Abstract
Background
Doxorubicin (DOX) is among the most widely employed antitumor agents, although its clinical applications have been largely hindered by severe cardiotoxicity. Earlier studies described an essential role of mitochondrial injury in the pathogenesis of DOX cardiomyopathy. PHB2 (Prohibitin 2) is perceived as an essential regulator for mitochondrial dynamics and oxidative phosphorylation (OXPHOS) although its involvement in DOX cardiomyopathy remains elusive.
Methods
To decipher the possible role of PHB2 in DOX cardiomyopathy, tamoxifen-induced cardiac-specific PHB2 conditional knockout mice were generated and subjected to DOX challenge. Cardiac function and mitochondrial profiles were examined. Screening of downstream mediators of PHB2 was performed using proteomic profiling and bioinformatic analysis, and was further verified using co-immunoprecipitation and pulldown assays.
Results
Our data revealed significantly downregulated PHB2 expression in DOX-challenged mouse hearts. PHB2CKO mice were more susceptible to DOX cardiotoxicity compared with PHB2flox/flox mice, as evidenced by more pronounced cardiac atrophy, interstitial fibrosis and decrease in left ventricular ejection fraction and fractional shortening. Mechanistically, PHB2 deficiency resulted in the impairment of mitochondrial bioenergetics and oxidative phosphorylation in DOX cardiotoxicity. Proteomic profiling and interactome analyses revealed that PHB2 interacted with NDUFV2 (NADH-ubiquinone oxidoreductase core subunit V2), a key subunit of mitochondrial respiratory Complex I to mediate regulatory property of PHB2 on mitochondrial metabolism. PHB2 governed the expression of NDUFV2 by promoting its stabilization, while PHB2 deficiency significantly downregulated NDUFV2 in DOX-challenged hearts. Cardiac overexpression of PHB2 alleviated mitochondrial defects in DOX cardiomyopathy both in vivo and in vitro.
Conclusions
Our study defined a novel role for PHB2 in mitochondrial dynamics and energetic metabolism through interacting with NDUFV2 in DOX-challenged hearts. Forced overexpression of PHB2 may be considered a promising therapeutic approach for patients with DOX cardiomyopathy.
Keywords: Doxorubicin cardiotoxicity, PHB2, NDUFV2, Mitochondrial complex I
1. Introduction
Chemotherapy remains a fundamental and indispensable option for various forms of cancers, despite the advances in targeted therapy and immune checkpoint inhibitors. Doxorubicin (DOX), one of the widely employed anthracyclines, is among the first-line chemotherapeutic agents for multiple hematologic malignancies and solid tumors [1]. Nonetheless, many patients with prolonged usage of DOX develop progressive and irreversible cardiotoxicity with a hallmark clinical presentation of left ventricular systolic dysfunction, with ultimate progression into heart failure [[2], [3], [4], [5]]. Over the past decades, several potential mechanisms were postulated for the pathogenesis of DOX cardiotoxicity, including oxidative stress, DNA damage, inflammation, alterations of mitochondrial cardiac energetics, and cell death [6,7]. However, there are little effective treatment options with relatively poor prognosis for DOX cardiotoxicity and only symptomatic therapies are currently available [2]. To identify novel therapeutic targets, a better understanding of underlying molecular mechanisms is pertinent, given the abundant patient populations suffering from this devastating myopathic anomaly.
Accumulating clinical evidence has depicted the involvement of mitochondrial dysfunction in adverse cardiac remodeling and heart failure in the onset and development of DOX cardiomyopathy [7]. The heart is one of the most metabolically active organs, with mitochondrial dynamics and bioenergetic capacity providing indispensable roles in the maintenance of cellular energy metabolism and cardiac function. Impaired mitochondrial bioenergetics, particularly deficiency in mitochondrial oxidative metabolism, results in contractile defects and heart failure in patients and experimental animals [[8], [9], [10]]. Evidence from in vivo and in vitro studies has indicated that DOX inhibits electron transport chain (ETC) complex I, II, and IV proteins and complex I activity, thus interrupting mitochondrial energy production [[11], [12], [13]]. A fine balance in mitochondrial dynamics (fusion and fission) is vital to meet the everchanging demands for energy production [14,15]. Ample earlier reports favor that DOX disrupts mitochondrial dynamics by inhibiting mitochondrial fusion and promoting mitochondrial fission through DRP1 (dynamin-related protein 1) phosphorylation at serine 616 [13,16,17]. Besides, DOX compromises autophagy and mitophagy clearance to provoke excessive oxidative stress and apoptosis [18]. Therefore, mitochondrial dynamics and bioenergetics are gaining intense attention as important therapeutic targets for DOX-induced cardiomyopathy.
