Abstract
Thiopurines, such as 6-mercaptopurine (6-MP), are widely used as cytotoxic agents and immunosuppressants for leukemia and autoimmune or inflammatory diseases. A nonsynonymous single nucleotide polymorphism (p.Arg139Cys; R139C) of the nucleoside diphosphate-linked moiety X-type motif 15 (NUDT15) gene causes the loss of thiopurine detoxification, inducing myelosuppression. To understand such hematotoxicity, we investigate the effects of NUDT15 R139C on hematopoietic stem cells (HSCs) upon thiopurine administration. Using previously established Nudt15R138C knock-in mice, which mimic myelosuppression in NUDT15R139C homozygous or heterozygous patients following thiopurine administration, we investigated the numerical changes of HSCs and hematopoietic progenitor cells following 6-MP administration using in vivo flowcytometry and ex vivo HSC expansion. Genes differentially expressed between Nudt15+/+ HSCs and Nudt15R138C/R138C HSCs were identified using RNA-sequencing before the emergence of 6-MP-induced HSC-damage. Gene Ontology (GO) and Transcriptional Regulatory Relationships Unraveled by Sentence-based Text Mining (TRRUST) analyses were performed to elucidate the molecular effects of 6-MP on HSCs. In Nudt15R138C/R138C mice, 6-MP induced exhaustion of HSCs faster than that of multipotent progenitors and as fast as that of myeloid-committed progenitors. Ex vivo-expanded Nudt15R138C/R138C HSCs were dose- and time-dependently damaged by 6-MP. GO analysis identified the DNA damage response and cell cycle process as the most strongly influenced processes in Nudt15R138C/R138C HSCs. TRRUST analysis revealed that the Trp53-regulated transcriptional regulatory network is influenced prior to HSC exhaustion in Nudt15R138C/R138C HSCs. The loss of NUDT15 thiopurine detoxification enhances thiopurine-mediated DNA damage via the Trp53 networks in HSCs. Therefore, caution is required in long-term thiopurine use in patients with NUDT15 R139C in view of its adverse effects on HSCs in the form of DNA damage.
Subject terms: Cell biology, Cell death, Cell growth, Stem cells, Haematopoietic stem cells
Introduction
Thiopurines, 6-mercaptopurine (6-MP) and azathioprine (AZA), are widely used as cytotoxic agents for the management of many cancers, especially acute lymphoblastic leukemia (ALL)1,2, and as immunosuppressants for autoimmune or inflammatory diseases, especially inflammatory bowel disease (IBD)3. Despite their clinical usefulness, thiopurines frequently have serious adverse effects4, the most common being myelosuppression, which can force patients to discontinue treatment5. A nonsynonymous single nucleotide polymorphism (SNP), rs116855232 (the c.415C > T variant), which gives rise to the p.Arg139Cys (R139C), in the nucleoside diphosphate-linked moiety X-type motif 15 (NUDT15) gene, has recently been identified as a strong risk factor for thiopurine-induced severe leukopenia in patients with IBD and ALL 6–9. The allele with this SNP is present in ~ 25% of the Japanese9 and 20% of the East Asians, but rare in Caucasians and Africans10. NUDT15 is a nucleotide diphosphatase that converts thio-guanosine-5′-triphosphate, an active thiopurine metabolite that functions by being incorporated into RNA or DNA, into thio-guanosine-5′-monophosphate, an inactive thiopurine metabolite, thereby detoxifying the thiopurine. NUDT15 R139C is reported to exhibit strong cytotoxicity as a result of the almost complete loss of this enzymatic activity10. The number of NUDT15R139C alleles present correlates with myelosuppression strength, and thiopurines cannot be administered to NUDT15R139C homozygous patients with non-malignant diseases such as IBD9.
We have previously established knock-in mice harboring a p.Arg138Cys mutation (Nudt15R138C) that corresponds to NUDT15R139C in humans. Nudt15R138C/R138C mice exhibit myelosuppression and leukopenia, with approximately tenfold higher sensitivity to 6-MP than Nudt15+/+ mice; thiopurine administration is consequently lethal in Nudt15R138C/R138C mice11. At 5 days after peritoneal administration with 5 mg/kg 6-MP, the number of hematopoietic stem cells (HSCs) in Nudt15R138C/R138C mice was reduced by 70%. HSCs are generally thought to be resistant to antimetabolites that interfere with DNA synthesis and repair, such as 5-fluorouracil (5-FU) and hydroxycarbamide, owing to their own relative quiescence12–14. While HSCs maintain dormancy during homeostasis, their self-renewal capacity is activated in response to anticancer drugs, to rescue the injured hematopoietic system15. However, it is unclear what damage HSCs undergo when they are remarkably sensitive to antimetabolic cytotoxic agents due to abnormal drug metabolism. To elucidate these dynamics, we examine how the loss of Nudt15 thiopurine detoxification affects HSCs upon thiopurine exposure.
