Abstract
Drug response prediction is important to establish personalized medicine for cancer therapy. Model construction for predicting drug response (i.e., cell viability half-maximal inhibitory concentration [IC50]) of an individual drug by inputting pharmacogenomics in disease models remains critical. Machine learning (ML) has been predominantly applied for prediction, despite the advent of deep learning (DL). Moreover, whether DL or traditional ML models are superior for predicting cell viability IC50s has to be established. Herein, we constructed ML and DL drug response prediction models for 24 individual drugs and compared the performance of the models by employing gene expression and mutation profiles of cancer cell lines as input. We observed no significant difference in drug response prediction performance between DL and ML models for 24 drugs [root mean squared error (RMSE) ranging from 0.284 to 3.563 for DL and from 0.274 to 2.697 for ML; R2 ranging from −7.405 to 0.331 for DL and from −8.113 to 0.470 for ML]. Among the 24 individual drugs, the ridge model of panobinostat exhibited the best performance (R2 0.470 and RMSE 0.623). Thus, we selected the ridge model of panobinostat for further application of explainable artificial intelligence (XAI). Using XAI, we further identified important genomic features for panobinostat response prediction in the ridge model, suggesting the genomic features of 22 genes. Based on our findings, results for an individual drug employing both DL and ML models were comparable. Our study confirms the applicability of drug response prediction models for individual drugs.
Subject terms: Computational biology and bioinformatics, Machine learning
Introduction
Drug response prediction is crucial to identify appropriate therapies for patients with cancer. However, extensive clinical trials to predict drug responses in diverse cancers remain impracticable, accompanied by unaffordable costs1–4. Consequently, cancer cell lines have been established as disease models to overcome these limitations, resulting in the emergence of drug response research for developing pharmacogenomic databases using cancer cell lines1. Large-scaled pharmacogenomic databases for cancer cell lines, such as Cancer Cell Line Encyclopedia (CCLE) and Genomics of Drug Sensitivity in Cancer (GDSC), have enabled the construction of drug response prediction models using artificial intelligence2,3.
To date, several studies have attempted to construct drug response prediction models using these pharmacogenomic databases as training data5–19. In these studies, to predict the drug response (i.e., cell viability half-maximal inhibitory concentration [IC50]) of various individual drugs on a cancer cell line, the genomic profile (e.g., mutations and gene expression profiles) of a cancer cell line was used as input data6–14. However, these studies were primarily based on machine-learning (ML) method. Importantly, the superiority of ML or deep learning (DL) models for predicting responses of an individual drug is yet to be determined.
Furthermore, genomics features affecting the values predicted in the drug response prediction model need to be derived. Consequently, explainable artificial intelligence (XAI) technique20 was introduced in predicting models. The XAI technique enables the introduction of important features that affect the predicted values in the model. However, few studies have explored the application of XAI for constructing drug response prediction models, especially considering data from patients with cancer. Hence, it is critical to establish a drug response prediction model for individual drugs and identify important genomic features using XAI.
To address these limitations, we constructed two datasets by combining the drug response data from CCLE and gene expression and mutation profiles from CCLE and GDSC, respectively. Next, we established two input settings for the drug response prediction models for 24 individual drugs (a model for each drug). We compared the prediction performance of DL and ML models in the two input settings. Additionally, we identified the major genomic features affecting drug sensitivity by applying XAI to the best model based on the performance comparison.
Results
Overview
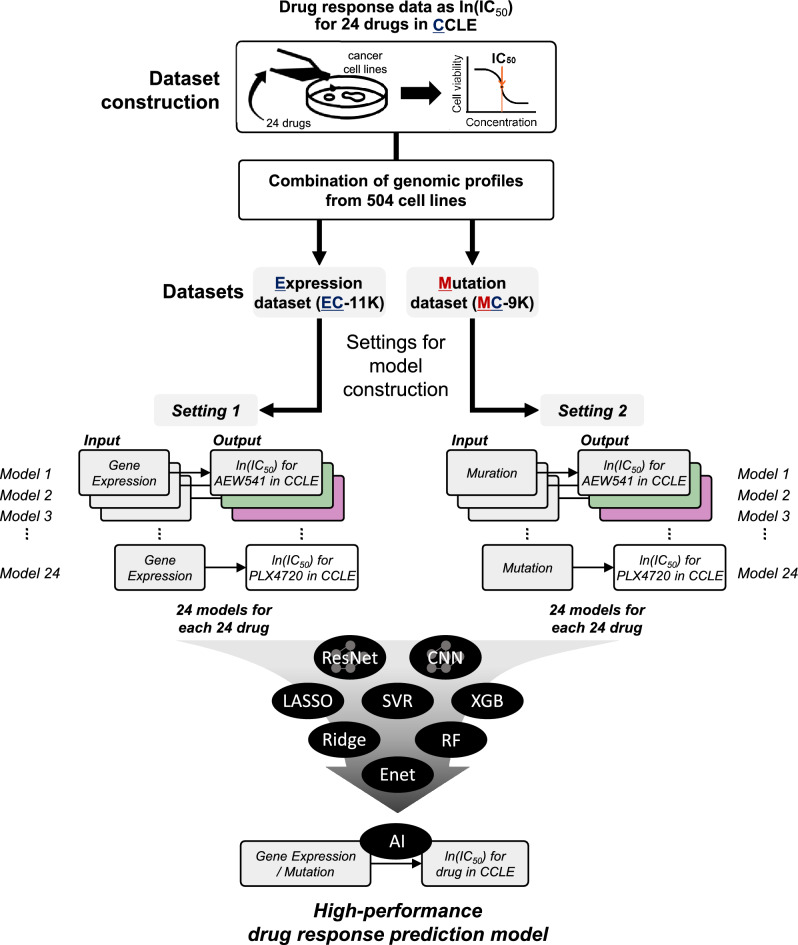
Herein, we first constructed two datasets, a gene expression dataset and a mutation dataset, to establish a drug response prediction model. Each dataset was named to describe the data (Fig. 1) and the number of cases in Supplementary Table S1. For example, one combination was named EC-11K, representing a combined dataset of gene expression data (denoted as “E”), and ln(IC50)s as drug response measurements from the CCLE (denoted as “C”), consisting of a total of ~ 11,000 (11K) cases. The other dataset was named MC-9K, including mutation statuses (denoted as “M”) and ln(IC50)s from the CCLE, comprising ~ 9000 (9K) cases (Supplementary Table S1).