As a highly conserved protein present in the nucleus, cytosol, and mitochondria, prohibitin 2 (PHB2) plays multiple important roles in physiological and pathophysiological processes, including cancer, aging, neuromuscular diseases, and cardiovascular diseases [19]. Of note, PHB2 forms a large multimeric scaffold complex with prohibitin 1 (PHB1) in the inner mitochondrial membrane (IMM), with manifold properties including chaperone function in the stabilization and assembly of the mitochondrial respiratory chain, mitochondrial biogenesis, maintenance of mtDNA stability, mitochondrial cristae morphogenesis and dynamics, and regulation of membrane protein degradation through interaction with mitochondrial proteases [[19], [20], [21], [22]]. PHB1 and PHB2 usually function as a heterodimeric complex, albeit with distinct roles in mitochondria [23]. PHB2 localized in IMM is essential for the maintenance of mitochondrial dynamics and mitophagy, normal metabolic processes, and bioenergetics in mitochondria. More importantly, PHB2 serves as an IMM mitophagy receptor to bind with LC3 upon mitochondrial depolarization and outer membrane rupture, to regulate cellular properties including mitochondrial dynamics and energy metabolism [19,24,25]. Besides, PHB2 recruits Parkin to mitochondria to engage PINK1/Parkin-dependent mitophagy [26]. Tissue-specific PHB2 knockout mice were utilized to evaluate its functions in different organs given the embryonical lethality of global PHB2 deletion. Of note, cardiac-specific PHB2 knockout mice exhibited cardiac systolic dysfunction and severe heart failure at 8 weeks of age, with disturbed fatty acid oxidation (FAO) and mitochondrial function [27]. On the other hand, vascular smooth muscle cells (VSMCs)-specific PHB2 knockout was more prone to postinjury VSMC proliferation and neointimal formation courtesy of their role in energy metabolism [28]. Hepatocyte-specific PHB2 conditional knockout mice exhibited defective gluconeogenesis, severe hypoglycemia, and lipid accumulation with profound mitochondrial fragmentation in the liver [29]. PHB2 deficiency led to mitochondrial fragmentation through excessive cleavage of L-OPA1. Overexpression of L-OPA1 in the liver was insufficient to rescue metabolic disorders in hepatocyte-specific PHB2 knockout mice since it only restored mitochondrial morphology without ameliorating oxidative phosphorylation (OXPHOS) function [29].
To this end, our current study was designed to explore the role of PHB2 in the pathogenesis of DOX cardiomyopathy and the underlying mechanisms. Taking advantage of cardiomyocyte-specific PHB2 knockout mice conditionally induced by tamoxifen, PHB2 was found to protect against DOX cardiotoxicity by maintaining mitochondrial dynamics and energetic production, thus revealing the therapeutic potential of PHB2 as a target in DOX cardiomyopathy.
2. Materials and methods
2.1. Experimental animal models
All animal procedures were performed according to the guidelines of the Care and Use of Laboratory Animals (National Institutes of Health Publication) and were approved by the Animal Welfare and Ethics Committee of Fudan University (approval number: 202209028Z). All mice were fed in a controlled environment (12 h day-night cycle) with free access to food and sterilized water. Male mice (8-10-week-old) were used. PHB2flox/flox mice were generated using a CRISPR/Cas9-mediated genome engineering system. Briefly, exon 4 of the mouse PHB2 gene was inserted into two LoxP sites (Fig. S1A). Conditionally cardiomyocyte-specific PHB2 knockout mice (PHB2flox/floxMyh6-CreERT2) were generated by crossing PHB2flox/flox mice with Myh6-CreERT2 mice (Cyagen Biosciences Co., Ltd., Guangzhou, Guangdong, China). Target gene manipulation was induced by intraperitoneal injection of tamoxifen (10540-29-1; Sigma-Aldrich, St. Louis, MO, USA) with a dosage of 50 mg/kg body weight once daily for 3 consecutive days [30,31]. PHB2flox/floxMyh6-Cre littermates without tamoxifen administration (receiving the solvent corn oil) were used as normal controls. Mice were genotyped using PCR with mouse tail DNA and primers as shown in Table S1.