Methods
Mice
We used Nudt15R138C knock-in mice, previously established via CRISPR/Cas9 gene editing using oligo DNA to induce the c.412C > T transition11. This line was maintained via mating with wild-type C57BL/6 mice. Genotype sequencing was conducted using the forward primer GGCATCTAGCCTGTAATATAGACAT, reverse primer CAGAGGTAGGTAGGCAGATCTGAG, and sequencing primer CCCGGCCTGCAGGTCTATGCCACCAGGACAATTCAG. For analysis, 8–12-week-old male and female mice were used. 6-MP (Sigma-Aldrich, St. Louis, MO) was dissolved in DMSO with each injection and administered peritoneally at 5 mg/kg once daily. A dose of 5 mg /kg in mice is equivalent to 0.4 mg/kg in an adult human16. The recommended initial dose of 6-MP in adult Japanese IBD patients is 30 mg/day9. All animal experiments were approved by the Animal Research Committee of Shiga University of Medical Science (permission number 2021-5-7) and performed under specific pathogen-free conditions at the Research Center for Animal Life Science, Shiga University of Medical Science.
Fluorescence-activated cell sorting (FACS) analysis
Bone marrow cells were isolated from the tibia, femur, and pelvis of mice and stained with the antibodies. We used antibodies directed against CD3e [145-2C11], CD4 [GK1.5], CD8a [53-6.7], CD19 [6D5], Mac1 [M1/70], Gr-1 [RB6-8C5], B220 [RA3-6B2], Ter119 [TER-119], CD127 [A7R34], CD34 [RAM34], CD16/32 [clone 93], CD135 [A2F10], c-kit [2B8], CD48 [HM-48-1], CD150 [TC15-12F12.2], Sca-1 [D7], CD45.1 [clone A20], and CD45.2 [clone 104] (Additional File 1, Supplementary Table 1). Antibodies were purchased from Thermo Fisher Scientific (Waltham, MA) or BioLegend (San Diego, CA). Lineage markers used in analysis of hematopoietic stem and progenitor cells (HSPCs) included CD3e, CD4, CD8a, CD19, Mac1, Gr-1, B220, and Ter119. The HSPC populations have been defined elsewhere15 (Additional File 1, Supplementary Fig. 1): HSC (CD34−/lowCD135−CD48−CD150+Lineage−Sca-1+c-Kit+); multipotent progenitor 1–4 (MMP1–4, CD34+CD135−CD48−CD150+Lineage−Sca-1+c-Kit+; CD34+CD135−CD48+CD150+Lineage−Sca-1+c-Kit+; CD34+CD135−CD48+CD150−Lineage−Sca-1+c-Kit+; CD34+CD135+CD48+CD150−Lineage−Sca-1+c-Kit+); common myeloid progenitor (CMP, Lineage−Sca-1−c-Kit+CD16/32lowCD34low); granulocyte-monocyte progenitor (GMP, Lineage−Sca-1−c-Kit+CD16/32+CD34+); and megakaryocyte-erythrocyte progenitor (MEP, Lineage−Sca-1−c-Kit+CD16/32−CD34−). Flow-cytometric data acquisition and cell sorting were performed using a FACSCanto™ II or FACSAriaFusion system (BD Biosciences, San Jose, CA). The data were analyzed using FACSDiva and FlowJo (BD Biosciences).
Ex vivo HSC expansion and Cell viability assay
CD48−CD150+CD127−Lineage−Sca-1+c-Kit+ cells were sorted as HSCs after pre-enrichment of c-Kit-positive cells using MACS (Miltenyi Biotec, Bergisch Gladbach, Germany) were directly purified onto a plate well. and expanded17. Briefly, 50 HSC cells were directly purified onto a fibronectin-coated plate well containing F-12 media (Nacalai Tesque, Kyoto, Japan) supplemented with 10 mM HEPES, 1 × Penicillin–Streptomycin-Glutamine (Thermo Fisher Scientific), 1 × Insulin-Transferrin-Selenium-ethanolamine (Thermo Fisher Scientific), 10 ng/ml stem cell factor (Peprotech, Cranbury, NJ), 100 ng/ml thrombopoietin (Peprotech), and 1 mg/ml polyvinyl alcohol (Sigma-Aldrich). HSCs were cultured in a humidified tissue culture 37 °C incubator with 5% CO2. After expansion for 28 d with half medium change every few days, 5000 cells were reseeded into a well and incubated with 0.01, 0.1, or 1 µM 6-MP. DMSO was used as a control. Cell viability was assessed using a Cell Count Reagent SF (Nacalai Tesque) according to the manufacturer’s instructions. The data were obtained using an Infinite M200 Plate Reader (Tecan, Männedorf, Switzerland).
