Abstract
Background
Morphoea can have a significant disease burden. Aetiopathogenesis remains poorly understood, with very limited existing genetic studies. Linear morphoea (LM) may follow Blascho’s lines of epidermal development, providing potential pathogenic clues.
Objective
The first objective of this study was to identify the presence of primary somatic epidermal mosaicism in LM. The second objective was tTo explore differential gene expression in morphoea epidermis and dermis to identify potential pathogenic molecular pathways and tissue layer cross-talk.
Methodology
Skin biopsies from paired affected and contralateral unaffected skin were taken from 16 patients with LM. Epidermis and dermis were isolated using a 2-step chemical-physical separation protocol. Whole Genome Sequencing (WGS; n = 4 epidermal) and RNA-seq (n = 5-epidermal, n = 5-dermal) with gene expression analysis via GSEA-MSigDBv6.3 and PANTHER-v14.1 pathway analyses, were performed. RTqPCR and immunohistochemistry were used to replicate key results.
Results
Sixteen participants (93.8% female, mean age 27.7 yrs disease-onset) were included. Epidermal WGS identified no single affected gene or SNV. However, many potential disease-relevant pathogenic variants were present, including ADAMTSL1 and ADAMTS16. A highly proliferative, inflammatory and profibrotic epidermis was seen, with significantly-overexpressed TNFα-via-NFkB, TGFβ, IL6/JAKSTAT and IFN-signaling, apoptosis, p53 and KRAS-responses. Upregulated IFI27 and downregulated LAMA4 potentially represent initiating epidermal ‘damage’ signals and enhanced epidermal-dermal communication. Morphoea dermis exhibited significant profibrotic, B-cell and IFN-signatures, and upregulated morphogenic patterning pathways such as Wnt.
Conclusion
This study supports the absence of somatic epidermal mosaicism in LM, and identifies potential disease-driving epidermal mechanisms, epidermal-dermal interactions and disease-specific dermal differential-gene-expression in morphoea. We propose a potential molecular narrative for morphoea aetiopathogenesis which could help guide future targeted studies and therapies.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00403-023-02541-5.
Keywords: Morphoea, Linear morphoea, Localised scleroderma, Aetiopathogenesis, Next-generation sequencing, Genomics, Transcriptomics, Gene expression
Introduction
Morphoea is characterised by fibrosis of the skin and/or underlying connective tissues, with the potential for significant functional and psychological impact. It is suggested that environmental triggers [1–3], occurring in a genetically susceptible individual, underpin the inflammation and deregulated tissue injury response in morphoea [4]. However, precise genetic susceptibility factors, inciting and propagating molecular mechanisms, remain unclear.
Linear morphoea (LM) may follow Blaschko’s lines of epidermal development, and hence may represent epidermal somatic mosaicism for a mutation conferring increased risk of disease at specific sites [5–9]. Accordingly, keratinocyte-derived signals and epidermal-dermal communication pathways vital to normal skin development and wound repair, are also key to pathological skin fibrosis and highly active, proliferative keratinocytes are seen in systemic sclerosis (SSc) [4, 10, 11].
However, LM is a non-congenital and morphologically heterogeneous dermal pathology, potentially suggesting more complex underlying aetiopathogenic mechanisms. Correspondingly, non-linear morphoea subtypes show alternative, but often symmetrical and somewhat predictably patterned skin involvement. As such, dermal fibroblasts have site-specific gene expression, known as positional identity (PI). Many molecular pathways instrumental in developmental patterning, regional-specific mesenchymal differentiation and epidermal fate, such as FGFs, TGF-β and Wnt [12, 13], are also involved in pathogenic fibrosis and SSc [14, 15]. Similarly, morphogenic and epidermal–dermal signaling pathways, including Wnt, Hedgehog [14, 16] and Notch [14, 17, 18], are deregulated in fibrosis and SSc [17, 19–23].
Morphoea’s morphological heterogeneity, clinical symmetry, patterning and possibly Blaschkoid distribution, may therefore provide clinical clues to potential underlying epidermal and dermal genetic aetiopathogenic and disease-driving mechanisms [4].
The goals of this study were to identify the presence or absence of primary somatic epidermal genomic variation (as a common single nucleotide variant (SNV), or differing SNVs in a commonly affected gene, across all study samples) in LM, and to explore differential gene expression (DGE) in isolated epidermal and dermal site-matched tissue pairs, to identify potential inciting and pathogenic pathways in the epidermis and dermis. We aimed to correlate our data with the very limited current genetic data in morphoea, to propose a possible genetic and molecular narrative underlying morphoea aetiopathogenesis and hence identify potential future study and therapeutic targets.
Methodology
This study was approved by the National Research Ethics Service (London-Hampstead, MREC Reference 6398). Tissue specimens were obtained with written informed consent as part of an ongoing programme of research into the pathogenesis of scleroderma.
Specimen source
Patients with LM involving the limb(s) and/or trunk identified from our previously characterised morphoea cohort were eligible for specimen collection [24]. A total of 16 patients were enrolled (Table 1). Details regarding sample selection for each molecular (DNA/RNA) and tissue layer (epidermal/dermal) dataset are described in the Supplemental Methods section.
Table 1.
Study cohort; experimental studies and clinical characteristic
| Study no | Sex, age onset (yrs) | Epidermal WGS | Epidermal/dermal RNA-seq | Validation studies | Disease status | Biopsy site activity | Site and phenotype biopsied | Cutaneous symptoms | Current treatment |
|---|---|---|---|---|---|---|---|---|---|
| 1 | F, 26 | Yes | Epidermal*, dermal | Epidermal RT-qPCR | Stable | Yes | Upper limb; inflammatory, sclerotic | Pruritus, tingling | Topical |
| 2 | F, 18 | Epidermal | Stable | No | Lower limb; inflammatory, sclerotic | Pruritus | Systemic | ||
| 3 | F, 19 | Yes | Epidermal*, dermal | Epidermal RT-qPCR | Active | Yes | Upper limb; inflammatory | Pruritus | Topical |
| 4 | F, 19 | Yes | Epidermal, dermal | Active | Yes | Upper limb; inflammatory, sclerotic | Tingling | Systemic | |
| 5 | F, 51 | Epidermal | Active | No | Lower limb; atrophic, pigmented | Nil | Nil; treatment naive | ||
| 6 | F, 32 | Epidermal | Stable | No | Lower limb; atrophic, pigmented | Pain | Systemic | ||
| 7 | F, 21 | Yes; failed sequencing | Epidermal, dermal | Active | Yes | Upper limb; inflammatory, sclerotic | Pruritus, pain | Systemic | |
| 8 | F, 29 | Yes | Epidermal*, dermal | Epidermal RT-qPCR | Active | Yes | Upper limb; inflammatory | Tingling | Systemic |
| 9 | F, 54 | Epidermal RT-qPCR | Remission | No | Trunk; atrophic, pigmented | Nil | Nil; previous systemic | ||
| 10 | F, 26 | Epidermal RT-qPCR | Remission | No | Lower limb; atrophic, pigmented | Pain | Nil, previous topical and systemic | ||
| 11 | F, 45 | Epidermal RT-qPCR | Remission | No | Lower limb; atrophic, pigmented | Tingling | Nil, previous systemic | ||
| 12 | F, 12 | Whole skin IHC | Active | Yes | Lower limb; pigmented | Pruritus | Topical, systemic | ||
| 13 | M, 8 | Whole skin IHC | Stable | No | Sclerotic | Pain | Systemic | ||
| 14 | F, 10 | Whole skin IHC | Active | Yes | Upper limb; sclerotic, pigmented | Nil | Systemic | ||
| 15 | F, 32 | Whole skin IHC | Active | Yes | Lower limb; pigmented | Pruritus | Topical, systemic | ||
| 16 | F, 14 | Whole skin IHC | Active | Yes | Trunk; sclerotic | pruritus, tingling | Topical, systemic |
*Failed quality control with Beijing Genomics Institute for RNA-seq, alternative epidermal samples for RNA-seq selected (Study No. 2, 5 and 6)
Paired 4 mm whole skin punch-biopsies were taken from each participant; one or two from morphoea affected (lesional) skin, and one or two from site-matched contralateral unaffected skin. For tissues samples utilised for DNA/RNA isolation, epidermis was immediately chemically separated from the dermis utilising 3.8% ammonium thiocyanate (Sigma-Alrich USA) in Dulbecco's phosphate-buffered saline pH 7.4 at room temperature for 25 min. Residual epidermal tissue was gently curetted off the superficial dermal surface using a scalpel blade (no. 15) [25].
DNA isolation, whole genome sequencing and analysis; epidermis
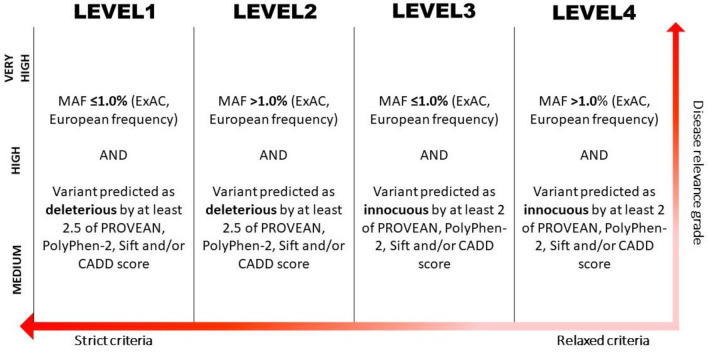
DNA was isolated from paired epidermal tissue and four selected paired samples underwent WGS. All identified genes with SNVs underwent network analysis utilising STRING online database (v11). Identified SNVs were then classified; graded according to disease relevance and sub-classified according to MAF (using ExAC) and pathogenicity (according to PolyPhen-2, PROVEAN, SIFT and CADD scores) (Supplemental Methods and Fig. 1).
Fig. 1.
Classification strategy for disease relevant gene candidates (graded as very high, high or medium according to functional relevance to morphoea aetiopathogenesis; vertical grading) and for pathogenicity (according to allele frequency and pathogenicity criteria; horizontal classification ranking)
RNA isolation, sequencing and analysis; epidermis and dermis
Total RNA was isolated from paired epidermal and dermal tissue, and selected samples underwent RNA-seq. Epidermal and dermal differentially expressed genes (DEG) were further analysed via Gene Set Enrichment Analysis (GSEA), using MSigDB Hallmark gene sets [26, 27]. Enrichment was reported as significant if the false discovery rate (FDR) was less than 0.25 [28] and each GSEA set was ranked according to log2 fold change (log2FC).
