandrini F., Tagliabracci A., Sabbatini A., Bracci M., et al. Mechanism of miR-222 and miR-126 regulation and its role in asbestos-induced malignancy. Int. J. Biochem. Cell Biol. 2020;121 doi: 10.1016/j.biocel.2020.105700. [DOI] [PubMed] [Google Scholar]
- 95.Frontini F., Bononi I., Torreggiani E., Di Mauro G., Mazzoni E., Stendardo M., et al. Circulating microRNA-197-3p as a potential biomarker for asbestos exposure. Sci. Rep. 2021;11(1):1–9. doi: 10.1038/s41598-021-03189-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bononi I., Comar M., Puozzo A., Stendardo M., Boschetto P., Orecchia S., et al. Circulating microRNAs found dysregulated in ex-exposed asbestos workers and pleural mesothelioma patients as potential new biomarkers. Oncotarget. 2016;7(50) doi: 10.18632/oncotarget.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Okazaki Y., Chew S.H., Nagai H., Yamashita Y., Ohara H., Jiang L., et al. Overexpression of miR‐199/214 is a distinctive feature of iron‐induced and asbestos‐induced sarcomatoid mesothelioma in rats. Cancer Sci. 2020;111(6):2016–2027. doi: 10.1111/cas.14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ding E., Guo J., Bai Y., Zhang H., Liu X., Cai W., et al. MiR-92a and miR-486 are potential diagnostic biomarkers for mercury poisoning and jointly sustain NF-κB activity in mercury toxicity. Sci. Rep. 2017;7(1):1–10. doi: 10.1038/s41598-017-13230-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tunçdemir M.T., YERLİKAYA F.H. The relationship between plasma MicroRNAs and serum mercury levels in patients with amalgam filling and dentists. Selcuk Dental J. 2021;8(3):736–743. [Google Scholar]
- 100.Pallocca G., Fabbri M., Nerini-Molteni S., Pistollato F., Zagoura D., Sacco M.G., et al. Changes in miRNA expression profiling during neuronal differentiation and methyl mercury-induced toxicity in human in vitro models. Toxics. 2014;2(3):443–463. [Google Scholar]
- 101.Wang L., Bayanbold K., Zhao L., Wang Y., Adamcakova-Dodd A., Thorne P.S., et al. Redox sensitive miR-27a/b/Nrf2 signaling in Cr (VI)-induced carcinogenesis. Sci. Total Environ. 2022;809 doi: 10.1016/j.scitotenv.2021.151118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pratheeshkumar P., Son Y.-O., Divya S.P., Turcios L., Roy R.V., Hitron J.A., et al. Hexavalent chromium induces malignant transformation of human lung bronchial epithelial cells via ROS-dependent activation of miR-21-PDCD4 signaling. Oncotarget. 2016;7(32) doi: 10.18632/oncotarget.9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang Q., Sun X., Xiao X., Zheng J., Li M., Yu M., et al. Maternal chromium restriction induces insulin resistance in adult mice offspring through miRNA. Int. J. Mol. Med. 2018;41(3):1547–1559. doi: 10.3892/ijmm.2017.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dioni L., Sucato S., Motta V., Iodice S., Angelici L., Favero C., et al. Urinary chromium is associated with changes in leukocyte miRNA expression in obese subjects. Eur. J. Clin. Nutr. 2017;71(1):142–148. doi: 10.1038/ejcn.2016.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang Z., Lin H.-P., Li Y., Tao H., Yang P., Xie J., et al. Chronic hexavalent chromium exposure induces cancer stem cell-like property and tumorigenesis by increasing c-Myc expression. Toxicol. Sci. 2019;172(2):252–264. doi: 10.1093/toxsci/kfz196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Y., Li P., Yu S., Zhang J., Wang T., Jia G. miR-3940-5p associated with genetic damage in workers exposed to hexavalent chromium. Toxicol. Lett. 2014;229(1):319–326. doi: 10.1016/j.toxlet.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 107.Liu Y.P., Cai Y., Lei Y.D., Yuan X.Y., Wang Y., Yi S., et al. Circular RNA expression profiles in human bronchial epithelial cells treated with beryllium sulfate. Toxicol. Res. 2021;10(5):1013–1021. doi: 10.1093/toxres/tfab086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yi S., Liu Y.P., Li X.Y., Yuan X.Y., Wang Y., Cai Y., et al. The expression profile and bioinformatics analysis of microRNAs in human bronchial epithelial cells treated by beryllium sulfate. J. Appl. Toxicol. : JAT. 2021;41(8):1275–1285. doi: 10.1002/jat.4116. [DOI] [PubMed] [Google Scholar]
- 109.Parada-Cruz B., Aztatzi-Aguilar O.G., Ramírez-Martínez G., Jacobo-Estrada T.L., Cárdenas-González M., Escamilla-Rivera V., et al. Inflammation- and cancer-related microRNAs in rat renal cortex after subchronic exposure to fluoride. Chem. Biol. Interact. 2023;379 doi: 10.1016/j.cbi.2023.110519. [DOI] [PubMed] [Google Scholar]