Figure 1.
Dataset and model description. Considering the drug response data as learning data for our prediction target (ln[IC50] values), we combined two types of data, including genomic information (gene expression and mutation profiles). This yielded expression (EC-11K) and mutation (MC-9K) datasets for the drug-response prediction model. We set two input settings to construct drug response, prediction models. Settings 1 and 2 handle gene expression profiles (mutation profiles for setting 2) to predict ln(IC50) values for an individual drug in one model, such that settings 1 and 2 had a total of 24 models (for prediction of drug response for 24 drugs). We have used three abbreviations: E (expression), M (mutation), and C (drug response of CCLE cell lines, ln[IC50]). CCLE, Cancer Cell Line Encyclopedia.
Subsequently, we constructed two input settings (settings 1 and 2) to predict the response of an individual drug using two types of datasets. Setting 1 was the construction of 24 drug response prediction models for 24 individual drugs, considering the gene expression dataset (EC-11K) as the input. Setting 2 employed the same approach as in setting 1 using the mutation dataset (MC-9K) (Fig. 1, Supplementary Fig. S1, Supplementary Tables S2, and S3). For each setting, we constructed DL models, using architecture of convolutional neural network (CNN) (Fig. 2a) and ResNet (Fig. 2b), and ML models [lasso, ridge, support vector regression (SVR), random forest (RF), extreme gradient boosting (XGBoost), and ElasticNet (Enet)] to predict ln(IC50) values. We adopted ‘CDRscan master’ model (henceforth, CDRscan) as CNN15.
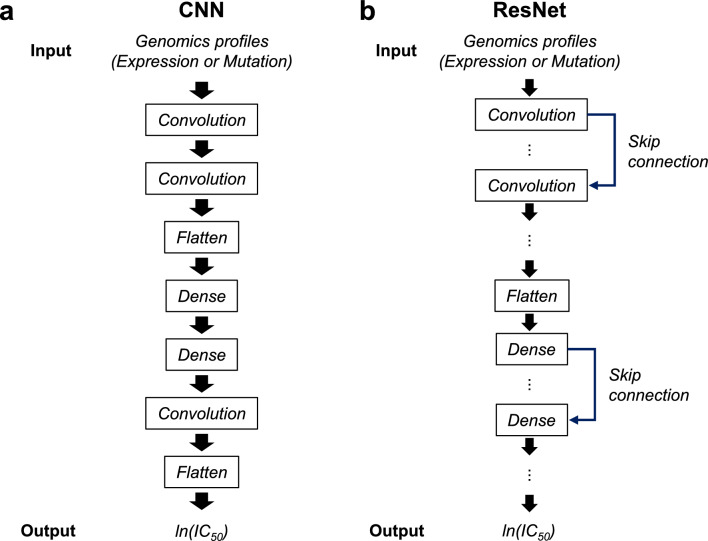
Figure 2.
Conceptual architecture of ResNet and CNN. (a) ResNet features usage of skip connection (b) CNN architecture adopted from CDRscan, except for the convolutional neural network layers for a drug.
Setting 1: model construction for individual drugs, considering expression profiles (EC-11K) as input for prediction of ln(IC50)s
Under setting 1, with gene expression profiles as the input, we constructed an ln(IC50) drug responsiveness prediction model for individual drugs (Supplementary Fig. S1a). In the aforementioned instances, a few hundred ln(IC50) measurements were available for individual drugs in diverse cancer cell lines (Supplementary Table S2). For each method (CNN, ResNet, lasso, ridge, SVR, RF, and XGBoost, Enet), we constructed drug response prediction models using gene expression profiles for individual drugs, using the training set from EC-11K (Supplementary Table S2).
Using test sets, the prediction performances of 24 drugs using DL and ML models were described using the root mean squared error (RMSE) and R-squared value (R2) (Supplementary Tables S4, S5, S6, S7, S8, S9, S10, and S11; Supplementary Figs. S2, S3, S4, S5, S6, S7, S8, and S9).
Accordingly, the ridge prediction model for panobinostat showed the best performance (R2: 0.470 and RMSE: 0.623) when compared with all models for other drugs (Fig. 3a, Supplementary Figure S3, and Supplementary Table S5). Notably, we detected no significant difference in drug response prediction performance between the DL and ML models for 24 drugs (RMSE ranging from 0.284 to 3.563 for DL and from 0.274 to 2.564 for ML; R2 ranging from −2.763 to 0.331 for DL and from −8.113 to 0.470 for ML).
Figure 3.
Integrated heatmap with dot plot for performance comparisons in settings 1 and 2. The R2 value is depicted as a size of the circle. Red and blue colors indicate RMSE values. The model with higher R2 and lower RMSE value is considered to have good performance. Note that R2 with a 0.0 value indicates that the R2 values of each model exhibit negative values or zero. (a) In setting 1, the ridge model for panobinostat shows the best performance among other models (b) In setting 2, no model outperformed the ridge model for panobinostat in setting 1. R2 R-squared value, RMSE root mean squared error.