For dermal RNA-seq data, further complimentary analysis via PANTHER (PANTHER Gene Ontology (GO)-Slim Biological Process) [29] was completed. An adjusted P-value was calculated using Bonferroni correction, with a statistical significance cut-off of < 0.05. STRING database was utilised to review protein–protein interactions between products of particular DEGs of interest. (Supplemental Methods).
RT-qPCR and IHC of selected epidermal and dermal gene candidates derived from epidermal RNA-seq
Details can be found in the relevant Supplemental Methods sections.
Results
Epidermal protein coding single nucleotide variants
861 SNVs were identified in morphoea-affected epidermis, but absent in paired unaffected epidermis. Of these, 119 were protein-coding exonic and 72 nonsynonymous. No single common SNV or commonly affected gene was identified across all four sequenced epidermal tissue pairs.
A number of nonsynonymous protein-coding SNVs had high CADD scores (> 20) and pathogenicity rated as damaging or possibly damaging by at least two of PolyPhen-2, PROVEAN and SIFT algorithms, including; ADAMTS16, ADAMTSL1 and CBX2 (Table 2). STRING network analyses of these variants yielded no noteworthy gene clusters.
Table 2.
All protein coding, nonsynonymous genomic variants (alphabetised, CADD scores rounded to the nearest whole number)
| Gene symbol | Variant | Study participant | PolyPhen- 2 | PolyPhen-2 | PROVEAN | PROVEAN | SIFT | SIFT | CADD score | ExAC European frequency (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| ADAMTS16 | p.C1206V | 4 | 0.999 | Damaging | − 9.08 | Damaging | 0 | Damaging | 30 | 0 |
| ADAMTSL1 | p.A322T | 3 | 0.999 | Damaging | − 2.78 | Damaging | 0.005 | Damaging | 33 | 0 |
| C6orf15 | p.R27Q | 8 | 0.955 | Damaging | − 3.71 | Damaging | 0 | Damaging | 26 | 0.001522 |
| CACNA1D | p.K776R, p.K796R | 1 | 0.023 | Benign | − 1.91 | Neutral | 0.064 | Tolerated | 24 | 0 |
| CAD | p.E1420K, p.E1483K | 1 | 0.190 | Benign | − 2.58 | Damaging | 0.197 | Tolerated | 27 | 0 |
| CBX2 | p.G367R | 8 | 0.561 | Possibly damaging | − 2.21 | Neutral | 0 | Damaging | 24 | 0 |
| CBX2 | p.G367E | 8 | 0.360 | Benign | − 1.98 | Neutral | 0 | Damaging | 22 | 0 |
| CNTNAP3 | p.G1195R | 3 | 0.999 | Damaging | − 5.11 | Damaging | 0.015 | Damaging | 23 | 0 |
| CNTNAP3B | p.D281H | 1 | N/A | NA | N/A | N/A | N/A | N/A | N/A | 0 |
| CNTNAP3B | p.V275I | 1 | 0.191 | Benign | − 0.80 | Neutral | 0.020 | Damaging | 6 | 0 |
| DEF8 | p.P71L, p.P131L, p.P121L, p.P192L | 1 | 0.264 | Benign | − 1.57 | Neutral | 0.262 | Tolerated | 24 | 0 |
| DEF8 | p.P71S, p.P131S, p.P121S, p.P192S | 1 | 0.692 | Possibly damaging | − 2.60 | Damaging | 0.029 | Damaging | 25 | 0 |
| DENND1C | p.R515H | 3 | 0.001 | Benign | − 0.43 | Neutral | 0.545 | Tolerated | 7 | 0 |
| EFCC1 | p.A165T | 4 | 0.118 | Benign | N/A | N/A | N/A | N/A | 22 | 0 |
| FAM186A | p.T1377P | 3 | 0 | Benign | 2.78 | Neutral | 1.000 | Tolerated | < 1 | 0 |
| FAM231B | p.S38T | 3 | N/A | N/A | N/A | N/A | N/A | N/A | < 1 | 2.20 |
| FAN1 | p.F866S | 4 | 0.007 | Benign | − 0.79 | Neutral | 0.457 | Tolerated | 3 | 0 |
| GOLGA6B | p.G648D | 3 | 0.017 | Benign | 1.40 | Neutral | 1.000 | Tolerated | 1 | 0 |
| HCFC1 | p.A934T | 8 | 0.995 | Damaging | − 1.68 | Neutral | 0.001 | Damaging | 32 | 0.0021 |
| HES6 | p.R49Q | 8 | 0.737 | Possibly damaging | − 2.68 | Damaging | 0.008 | Damaging | 24 | 0 |
| HRNR | p.L1722S | 8 | 0 | Benign | 1.80 | Neutral | 0.125 | Tolerated | 2 | 0.11 |
| HS6ST1 | p.V114G | 3 | 0.679 | Possibly damaging | 0.32 | Neutral | 0.262 | Tolerated | 23 | 26.27 |
| IGSF3 | p.660Q, p.R680Q | 8 | 0.345 | Benign | − 1.71 | Neutral | 0.095 | Tolerated | 16 | 4.97 |
| IMPG2 | p.G2386A | 4 | 0.109 | Benign | − 1.73 | Neutral | 0.01 | Damaging | 20 | 0 |
| KIF21B | p.R1371W, p.R1384W | 1 | 0.993 | Damaging | − 5.51 | Damaging | 0.001 | Damaging | 33 | 0 |
| KRT8 | p.S31A, p.S59A | 8 | 0.001 | Benign | 0.27 | Neutral | 1.000 | Tolerated | < 1 | 0.03 |
| MST1L | p.R483C | 3 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 0.02 |
| MUC12 | p.T3428I | 8 | N/A | N/A | − 2.00 | Neutral | 0.006 | Damaging | 4 | 0 |
| MUC20 | p.S182G | 8 | 0.475 | Possibly damaging | − 0.97 | Neutral | 0.411 | Tolerated | < 1 | 0.03 |
| MUC4 | p.I2761V | 4 | 0.001 | Benign | − 0.12 | Neutral | 1.000 | Tolerated | < 1 | 4.30 |
| MUC5B | p.M2869T | 1 | 0 | Benign | 1.03 | Neutral | 1.000 | Tolerated | < 1 | 15.68 |
| NBPF20 | p.D3013E | 4 | N/A | N/A | N/A | N/A | N/A | N/A | < 1 | 0 |
| NDST2 | p.R464C | 4 | 0.969 | Damaging | − 5.99 | Damaging | 0.001 | Damaging | 32 | 0.0015 |
| NOS1AP | p.A31V, p.A321V, p.A326V | 3 | 0.511 | Possibly damaging | − 2.35 | Neutral | 0.017 | Damaging | 30 | 0.00301 |
| NR2F2 | p.Y179S, p.Y159S, p.Y312S | 4 | 0.944 | Damaging | − 7.31 | Damaging | 0 | Damaging | 24 | 0 |
| OR11H12 | p.W68R | 8 | 0 | Benign | 2.67 | Neutral | 0.475 | Tolerated | < 1 | 0.001648 |
| OR2T6 | p.G151S | 1 | 0.971 | Damaging | − 1.61 | Neutral | 0.032 | Damaging | 23 | 0.001502 |
| PACS1 | p.Q35P | 8 | N/A | N/A | 0.02 | Neutral | 0.364 | Tolerated | < 1 | 0.06 |
| PARG | p.R377W, p.R403W, p.R485W | 8 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| PAX2 | p.Q255R, p.Q286R, p.Q278R | 4 | 0.104 | Benign | − 1.49 | Neutral | 0.149 | Tolerated | 14 | 0 |
| PAX3 | p.G15D | 1 | 0.025 | Benign | − 1.37 | Neutral | 0.002 | Damaging | 24 | 0 |
| PAX3 | p.G15D | 1 | 0.001 | Benign | 0.05 | Neutral | 0.251 | Tolerated | 22 | 0.006088 |
| PRAMEF10 | p.N459T | 3 | 0 | Benign | − 1.45 | Neutral | 0.254 | Tolerated | < 1 | 0.007823 |
| PRAMEF6 | p.N381T | 8 | 0 | Benign | 1.42 | Neutral | 1.000 | Tolerated | < 1 | 0 |
| PRAMEF6 | p.S375N | 8 | 0.996 | Damaging | − 2.22 | Neutral | 0.017 | Damaging | 4 | 0 |
| PRDM9 | p.T713R | 4 | 0.513 | Possibly damaging | − 4.51 | Damaging | 0.001 | Damaging | 23 | 0.001675 |
| RFPL4A | p.V179E | 1 | 0.018 | Benign | 1.37 | Neutral | 0.910 | Tolerated | < 1 | 17.13 |
| RGPD5;RGPD8 | p.R952S | 3 | 0 | Benign | 2.33 | Neutral | 1.000 | Tolerated | < 1 | 0 |
| RMDN3 | p.K285R | 1 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| RYR1 | p.D1377E | 3 | 0.231 | Benign | − 2.08 | Neutral | 0.193 | Tolerated | 23 | 0 |
| SAA2;SAA2-SAA4 | p.S156 | 8 | 0 | Benign | 2.70 | Neutral | 1.000 | Tolerated | 5 | 0 |
| SDR39U1 | p.D115Y, p.D89Y, p.D197Y | 8 | 1.000 | Damaging | − 8.85 | Damaging | 0 | Damaging | 32 | 0 |
| SGIP1 | p.G427R, p.G431R | 8 | 0.999 | Damaging | − 2.06 | Neutral | 0.031 | Damaging | 25 | 0 |
| SLC17A7 | p.F8V | 1 | 0.002 | Benign | − 0.53 | Neutral | 0.610 | Tolerated | 14 | 0 |
| SMG1 | p.I612K | 8 | 0 | Benign | − 3.00 | Damaging | 0.028 | Damaging | 17 | 3.57 |
| SPATA31D1 | p.A192P | 3 | 0 | Benign | 3.61 | Neutral | 1.000 | Tolerated | < 1 | 0.004496 |
| SPTBN1 | p.R1741H, p.R1754H | 3 | 0.987 | Damaging | − 4.60 | Damaging | 0.001 | Damaging | 31 | 0 |
| SYNE1 | p.V5268I, p.V5339I | 1 | 0.006 | Benign | 0.36 | Neutral | 1.000 | Tolerated | 13 | 0 |
| TBC1D3B;TBC1D3D;TBC1D3G;TBC1D3H;TBC1D3I;TBC1D3L | p.I117T | 3 | 0.349 | Benign | N/A | N/A | N/A | N/A | 9 | 0 |
| TBC1D3D;TBC1D3H;TBC1D3I | p.R399W | 3 | 0 | Benign | N/A | N/A | N/A | N/A | 12 | 0 |
| TCP10 | p.A256S | 1 | 0 | Benign | 0.69 | Neutral | 0.807 | Tolerated | < 1 | 5.39 |
| TCP10 | p.R262W | 8 | 0.035 | Benign | 0.52 | Neutral | 0.078 | Tolerated | 12 | 0.66 |
| TNS3 | p.S120Y | 8 | 0.997 | Damaging | − 3.34 | Damaging | 0 | Damaging | 31 | 0 |
| UFSP2 | p.E440K | 8 | 0.074 | Benign | − 1.13 | Neutral | 0.244 | Tolerated | 24 | 0 |
| URB1 | p.H967Y | 1 | 0.469 | Possibly damaging | − 0.49 | Neutral | 0.050 | Damaging | 25 | 0 |
| USP22 | p.F428S | 3 | 0.998 | Damaging | − 7.53 | Damaging | 0 | Damaging | 34 | 0 |
| WWC3 | pQ827K | 1 | 0.108 | Benign | − 0.73 | Neutral | 0.421 | Tolerated | 19 | 0 |
| ZNF608 | p.S1287L | 1 | 0.716 | Possibly damaging | − 1.31 | Neutral | 0.011 | Damaging | 23 | 0.001498 |
| ZNF614 | p.I201T | 3 | 0.005 | Benign | − 1.24 | Neutral | 0.275 | Tolerated | < 1 | 0 |
| ZNF705E | p.Q67R | 4 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | 0 |
| ZNF862 | p.R923K | 4 | 0.001 | Benign | 0.35 | Neutral | 0.463 | Tolerated | < 1 | 0 |
| ZP3 | p.s264P | 4 | 0 | Benign | 0.71 | Neutral | 1.000 | Tolerated | < 1 | 54.05 |
Disease relevance of epidermal genomic variants
No protein coding nonsynonymous SNVs were graded as very high for disease relevance. Variants in the genes ADAMTS16 and ADAMTSL1 were graded as high for disease relevance and Level 1 for potential pathogenicity and rarity. All other protein-coding nonsynonymous variants were graded as medium disease relevance (Table 3).