To overexpress PHB2 in mouse hearts, PHB2 exons were inserted into the vector, and sequences were cloned into the recombinant adeno-associated virus serotype 9 (AAV9) frame vector containing mCherry fluorescence sequence, which was then amplified in HEK293 cells (Obio Technology Co., Ltd., Shanghai, China). For cardiac delivery of AAV9, 5 × 1011 particles of 100 μl (diluted in sterile PBS) were injected into C57BL/6 mice aged 8 weeks through tail veins. Three weeks later, PHB2 overexpression efficiency and reporter fluorescence were examined in mouse heart tissues. C57BL/6 mice were obtained from Shanghai Jihui Laboratory Animal Care Co., Ltd.
DOX (S1208; Selleck Chemicals, Houston, TX, USA) dissolved in saline (0.9% sterile sodium chloride) was administrated through intraperitoneal injection 2 weeks following tamoxifen injection or 3 weeks after AAV9 delivery. DOX-free mice received an equal volume of saline. A sub-chronic model of DOX was utilized with a dose of 4 mg/kg body weight once a week for 5 weeks (with an accumulated dose of 20 mg/kg) [32].
2.2. Echocardiographic assessment of myocardial function
Echocardiography was performed one week following the final DOX administration using a two-dimension guided M-mode echocardiography (Vevo 2100; VisualSonics, Toronto, ON, Canada). Mice were continuously anesthetized by 2–3% isoflurane inhalation. Parameters of cardiac function and ventricular volume were calculated using Vevo computer algorithms, including interventricular septum (IVS) thickness, left ventricular posterior wall (LVPW) thickness, left ventricular end-diastolic dimension (LVEDD), end-systolic dimension (LVESD), end-diastolic volume (LVEDV), end-systolic volume (LVESV), ejection fraction (EF), fractional shortening (FS) and heart rate (HR) [33]. Cardiac output (CO) was calculated using the following formula: CO = (LVEDV-LVESV) × HR [34].
2.3. Histological examination and immunohistochemistry staining
Following fixation in 4% paraformaldehyde, mouse heart tissues were embedded in paraffin and cut cross-sectionally into 5 μm-slides. After continuous deparaffinization and rehydration, paraffin sections were subjected to hematoxylin and eosin (H&E) staining to evaluate general morphology, including Alexa Fluor 488 labeled wheat germ agglutinin (WGA) staining to evaluate myocardial atrophy, and picrosirius Red (PSR) staining to assess collagen content. For WGA staining, five to six images were taken and nearly 200–300 cells were measured for each heart. An average value was calculated for each mouse heart. For picrosirius Red staining fibrosis, the collagen volume fraction was determined using the percentage of positively stained area normalized to the entire field [32]. Images were taken using a Leica microscope (Leica Microsystems, Wetzlar, Germany) at a 20 × objective, and cross-sectional area and collagen volume fraction were analyzed using the Image J software (National Institutes of Health, Bethesda, MD, USA).
For immunohistochemistry staining, following the antigen retrieval process, paraffin-embedded sections were incubated with primary antibodies against PHB2 (12295-1-AP; Proteintech, Wuhan, Hubei, China) overnight at 4 °C and were then incubated with biotin-conjugated secondary antibodies. Positive antibody binding was visualized using a 3,3′-diaminobenzidine (DAB) peroxidase substrate kit, and nuclei were stained using hematoxylin. Images were taken using a Leica microscope.