We also inspected whether fine-tuning (i.e., feature selection) improved the performances of CNN and ridge for panobinostat. For the feature section, lasso was used in the training set. The numbers of features selected by lasso were 2000, 4000, 6000, 8000, 10,000, 12,000, 14,000, 16,000, and 18,988 (i.e., whole features). Given the features, we trained CNN and ridge, and measured the performances of the two models. As a result, the best prediction model in the two models was the ridge model using whole features (Supplementary Fig. S10). We attempted two more feature engineering techniques [i.e., non-linear feature generation21 and feature selection by ‘lasso with lars’ (least angle regression and shrinkage)22] using the Autofeat library of Python. As a result, the R2 values for the two feature engineering techniques were 0.218 and 0.409, respectively, which were not improved.
Setting 2: model construction for individual drugs, taking mutation profiles (MC-9K) as input for prediction of ln(IC50)s
For setting 2, similar to setting 1, 24 individual drug prediction models were constructed using mutation profiles using the training set from MC-9K (Supplementary Fig. S1b and Supplementary Table S3). Comparing the performance using test sets, all models failed to display strong positive correlations between predicted and actual ln(IC50) values for any drug (Fig. 3b; Supplementary Figs. S11, S12, S13, S14, S15, S16, S17, and S18; Supplementary Tables S12, S13, S14, S15, S16, S17, S18 and S19).
Application of ridge for panobinostat from setting 1 to gastric cancer (GC) cell lines and patient datasets
As mentioned in setting 1, the ridge model for panobinostat exhibited better performance than all the models for other drugs. Panobinostat is a histone deacetylase (HDAC) inhibitor. HDAC2 was overexpressed in gastric cancer (GC) cell lines to utilize panobinostat for GC treatment23.
In addition, given that GC tumors are highly heterogeneous24, we applied the ridge model of panobinostat to GC cell lines and patients with GC. We obtained four datasets of gene expression profiles of GC from CCLE2, GDSC3,25, GSE11891626, and patients with GC (n = 450) from The Cancer Genome Atlas (TCGA)27. We then entered the gene expression status vectors from the four datasets into the ridge model. For each dataset, we obtained a predicted ln(IC50) value for each cell line (or patient) for panobinostat (Fig. 4a).
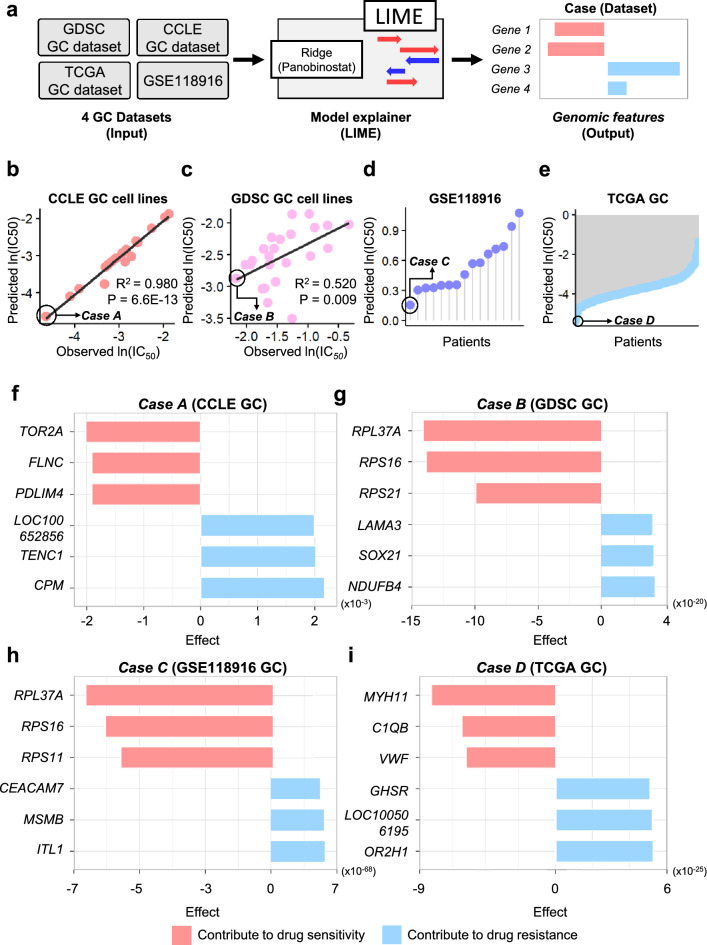
Figure 4.
Application of the ridge model for panobinostat in setting 1 to the GC cell line (CCLE and GDSC) and patient datasets (GSE118916 and TCGA). (a) Description of XAI application. (b) CCLE and (c) GDSC GC cell lines show a good correlation between predicted and observed ln(IC50) values. (d) GSE118916 and (e) TCGA datasets for patients with GC show a broad distribution of predicted ln(IC50) values. Furthermore, we selected a sensitive case for panobinostat in each GC dataset, and cases A through D are indicated in the circle. For case A and B, the predicted IC50 values of the two cases agree with their observed IC50 values. Considering case C and D, their datasets did not have patient information on the response, and the prediction cannot be compared. Considering prediction, the four cases are sensitive to the panobinostat when compared with the other samples. (f–i) To identify major genes affecting drug response, we performed XAI analysis with the four cases (case A through D). The six genes affecting drug sensitivity (red bar) and drug resistance (blue bar) were detected in (f) case A, (g) case B, (h) case C, and (i) case D, respectively. CCLE Cancer Cell Line Encyclopedia, GDSC genomics of drug sensitivity in cancer, GC gastric cancer, TCGA The Cancer Genome Atlas, XAI explainable artificial intelligence.