RNA-sequencing
Twenty-four hours after peritoneal administration of 5 mg/kg 6-MP to Nudt15+/+ female mice (N = 3–6) and Nudt15R138C/R138C female mice (N = 2–9), HSCs were isolated and pooled as each one sample. This process was repeated 3 times and finally 3 samples each for Nudt15+/+ HSCs and Nudt15R138C/R138C HSCs were prepared for analysis. Total RNA was extracted from the isolated HSCs using a RNeasy Micro kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. After checking the quality using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA), cDNA were synthesized using the SMART-Seq v. 4 Ultra Low Input RNA Kit for Sequencing (Takara Bio USA, San Jose, CA). The libraries were prepared using the Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA), and were sequenced on a NovaSeq 6000 platform (Illumina). Transcriptome sequence data were mapped to the GRCm38 mouse genome (GCA_000001635.2), and expression levels were normalized as transcripts per kilobase million using the DRAGEN Bio-IT platform v. 3.7.5 (Illumina). Differentially expressed genes were identified at cutoffs of log2 fold change >|1| and p < 0.05, using the bioconductor R package edgeR18. Gene Ontology (GO) analysis were performed using Metascape19 (http://metascape.org). The Transcriptional Regulatory Relationships Unraveled by Sentence-based Text Mining (TRRUST) database, containing 6552 TF–target interactions for 828 mouse TFs (https://www.grnpedia.org/trrust/)20, was used on Metascape to examine the activated transcriptional regulatory networks.
Statistics
ANOVA followed by Šidák tests was adopted for multiple comparison analysis using GraphPad Prism v. 9 (GraphPad, La Jolla, CA). All tests were two-sided, with p < 0.05 considered significant.
The number of experiments and replicates are descried in methods and figure legends.
Ethics
All methods were performed and reported in accordance with the relevant guidelines and regulations including ARRIVE guidelines.
Results
6-MP induces exhaustion of HSCs faster than that of multipotent progenitors and as fast as that of myeloid-committed progenitors in Nudt15R138C/R138C mice
To investigate whether the reduction of HSCs follows damage of hematopoietic progenitor cells (HPCs) or more mature blood cells in Nudt15R138C/R138C mice following thiopurine administration, we counted total bone marrow cells as well as each population of HSPCs, including HSCs, MPPs, CMPs, GMPs, MEPs, in mouse bone marrow every day after 6-MP administration. The total number of bone marrow cells was significantly reduced in Nudt15R138C/R138C mice, to approximately half of that in Nudt15+/+ mice at 72 h, but negligibly at 48 h, after 6-MP administration (Fig. 1). Numbers of myeloid-committed HPCs, such as GMPs and MEPs, were significantly reduced to approximately 70% at 48 h, and < 20% at 72 h after 6-MP administration. The number of CMPs, earlier step in myeloid-committed HPCs than GMPs and MEPs, was significantly reduced, to < 20% at 72 h, but not at all at 48 h, after 6-MP administration. The number of MPP1, MPP3, and MPP4, which are earlier steps than CMPs, GMPs, and MEPs was reduced by 40–70% at 72 h, but not at all at 48 h, after 6-MP administration. These data suggest that GMPs and MEPs are the most damaged by 6-MP, followed by reductions in the more mature blood cells that they supply, as well as in the CMPs and MPPs that supply them. By contrast, the number of HSCs was significantly reduced to 70% at 48 h and 20% at 72 h after 6-MP administration. This earlier reduction in HSCs than in MPPs after 6-MP administration in Nudt15R138C/R138C mice suggests that increased 6-MP intracellular toxicity may directly affect HSCs prior to the injury-induced reduction in differentiated blood cells.
Figure 1.
Thiopurine-induced damage of bone marrow cells by Nudt15 R138C. Bone marrow of Nudt15+/+ mice and Nudt15R138C/R138C mice was analyzed at 0, 24, 48, and 72 h after peritoneal administration of 5 mg/kg 6-MP. Data are presented as the mean ± SD (N = 5 each condition). The experiment with one or two mice in each condition was performed independently four times. *p < 0.05, **p < 0.01, and ***p < 0.001. HSC, hematopoietic stem cell; MPP, multipotent progenitor; CMP, common myeloid progenitor; GMP, granulocyte–macrophage progenitor; MEP, megakaryocyte-erythroid progenitor.