2.4. Transmission electron microscopy (TEM)
TEM was used to evaluate mitochondrial ultrastructure in cardiomyocytes. After fixation with glutaraldehyde and trimming into tiny blocks, mouse heart tissues were cut into slides and were collected onto 200-mesh copper grids for staining. Sections were then visualized using an 80-kV transmission electron microscope (JEM-1230; JOEL Ltd., Tokyo, Japan). At least 3 fields per slice were observed at low magnification (5000 ×) for assessment of mitochondrial ultrastructure and 6 independent experiments were performed for each group. The examiner was blinded for TEM sample collection and subsequent data analysis. Mitochondrial size, and major and minor axis lengths were measured using an Image J software (National Institutes of Health, Bethesda, MD, USA) [33].
2.5. Isolation of primary neonatal rat cardiomyocytes (NRCM)
Hearts were harvested from neonatal Sprague-Dawley rats (1–2 days old) and excised into small pieces. Tissues were digested with 0.08% type I Collagenase (LS004196; Worthington Biochemical Corporation, Lakewood, NJ, USA), and the supernatant was neutralized in a complete Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). After digestion, the supernatant was centrifuged at 1000 rpm for 5 min, and pellets were resuspended in the complete DMEM medium, followed by pre-plating for 90 min to remove fibroblasts. Then the supernatant was plated and cultured for 48 h before transfection and further treatment in 37 °C incubators with 95% oxygen and 5% CO2.
2.6. Isolation of primary adult mouse cardiomyocytes (AMCM), cell shortening and relengthening
Adult mice (8 weeks old) were anesthetized and fully exposed. After trimming off the inferior vena cava, an EDTA buffer was immediately delivered into the right ventricle. Hearts were excised from ascending aorta and were digested with type II and type IV collagenase (LS004176 and LS004188; Worthington Biochemical Corporation, Lakewood, NJ, USA) after sequential perfusion of EDTA and perfusion buffers. Digested tissues were gently separated into small pieces using forceps, and cells were collected using gravity settling, followed by gradient calcium re-introduction. At least 80%–90% of rod-shaped AMCMs were deemed successful [35]. AMCMs were resuspended with a contractile buffer for electrical stimulation, or cultured in M199 medium (supplemented with 1% penicillin-streptomycin, 1% ITS, 1% CD lipid, 0.1% BSA, and 0.1% BDM) for further treatment.
A SoftEdge MyoCam system (IonOptix, Milton, MA, USA) with an Olympus IX-70 microscope was employed to monitor the mechanical properties of adult mouse cardiomyocytes. Cells were resuspended with a contractile buffer and were stimulated to contract at a frequency of 0.5 Hz. Isotonic contraction of AMCMs was recorded using an IonOptix SoftEdge device [33].
2.7. Constructions and infections of adenoviruses, and DOX treatment in primary cardiomyocytes
To overexpress PHB2 in primary cardiomyocytes, PHB2 exons were inserted into a pAdeno-MCMV-T2A vector to create the pAdeno-MCMV-Phb2-T2A vector (Obio Technology Co., Ltd., Shanghai, China). To knockdown target genes, siRNAs were deigned and synthesized (Genecefe Biotechnology Co., Ltd., Wuxi, Jiangsu, China) using sequences listed in Table S2. Then shRNA target and negative control sequences were cloned to pADV-U6-shRNA-CMV-MCS vectors. All constructs were verified using sequencing. Finally, these vectors were cloned into the AdMax System (MicroBix Biosystems, Mississauga, ON, Canada) following the manufacturer's instructions. The viral transfection was verified using Western blot.
Primary cardiomyocytes including NRCMs and AMCMs were infected with adenoviruses for 24–48 h followed by DOX (1 μM) treatment for another 24 h before cell harvest. Cells in control groups were treated with saline for an equivalent period [32].
2.8. Culture of cell lines and plasmid transfection
HEK293T cells were cultured in DMEM with 10% FBS and 1% penicillin-streptomycin. Plasmids encoding wild-type and mutated PHB2 and NDUFV2 (Genecefe Biotechnology Co., Ltd., Wuxi, Jiangsu, China) were transfected into HEK293T cells using the Lipofectamine 2000 reagent (11668030; Invitrogen, Carlsbad, CA, USA) for 48 h before cell lysis.