Using ln(IC50) prediction values from each dataset, we first compared the observed ln(IC50)s of the drugs in CCLE and GDSC GC cell lines to predict the drug ln(IC50)s. Both CCLE and GDSC GC cell lines presented a strong positive correlation (R2 for CCLE GC: 0.980 and R2 for GDSC GC: 0.520) between predicted and observed ln(IC50) values (Fig. 4b, c).
Second, we attempted to predict ln(IC50) values from patients with GC (GSE118916 and TCGA GC). In particular, the patient dataset yielded results revealing a broad distribution, which indicated that the drug response of panobinostat differed from the gene expression profile of each patient, thereby implying that the ridge model considers the heterogeneity of GC tumors27 (Fig. 4d, e).
Inspection of major genomic features affecting drug response prediction using local interpretable model-agnostic explanation (LIME) analysis
We investigated ln(IC50) predictions by inputting each of the four GC datasets into the ridge (panobinostat) model in setting 1. We confirmed that the drug response of each GC cell line (or patient) to panobinostat was predicted distinctly according to the gene expression vector. Accordingly, by utilizing an XAI approach such as LIME, we extracted significant genomic features in the ridge model of panobinostat in setting 1. Subsequently, we selected a sensitive case for panobinostat in each dataset, with case A for the CCLE GC dataset, case B for the GDSC GC dataset, case C for the GSE118916 GC dataset, and case D for the TCGA GC dataset, denoted in Fig. 4b–e, respectively. Given that the two datasets (GDSC and CCLE) contained actual IC50s derived from cell viability assays, the predicted IC50s of the two cases (cases A and B) were compared with their observed IC50s to establish confirmation between the prediction and the observation. Furthermore, the four cases (A, B, C, and D) were predicted to be more sensitive to panobinostat when compared with that of the other samples. Then, by applying LIME to the ridge model, we inspected the top three explainable genes affecting drug response and the top three genes affecting drug response in each case (cases A through D).
The top three genes that most affected drug response were TOR2A, FLNC, and PDLIM4 in case A; RPL37A, RPS16, and RPS21 in case B; RPL37A, RPS16, and RPS11 in case C; and MYH11, C1QB, and VWF in case D (red bars in Fig. 4f–i). Conversely, the top three genes impacting drug resistance were LOC100652856, TENC1, and CPM in case A; LAMA3, SOX21, and NDUFB4 in case B; CEACAM7, MSMB, and ITLN1 in case C; GHSR, LOC100506195, and OR2H1 in case D (blue bars in Fig. 4f–i).
Next, we inspected whether application of XAI to the ridge (panobinostat) model in setting 1 could reveal novel features other than the features selected by the ridge model in setting 1. For the purpose, we compared the features by the XAI with those by the ridge (panobinostat) model. As a result, the gene features selected by XAI were found to overlap with only three genes of the gene features selected in the panobinostat ridge model. It indicates that XAI selected novel gene features (Supplementary Fig. S19).
Since XAI is usually applied to DL, we applied LIME to CNN (i.e., CDRscan) in setting 1. For case D, the same important features were selected in both the panobinostat ridge model and CNN. For cases A, B, and C, no overlapped selected features with the panobinostat ridge model and CNN were revealed (Supplementary Fig. S20).
Discussion
Using diverse input settings, we compared the performance of ML and DL models in predicting IC50 cell viability values as drug responses based on pharmacogenomic databases by employing gene expression and mutation profiles.
Following this scheme, we also constructed AI and ML models, for individual drug species, for the EC-11K and MC-9K datasets (settings 1 and 2). Considering visual inspection (Supplementary Figs. S2, S3, S4, S5, S6, S7, S8, S9, S11, S12, S13, S14, S15, S16, S17, and S18), R2, and RMSE (Supplementary Tables S4, S5, S6, S7, S8, S9, S10, S11, S12, S13, S14, S15, S16, S17, S18, and S19), we noted that DL and ML models did not exhibit substantial differences in drug response prediction performance for 24 drugs (RMSE ranging from 0.284 to 3.563 for DL and from 0.274 to 2.697 for ML; R2 ranging from -7.405 to 0.331 for DL and from -8.113 to 0.470 to ML). Notably, the panobinostat ridge prediction model outperformed other models (Fig. 3a).
On applying the ridge (panobinostat) model in setting 1 to patients with GC, CCLE and GDSC GC revealed a positive correlation between the predicted and observed ln(IC50)s (Fig. 4b, c). However, we failed to detect any correlation between predicted and observed drug responses in GSE118916 and TCGA GC, given the lack of drug response data for panobinostat. Nevertheless, we observed that patients with GC exhibited a heterogeneous drug response to panobinostat (Fig. 4d, e).
In addition, we performed XAI analysis using LIME to identify the major genes that affect drug responsiveness prediction (drug sensitivity and resistance) in patients with GC (Fig. 4f–i). Using XAI, we confirmed that the genes affecting drug response and resistance in each case were related to the onset of GC and other cancers4,7,15,26,28–56. Interestingly, among genes affecting drug response prediction, CPM was found to contribute to chemoresistance in GC49, whereas RPL37A was a potential biomarker for response to neoadjuvant chemotherapy (NCT) against non-metastatic locally advanced breast carcinoma (LABC)41. Furthermore, we observed that the upregulation of MYH11 inhibited tumor growth in GC45, and genes encoding ribosomal proteins (RPL16, RPS11, and RPS21) were related to ulcerative colitis and GC. Accordingly, XAI analysis can reveal important genes that affect drug responses based on the genomic profile of each patient. Additionally, it can be used as a personalized medical strategy to overcome the heterogeneity observed in patients with cancer.