Ex vivo expansion of Nudt15R138C/R138C HSCs is dose- and time-dependently damaged by 6-MP
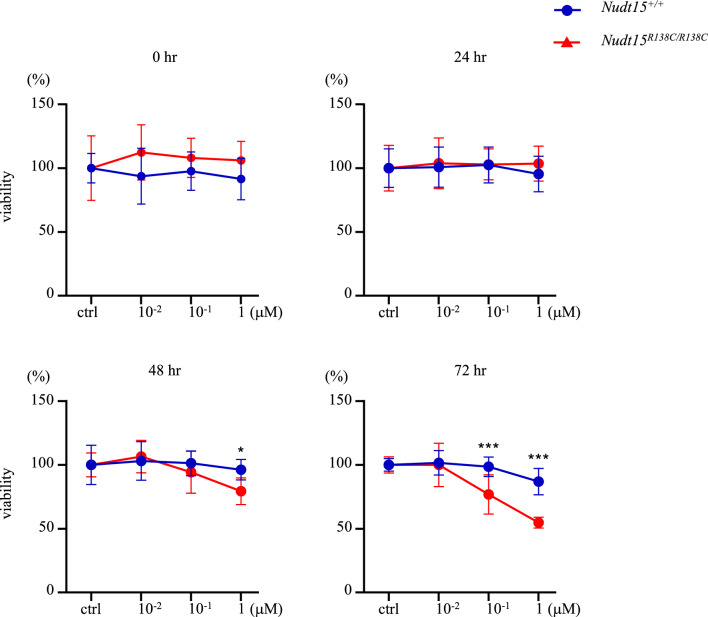
To investigate the direct effects of thiopurine on Nudt15R138C/R138C HSCs, we used the ex vivo HSC expansion. CD48−CD150+CD127−Lineage−Sca-1+c-Kit+ cells were sorted and expanded as HSCs from Nudt15R138C/R138C or Nudt15+/+ mice (Additional File 1, Supplementary Fig. 2). The ex vivo HSCs were verified to comparably engraft in mice and reproduce blood cells (Additional File 1, Supplementary Fig. 3). Using these cells, we performed a cell viability assay. Nudt15R138C/R138C ex vivo HSCs exhibited significantly lower viability after 48 h and 72 h of culture with 1 μM 6-MP and after 72 h culture with 0.1 μM 6-MP relative to Nudt15+/+ HSCs (Fig. 2). This provides evidence that enhanced 6-MP intracellular toxicity by Nudt15 R138C may be linked to direct damage of HSCs.
Figure 2.
Thiopurine-induced damage of Nudt15R138C/R138C ex vivo-expanded HSCs. The ex vivo-expanded HSCs were incubated with DMSO (control) or 0.01, 0.1, or 1 µM 6-MP and analyzed using the cell viability assay. Viability (%) is relative to that in the control. This experiment was performed independently twice with quadruplicates. Data are presented as the mean ± SD. *p < 0.05, **p < 0.01, and ***p < 0.001.
DNA damage response and cell cycle process are affected in Nudt15R138C/R138C HSCs prior to HSC exhaustion
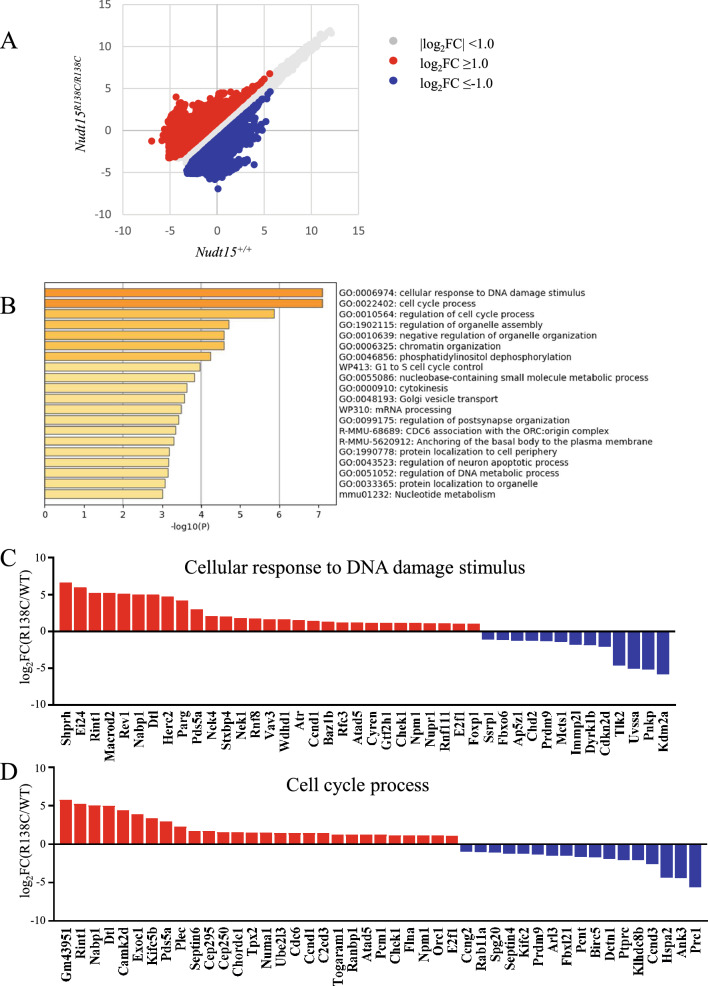
To understand the molecular mechanisms that occur before the reduction in the number of Nudt15R138C/R138C HSCs following 6-MP administration, we compared the gene expression profiles in HSCs directly isolated from Nudt15R138C/R138C and Nudt15+/+ mice 24 h after 6-MP administration (Fig. 3A): 289 genes were upregulated and 235 genes were downregulated in Nudt15R138C/R138C HSCs (Additional File 1, Supplementary Table 2). GO analysis revealed that these genes are enriched in cellular response to DNA damage and in the cell cycle process (Fig. 3B). DNA repair genes that protect against mutagenesis and chromosomal instability, such as Shprh21 and Macrod222, were upregulated, whereas those that support cell survival, cancer development, and