Table 3.
Potential gene candidates from epidermal whole genome sequencing as selected by network analyses and disease relevance; graded by potential relevance to morphea pathogenesis, and sub-categorised by Level, based on potential pathogenicity
| Disease/functional relevance grade | Level 1 | Level 2 | Level 3 | Level 4 | Non-coding variants |
|---|---|---|---|---|---|
| Very high | CCL5, FGF9, HBEGF, SMAD4, SMAD6 | ||||
| High | ADAMTS16, ADAMTSL1 | ACTN4, ADAM9, ADAMTS14, ADAMTS6, DTX2, FLRT2, ITGB1, LTBP1, MAP3K7, MAP3K13, MTOR, NANOG, NFE2L2, PIAS1, PIK3CA, POU5F1, PTEN, RB1CC1, ROCK1, SPRTN | |||
| Medium | C6orf15, CBX2 (p.G367R), HES6, CNTNAP3, DEF8*, HCFC1, NDST2, NOS1AP, NR2F2, OR2T6, PRDM9, SDR39U1, SGIP1, SMG1, SPTBN1, TNS3, URB1, USP22, ZNF608 | CAD, CBX2 (G367E), CNTNAP3B, DEF8^, DENND1C, EFCC1, FAM186A, FAN1, GOGLA6B, HRNR, MUC4, MUC20, NBPF20, OR11H12, PACS1, PARG, PAX2, PAX3, PRAMEF10, PRAMEF6, RGPDS;RGPD8, RYR1, SAA2;SAA2-SAA4, SLC17A7, SPATA31D1, SYNE1, TBC1D3B;TBC1D3D;TBC1D3G;TBC1D3H;TBC1D3I;TBC1D3L, TBC1D3D;TBC1D3H;TBC1D3I, WWC3, ZNF614, ZNF705E, ZNF862 | FAM231B, HS6ST1, MST1L, MUC5B, MUC12, RFP44A, ZP3 | ATR, BCL2L11, BMF, CBL, CRTAP, CTBP2, EHMT1, EPS1SL1, ERBIN, FBXO27, FBXW8, GNAQ, IGF1, IGF2, MAGI1, MAGI3, MOB1A, MOB1B, NEURL, VCL, VPS37C |
*p.P71S, p.P131S, p.P121S, p.P192S; ^p.P71L, p.P131L, p.P121L, p.P192L
Epidermal gene expression
Only three gene transcripts were significantly upregulated, including gene paralogs SPRR4 (FDR = 0.011, Log2FC 1.266) and SPRR1B (FDR = 0.026, log2FC 1.252), and four were significantly downregulated including LAMA4 (FDR = 0.026, log2FC −1.263) and PAX8 (FDR = 0.029, log2FC −0.785). Despite FDR > 0.05, IFI27 (log2FC 1.565) and WNT2 (log2FC 1.351) were noted with log2FC > 1.
Epidermal gene signatures; gene set enrichment analysis
Thirty-six Hallmark gene sets had significant enrichment; 16 with positive and 20 with negative enrichment. TNF-α signalling via NFkB (NES = 2.514, FDR = < 0.001), TGF-β signalling (NES = 2.006, FDR = 0.001) and IL-6/JAKSTAT3 signalling (NES = 1.961, FDR = 0.001) were the most strongly positively enriched (Fig. 2 and Table 4).
Fig. 2.
Enrichment of disease relevant Hallmark gene sets on GSEA, comparing epidermal and dermal datasets. An asterix (*) denotes significantly enriched sets (FDR < 0.25). Dermal Wnt signaling and epidermal Notch signaling were not in the top 20 differentially expressed Hallmark sets within their respective dataset and hence are not displayed graphically
Table 4.
Epidermal RNA sequencing: Hallmark gene sets with significant positive or negative enrichment on GSEA, listed by NES
| Hallmark gene set | NES | FDR |
|---|---|---|
| Positively enriched sets | ||
| TNF-α signaling via NFkB | 2.514 | < 0.001 |
| TGF-β signaling | 2.006 | 0.001 |
| IL-6/JAKSTAT3 signaling | 1.961 | 0.001 |
| IFNα response | 1.942 | 0.001 |
| Inflammatory response | 1.874 | 0.002 |
| Androgen response | 1.821 | 0.002 |
| Early estrogen response | 1.800 | 0.003 |
| Protein secretion | 1.664 | 0.009 |
| IFNγ response | 1.591 | 0.014 |
| Heme metabolism | 1.564 | 0.016 |
| KRAS signaling ↑ | 1.515 | 0.022 |
| Complement | 1.456 | 0.032 |
| p53 pathway | 1.451 | 0.031 |
| Late estrogen response | 1.438 | 0.032 |
| Apoptosis | 1.268 | 0.109 |
| mTOR-C1 signaling | 1.178 | 0.191 |
| Negatively enriched sets | ||
| E2F targets | -2.596 | < 0.001 |
| G2M check point | -2.375 | < 0.001 |
| Myogenesis | -1.800 | 0.005 |
| Epithelial to mesenchymal transition | -1.796 | 0.005 |
| MYC targets-V2 | -1.754 | 0.006 |
| Angiogenesis | -1.732 | 0.006 |
| KRAS signaling ↓ | -1.724 | 0.006 |
| MYC targets-V1 | -1.671 | 0.010 |
| Glycolysis | -1.606 | 0.017 |
| Apical surface | -1.581 | 0.020 |
| DNA repair | -1.580 | 0.018 |
| Hedgehog signaling | -1.571 | 0.018 |
| Spermatogenesis | -1.506 | 0.030 |
| Hypoxia | -1.497 | 0.030 |
| Wnt-β-catenin signaling | -1.424 | 0.054 |
| Mitotic spindle | -1.298 | 0.141 |
| Apical junction | -1.268 | 0.167 |
| Coagulation | -1.267 | 0.159 |
| Oxidative phosphorylation | -1.205 | 0.232 |
| Xenobiotic metabolism | -1.189 | 0.245 |
Dermal gene expression
Ninety-three gene transcripts were significantly upregulated, 263 downregulation and 15,206 had nonsignificant differential expression (DE). A number of immunoglobulin-related genes were amongst the most strongly DEGs [(all FDR < 0.001, log2FC > 2.927). Other genes with significant positive DE included SFRP4 (log2FC 3.277), CXCL9 (log2FC 2.709), COMP (log2FC 1.664), WNT16 (log2FC 0.742), CCL2 (log2FC 0.701), WNT2B (log2FC 0.576), NOTCH4 (log2FC 0.500)]; while MMP7 (log2FC −2.861) and NR4A1 (log2FC −0.630) were negatively expressed.
Dermal gene signatures; gene set enrichment analysis and PANTHER statistical enrichment analysis
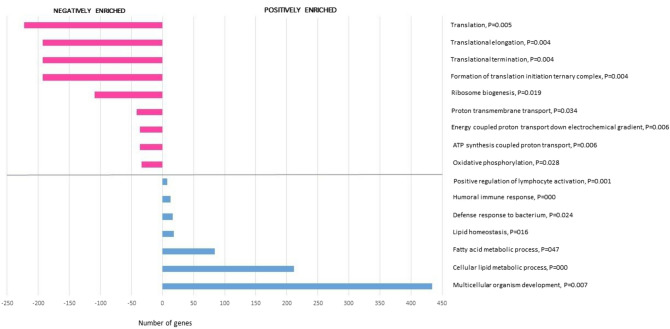
Seventeen GSEA Hallmark gene sets were significantly enriched; 9 with positive and 8 with negative enrichment (Fig. 2 and Table 5). Sixteen biological processes were statistically enriched on PANTHER statistical enrichment testing; 7 with positive and 9 with negative enrichment (Fig. 3).
Table 5.