The limitations of the present study need to be addressed. First, the drug response prediction models were trained using one pharmacogenomic database (CCLE), and we attempted to overcome this drawback by applying multiple GC datasets to the prediction model. Second, to explore the genomic features affecting the sensitivity to various drugs, numerous drug prediction models should be incorporated into a one-model approach for individual drugs. Third, genomic profiles may not have local patterns and genomic profile data may not be suitable for CNN model using stride. However, a recent study demonstrated that a CNN model using strides in unstructured genomic profile data improved the performance of a cancer type prediction model57. In this sense, CDRscan is a CNN model with strides. Fourth, some models have low correlation coefficients between the actual and predicted values, explainable gene features should be carefully interpreted.
Regarding modeling individual cancer drug response dataset and modeling the entire drug response dataset14,58, the two kinds of modeling serve different purposes, and the evaluation of the modeling results can vary depending on their intended usages. Modeling individual cancer drug response helps to identify gene features specific to a drug58. On the other hand, modeling the entire dataset helps to identify the most responsive drug in a patient among diverse drugs14. Therefore, modeling individual cancer drug response can be useful in identifying drug-specific informative genes58.
Conclusion
Our research offers a useful guide for constructing a drug response prediction model using a one-model approach for individual drugs by integrating with XAI.
Methods
Data collection
Using CCLE2, we collected drug screening data as cell viability IC50, which is the half-maximal inhibitory concentrations, from cell viability assays in cancer cell lines (Fig. 1). For 24 drugs, IC50 data were available for 504 cancer cell lines in CCLE (version 24 Feb 2015). The screening concentrations for CCLE ranged between 0.0025 and 8.0 μM. In addition, we obtained mutation profile data from GDSC3,25. The mutation profile data consisted of 21,213 mutation sites in 1001 cell lines. The mRNA expression profiles of 18,988 genes in 1,037 cell lines were obtained from CCLE.
To generate combination datasets (Supplementary Table S1), we used cell line expression profiles (denoted as “E”), cell line mutation statuses (denoted as “M”), and drug response measurements as ln(IC50)s from CCLE (denoted as “C”)2.
The sources of the downloaded datasets are described in Supplementary Method S1.
Setting 1: construction of a one-model approach for an individual drug, considering expression profiles (EC-11K) as input for prediction of ln(IC50)s
The EC-11K dataset for setting 1 consisted of the expression profiles of 504 CCLE cancer cell lines as input and ln(IC50) values of 24 drugs examined in cancer cell lines as output (Supplementary Table S1). Accordingly, 11,360 (~ 11K) ln(IC50) measurements were available for cell line-drug treatment pairs from CCLE. In the dataset, the input vector had z-normalized expression elements for 18,988 genes (“EC-11K”, Supplementary Table S1). We then obtained 24 data matrices for 24 drugs from the EC-11K. Each data matrix was randomly divided into training and test sets at a ratio of 8 to 2 (Supplementary Fig. S1a and Supplementary Table S2).
Setting 2: construction of one-model approach, considering mutation as input for predicting ln(IC50)s, from dataset MC-9K
MC-9K, the combined dataset, comprised mutational statuses for 504 cancer cell lines, amounting to 21,213 mutation positions (point mutations) as inputs and ln(IC50) values of 24 drugs as outputs (Supplementary Table S1). For each cancer cell line, the mutational statuses for 21,213 mutation sites were binarized to either 1 (presence) or 0 (absence). As a result, the number of features (mutations) in an the input vector was 21,213 for the dataset of 8727 (~ 9K) ln(IC50) measurements (“MC-9K”, Supplementary Table S1).
Likewise to setting 1, in the MC-9K dataset, 24 data matrices for 24 drugs were obtained, and each data matrix was randomly split into training and test sets at a ratio of 8 to 2 (Supplementary Fig. S1b and Supplementary Table S3).
Construction of DL and ML models
We adopted CNN and ResNet architectures for regression-based DL models. For CNN architecture, we modified the architecture of the ‘CDRscan master’ model15 by eliminating CNN layers for drugs (Fig. 2a, Supplementary Fig. S21, and Supplementary Tables S20 and S21).
ResNet architecture (Fig. 2b, Supplementary Tables S22 and S23) was adopted from ResNetIC50, as previously reported14,35. The ResNet had 30 layers, including 9 skip connections (Fig. 2b and Supplementary Fig. S22).
For both the CNN and ResNet architectures, the loss function was the mean square error, while the rectified linear activation (ReLU) or the hyperbolic tangent function was used as the activation function. The CNN and ResNet parameters are indicated in Supplementary Tables S24 and S25.
We employed lasso, ridge, RF, SVR, and XGBoost for ML models. In all settings, the hyperparameter was set to alpha 0.001 for ridge and lasso, C 0.01 for SVR, and default options for RF and XGBoost (Supplementary Tables S24 and S25).
Model performance comparisons
For model performance comparisons, we calculated R2 and RMSE values between the predicted and the observed ln(IC50) values in the test set. The formulas are as follows:
| 1 |
| 2 |
where N is the number of cell lines in the test set; yi is the ith observed ln(IC50); and fi is the predicted ln(IC50) for the ith case. Thus, indicates the average of all y values. A scatter plot was adopted to visualize predicted and observed ln(IC50) values in the test set.