2.9. Probes for mitochondrial detection
For measurements of mitochondrial membrane potential, primary adult murine cardiomyocytes were stained with the mitochondrial membrane potential indicator tetramethylrhodamine methyl ester (TMRM) reagent (I34361; Invitrogen, Carlsbad, CA, USA) and MitoTracker Green FM (M7514; Invitrogen) following the manufacturer's instructions. The level of mitochondrial ROS production was determined using MitoSOX Red mitochondrial superoxide indicator (M36008; Invitrogen). The morphology of mitochondria was assessed using MitoTracker Red CMXRos staining (M7512; Invitrogen) in neonatal primary cardiomyocytes. Nuclei were counterstained with Hoechst 33342 (R37605; Invitrogen). Finally, cells were observed using a laser scanning confocal microscope (SP8; Leica Microsystems, Wetzlar, Germany) at a 63 × objective [33,36].
Following MitoTracker staining, neonatal primary cardiomyocytes were categorized into those with predominantly fragmented mitochondria (fragmented mitochondria >50%) and those with predominantly elongated mitochondria (elongated mitochondria >50%). Approximately 20 cells were evaluated for each independent experiment and 4 independent experiments were carried out for each group. The percentage of cells with fragmented mitochondria was calculated, and the examiner was blinded to the group identity during data collection and analysis [37].
2.10. Measurements of cell bioenergetics
Oxygen consumption rate (OCR) and adenosine triphosphate (ATP) production were measured using a Seahorse XF96 analyzer (Agilent, Santa Clara, CA, USA) per the manufacturer's instructions. NRCMs were seeded into XFe96 plates at a proper density. Measurement of OCR was performed using the Seahorse XF Cell Mitochondrial Stress Test (103015–100; Agilent). Following baseline OCR recording, oligomycin, trifluoromethoxy carbonylcyanide phenylhydrazone (FCCP), and rotenone along with antimycin A were sequentially added to measure ATP-coupled respiration, maximal respiration, and non-mitochondrial respiration [38]. Values of OCR and ATP production rate were normalized to protein concentrations.
2.11. Mitochondria isolation and mitochondrial complex activity
Mitochondrial fractions were extracted using the mitochondrial isolation kit for cultured cells (KTP4003; Abbkine, Wuhan, Hubei, China). Briefly, NRCMs were collected with PBS followed by centrifugation at 500×g for 5 min and washed twice in a cold PBS buffer. Cell pellets were resuspended with cold lysis buffer A, B, and C, followed by centrifugation at 600×g for 10 min to remove unwanted sediment and centrifugation at 11,000×g for another 10 min for mitochondrial fractionation. Activities of mitochondrial complex I–V were measured by mitochondrial respiratory chain complex activity assay kits (Solarbio, Beijing, China) using isolated mitochondria from NRCMs per the manufacturer's instructions [38].
2.12. Proteomics
Quantitative proteomics was performed from mouse heart tissues using TMTpro 16 labeling (Thermo Fisher Scientific, Waltham, MA, USA) in combination with liquid chromatography-mass spectrometry (LC-MS) technology (Shanghai Lu-Ming Biotech Co., Ltd., Shanghai, China). After database searching using Proteome Discover 2.4 (Thermo Fisher Scientific, Waltham, MA, USA) and protein quantification, only proteins with false discovery rate (FDR) < 1% and unique peptides ≥1 were considered for the protein listing and further downstream analysis.
2.13. Co-immunoprecipitation (Co-IP) and mass spectrometry analysis
Mouse heart tissues and cells were lysed with a lysis buffer containing 0.5% NP-40, 150 mM NaCl, 50 mM Tris-HCl, and a protease inhibitor cocktail at pH 7.4. Lysates were centrifuged and incubated with primary antibodies including anti-Flag M2 Magnetic Beads (M8823; Millipore, Sigma-Aldrich, St. Louis, MO, USA), or mouse anti-PHB2 (sc-133094; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and rabbit anti-NDUFV2 (ab183715; Abcam, Boston, MA, USA) antibodies for overnight incubation on a rotary shaker at 4 °C. Lysates incubated with mouse IgG (5415; Cell Signaling Technology, Danvers, MA, USA) or rabbit IgG (3900; Cell Signaling Technology) were used as a negative control. Protein G beads were added into the immune complexes for 2 h at 4 °C. Beads were then rinsed 3–5 times and were eluted for further examinations.