Dermal RNA sequencing: Hallmark gene sets with significant positive or negative enrichment on GSEA, listed by NES
| Hallmark gene set | NES | FDR |
|---|---|---|
| Positively enriched sets | ||
| Bile acid metabolism | 1.617 | 0.095 |
| Adipogenesis | 1.699 | 0.098 |
| Epithelial to mesenchymal transition | 1.536 | 0.125 |
| Xenobiotic metabolism | 1.464 | 0.131 |
| Cholesterol metabolism | 1.389 | 0.136 |
| IFNγ response | 1.402 | 0.145 |
| Angiogenesis | 1.422 | 0.147 |
| IFNα response | 1.465 | 0.162 |
| Peroxisome | 1.292 | 0.227 |
| Negatively enriched sets | ||
| Androgen response | -1.760 | 0.052 |
| Oxidative phosphorylation | -1.675 | 0.071 |
| Early estrogen response | -1.539 | 0.071 |
| Protein secretion | -1.549 | 0.075 |
| MYC targets, V1 | -1.571 | 0.076 |
| KRAS signaling (down) | -1.574 | 0.094 |
| G2M checkpoint | -1.592 | 0.108 |
| Late estrogen response | -1.468 | 0.113 |
Fig. 3.
PANTHER Gene Ontology biological processes with significant positive and negative enrichment according to PANTHER enrichment test (Bonferroni correction, adjusted P-values listed next to biological process name)
Two distinct gene expression clusters were evident from analyses; inflammatory [GSEA: IFNα response (NES = 1.465, FDR = 0.162) and IFNγ response (NES = 1.402, FDR = 0.145), and PANTHER: Humoral immune response (P < 0.001) and Positive regulation of lymphocyte reactivation (P = 0.001) see Table 6], and; profibrotic, morphogenic signatures [GSEA: Epithelial to mesenchymal transition (NES = 1.536, FDR = 0.125) and Angiogenesis (NES = 1.422, FDR = 0.147), as well as nonsignificant positive enrichment of Hedgehog signalling (NES = 1.217, FDR = 0.291), Notch signalling (NES = 0.981, FDR = 0.655) and Wnt signalling (NES = 0.453, FDR = 0.999), and PANTHER: Multicellular organism development (P = 0.007); 434 contributory genes including WNT (WNT16, WNT10B, WNT2B), hedgehog (HHAT, HHATL), disheveled (DVL1, DVL2, DVL3) and frizzled (SMO), HOX (HOXA1a HOXA3, HOXA4, HOXA5, HOXA6, HOXA7, HOXA13, HOXB3, HOXB4, HOXB5, HOXB6, HOXB7, HOXC4, HOXC6, HOXC13) and PAX (PAX3, PAX6, PAX8)] (Fig. 4).
Table 6.
Dermal RNA sequencing: transcripts contributing to the three key selected positively enriched PANTHER GO-Slim Biological Processes (multicellular organism development, humoral immune response and positive regulation of lymphocyte activation) with significant upregulation
| Gene symbol | Description | FDR | Log2FC | Log2CPM |
|---|---|---|---|---|
| IGHG2 | Immunoglobulin heavy constant gamma 2 (G2m marker) | < 0.001 | 5.508 | 4.426 |
| IGHG1 | Immunoglobulin heavy constant gamma 1 (G1m marker) | < 0.001 | 5.162 | 7.118 |
| IGLC2 | Immunoglobulin lambda constant 2 | < 0.001 | 4.302 | 4.821 |
| IGHG4 | Immunoglobulin heavy constant gamma 4 (G4m marker) | 0.037 | 4.112 | 2.760 |
| IGHM | Immunoglobulin heavy constant mu | < 0.001 | 4.027 | 5.798 |
| IGHA1 | Immunoglobulin heavy constant alpha 1 | < 0.001 | 3.794 | 6.702 |
| IGLC3 | Immunoglobulin lambda constant 3 (Kern-Oz marker) | < 0.001 | 3.215 | 4.507 |
| IGHA2 | Immunoglobulin heavy constant alpha 2 (A2m marker) | < 0.001 | 2.927 | 4.098 |
| CXCL9 | C-X-C motif chemokine ligand 9 | < 0.001 | 2.709 | 3.880 |
| SULF1 | Sulfatase 1 | < 0.001 | 0.976 | 5.124 |
| WNT10B | Wnt family member 10B | 0.024 | 0.895 | 2.714 |
| WNT16 | Wnt family member 16 | 0.001 | 0.742 | 5.145 |
| COL14A1 | Collagen type XIV alpha 1 chain | 0.003 | 0.723 | 7.332 |
| TENM4 | Teneurin transmembrane protein 4 | 0.032 | 0.668 | 5.754 |
| JCAD | Junctional cadherin 5 associated | 0.028 | 0.655 | 6.112 |
| NREP | Neuronal regeneration related protein | 0.017 | 0.613 | 5.547 |
| WNT2B | Wnt family member 2B | 0.048 | 0.576 | 5.703 |
| SULF2 | Sulfatase 2 | 0.006 | 0.546 | 7.069 |
Fig. 4.
Interactions between leading edge genes within inflammatory gene sets IFN-signaling (α and γ), and developmental related gene sets of epithelial to mesenchymal transition, Angiogenesis and Hedgehog signaling, demonstrating clustering and inter-pathway interactions. Default STRING criteria used: nodes linked by evidence, with medium confidence level of 0.4
Many HOX, PAX, SOX and CBX genes were impacted across all three epidermal/dermal and genomic/transcriptomic datasets (Fig. 5).
Fig. 5.
STRING network diagram demonstrating multiple strong and overlapping interactions between PAX, HOX, SOX and CBX genes with protein or non-protein coding epidermal SNVs on WGS and/or differential epidermal or dermal expression on RNA-seq. Nodes linked by evidence with medium confidence level of 0.4 (default STRING criteria)
Thirty-two members of the ADAM, ADAMTS and ADAMTSL super-family were nonsignificantly DE in the dermis (13 downregulated and 19 upregulated) and 12 in the epidermis (6 upregulated and 6 downregulated). Overall, 50 ADAM/ADAMTS-family genes were affected across all three datasets, including the potentially highly pathogenic (according to criteria described in Fig. 1) nonsynonymous SNVs in ADAMTS16 and ADAMSTL1 (Fig. 6).
Fig. 6.
STRING network diagram of all ADAM, ADAMTS and ADAMTSL proteases with epidermal SNVs and/or epidermal and/or dermal differential RNA expression. Nodes linked by evidence, with medium confidence level of 0.4 (default STRING criteria). Further genes with strong links to the ADAM, ADAMTS and/or ADAMTSL proteins were also included (via STRING extended analysis); two of which were the ‘delta like canonical notch ligands’ (1 and 4); linking the ADAM, ADAMTS and ADAMTSL proteins, to notch signalin
Candidate genes and pathways based on epidermal genomic and epidermal and dermal transcriptomic profiles
Based on the WGS and RNA-seq results, a number of gene candidates were selected; some for further study. Selected epidermal candidate genes included ADAMTS16, ADAMTSL1 and the inflammatory and profibrotic TGF-β1 and JUNB. Selected dermal candidates included members of some developmental and morphogenic signaling pathways; SFRP4, SIX1, WNT2 and NOTCH4. Key characteristics of these genes and justification for their selection as candidates are detailed in Table 7.
Table 7.
Descriptive and statistical characteristics of selected gene candidates in epidermal and dermal tissue
| Gene symbol | Description | FDR | Log2FC | Log2CPM | Notes/data related Justification |
|---|---|---|---|---|---|
| Epidermal candidates | |||||
| ADAMTS16 | ADAM Metallopeptidase With Thrombospondin Type 1 Motif 16 | N/A | N/A | N/A |
WGS data: Novel variant Denoted deleterious by PolyPhen2,PROVENA and SIFT scores. CADD score 33 Only variants graded as High and subcategorised as Level 1 for disease relevance and pathogenicity Known links to fibrosis |
| ADAMTSL1 | ADAMTS Like 1 | N/A | N/A | N/A |
WGS data only: Novel variant Denoted deleterious by PolyPhen2,PROVENA and SIFT scores. CADD score 30 Only variants graded as High and subcategorised as Level 1 for disease relevance and pathogenicity Known links to fibrosis |
| LAMA4 | Laminin subunit alpha 4 | 0.026 | − 1.26 | 2.21 |
RNA-seq data: Significant FDR, log2FC < -1 Known links to fibrosis in other organs Plausible involvement in epidermal-dermal interactions in pathogenic mechanisms |
| IFI27 | Interferon Alpha Inducible Protein 27 | 0.952 | 1.565 | 5.721 |
Only epidermal transcript with log2FC > 1.5 Epidermal GSEA, Hallmark gene set leading edge gene: IFNα signaling (NES = 1.924, FDR = 0.0011) IFNγ signaling (NES = 1.591, FDR = 0.014) Plausible epidermal early ‘damage’ signal, with links to downregulation of NR4A1 |
| TGF-β1 | Transforming Growth Factor Beta 1 | 0.990 | -0.036 | 5.362 |
Key initiator and mediator of fibrosis Epidermal expression never specifically investigated in morphoea Overall signaling (TGF-β signaling Hallmark set) strongly positively enriched via GSEA analysis (NES = 2.006, FDR = 0.001) |
| JUNB | JunB Proto-Oncogene, AP-1 Transcription Factor Subunit | 0.952 | 0.424 | 7.939 |
Relatively high log2CPM of 7.939 Epidermal GSEA, Hallmark gene set leading edge gene in TGF-β signaling Hallmark set (NES = 2.006, FDR = 0.001) |
| PAX3 | Paired box gene 3 | N/A | N/A | N/A |
Epidermal WGS: nonsynonymous protein coding deleterious SNV Links to epidermal upregulation of PAX8 as well as many other PAX, HOX, SOX and CBX genes in both epidermal and dermal datasets; many with links to fibrosis and SSc |
| Dermal candidates | |||||
| SFRP4 | Secreted Frizzled Related Protein 4 | < 0.001 | 3.277 | 5.582 |
Frizzled related protein with significant differential expression and log2FC > 3 Dermal GSEA, Hallmark gene set leading edge gene: Epithelial to mesenchymal transition (NES = 1.536, FDR = 0.125), highest ranked leading edge gene |
| SIX1 | SIX Homeobox 1 | 0.641 | 2.333 | 2.529 | Homeobox gene with the highest log2FC |
| WNT2 | Wnt Family Member 2 | 0.061 | 1.793 | 2.283 |
Only Wnt signaling with log2FC > 1.5 Differential expression approaching significance Dermal GSEA, Hallmark gene set leading edge gene: Notch signaling, top 20 positively enriched sets (NES = 0.980, FDR = 0.655), highest ranked leading edge gene PANTHER statistical enrichment test: Present within the significantly enriched Multicellular organism development gene set (PANTHER GO-Slim Biological Process), P = 0.007 |
| NOTCH4 | Notch Receptor 4 | 0.008 | 0.500 | 5.631 |
Only significantly differentially expressed NOTCH gene Relatively high log2CPM |
| NR4A1 | Nuclear Receptor Subfamily 4 Group A Member 1 | 0.003 | −0.63 | 4.81 |
Significant dermal downregulation Downregulated by IFI27 (see above) Endogenous regulator of TGF-β1 signaling and known involvement in fibrotic processes |
| CXCL9 | C-X-C Motif Chemokine Ligand 9 | < 0.001 | 2.71 | 3.88 |
Inflammatory IFN response related gene with significant and strong differential expression Dermal (and epidermal) GSEA, Hallmark gene set leading edge gene: Contribution to the leading edge gene profile for IFNγ signaling in both the dermis and epidermis Suggested as a biomarker in morphoea |
| CCL2 | C-C Motif Chemokine Ligand 2 | 0.034 | 0.7 | 4.34 |
Inflammatory IFN response related gene with significant differential expression Dermal (and epidermal) GSEA, Hallmark gene set leading edge gene: Contribution to the leading edge gene profile for IFNγ signaling in both the dermis and epidermis Over-expressed amongst morphoea patients included in the Milano et al. ‘intrinsic gene subset’ scleroderma study and has been isolated to dermal macrophages in morphoea |
RT-qPCR and immunohistochemistry validation of selected epidermal and dermal gene candidates
Two key candidate genes were validated by RT-pPCR in this study; TGF-β1 and JUNB. These were from the strongly over-expressed and highly disease-relevant TGF-β signaling gene set. TGF-β1 is the recognised orchestrator of fibrosis and the role of its epidermal production and expression have not been specifically investigated in morphoea. JUNB is also a key player in TGF-β signaling and hence with its relatively high log2CPM, JUNB was selected as the second validation candidate, keeping both genes for qPCR from the TGF-β signaling gene set (NES = 2.006, FDR = 0.001).