Application of ridge for panobinostat from setting 1 to GC cell lines and patient datasets
Considering the performance comparisons, we confirmed that the ridge model for panobinostat in setting 1 exhibited superior performance to other models in all settings. Then, to confirm the applicability of this model, we obtained gene expression profiles of GC cell lines from CCLE (n = 18)2 and GDSC (n = 24)3,25.
As cell lines are recognized as potential proxies for drug development2,10,59,60, we assumed that patients’ drug response correlated with cell line-drug response.
For this purpose, we also obtained gene expression profiles of patients with GC from GSE118916 (n = 15)26 and The Cancer Genome Atlas of Stomach Adenocarcinoma (TCGA-STAD) from UCSC XENA (IlluminaHiSeq pancan normalized [n = 450])27,61. The gene expression profiles from the four GC datasets were also z-normalized; then, the predicted ln(IC50)s for panobinostat were obtained by inputting the z-normalized gene expression profiles into the ridge model for panobinostat. Using predicted ln(IC50)s values in the four GC datasets, we selected the four sensitive cases for panobinostat from the GC datasets, respectively (case A for CCLE GC, case B for GDSC GC, case C for GSE118916 GC, and case D for TCGA GC).
For CCLE and GDSC GC cell lines, we performed pearson’s correlation analysis between the predicted ln(IC50) values and the observed ln(IC50) values to investigate the applicability of the ridge model for panobinostat in setting 1 to GC cell lines and patients.
Inspection of the major genomic features affecting drug response prediction using LIME analysis
To explore explainable genomic features of the ridge model for panobinostat in setting 1, we adopted LIME for an XAI method62 to the ridge model for panobinostat, obtaining important genes in the model using the four selected GC cases (cases A through D). For LIME, the python lime package62 was used with default parameter settings, with 18,988 explainable features. Then, based on the LIME analysis, the top three explainable biological (or genomic) features affecting drug response and the top three features affecting drug response were yielded in each case (cases A through D).
Also, we compared the gene features selected by application of XAI to the panobinostat ridge model (setting 1) in the four cases (cases A through D) with those selected in the panobinostat ridge model (setting 1). The top 100 gene features by XAI in each case were merged to obtain 374 non-redundant gene features. Subsequently, the non-redundant gene features selected by XAI were compared with the 100 gene features selected in the panobinostat ridge model.
Supplementary Information
Author contributions
Conceptualization, S.N.; methodology, S.N., A.P and Y.L..; formal analysis, A.P. and Y.L.; investigation, A.P and Y.L..; data curation, A.P.; writing—original draft preparation, A.P.; writing—review and editing, S.N. and Y.L.; visualization, A.P. and Y.L.; supervision, S.N.; funding acquisition, S.N. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Gachon University research fund of 2020 (GCU-202008430003 to S.N.); the Industrial Technology Innovation program (NO. 20016417, AI prediction platform development for lung and gastric cancer with Korean genetic data and its servitization) funded by the Ministry of Trade, Industry & Energy (MOTIE (KATS)/KEIT, Korea); and the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2020R1F1A1069206 to S.N.).
Data availability
The data underlying this article are available on GitHub at https://github.com/labnams/IC50_individual_drug.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-39179-2.
References
- 1.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 2.Barretina J, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W, et al. Genomics of drug sensitivity in cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baptista D, Ferreira PG, Rocha M. Deep learning for drug response prediction in cancer. Brief Bioinform. 2020 doi: 10.1093/bib/bbz171. [DOI] [PubMed] [Google Scholar]
- 5.Daemen A, et al. Modeling precision treatment of breast cancer. Genome Biol. 2013;14:R110. doi: 10.1186/gb-2013-14-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costello JC, et al. A community effort to assess and improve drug sensitivity prediction algorithms. Nat. Biotechnol. 2014;32:1202–1212. doi: 10.1038/nbt.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong Z, et al. Anticancer drug sensitivity prediction in cell lines from baseline gene expression through recursive feature selection. BMC Cancer. 2015;15:489. doi: 10.1186/s12885-015-1492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakellaropoulos T, et al. A deep learning framework for predicting response to therapy in cancer. Cell Rep. 2019;29:3367–3373. doi: 10.1016/j.celrep.2019.11.017. [DOI] [PubMed] [Google Scholar]
- 9.Wei D, Liu C, Zheng X, Li Y. Comprehensive anticancer drug response prediction based on a simple cell line-drug complex network model. BMC Bioinform. 2019;20:44. doi: 10.1186/s12859-019-2608-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu YC, et al. Predicting drug response of tumors from integrated genomic profiles by deep neural networks. BMC Med. Genomics. 2019;12:18. doi: 10.1186/s12920-018-0460-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.GuvencPaltun B, Mamitsuka H, Kaski S. Improving drug response prediction by integrating multiple data sources: Matrix factorization, kernel and network-based approaches. Brief. Bioinform. 2019 doi: 10.1093/bib/bbz153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurilov R, Haibe-Kains B, Brors B. Assessment of modelling strategies for drug response prediction in cell lines and xenografts. Sci. Rep. 2020;10:2849. doi: 10.1038/s41598-020-59656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koras K, et al. Feature selection strategies for drug sensitivity prediction. Sci. Rep. 2020;10:9377. doi: 10.1038/s41598-020-65927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park A, et al. A comprehensive evaluation of regression-based drug responsiveness prediction models, using cell viability inhibitory concentrations (IC50 values) Bioinformatics. 2022;38:2810–2817. doi: 10.1093/bioinformatics/btac177. [DOI] [PubMed] [Google Scholar]
- 15.Chang Y, et al. Cancer drug response profile scan (CDRscan): A deep learning model that predicts drug effectiveness from cancer genomic signature. Sci. Rep. 2018;8:8857. doi: 10.1038/s41598-018-27214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, et al. FN1, SPARC, and SERPINE1 are highly expressed and significantly related to a poor prognosis of gastric adenocarcinoma revealed by microarray and bioinformatics. Sci. Rep. 2019;9:1–9. doi: 10.1038/s41598-019-43924-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rampasek, L., Hidru, D., Smirnov, P., Haibe-Kains, B. & Goldenberg, A. Dr.VAE: Improving drug response prediction via modeling of drug perturbation effects. Bioinformatics35, 3743–3751. 10.1093/bioinformatics/btz158 (2019). [DOI] [PMC free article] [PubMed]
- 18.Sharifi-Noghabi H, Zolotareva O, Collins CC, Ester M. MOLI: Multi-omics late integration with deep neural networks for drug response prediction. Bioinformatics. 2019;35:i501–i509. doi: 10.1093/bioinformatics/btz318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bomane A, Goncalves A, Ballester PJ. Paclitaxel response can be predicted with interpretable multi-variate classifiers exploiting DNA-methylation and miRNA data. Front. Genet. 2019;10:1041. doi: 10.3389/fgene.2019.01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Lent, M., Fisher, W. & Mancuso, M. Proceedings of the National Conference on Artificial Intelligence. 900–907 (AAAI Press/MIT Press, 1999).