resistance to DNA damage agents, such as Kdm2a23,24, Tlk225, and Uvssa26, were downregulated (Fig. 3C). Among those genes involving cell cycle process, Cdc6 and Orc127, which participate in initiating DNA replication, and Ccnd1 and E2f128,29, which have important roles in G1/S transition and DNA repair and apoptosis, were upregulated. Cdkn2, which induces G1 phase arrest, was downregulated30. Chk1, a key regulator of checkpoint signaling in both the unperturbed cell cycle and DNA damage response31, was upregulated (Fig. 3D). These results suggest that the DNA repair system and the cell cycle regulation system are closely related and simultaneously altered in HSCs prior to the reduction in the number of HPCs.
Figure 3.
Gene expression profiling via RNA-seq and GO analyses. HSCs were sorted 24 h after peritoneal administration of 5 mg/kg 6-MP and were analyzed via RNA-seq. (A) Scatter plot comparing Nudt15+/+ HSC (WT, N = 3) and Nudt15R138C/R138C HSC (R138C, N = 3). (B) GO analysis. Differentially expressed genes (log2FC >|1|, p < 0.05) were analyzed using Metascape (http://metascape.org). The top 20 GO terms are presented. (C) Change in expression of genes listed under cellular response to DNA damage stimulus. (D) Change in expression of genes listed under the cell cycle process.
Trp53-regulated transcriptional regulatory network is affected in Nudt15R138C/R138C HSCs before emergence of HSC exhaustion
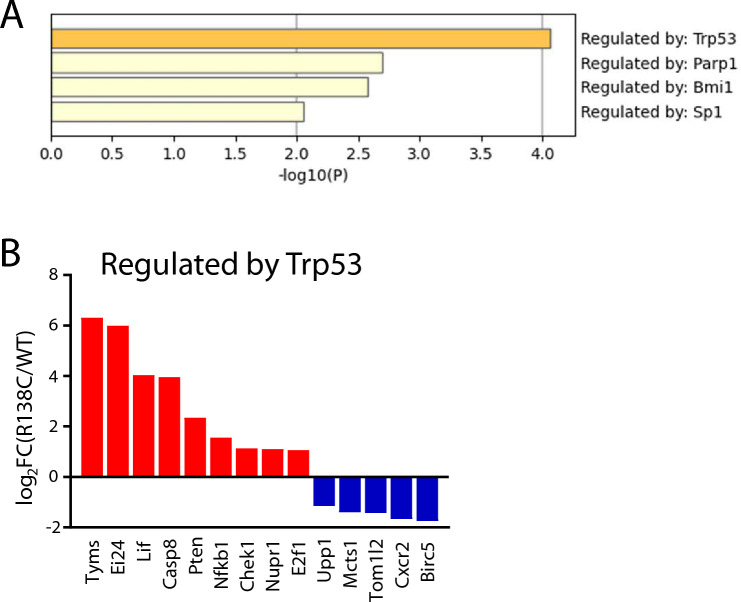
TRRUST analysis20 revealed that the Trp53-regulated transcriptional regulatory network was the most highly activated (Fig. 4A). Among the genes in the Trp53 network, Pten and Lif, which maintain HSC function32–34, were upregulated (Fig. 4B). This is consistent with the enrichment of phosphatidylinositol dephosphorylation, revealed via GO analysis. Ei24 and Casp8, involved in apoptosis35,36, were upregulated, suggesting that excessive DNA damage accumulates in HSCs. These results suggest that the loss of thiopurine detoxification due to Nudt15 R138C could activate the Trp53-regulated transcriptional network, inducing cell-cycle dysregulation and increasing DNA damage response, thus impairing the HSCs following 6-MP exposure.