Expression of TGF-β1 and JUNB was higher in morphoea affected epidermis compared to the contralateral site-matched unaffected epidermis in all samples, but this trend was not significant (TGF-β1; P = 0.476, JUNB; P = 0.105, Fig. 7).
Fig. 7.
RT-qPCR validation for key epidermal upregulated TGF-β signaling genes, mean expression levels as normalised copy number; A TGF-β1, B JUNB
WNT2 was selected for validation via IHC on formalin-fixed, wax-embedded paraffin whole skin sections. WNT2 was highlighted by dermal transcriptomic profiling, subsequent pathway analysis and is a member of the developmental morphogenic pathways which are of particular relevance to the anatomical patterning in morphoea and its pathogenesis. Of note, WNT2 was also highlighted by epidermal RNA-seq.
In the dermis, WNT2 was the only Wnt signaling gene with log2FC > 1.5 (log2FC = 1.79), its FDR approached significance (FDR = 0.061), it was a leading edge gene (highest ranked) within the positively enriched Notch signaling Hallmark gene set within dermal GSEA data and was also present within the significantly enriched Multicellular organism development gene set (PANTHER GO-Slim Biological Process; P = 0.007).
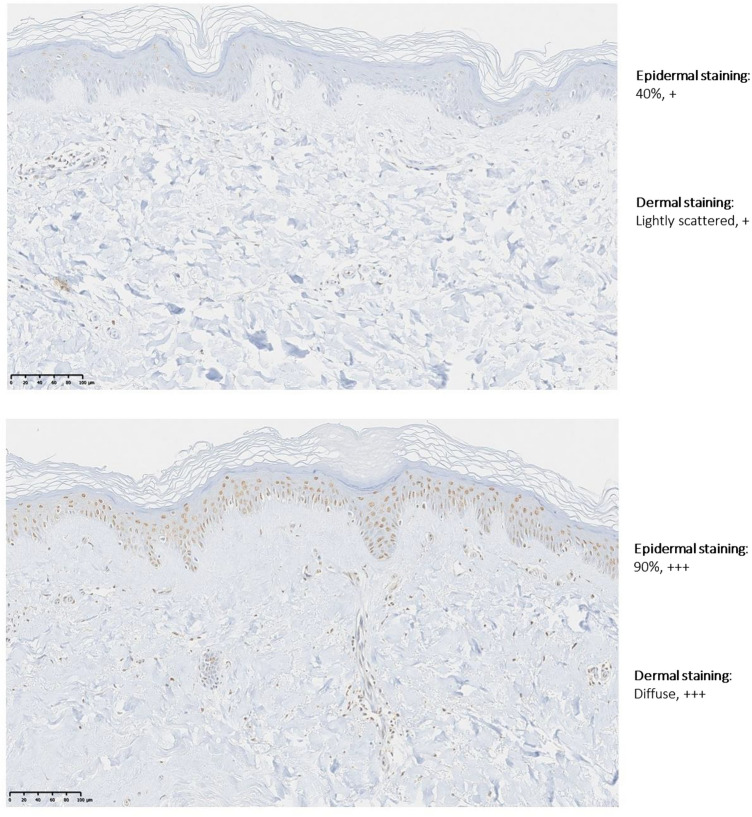
WNT2 staining demonstrated discernible staining differences between morphpea-affected and unaffected control skin in both epidermis (4 of 5) and dermis (3 of 5) (Fig. 8).
Fig. 8.
High power images of immunohistochemical staining with WNT2 antibody; unaffected control skin (above) and morphoea affected contralateral site-matched skin (below); study participant 15
Discussion
In this study, WGS did not identify a single common somatic mutation occurring in all four epidermal samples taken from LM-affected skin, or a commonly affected gene across all study samples. To our knowledge, this is the first study to investigate the presence of primary genomic variation in morphoea skin. This critical finding provides robust evidence against primary genomic epidermal segmental mosaicism-related aetiology in adult-onset LM. There are several clinical complexities of LM supporting more multifaceted aetiopathogenesis. LM may not be truly Blaschkoid [8], morphoea is a dermal pathology, has vast clinical heterogeneity with complex patterning and morphology [4, 30] and is not congenital.
Accordingly, we identified 861 epidermal SNVs, including 119 protein-coding variants, many with medium to high disease relevance and potential pathogenicity, providing possible support for complex polygenic epidermal mosaicism in LM [31, 32].
The ADAM/ADAMTS-family genes were widely affected across all three datasets, including potentially highly pathogenic nonsynonymous SNVs in ADAMTS16 and ADAMSTL1, possibly pointing to their pathogenic role in morphoea. These proteins/proteases are ECM-regulators implicated in embryological morphogenesis, skin development, wound healing, fibrosis [33–36], rare primary fibrotic genetic disorders [37, 38], SSc and keiloidal morphoea [39, 40]. Using site-matched tissue-pair methodology, Badshah et.al. recently demonstrated upregulated ADAMTS8 in LM fibroblasts and whole-skin, hypothesising ADAMTS8’s role in tissue atrophy [41]. Whilst links between the ADAMTS/ADAMTSL’s and their precise functions in morphoea are unclear, their possible role in LM is further supported by our findings.
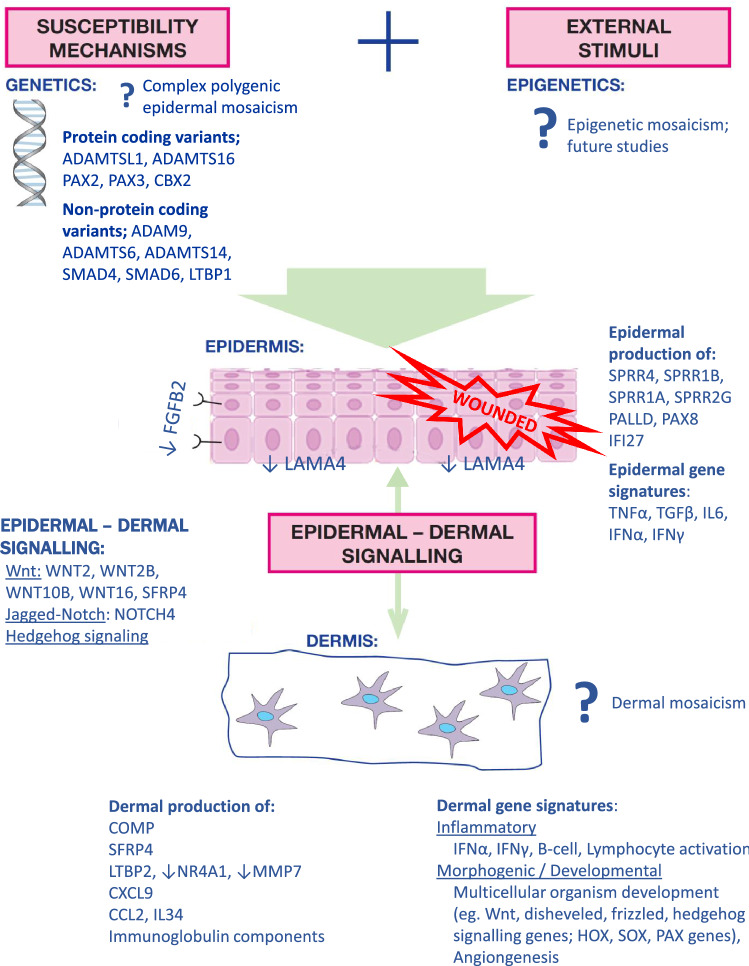
Corroborating the potential key role of the epidermis in morphoea pathogenesis, we demonstrated a structurally active, proliferative and differentiating epidermis, with significant overexpression of SPPRs, PALLD, WNT2, other cell cycle/cell division (such as p53 and KRAS signalling) and apoptosis-related gene pathways, along with significant down-regulation of checkpoint and DNA repair-related genes (such as G2M DNA checkpoint and E2F targets) (Fig. 9).
Fig. 9.
Multicomponent morphea etiopathogenesis; summary of key epidermal and dermal genes involved in morphea, as highlighted by NGS of paired epidermal and dermal tissue samples in this study
We also demonstrated an inflammatory and profibrotic epidermal gene signature, which corresponds to the early inflammatory and profibrotic disease phases previously mapped by blood cytokine profiles [42–46]. A Th1 response (IL-2, TNF-α and IL-6) seen in the first year, is followed by a Th17 response (IL-1, IL-17, IL-22 and TGF-β) and Th2 cytokines (IL-4 and IL-13) [47]. Accordingly, the three Hallmark gene sets with the strongest significant positive enrichment in this study were TNF-α signalling via NFkB, TGF-β signalling and IL-6/JAKSTAT3 signalling; all suggesting early active inflammatory and fibrotic phase disease (Fig. 9). This was despite study samples being from LM of at least 3-years duration and not all demonstrating an inflammatory clinical phenotype; supporting an ongoing disease-driving role of the epidermis.