To explore potential interactors of PHB2, co-IP samples were separated through SDS-PAGE electrophoresis followed by Coomassie blue staining (P0017; Beyotime Biotechnology, Shanghai, China). Stained gels were excised and were further digested into peptides, which were processed to LC-MS (Shanghai Lu-Ming Biotech Co., Ltd., Shanghai, China) for protein identification [9].
2.14. Structure-based protein interaction interface analysis
The molecular structure of NDUFV2 was obtained from the RCSB PDB database (http://www.rcsb.org/) with the accession of 5XTB, chain O. Protein structure of PHB2, which was predicted using AlphaFold2, a highly accurate artificial intelligence-based computational structure modeling method, was retrieved from the UniProt database (http://www.uniprot.org/) [39]. Then the structures of these proteins were submitted to the PRISM tool (http://cosbi.ku.edu.tr/prism) to predict their potential interaction interface [40]. Finally, the prediction results were visualized using the PyMol tool (http://pymol.org).
2.15. Immunofluorescence
Following fixation with 4% paraformaldehyde, permeabilization with Triton X-100, and blocking with BSA, AMCMs were incubated with primary antibodies overnight at 4 °C, including mouse anti-PHB2 (sc-133094; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and rabbit anti-NDUFV2 (ab183715; Abcam, Boston, MA, USA) antibodies. Cells were then incubated with secondary antibodies Alexa Flour 488 (4408; Cell Signaling Technology, Danvers, MA, USA) and Alexa Flour 594 (8889; Cell Signaling Technology, Danvers, MA, USA) at 37 °C for 1 h. Nuclei were counterstained with DAPI (P0131; Beyotime Biotechnology, Shanghai, China). Images were taken using a laser scanning confocal microscope (SP8; Leica Microsystems, Wetzlar, Germany) at a 63 × objective [33].
2.16. GST pulldown
Human PHB2 fused with GST tag and Human NDUFV2 fused with His tag were cloned into pGEX-4T-1 vectors, before transformation into E. coli BL21 cells to generate recombinant PHB2 and NDUFV2 proteins. For in vitro protein interaction assay, GST-PHB2 or GST (as control) proteins were mixed with GST Resin (17075601; GE Healthcare Systems, Saint Louis, MO, USA) for 2-h incubation on the shaker at 4 °C. Following centrifugation, His-NDUFV2 proteins were added to the mixture and were incubated overnight at 4 °C. Products were precipitated using reduced l-Glutathione (G4251; Sigma-Aldrich, St. Louis, MO, USA) for Western blot analysis [41].
2.17. Cell viability assay
NRCMs were plated in 96-well plates for the determination of cell viability using the published protocol [32]. Following the addition of 10 μl Enhanced Cell Counting Kit 8 (CCK-8) solution (C0042; Beyotime Biotechnology, Shanghai, China) into each well, the plate was incubated for 1 h at 37 °C and absorbance was measured at 450 nm using a microplate reader (FlexStation 3, Molecular Devices, San Jose, CA, USA). CellTiter-Glo Luminescent Cell Viability Assay (G7570; Promega Corporation, Madison, WI, USA) was also used to determine cell viability according to the manufacturer's instructions. Lactate dehydrogenase (LDH) was determined using the CyQUANT LDH Cytotoxicity Assay (C20300; Invitrogen, Carlsbad, CA, USA), and cytotoxicity was calculated using the following formula:
2.18. Real-time quantitative PCR (real-time qPCR)
Total RNA was extracted from mouse heart tissues using FastPure Cell/Tissue Total RNA Isolation Kit (RC112-01; Vazyme Biotech Co., Ltd., Nanjing, Jiangsu, China) according to the manufacturer's instructions. Total RNA was reversed into cDNA using PrimerScript RT Master Mix (RR036A; Takara Bio Inc., Shiga, Japan), followed by real-time qPCR using TB Green Premix Ex TaqII kit (RR820A; Takara Bio Inc., Shiga, Japan) with the CFX Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). GAPDH was used as a control, and a relative quantification method 2−ΔΔCT was used to quantify the relative expression of genes. Gene-specific primers for Real-time qPCR (Sangon Biotech, Shanghai, China) are listed in Table S3.