Figure 4.
TRRUST analysis. Differentially expressed genes (log2FC >|1|, p < 0.05) analyzed using TRRUST (https://www.grnpedia.org/trrust/). (A) Four transcriptional regulatory networks from the list of differentially expressed genes are listed. (B) Change in expression of genes listed in the Trp53-regulated regulatory network.
Discussion
We have previously demonstrated both significant bone marrow suppression and a reduction in the number of HSCs following 5 days of 6-MP administration in Nudt15R138C/R138C mice. Here, we examined further how the loss of Nudt15 thiopurine detoxification affects HSCs upon thiopurine exposure. Our findings demonstrate that the loss of Nudt15 thiopurine detoxification impairs HSCs faster than or as fast as HPCs. Cells with greater proliferative capacity typically suffer greater DNA damage from antimetabolites that interfere with DNA synthesis and repair12–14. The steady state proliferative activity differs significantly different between HSCs and HPCs15. However, our findings show that HSC impairment coincides with that of myeloid-committed progenitor cells following 6-MP exposure and occurs earlier than that of MPPs in Nudt15R138C/R138C mice. According to previous reports with 5-FU treatment which is another antimetabolite, dormant HSCs were induced to exit their homeostatic quiescent status from the second day after treatment and to rapidly increase BrdU incorporation over a period of 24 h15. HSCs are reported to undergo DNA damage as a direct consequence of their exit from dormancy37. Our GO analysis revealed that abnormal thiopurine metabolism facilitates the induction of the molecular mechanism for cell cycle activation by upregulating positive regulators such as Ccnd1 and downregulating negative regulators such as Cdkn2d 24 h after 6-MP treatment in Nudt15R138C/R138C HSCs. Simultaneously, the molecular mechanisms for cellular response to DNA damage stimulus and chromatin organization were enhanced. These molecular mechanisms are activated prior to actual HPC damage, suggesting that HSC impairment due to the loss of Nudt15 thiopurine detoxification is directly induced as an intracellular abnormality rather than as a reactive event following bone marrow injury.
Among the top upregulated genes in the GO signature of cellular response to DNA damage stimulus, Shprh is reported to act in a DNA damage-specific manner and to prevent mutation21. Macrod2 is reported to participate in chromosomal stability and to be essential in the catalytic activity of Parp1, of which the network seems to be affected according to our TRRUST analysis22. Among the top downregulated genes, Kdm2a participates in cell survival or immortality23,24. Uvssa-knockout cells are sensitive to UV and cisplatin26. TLK2 inhibition suppresses the growth of several cancers38,39. These findings suggest that HSCs with the loss of Nudt15 thiopurine detoxification are preparing for cell death while vigorously defending themselves against mutagenesis by thiopurine.
Our TRRUST analysis revealed activation of the Trp53-regulated transcriptional regulatory network as an initial response to 6-MP in Nudt15R138C/R138C HSCs. Trp53 plays a central role in cell-cycle activation and DNA damage response40–42. DNA double-strand breaks activate the p53 pathway and DNA damage response in HSPCs43. Lif expression is upregulated during stress hematopoiesis and promotes maintenance of HSC self-renewal via JAK/STAT344,45. Pten, which negatively regulates PI3/AKT/mTOR signaling46, is important in regulating the cell cycle and DNA damage repair47 and is essential for maintaining HSCs32. The upregulation of Lif and Pten expression may protect HSCs from delayed thiopurine metabolism. However, the simultaneous upregulation of Ei24, Casp8, and Chk1 expression, in the background of the cytoprotective program, seems to reflect preparation for triggering cell death.
Although HSCs with the loss of Nudt15 thiopurine detoxification immediately launch defenses against DNA damage upon thiopurine exposure, the number of HSCs significantly decreases after 48 h. DNA damage due to cancer therapies has been reported to initiate clonal hematopoiesis that is related to the development of hematological malignancies and cardiovascular diseases48,49. Young IBD patients exposed to thiopurine have significantly higher levels of mosaic chromosomal alterations shaping a part of clonal hematopoiesis50. Future studies will be required to understand whether the loss of NUDT15 thiopurine detoxification could give rise to DNA damage-tolerant clones that outcompete other clones, leading to clonal hematopoiesis.
Conclusion
The loss of NUDT15 thiopurine detoxification enhances thiopurine-mediated DNA damage via the Trp53 networks in HSCs, leading to HSC exhaustion. Although low doses of thiopurine may attenuate acute myelosuppression in NUDT15R139C homozygous or heterozygous patients, long-term thiopurine use should be applied with caution, in view of its adverse effects on HSC dynamics due to DNA damage accumulation.