Importantly, in recently published work evaluating transcriptomic whole-skin profiles of pediatric-onset morphoea, healthy controls, active and inactive disease were compared, and JAK/STATs were highlighted as the most prevalent DE pathway [48]. By separating the epidermis and dermis, we have highlighted that this signature may originate from the epidermis, promoting ongoing dermal disease activity. These findings provide further support for future studies to better elucidate precise pathogenic JAK/STAT-related mechanisms in morphoea and the use of therapeutic JAK-inhibitors in sclerotic skin disease [49].
Finally, the epidermal molecular picture was also that of a ‘wounded epidermis’, similar to the epidermal phenotype demonstrated in SSc [10, 50, 51]. TGF-β is a key orchestrator of wound healing responses, also propagating pathological fibrosis [52]. Isolating a strongly enriched TGF-β signature in morphoea epidermis is unique, significant, and could provide impetus for further study of local TGF-β inhibition in appropriate clinical scenarios of superficial disease (e.g. with pirfenidone) [53]. However, precisely whether these signals are originating in the epidermis, or due to secondary unchecked positive feedback from the dermis, remains unclear.
Relevantly, epidermal IFI27 was upregulated (nonsignificant, but with the dataset’s highest log2FC). It is known to induce IFNγ-related epidermal apoptosis. We saw significant upregulation of the epidermal Apoptosis gene set, and epidermal and dermal IFNα and IFNγ responses. IFN-signalling has been widely implicated in SSc and morphoea [11, 48, 54]. IFNγ-related chemokines and their receptors may stimulate fibroblasts, including in morphoea [46, 48, 55]. CXCL9 was significantly upregulated in morphoea dermis in our study, and it has previously been suggested as a disease biomarker [46, 55].
Importantly, IFI27 negatively regulates NR4A1 [54], which was significantly downregulated in the dermal dataset. In turn, NR4A1 is an endogenous TGF-β inhibitor [56]. Fibrotic diseases appear to utilise this NR4A1-dependent mechanism to enable persistent TGF-β signaling and deregulated fibrosis and NR4A1 agonists inhibit laboratory-induced fibrosis of the skin, lung, liver, and kidney in mice [56, 57].
Clues to another potential inciting epidermal ‘damage’ signal in morphoea lie in the significant downregulation of LAMA4. Laminins are extracellular matrix (ECM) glycoproteins involved in differentiation, cell adhesion, signaling, migration, and form a key non-collagen component of the dermo-epidermal junction (DEJ) [54]. Related DEJ disruption could plausibly enhance epidermal-dermal communication and/or act as an initiating ‘damage’ signal, inciting proinflammatory and profibrotic dermal responses. Correspondingly, LAMA4-deficiency has been linked to cardiac [58–60] and renal fibrosis [61].
Individual dermal-genes demonstrated far greater DGE compared to the epidermis, suggesting dermal factors are more disease-specific in morphoea; in keeping with its predominantly dermal pathology. Two distinct DGE clusters were identified; inflammatory and profibrotic. The inflammatory signature, with significant upregulation of Humoral immunity, Lymphocyte activation and IFN-response-related genes, validates and adds to the limited morphoea gene expression data currently available [11, 48, 62]. This corroborated over-expression of IFN-signalling has an immediate foreseeable opportunity for potential therapeutic exploitation via anifrolimab, FDA-approved for systemic lupus erythematosus. Interestingly, KRAS-signalling has been identified as a potential biomarker for disease activity [48]. We demonstrated significant downregulation of inhibitory KRAS-signalling in the dermis and upregulated KRAS-signalling in the epidermis also. All our cases had disease activity as demonstrated by LoScAT-activity scores of greater than zero (progressive or stable disease activity) (Tables 1, 4 and 5).
In the profibrotic DGE cluster, upregulated genes involved in embryogenesis and oncogenesis was seen such as Wnt, Hedgehog, dishevelled, frizzled family, HOX and PAX. PAX and HOX genes were specifically highlighted by PANTHER pathway analysis of dermal RNA-seq data. These families of biologically and functionally related developmental genes were collectively impacted in all three data sets (epidermal WGS, epidermal RNA-seq and dermal RNA-seq). HOX genes are the key orchestrating genes involved in fibroblast PI [12, 13, 63–65]. Related location-specific gene signatures confer developmental patterning, position and help determine downstream differentiation of site-specific mesenchymal cells [13, 66]. The genetic origin of fibroblasts can also alter their crosstalk with overlying keratinocytes [67]. Several HOX genes have shown significant DE in affected SSc-skin compared to unaffected skin [68] and related SOX genes have also been implicated in fibrosis and SSc [23, 69]. Accordingly, one can deduce the feasible role HOX and related developmental and patterning genes could play in morphoea aetiopathogenesis and observed clinical patterning of non-linear subtypes. Indeed, their involvement in ‘dermal mosaicism’ has been suggested.
It is also suggested that via its regulation of dermal development, epidermal Wnt- signalling could account for the Blaschkoid distribution of dermal dermatoses, including Focal Dermal Hypoplasia [70]. Twelve Wnt-signalling genes contributed to the upregulation of the GO-Slim Biological Process of Multicellular organism development; WNT2B, WNT10B and WNT16 with significant DE. WNT2 was significantly upregulated in the epidermis, approached significance in the dermis (FDR = 0.061) and both these RNA-seq results were validated with IHC whole skin staining. Correspondingly, WNT2, WNT3A and β-catenin have previously demonstrated increased activity via IHC staining in both SSc and morphoea [71] and the role of Wnt-signalling in morphoea is established [20, 55, 71–75]. Dermal SFRP4 was also significantly upregulated and recent data demonstrated the upregulation of SFRP2 in morphoea dermal fibroblasts [55]. SFRPs are homologous to the Wnt-binding site on frizzled proteins and, therefore, modulate Wnt-signalling via direct interactions [54]. Interestingly, SFRP4 expression in the myocardium is associated with an apoptotic-related gene expression profile [54], feasibly associating its overexpression in morphoea to a disease-related damage signal.
Limitations of this study include its cross-sectional nature, small datasets and limited validation of transcriptomic data. It is also impossible to differentiate primary from secondary gene expression changes or to adjust for treatment effect.
In summary, despite the often assumed Blaschkoid distribution of LM, data from this study indicate the absence of a single epidermal developmental somatic mutation responsible for disease causation. Instead, this study’s molecular (genomic and transcriptomic) and tissue (epidermis and dermis) layered approach highlights possible polygenic epidermal mosaicism in initiating a complex multicomponent disease aetiopathogenesis. A wounded epidermal phenotype could, perhaps via Wnt-signalling, depletion of NR4A1 and other complex tissue layer crosstalk, contribute to the consequent inflammatory dermal fibrosis of morphoea, with its variable patterning possibly explained, at least in part, by the involvement of HOX, SOX, PAX and WNT developmental patterning genes (Fig. 9).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to sincerely thank Dr Chiara Bacchelli for her expert advice and collaboration with whole genome and RNA sequencing work and bioinformatics support. Dr Ioannis Papaioannou and Dr Markella Ponticos, for their tireless practical laboratory support. Korsa Khan and Francesca Launchbury for their assistance with histology slide preparation and immunohistochemical staining, as well as Drs Florence Deroide and Victoria Swale for their expert slide interpretation. Bahja Ahmed Abdi for logistical laboratory assistance. Dr Catherine Orteu for assistance in the early phases of this project.
Abbreviations
- BGI
Beijing Genomics Institute
- C
Control skin (denoting a skin sample taken from a site unaffected by morphoea; contralateral site-matched pair)
- CADD
Combined annotation-dependent depletion
- CCL
CC chemokine ligand
- CNS
Central nervous system
- COMP
Cartilage oligomeric matrix protein
- CPM
Counts per million
- CTGF
Connective tissue growth factor
- CXCL
Chemokine C-X-C (motif) ligand
- DE
Differentially expressed
- DGE
Differential gene expression
- DNA
Deoxyribonucleic acid
- ECM
Extracellular matrix/extracutaneous manifestations
- EMT
Epithelial to mesenchymal transition
- ES
Enrichment score
- ET-1
Endothelin 1
- ExAC
Exonerated aggregation consortium
- FC
Fold change
- FDR
False discovery rate
- FGF
Fibroblast growth factor
- Fli1
Friend leukaemia virus integration 1
- GSEA
Gene set enrichment analysis
- H&E
Haematoxylin and eosin
- IFN
Interferon
- IHC
Immunohistochemistry
- IL
Interleukin
- LoSCAT
Localised scleroderma cutaneous assessment tool
- LM
Linear morphoea
- LTBP
Latent transforming growth factor beta binding protein
- M
Morphoea-affected skin (denoting a sample from skin affected by morphoea)
- MAC
Morphoea in adults and children cohort
- MAF
Minor allele frequency
- mLoSDI
Modified localised scleroderma damage index
- mLoSSI
Modified localised scleroderma severity index
- MMF
Mycophenolate mofetil
- MMP
Matrix metalloproteinase
- mRSS
Modified Rodnan skin score
- MTX
Methotrexate
- NGS
Next-generation sequencing
- NES
Normalised enrichment score
- NFkB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- ng
Nanogram
- NLRP3
NACHT, LRR and PYD domains-containing protein 3
- PANTHER
Protein analysis through evolutionary relationships
- PCR
Polymerase chain reaction
- PDGF
Platelet-derived growth factor
- PI
Positional identity
- PPAR-γ
Peroxisome proliferator-activated receptor gamma
- PROVEAN
Protein variation effect analyser
- PUVA
Psoralen ultraviolet-A
- QC
Quality control
- qPCR
Quantitative polymerase chain reaction
- QoL
Quality of life
- RNA
Ribonucleic acid
- RNA seq
RNA sequencing
- RT-qPCR
Reverse transcriptase quantitative polymerase chain reaction
- SIFT
Sorting intolerant from tolerant
- SNP
Single nucleotide polymorphism
- SNV
Single nucleotide variant
- SSc
Systemic sclerosis
- TGF-β
Transforming growth factor beta
- TIMP
Tissue inhibitor of metalloproteinase
- TNF-α
Tumour necrosis factor alpha
- µL
Microlitre
- WES
Whole exome sequencing
- WGS
Whole genome sequencing
Author contributions
All authors contributed to the study’s conception and design. Material preparation, data collection and analysis were performed by AS, AG, GO, DK, CD and DA. The first draft of the manuscript was written by AS and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by funding from the following organisations; Rosetrees Trust (The Teresa Rosenbaum Golden Charitable Trust); Australasian College of Dermatologists (as part of the Scientific Research Fund); Royal Free Charity; Versus Arthritis; Dermatrust (Dermatitis and Allied Disease Research Trust); and Skin Health Institute of Victoria, Australia (as part of the Paul Eddington Scholarship). GOSgene is funded by the NIHR GOSH BRC. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Data availability
The data that support the findings of this study are available upon reasonable request to the corresponding author, after validation by co-authors.