- 21.Horn, F., Pack, R. & Rieger, M. Machine Learning and Knowledge Discovery in Databases: International Workshops of ECML PKDD 2019, Würzburg, Germany, September 16–20, 2019, Proceedings, Part I. 111–120 (Springer, 2019).
- 22.Zhang L, Li K. Forward and backward least angle regression for nonlinear system identification. Automatica. 2015;53:94–102. doi: 10.1016/j.automatica.2014.12.010. [DOI] [Google Scholar]
- 23.Regel I, et al. Pan-histone deacetylase inhibitor panobinostat sensitizes gastric cancer cells to anthracyclines via induction of CITED2. Gastroenterology. 2012;143:99–109.e110. doi: 10.1053/j.gastro.2012.03.035. [DOI] [PubMed] [Google Scholar]
- 24.Nam S, Kim JH, Lee DH. RHOA in gastric cancer: Functional roles and therapeutic potential. Front. Genet. 2019;10:438. doi: 10.3389/fgene.2019.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pozdeyev N, et al. Integrating heterogeneous drug sensitivity data from cancer pharmacogenomic studies. Oncotarget. 2016;7:51619–51625. doi: 10.18632/oncotarget.10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amini M, et al. GHSR DNA hypermethylation is a new epigenetic biomarker for gastric adenocarcinoma and beyond. J. Cell. Physiol. 2019;234:15320–15329. doi: 10.1002/jcp.28179. [DOI] [PubMed] [Google Scholar]
- 27.Cancer_Genome_Atlas_Research_Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature513, 202–209. 10.1038/nature13480 (2014). [DOI] [PMC free article] [PubMed]
- 28.Zheng L, et al. Aberrant expression of intelectin-1 in gastric cancer: Its relationship with clinicopathological features and prognosis. J. Cancer Res. Clin. Oncol. 2012;138:163–172. doi: 10.1007/s00432-011-1088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohnuma S, et al. Cancer-associated splicing variants of the CDCA1 and MSMB genes expressed in cancer cell lines and surgically resected gastric cancer tissues. Surgery. 2009;145:57–68. doi: 10.1016/j.surg.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J, et al. Dynamic expression of CEACAM7 in precursor lesions of gastric carcinoma and its prognostic value in combination with CEA. World J. Surg. Oncol. 2011;9:1–8. doi: 10.1186/1477-7819-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ii M, et al. Co-expression of laminin β3 and γ2 chains and epigenetic inactivation of laminin α3 chain in gastric cancer. Int. J. Oncol. 2011;39:593–599. doi: 10.3892/ijo.2011.1048. [DOI] [PubMed] [Google Scholar]
- 32.Caglayan D, Lundin E, Kastemar M, Westermark B, Ferletta M. Sox21 inhibits glioma progression in vivo by forming complexes with Sox2 and stimulating aberrant differentiation. Int. J. Cancer. 2013;133:1345–1356. doi: 10.1002/ijc.28147. [DOI] [PubMed] [Google Scholar]
- 33.Bizama C, et al. The low-abundance transcriptome reveals novel biomarkers, specific intracellular pathways and targetable genes associated with advanced gastric cancer. Int. J. Cancer. 2014;134:755–764. doi: 10.1002/ijc.28405. [DOI] [PubMed] [Google Scholar]
- 34.Qiao J, et al. Filamin C, a dysregulated protein in cancer revealed by label-free quantitative proteomic analyses of human gastric cancer cells. Oncotarget. 2015;6:1171. doi: 10.18632/oncotarget.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He, K., Zhang, X., Ren, S. & Sun, J. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 770–778 (2016).