Supplementary Information
Acknowledgements
We thank the Central Research Laboratory of Shiga University of Medical Science. In particular, Yasuhiro Mori provided technical support with FACS analyses and sorting, and Takefumi Yamamoto provided technical support with mouse irradiation.
Author contributions
N.Y. performed all experiments and wrote the paper. M.K. designed all experiments and wrote the paper. T.I. performed several experiments and wrote the paper. G.T. maintained the mouse strain and performed histology and cytology experiments. AA.-.N. performed several experiments. A.A. wrote and edited the paper. All authors read and approved the final manuscript.
Funding
This work was supported by AMED under Grant Number 21ek0410056 and 22ek0410091 (MK and AA) and JSPS KAKENHI under Grant Number JP19K08811 (MK).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. The datasets generated and analyzed during the current study are available in the Gene Expression Omnibus repository, accession number GSE214604.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-38952-7.
References
- 1.Karran P, Attard N. Thiopurines in current medical practice: Molecular mechanisms and contributions to therapy-related cancer. Nat. Rev. Cancer. 2008;8:24–36. doi: 10.1038/nrc2292. [DOI] [PubMed] [Google Scholar]
- 2.Toksvang LN, Lee SHR, Yang JJ, Schmiegelow K. Maintenance therapy for acute lymphoblastic leukemia: Basic science and clinical translations. Leukemia. 2022;36:1749–1758. doi: 10.1038/s41375-022-01591-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agrawal M, Spencer EA, Colombel J-F, Ungaro RC. Approach to the management of recently diagnosed inflammatory bowel disease patients: A user’s guide for adult and pediatric gastroenterologists. Gastroenterology. 2021;161:47–65. doi: 10.1053/j.gastro.2021.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chande N, Patton PH, Tsoulis DJ, Thomas BS, MacDonald JK. Azathioprine or 6-mercaptopurine for maintenance of remission in Crohn's disease. Cochrane Database Syst. Rev. 2015 doi: 10.1002/14651858.CD000067.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser AG. The efficacy of azathioprine for the treatment of inflammatory bowel disease: A 30 year review. Gut. 2002;50:485–489. doi: 10.1136/gut.50.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang S-K, et al. A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat. Genet. 2014;46:1017–1020. doi: 10.1038/ng.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang JJ, et al. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J. Clin. Oncol. 2015;33:1235–1242. doi: 10.1200/jco.2014.59.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asada A, et al. NUDT15 R139C-related thiopurine leukocytopenia is mediated by 6-thioguanine nucleotide-independent mechanism in Japanese patients with inflammatory bowel disease. J. Gastroenterol. 2016;51:22–29. doi: 10.1007/s00535-015-1142-4. [DOI] [PubMed] [Google Scholar]
- 9.Kakuta Y, et al. NUDT15 codon 139 is the best pharmacogenetic marker for predicting thiopurine-induced severe adverse events in Japanese patients with inflammatory bowel disease: A multicenter study. J. Gastroenterol. 2018;53:1065–1078. doi: 10.1007/s00535-018-1486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moriyama T, et al. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat. Genet. 2016;48:367–373. doi: 10.1038/ng.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatsumi G, et al. Thiopurine-mediated impairment of hematopoietic stem and leukemia cells in Nudt15R138C knock-in mice. Leukemia. 2020;34:882–894. doi: 10.1038/s41375-019-0583-9. [DOI] [PubMed] [Google Scholar]
- 12.Lerner C, Harrison DE. 5-Fluorouracil spares hemopoietic stem cells responsible for long-term repopulation. Exp. Hematol. 1990;18:114–118. [PubMed] [Google Scholar]
- 13.Venezia TA, et al. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biology. 2004;2:e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arai F, Suda T. Maintenance of quiescent hematopoietic stem cells in the osteoblastic niche. Ann. N. Y. Acad. Sci. 2007;1106:41–53. doi: 10.1196/annals.1392.005. [DOI] [PubMed] [Google Scholar]
- 15.Wilson A, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 16.Estimating the maximum safe starting dose in initial clinical trials for therapeutics in adult healthy volunteers. U.S. Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation and Research (CDER). (2005).