Declarations
Conflict of interest
AMS: has received honoraria from UCB outside the submitted work. DK: nil. GWO: nil. AG: nil. DJA: nil. CPD: reports personal fees or research grants to his institution from GlaxoSmithKline, Galapagos, Boehringer Ingelheim, Roche, CSL Behring, Corbus, Horizon, and Arxx Therapeutics outside the submitted work.
Ethics approval
This study was approved by the National Research Ethics Service (London-Hampstead, MREC Reference 6398). Tissue specimens were obtained with written informed consent as part of an ongoing programme of research into the pathogenesis of scleroderma.
Informed consent
Written informed consent to participate in the study and publication was obtained from all participants.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Prinz JCKZ, Weisenseel P, Poto L, Battyani Z, Ruzzicka T. Borrelia-associated early-onset morphea: a particular type of scleroderma in childhood and adolescence with high tire antinuclear antibodies? Results of a cohort analysis and presentation of three cases. J Am Acad Dermatol. 2009;60(2):8. doi: 10.1016/j.jaad.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 2.Fett N, Werth VP. Update on morphea: part II. Outcome measures and treatment. J Am Acad Dermatol. 2011;64(2):231–242. doi: 10.1016/j.jaad.2010.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fett N, Werth VP. Update on morphea: part I. Epidemiology, clinical presentation, and pathogenesis. J Am Acad Dermatol. 2011;64(2):217–228. doi: 10.1016/j.jaad.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 4.Saracino AM, Denton CP, Orteu CH. The molecular pathogenesis of morphoea: from genetics to future treatment targets. Br J Dermatol. 2016;177(1):34–46. doi: 10.1111/bjd.15001. [DOI] [PubMed] [Google Scholar]
- 5.Weibel L, Harper JI. Linear morphoea follows Blaschko's lines. Br J Dermatol. 2008;159(1):175–181. doi: 10.1111/j.1365-2133.2008.08647.x. [DOI] [PubMed] [Google Scholar]
- 6.Danarti R, Bittar M, Happle R, König A. Linear atrophoderma of Moulin: postulation of mosaicism for a predisposing gene. J Am Acad Dermatol. 2003;49(3):492–498. doi: 10.1067/s0190-9622(03)00895-8. [DOI] [PubMed] [Google Scholar]
- 7.Mukhopadhyay AK. Linear scleroderma following Blaschko's lines. Indian J Dermatol Venereol Leprol. 2005;71(6):421–422. doi: 10.4103/0378-6323.18949. [DOI] [PubMed] [Google Scholar]
- 8.Shmuylovich L, Paller AS, Kiguradze T, Anderson K, Sibbald C, Tollefson M, et al. 385 Patterning of linear morphea on the face and neck: Blaschkoid or not? J Investig Dermatol. 2019;139(5):S66. [Google Scholar]
- 9.Soma Y, Kawakami T, Yamasaki E, Sasaki R, Mizoguchi M. Linear scleroderma along Blaschko's lines in a patient with systematized morphea. Acta Derm Venereol. 2003;83(5):362–364. doi: 10.1080/00015550310013088. [DOI] [PubMed] [Google Scholar]
- 10.Aden N, Shiwen X, Aden D, Black C, Nuttall A, Denton CP, et al. Proteomic analysis of scleroderma lesional skin reveals activated wound healing phenotype of epidermal cell layer. Rheumatology (Oxford) 2008;47(12):1754–1760. doi: 10.1093/rheumatology/ken370. [DOI] [PubMed] [Google Scholar]
- 11.Milano A, Pendergrass SA, Sargent JL, George LK, McCalmont TH, Connolly MK, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS ONE. 2008;3(7):e2696. doi: 10.1371/journal.pone.0002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinn JL, Wang JK, Liu H, Montgomery K, van de Rijn M, Chang HY. A systems biology approach to anatomic diversity of skin. J Invest Dermatol. 2008;128(4):776–782. doi: 10.1038/sj.jid.5700986. [DOI] [PubMed] [Google Scholar]
- 13.Chang HY, Chi JT, Dudoit S, Bondre C, van de Rijn M, Botstein D, et al. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci USA. 2002;99(20):12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacharyya S, Wei J, Varga J. Understanding fibrosis in systemic sclerosis: shifting paradigms, emerging opportunities. Nat Rev Rheumatol. 2012;8(1):42–54. doi: 10.1038/nrrheum.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denton CP, Ong VH. Targeted therapies for systemic sclerosis. Nat Rev Rheumatol. 2013;9(8):451–464. doi: 10.1038/nrrheum.2013.46. [DOI] [PubMed] [Google Scholar]
- 16.Dees C, Zerr P, Tomcik M, Beyer C, Horn A, Akhmetshina A, et al. Inhibition of Notch signaling prevents experimental fibrosis and induces regression of established fibrosis. Arthritis Rheum. 2011;63(5):1396–1404. doi: 10.1002/art.30254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horn A, Kireva T, Palumbo-Zerr K, Dees C, Tomcik M, Cordazzo C, et al. Inhibition of hedgehog signalling prevents experimental fibrosis and induces regression of established fibrosis. Ann Rheum Dis. 2012;71(5):785–789. doi: 10.1136/annrheumdis-2011-200883. [DOI] [PubMed] [Google Scholar]
- 18.Gilbane AJ, Denton CP, Holmes AM. Scleroderma pathogenesis: a pivotal role for fibroblasts as effector cells. Arthritis Res Ther. 2013;15(3):215. doi: 10.1186/ar4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kavian N, Servettaz A, Mongaret C, Wang A, Nicco C, Chereau C, et al. Targeting ADAM-17/notch signaling abrogates the development of systemic sclerosis in a murine model. Arthritis Rheum. 2010;62(11):3477–3487. doi: 10.1002/art.27626. [DOI] [PubMed] [Google Scholar]
- 20.Wei J, Melichian D, Komura K, Hinchcliff M, Lam AP, Lafyatis R, et al. Canonical Wnt signaling induces skin fibrosis and subcutaneous lipoatrophy: a novel mouse model for scleroderma? Arthritis Rheum. 2011;63(6):1707–1717. doi: 10.1002/art.30312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayle J, Fitch J, Jacobsen K, Kumar R, Lafyatis R, Lemaire R. Increased expression of Wnt2 and SFRP4 in Tsk mouse skin: role of Wnt signaling in altered dermal fibrillin deposition and systemic sclerosis. J Invest Dermatol. 2008;128(4):871–881. doi: 10.1038/sj.jid.5701101. [DOI] [PubMed] [Google Scholar]
- 22.Lam AP, Flozak AS, Russell S, Wei J, Jain M, Mutlu GM, et al. Nuclear beta-catenin is increased in systemic sclerosis pulmonary fibrosis and promotes lung fibroblast migration and proliferation. Am J Respir Cell Mol Biol. 2011;45(5):915–922. doi: 10.1165/rcmb.2010-0113OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frost J, Estivill X, Ramsay M, Tikly M. Dysregulation of the Wnt signaling pathway in South African patients with diffuse systemic sclerosis. Clin Rheumatol. 2019;38(3):933–938. doi: 10.1007/s10067-018-4298-5. [DOI] [PubMed] [Google Scholar]
- 24.Saracino AM, George C, Nihtyanova SI, Denton CP. Comparing paediatric- and adult-onset linear morphoea in a large tertiary-referral scleroderma centre. J Scleroderma Relat Disord. 2021;6(1):102–108. doi: 10.1177/2397198320925684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clemmensen ATM, Clemmensen O, Tan Q, Kruse TA, Petersen TK, Andersen F, Andersen KE. Extraction of high-quality epidermal RNA after ammonium thiocyanate-induced dermo-epidermal separation of 4 mm human skin biopsies. Exp Dermatol. 2009;18(11):979–984. doi: 10.1111/j.1600-0625.2009.00921.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81(6):1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.PANTHER (2018). http://www.pantherdb.org/about.jsp. Gene Ontology Unifying Biology. Version 14.1
- 30.Saracino AM, Orteu CH. Morphoea management: current approach and future perspectives. In: Zaheri S, Ali I, editors. Recent advances in dermatology. London: Jaypee UK; 2018. [Google Scholar]
- 31.Happle R. The categories of cutaneous mosaicism: a proposed classification. Am J Med Genet A. 2016;170(2):452–459. doi: 10.1002/ajmg.a.37439. [DOI] [PubMed] [Google Scholar]
- 32.Happle R. The molecular revolution in cutaneous biology: era of mosaicism. J Invest Dermatol. 2017;137(5):e73–e77. doi: 10.1016/j.jid.2016.03.045. [DOI] [PubMed] [Google Scholar]
- 33.Apte SS, Parks WC. metalloproteinases: a parade of functions in matrix biology and an outlook for the future. Matrix Biol J Int Soc Matrix Bio. 2015;44–46:1–6. doi: 10.1016/j.matbio.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Kawaguchi M, Hearing VJ. The roles of ADAMs family proteinases in skin diseases. Enzyme Res. 2011;2011:1–9. doi: 10.4061/2011/482498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) family. Genome Biol. 2015;16:113. doi: 10.1186/s13059-015-0676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keating DT, Sadlier DM, Patricelli A, Smith SM, Walls D, Egan JJ, et al. Microarray identifies ADAM family members as key responders to TGF-beta1 in alveolar epithelial cells. Respir Res. 2006;7:114. doi: 10.1186/1465-9921-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faivre L, Gorlin RJ, Wirtz MK, Godfrey M, Dagoneau N, Samples JR, et al. In frame fibrillin-1 gene deletion in autosomal dominant Weill–Marchesani syndrome. J Med Genet. 2003;40(1):34–36. doi: 10.1136/jmg.40.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hubmacher D, Apte S. Genetic and functional linkage between ADAMTS superfamily proteins and fibrillin-1: a novel mechanism influencing microfibril assembly and function. Cell Mol Life Sci. 2011;68(19):3137–3148. doi: 10.1007/s00018-011-0780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terao M, Murota H, Kitaba S, Katayama I. Tumor necrosis factor-alpha processing inhibitor-1 inhibits skin fibrosis in a bleomycin-induced murine model of scleroderma. Exp Dermatol. 2010;19(1):38–43. doi: 10.1111/j.1600-0625.2009.00973.x. [DOI] [PubMed] [Google Scholar]