2.19. Western blot analysis
Cardiac tissues were immediately frozen in liquid nitrogen after euthanasia and were stored at −80 °C for further examination. Heart tissues and cells were lysed using RIPA lysis buffer (P0013; Beyotime Biotechnology, Shanghai, China) with protease inhibitor cocktails, and then protein extractions were separated by SDS-PAGE and transferred onto membranes. After incubation with primary antibodies at 4 °C overnight, membranes were then incubated with respective secondary antibodies. Signals were imaged using a ChemiDoc Touch Imaging System (Bio-Rad, Hercules, CA, USA), and quantification was performed with the Image J software (National Institutes of Health, Bethesda, MD, USA) [33]. Catalog numbers of all antibodies used for western blotting are shown in Table S4.
2.20. Availability of other data and materials
All other materials not mentioned here are made available in Table S5.
2.21. Statistical analysis
All data were shown as mean ± SEM. Statistical analysis was performed using the GraphPad Prism 9.4.1 software (GraphPad Software, La Jolla, CA, USA). Comparison between the two groups was conducted using unpaired student's t-test (normal distribution and equal variances). Multiple group comparisons were performed using one-way or two-way ANOVA (normal distribution) followed by Tukey's multiple comparison test. Unless otherwise indicated, p < 0.05 was deemed as the threshold for statistical significance. In certain cases, raw values were normalized to the control group values.
3. Results
3.1. Cardiac-specific PHB2 deficiency aggravated DOX-induced cardiomyopathy in mice
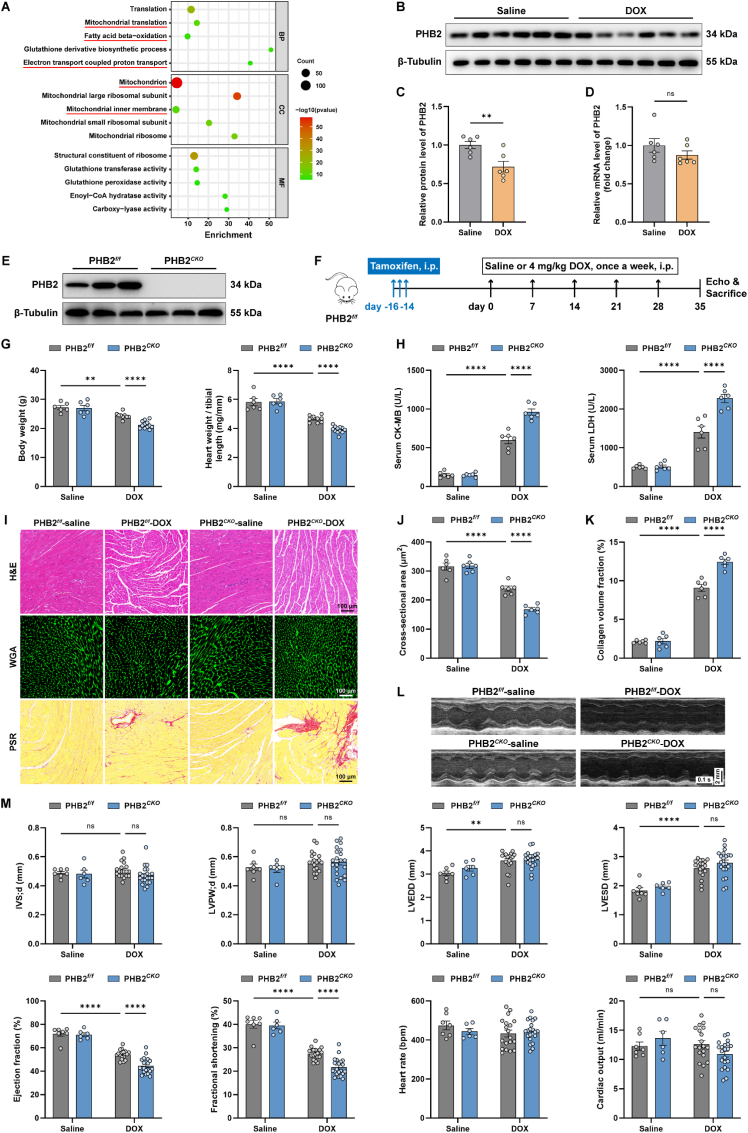
A sub-chronic mouse model of DOX cardiomyopathy was employed. TMT (Tandem Mass Tags) labeled quantitative proteomic profiling was performed using mouse heart lysates from DOX and saline control mice. Gene ontology (GO) analysis for biological process (BP) showed that translation, mitochondrial translation, fatty acid β-oxidation and electron transport coupled proton transport were among the top enriched biological processes, while GO analysis for cellular component (CC) noted mitochondrion, mitochondrial inner membrane (IMM), and mitochondrial ribosome as differentially expressed cellular components (Fig. 1A). PHB2 is an essential regulator localized in IMM, with an important role in the maintenance of mitochondrial cristae and dynamics, as well as the stabilization of mitochondrial respiratory chain and oxidative phosphorylation. Evaluation of PHB2 level under DOX challenge revealed overtly downregulated protein