- 17.Wilkinson AC, Ishida R, Nakauchi H, Yamazaki S. Long-term ex vivo expansion of mouse hematopoietic stem cells. Nat. Protoc. 2020;15:628–648. doi: 10.1038/s41596-019-0263-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MD, Mccarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019 doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han H, et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380–D386. doi: 10.1093/nar/gkx1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin JR, Zeman MK, Chen JY, Yee MC, Cimprich KA. SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Molecular Cell. 2011;42:237–249. doi: 10.1016/j.molcel.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakthianandeswaren A, et al. MACROD2 haploinsufficiency impairs catalytic activity of PARP1 and promotes chromosome instability and growth of intestinal tumors. Cancer Discov. 2018;8:988–1005. doi: 10.1158/2159-8290.cd-17-0909. [DOI] [PubMed] [Google Scholar]
- 23.Pfau R, et al. Members of a family of JmjC domain-containing oncoproteins immortalize embryonic fibroblasts via a JmjC domain-dependent process. Proc. Natl. Acad. Sci. 2008;105:1907–1912. doi: 10.1073/pnas.0711865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heltemes-Harris LM, et al. Sleeping Beauty transposon screen identifies signaling modules that cooperate with STAT5 activation to induce B-cell acute lymphoblastic leukemia. Oncogene. 2016;35:3454–3464. doi: 10.1038/onc.2015.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortuza GB, et al. Molecular basis of Tousled-Like Kinase 2 activation. Nat. Commun. 2018 doi: 10.1038/s41467-018-04941-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kordon MM, et al. The UVSSA protein is part of a genome integrity homeostasis network with links to transcription-coupled DNA repair and ATM signaling. Proc. Natl. Acad. Sci. 2022 doi: 10.1073/pnas.2116254119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding Q, Koren A. Positive and negative regulation of DNA replication initiation. Trends Genet. 2020;36:868–879. doi: 10.1016/j.tig.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JK, Diehl JA. Nuclear cyclin D1: An oncogenic driver in human cancer. J. Cell. Physiol. 2009;220:292–296. doi: 10.1002/jcp.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, et al. CHREBP suppresses gastric cancer progression via the cyclin D1-Rb-E2F1 pathway. Cell Death Discov. 2022 doi: 10.1038/s41420-022-01079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felisiak-Golabek A, et al. p19(INK4d) mRNA and protein expression as new prognostic factors in ovarian cancer patients. Cancer Biol. Ther. 2013;14:973–981. doi: 10.4161/cbt.25966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patil M, Pabla N, Dong Z. Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell. Mol. Life Sci. 2013;70:4009–4021. doi: 10.1007/s00018-013-1307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 33.Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 34.Hall J, et al. Oct4 and LIF/Stat3 additively induce Krüppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Gu Z, Flemington C, Chittenden T, Zambetti GP. ei24, a p53 response gene involved in growth suppression and apoptosis. Mol. Cell. Biol. 2000;20:233–241. doi: 10.1128/mcb.20.1.233-241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez KE, Bouchier-Hayes L. Lethal and non-lethal functions of caspases in the DNA damage response. Cells. 2022;11:1887. doi: 10.3390/cells11121887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter D, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520:549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- 38.Kim J-A, et al. Comprehensive functional analysis of the tousled-like kinase 2 frequently amplified in aggressive luminal breast cancers. Nat. Commun. 2016;7:12991. doi: 10.1038/ncomms12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin M, Yao Z, Zhao N, Zhang C. TLK2 enhances aggressive phenotypes of glioblastoma cells through the activation of SRC signaling pathway. Cancer Biol. Ther. 2019;20:101–108. doi: 10.1080/15384047.2018.1507257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams AB, Schumacher B. p53 in the DNA-Damage-Repair Process. Cold Spring Harbor Perspect. Med. 2016;6:a026070. doi: 10.1101/cshperspect.a026070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hernández Borrero LJ, El-Deiry WS. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta Rev. Cancer. 2021;1876:188556. doi: 10.1016/j.bbcan.2021.188556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schiroli G, et al. Precise gene editing preserves hematopoietic stem cell function following transient p53-mediated DNA damage response. Cell Stem Cell. 2019;24:551–565.e558. doi: 10.1016/j.stem.2019.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cartwright P, et al. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 45.Ropa J, Cooper S, Broxmeyer HE. Leukemia inhibitory factor promotes survival of hematopoietic progenitors ex vivo and is post-translationally regulated by DPP4. Stem Cells. 2022;40:346–357. doi: 10.1093/stmcls/sxac004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stambolic V, et al. Regulation of PTEN transcription by p53. Mol. Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 47.Brandmaier A, Hou S-Q, Shen WH. Cell cycle control by PTEN. J. Mol. Biol. 2017;429:2265–2277. doi: 10.1016/j.jmb.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019 doi: 10.1126/science.aan4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolton KL, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat. Genet. 2020;52:1219–1226. doi: 10.1038/s41588-020-00710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kakuta Y, et al. Crohn’s disease and early exposure to thiopurines are independent risk factors for mosaic chromosomal alterations in patients with inflammatory bowel diseases. J. Crohns Colitis. 2022;16:643–655. doi: 10.1093/ecco-jcc/jjab199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The datasets generated and analyzed during the current study are available in the Gene Expression Omnibus repository, accession number GSE214604.