- 40.Bohgaki T, Amasaki Y, Nishimura N, Bohgaki M, Yamashita Y, Nishio M, et al. Up regulated expression of tumour necrosis factor alpha converting enzyme in peripheral monocytes of patients with early systemic sclerosis. Ann Rheum Dis. 2005;64(8):1165–1173. doi: 10.1136/ard.2004.030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Badshah II, Brown S, Weibel L, Rose A, Way B, Sebire N, et al. Differential expression of secreted factors SOSTDC1 and ADAMTS8 cause profibrotic changes in linear morphoea fibroblasts. Br J Dermatol. 2019;180(5):1135–1149. doi: 10.1111/bjd.17352. [DOI] [PubMed] [Google Scholar]
- 42.Ihn H, Sato S, Fujimoto M, Kikuchi K, Takehara K. Demonstration of interleukin-2, interleukin-4 and interleukin-6 in sera from patients with localized scleroderma. Arch Dermatol Res. 1995;287(2):193–197. doi: 10.1007/BF01262331. [DOI] [PubMed] [Google Scholar]
- 43.Hasegawa M, Sato S, Nagaoka T, Fujimoto M, Takehara K. Serum levels of tumor necrosis factor and interleukin-13 are elevated in patients with localized scleroderma. Dermatology (Basel, Switzerland) 2003;207(2):141–147. doi: 10.1159/000071783. [DOI] [PubMed] [Google Scholar]
- 44.Arkachaisri T, Fertig N, Pino S, Medsger TA., Jr Serum autoantibodies and their clinical associations in patients with childhood- and adult-onset linear scleroderma. A single-center study. J Rheumatol. 2008;35(12):2439–2444. doi: 10.3899/jrheum.080098. [DOI] [PubMed] [Google Scholar]
- 45.Oriente A, Fedarko NS, Pacocha SE, Huang SK, Lichtenstein LM, Essayan DM. Interleukin-13 modulates collagen homeostasis in human skin and keloid fibroblasts. J Pharmacol Exp Ther. 2000;292(3):988–994. [PubMed] [Google Scholar]
- 46.O'Brien JC, Rainwater YB, Malviya N, Cyrus N, Auer-Hackenberg L, Hynan LS, et al. Transcriptional and cytokine profiles identify CXCL9 as a biomarker of disease activity in morphea. J Invest Dermatol. 2017;137(8):1663–1670. doi: 10.1016/j.jid.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurzinski K, Torok KS. Cytokine profiles in localized scleroderma and relationship to clinical features. Cytokine. 2011;55(2):157–164. doi: 10.1016/j.cyto.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mirizio ELC, Yan Q, Waltermire J, Mandel R, Schollaert KL, Konnikova L, Wang X, Chen W, Torok KS. Genetic signatures from RNA sequencing of pediatric localized scleroderma skin. Front Pediatr. 2021 doi: 10.3389/fped.2021.669116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGaugh S, Kallis P, De Benedetto A, Thomas RM. Janus kinase inhibitors for treatment of morphea and systemic sclerosis: a literature review. Dermatol Ther. 2022;35(6):e15437. doi: 10.1111/dth.15437. [DOI] [PubMed] [Google Scholar]
- 50.Arwert EN, Hoste E, Watt FM. Epithelial stem cells, wound healing and cancer. Nat Rev Cancer. 2012;12(3):170–180. doi: 10.1038/nrc3217. [DOI] [PubMed] [Google Scholar]
- 51.Aden N, Nuttall A, Shiwen X, de Winter P, Leask A, Black CM, et al. Epithelial cells promote fibroblast activation via IL-1alpha in systemic sclerosis. J Invest Dermatol. 2010;130(9):2191–2200. doi: 10.1038/jid.2010.120. [DOI] [PubMed] [Google Scholar]
- 52.Bhattacharyya S, Sargent JL, Du P, Lin S, Tourtellotte WG, Takehara K, et al. Egr-1 induces a profibrotic injury/repair gene program associated with systemic sclerosis. PLoS ONE. 2011;6(9):e23082. doi: 10.1371/journal.pone.0023082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodríguez-Castellanos M, Tlacuilo-Parra A, Sánchez-Enríquez S, Vélez-Gómez E, Guevara-Gutiérrez E. Pirfenidone gel in patients with localized scleroderma: a phase II study. Arthritis Res Ther. 2015;16(6):510. doi: 10.1186/s13075-014-0510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.GeneCardsSuite (2019) GeneCards human gene database. Weizman Institute of Science, Life Map Sciences
- 55.Mirizio ETT, Sun T, Schollaert-Fitch K, Chen W, Lafyatis R, Torok K (2019) Defining the transcriptional profile of the skin in pediatric localized scleroderma (LS). In: 16th International workshop on scleroderma research, abstract book. 2019 (abstract number 52):13
- 56.Palumbo-Zerr K, Zerr P, Distler A, Fliehr J, Mancuso R, Huang J, et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-[beta] signaling and fibrosis. Nat Med. 2015;21(2):150–158. doi: 10.1038/nm.3777. [DOI] [PubMed] [Google Scholar]
- 57.Domagalski K, Pawlowska M, Kozielewicz D, Dybowska D, Tretyn A, Halota W. The impact of IL28B genotype and liver fibrosis on the hepatic expression of IP10, IFI27, ISG15, and MX1 and their association with treatment outcomes in patients with chronic hepatitis C. PLoS ONE. 2015;10(6):e0130899. doi: 10.1371/journal.pone.0130899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Hoshijima M, Lam J, Zhou Z, Jokiel A, Dalton N, et al. Cardiomyopathy associated with microcirculation dysfunction in laminin alpha 4 chain-deficient mice. J Biol Chem. 2006;281(1):213–220. doi: 10.1074/jbc.M505061200. [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Pavia P, Cobo-Marcos M, Guzzo-Merello G, Gomez-Bueno M, Bornstein B, Lara-Pezzi E, et al. Genetics in dilated cardiomyopathy. Biomark Med. 2013;7(4):517–533. doi: 10.2217/bmm.13.77. [DOI] [PubMed] [Google Scholar]
- 60.Garcia C. Insights from human genetic studies of lung and organ fibrosis. J Clin Investig. 2018;128(1):36–44. doi: 10.1172/JCI93556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abrass C, Hansen K, Patton B. Laminin alpha 4-null mutant mice develop chronic kidney disease with persistent overexpression of platelet-derived growth factor. Am J Pathol. 2010;176(2):839–849. doi: 10.2353/ajpath.2010.090570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schutt C, Mirizio E, Salgado C, Reyes-Mugica M, Wang X, Chen W, et al. Transcriptomic evaluation of juvenile localized scleroderma skin with histologic and clinical correlation. Arthritis Rheumatol. 2021;73(10):1921–1930. doi: 10.1002/art.41758. [DOI] [PubMed] [Google Scholar]
- 63.Rinn JL, Wang JK, Allen N, Brugmann SA, Mikels AJ, Liu H, et al. A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev. 2008;22(3):303–307. doi: 10.1101/gad.1610508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006;2(7):e119. doi: 10.1371/journal.pgen.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2(2):E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Picchi J, Trombi L, Spugnesi L, Barachini S, Maroni G, Brodano GB, et al. HOX and TALE signatures specify human stromal stem cell populations from different sources. J Cell Physiol. 2013;228(4):879–889. doi: 10.1002/jcp.24239. [DOI] [PubMed] [Google Scholar]
- 67.Hausmann C, Zoschke C, Wolff C, Darvin M, Sochorová M, Kováčik A, et al. Fibroblast origin shapes tissue homeostasis, epidermal differentiation, and drug uptake. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-39770-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Makowska Z, Buttgereit A, Babaei S, Limaye N, Galant C, Houssiau F, et al. FRI0424 Comparative analysis of clinically affected and unaffected skin biopsies from scleroderma patients based on rna-sequencing. Ann Rheum Dis. 2018;77(s2):742. [Google Scholar]
- 69.Makino T, Jinnin M. Genetic and epigenetic abnormalities in systemic sclerosis. J Dermatol. 2016;43(1):10–18. doi: 10.1111/1346-8138.13221. [DOI] [PubMed] [Google Scholar]
- 70.Paller AS. Wnt signaling in focal dermal hypoplasia. Nat Genet. 2007;39(7):820–821. doi: 10.1038/ng0707-820. [DOI] [PubMed] [Google Scholar]
- 71.Liu J, Liu T. [Role of Wnt 2, Wnt 3a and beta-catenin in skin lesions of patients with scleroderma] Nan fang yi ke da xue xue bao J South Med Univ. 2012;32(12):1781–1786. [PubMed] [Google Scholar]
- 72.Carthy JM, Garmaroudi FS, Luo Z, McManus BM. Wnt3a induces myofibroblast differentiation by upregulating TGF-beta signaling through SMAD2 in a beta-catenin-dependent manner. PLoS ONE. 2011;6(5):e19809. doi: 10.1371/journal.pone.0019809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang W, Wei K, Jacobs SS, Upadhyay D, Weill D, Rosen GD. SPARC suppresses apoptosis of idiopathic pulmonary fibrosis fibroblasts through constitutive activation of beta-catenin. J Biol Chem. 2010;285(11):8196–8206. doi: 10.1074/jbc.M109.025684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thorne CA, Hanson AJ, Schneider J, Tahinci E, Orton D, Cselenyi CS, et al. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1alpha. Nat Chem Biol. 2010;6(11):829–836. doi: 10.1038/nchembio.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hamburg-Shields E, DiNuoscio GJ, Mullin NK, Lafyatis R, Atit RP. Sustained beta-catenin activity in dermal fibroblasts promotes fibrosis by up-regulating expression of extracellular matrix protein-coding genes. J Pathol. 2015;235(5):686–697. doi: 10.1002/path.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available upon reasonable request to the corresponding author, after validation by co-authors.