- 110.Zhang L., Li Y., Tao D., Yang L., Zhang Y., Zhang H., et al. The miR-34b-5p-negative target Gnai2 aggravates fluorine combined with aluminum-induced apoptosis of rat offspring hippocampal neurons and NG108-15 cells. Environ. Sci. Pollut. Res. Int. 2023;30(25):66822–66839. doi: 10.1007/s11356-023-27135-6. [DOI] [PubMed] [Google Scholar]
- 111.Zhao Y., Yu Y., Ommati M.M., Xu J., Wang J., Zhang J., et al. Multiomics analysis revealed the molecular mechanism of miRNAs in fluoride-induced hepatic glucose and lipid metabolism disorders. J. Agric. Food Chem. 2022;70(44):14284–14295. doi: 10.1021/acs.jafc.2c03049. [DOI] [PubMed] [Google Scholar]
- 112.Beck R., Chandi M., Kanke M., Stýblo M., Sethupathy P. Arsenic is more potent than cadmium or manganese in disrupting the INS-1 beta cell microRNA landscape. Arch. Toxicol. 2019;93(11):3099–3109. doi: 10.1007/s00204-019-02574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.María E.R.M.M.-P., Rodríguez-Sastre A., Valverde M. As-Cd-Pb mixture induces cellular transformation via post-transcriptional regulation of Rad51c by miR-222. Cell. Physiol. Biochem. 2019;53:910–920. doi: 10.33594/000000181. [DOI] [PubMed] [Google Scholar]
- 114.Sanders A.P., Burris H.H., Just A.C., Motta V., Amarasiriwardena C., Svensson K., et al. Altered miRNA expression in the cervix during pregnancy associated with lead and mercury exposure. Epigenomics. 2015;7(6):885–896. doi: 10.2217/epi.15.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mumtaz F., Albeltagy R.S., Diab M.S., Abdel Moneim A.E., El-Habit O.H. Exposure to arsenite and cadmium induces organotoxicity and miRNAs deregulation in male rats. Environ. Sci. Pollut. Control Ser. 2020;27(14):17184–17193. doi: 10.1007/s11356-020-08306-1. [DOI] [PubMed] [Google Scholar]
- 116.Alshammari G.M., Al‐Qahtani W.H., AlFaris N.A., Alzahrani N.S., Alkhateeb M.A., Yahya M.A. Quercetin prevents cadmium chloride‐induced hepatic steatosis and fibrosis by downregulating the transcription of miR‐21. Biofactors. 2021;47(3):489–505. doi: 10.1002/biof.1724. [DOI] [PubMed] [Google Scholar]
- 117.Chen H., Li P., Shen Z., Wang J., Diao L. Protective effects of selenium yeast against cadmium-induced necroptosis through mir-26a-5p/pten/pi3k/akt signaling pathway in chicken kidney. Ecotoxicol. Environ. Saf. 2021;220 doi: 10.1016/j.ecoenv.2021.112387. [DOI] [PubMed] [Google Scholar]
- 118.Shi Q., Jin X., Fan R., Xing M., Guo J., Zhang Z., et al. Cadmium-mediated miR-30a-GRP78 leads to JNK-dependent autophagy in chicken kidney. Chemosphere. 2019;215:710–715. doi: 10.1016/j.chemosphere.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 119.Chen Z., Gu D., Zhou M., Shi H., Yan S., Cai Y. Regulatory role of miR-125a/b in the suppression by selenium of cadmium-induced apoptosis via the mitochondrial pathway in LLC-PK1 cells. Chem. Biol. Interact. 2016;243:35–44. doi: 10.1016/j.cbi.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 120.Albeltagy R.S., Mumtaz F., Abdel Moneim A.E., El-Habit O.H. N-acetylcysteine reduces miR-146a and NF-κB p65 inflammatory signaling following cadmium hepatotoxicity in rats. Biol. Trace Elem. Res. 2021;199(12):4657–4665. doi: 10.1007/s12011-021-02591-8. [DOI] [PubMed] [Google Scholar]
- 121.Hao R., Ge J., Ren Y., Song X., Jiang Y., Sun-Waterhouse D., et al. Caffeic acid phenethyl ester mitigates cadmium-induced hepatotoxicity in mice: role of miR-182-5p/TLR4 axis. Ecotoxicol. Environ. Saf. 2021;207 doi: 10.1016/j.ecoenv.2020.111578. [DOI] [PubMed] [Google Scholar]
- 122.Zhang J., Zheng S., Wang S., Liu Q., Xu S. Cadmium-induced oxidative stress promotes apoptosis and necrosis through the regulation of the miR-216a-PI3K/AKT axis in common carp lymphocytes and antagonized by selenium. Chemosphere. 2020;258 doi: 10.1016/j.chemosphere.2020.127341. [DOI] [PubMed] [Google Scholar]
- 123.Hao R., Ge J., Li M., Song X., Jiang Y., Li F., et al. Caffeic acid phenethyl ester mitigates cadmium‐induced damage via the Hsa_circ_0010039/miR‐661/Caspase9 axis–mediated apoptosis. Food Frontiers. 2021;2(4):537–546. [Google Scholar]
- 124.Atteia H.H., Arafa M.H., Prabahar K. Selenium nanoparticles prevents lead acetate-induced hypothyroidism and oxidative damage of thyroid tissues in male rats through modulation of selenoenzymes and suppression of miR-224. Biomed. Pharmacother. 2018;99:486–491. doi: 10.1016/j.biopha.2018.01.083. [DOI] [PubMed] [Google Scholar]
- 125.Lin L.-T., Liu S.-Y., Leu J.-D., Chang C.-Y., Chiou S.-H., Lee T.-C., et al. Arsenic trioxide-mediated suppression of miR-182-5p is associated with potent anti-oxidant effects through up-regulation of SESN2. Oncotarget. 2018;9(22) doi: 10.18632/oncotarget.24678. [DOI] [PMC free article] [PubMed] [Google Scholar]