- 36.Mao Q, et al. iTRAQ-based proteomic analysis of Ginsenoside F2 on human gastric carcinoma cells SGC7901. Evid.-Based Complem. Altern. Med. 2016;2016:1–21. doi: 10.1155/2016/2635483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sotgia F, Lisanti MP. Mitochondrial biomarkers predict tumor progression and poor overall survival in gastric cancers: Companion diagnostics for personalized medicine. Oncotarget. 2017;8:67117. doi: 10.18632/oncotarget.19962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin T-C, Hsiao M. Ghrelin and cancer progression. Biochim. Biophys. Acta (BBA) Rev. Cancer. 2017;1868:51–57. doi: 10.1016/j.bbcan.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Yoo J-Y, et al. Pdlim4 is essential for CCR7-JNK–mediated dendritic cell migration and F-actin-related dendrite formation. FASEB J. 2019;33:11035–11044. doi: 10.1096/fj.201901031. [DOI] [PubMed] [Google Scholar]
- 40.Kravchenko DS, Ivanova AE, Podshivalova ES, Chumakov SP. PDLIM4/RIL-mediated regulation of Src and malignant properties of breast cancer cells. Oncotarget. 2020;11:22. doi: 10.18632/oncotarget.27410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carrara GFA, et al. Analysis of RPL37A, MTSS1, and HTRA1 expression as potential markers for pathologic complete response and survival. Breast Cancer. 2021;28:307–320. doi: 10.1007/s12282-020-01159-z. [DOI] [PubMed] [Google Scholar]
- 42.El Khoury, W. & Nasr, Z. Deregulation of ribosomal proteins in human cancers. Biosci. Rep.41, BSR20211577 (2021). [DOI] [PMC free article] [PubMed]
- 43.Chu J, et al. Bayesian hierarchical lasso Cox model: A 9-gene prognostic signature for overall survival in gastric cancer in an Asian population. PLoS ONE. 2022;17:e0266805. doi: 10.1371/journal.pone.0266805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elhamamsy, A. R., Metge, B. J., Alsheikh, H. A., Shevde, L. A. & Samant, R. S. Ribosome biogenesis: A central player in cancer metastasis and therapeutic resistance. Cancer Res. (2022). [DOI] [PMC free article] [PubMed]
- 45.Lee I-S, et al. A blood-based transcriptomic signature for noninvasive diagnosis of gastric cancer. Br. J. Cancer. 2021;125:846–853. doi: 10.1038/s41416-021-01461-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang J, et al. Identification of TYROBP and C1QB as two novel key genes with prognostic value in gastric cancer by network analysis. Front. Oncol. 2020;10:1765. doi: 10.3389/fonc.2020.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salmikangas S, et al. Tensin2 is a novel diagnostic marker in GIST, associated with gastric location and non-metastatic tumors. Cancers. 2022;14:3212. doi: 10.3390/cancers14133212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu H, Wang G, Zhu H, Xu A. MTFR2, a potential biomarker for prognosis and immune infiltrates, promotes progression of gastric cancer based on bioinformatics analysis and experiments. J. Cancer. 2021;12:3611. doi: 10.7150/jca.58158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang L, et al. Circular CPM promotes chemoresistance of gastric cancer via activating PRKAA2-mediated autophagy. Clin. Transl. Med. 2022;12:e708. doi: 10.1002/ctm2.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Razavi H, Katanforosh A. Identification of novel key regulatory lncRNAs in gastric adenocarcinoma. BMC Genomics. 2022;23:1–14. doi: 10.1186/s12864-022-08578-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moradi K, et al. High potential of SOX21 gene promoter methylation as an epigenetic biomarker for early detection of colorectal cancer. Indian J. Cancer. 2020;57:166. doi: 10.4103/ijc.IJC_542_18. [DOI] [PubMed] [Google Scholar]
- 52.Raj D, et al. CEACAM7 is an effective target for CAR T-cell therapy of pancreatic ductal adenocarcinoma CEACAM7-directed CAR T-cell therapy of pancreatic cancer. Clin. Cancer Res. 2021;27:1538–1552. doi: 10.1158/1078-0432.CCR-19-2163. [DOI] [PubMed] [Google Scholar]
- 53.Paval DR, Di Virgilio TG, Skipworth RJ, Gallagher IJ. The emerging role of intelectin-1 in cancer. Front. Oncol. 2022;12:767859. doi: 10.3389/fonc.2022.767859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hodkinson BP, et al. Biomarkers of response to ibrutinib plus nivolumab in relapsed diffuse large B-cell lymphoma, follicular lymphoma, or Richter's transformation. Translat. Oncol. 2021;14:100977. doi: 10.1016/j.tranon.2020.100977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin, A. L. et al. Olfactory Receptor OR2H1 is an effective target for CAR T cells in human epithelial tumors. Mol. Cancer Ther. (2022). [DOI] [PMC free article] [PubMed]
- 56.Qu C, et al. Tumor buster-where will the CAR-T cell therapy ‘missile’go? Mol. Cancer. 2022;21:1–53. doi: 10.1186/s12943-022-01669-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mostavi M, Chiu YC, Huang Y, Chen Y. Convolutional neural network models for cancer type prediction based on gene expression. BMC Med. Genomics. 2020;13:44. doi: 10.1186/s12920-020-0677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parca L, et al. Modeling cancer drug response through drug-specific informative genes. Sci. Rep. 2019;9:15222. doi: 10.1038/s41598-019-50720-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Francies HE, McDermott U, Garnett MJ. Genomics-guided pre-clinical development of cancer therapies. Nat. Cancer. 2020;1:482–492. doi: 10.1038/s43018-020-0067-x. [DOI] [PubMed] [Google Scholar]
- 60.Wilding JL, Bodmer WF. Cancer cell lines for drug discovery and development. Cancer Res. 2014;74:2377–2384. doi: 10.1158/0008-5472.CAN-13-2971. [DOI] [PubMed] [Google Scholar]
- 61.Gao, G. F. et al. Before and after: Comparison of legacy and harmonized TCGA genomic data commons' data. Cell Syst.9, 24–34 e10. 10.1016/j.cels.2019.06.006 (2019). [DOI] [PMC free article] [PubMed]
- 62.Ribeiro, M. T., Singh, S. & Guestrin, C. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 1135–1144 (Association for Computing Machinery).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available on GitHub at https://github.com/labnams/IC50_individual_drug.