expression but not mRNA level of PHB2 (Fig. 1B-D). To discern the precise role of PHB2 in DOX cardiomyopathy, a tamoxifen-induced cardiac-specific PHB2 knockout mouse model driven by the Myh6 promoter was generated. The efficacy of PHB2 knockout was verified in mouse hearts several weeks following tamoxifen delivery (Fig. 1E and S1B). Cardiac-specific knockout of PHB2 did not result in higher mortality or cardiac dysfunction at baseline (data not shown). PHB2flox/flox and PHB2CKO mice were subjected to DOX challenge and were followed up as illustrated (Fig. 1F). DOX resulted in significant weight loss and cardiac atrophy, as indicated by decreased body weight and heart weight-to-tibial length ratio, the effect of which was greatly aggravated by cardiac-specific PHB2 ablation (Fig. 1G). DOX-induced elevation of creatine kinase-muscle/brain (CK-MB) and LDH in mouse serum was further exacerbated by PHB2 knockout (Fig. 1H). In addition, histological analysis showed that PHB2CKO mouse heart displayed decreased cardiomyocyte size and increased myocardial fibrosis compared with PHB2flox/flox mice in the face of DOX insult (Fig. 1I-K). Echocardiographic findings indicated that DOX-induced systolic dysfunction was aggravated by PHB2 knockout, as evidenced by left ventricular ejection fraction and fractional shortening (Fig. 1L and M). Little changes were noted for left ventricular wall thickness, left ventricular dimension, and cardiac output between PHB2flox/flox and PHB2CKO mice with DOX challenge (Fig. 1M). To consolidate the effect of PHB2 knockout on the contractile properties of cardiomyocytes, the mechanical properties of primary adult mouse cardiomyocytes (AMCMs) were evaluated. DOX exposure evoked significant mechanical abnormalities as evidenced by decreased peak shortening, maximal velocity of shortening (+ dL/dt) and relengthening (- dL/dt) in cardiomyocytes. Although PHB2 knockout itself failed to affect these mechanical indices, it accentuated DOX-induced cardiomyocyte anomalies (Fig. S1C). In this context, assessment of cardiac morphology and function showed that cardiac-specific PHB2 knockout aggravated DOX cardiomyopathy in vivo and in vitro.
Fig. 1.
Cardiac-specific PHB2 deficiency aggravated DOX-induced cardiomyopathy in mice.
(A) Top 5 GO hits for biological process (BP), cellular component (CC), and molecular function (MF) analysis using differentially expressed proteins in mouse heart lysates from DOX and saline groups. (B and C) Representative immunoblotting images and quantification of PHB2 in mouse heart lysates with and without DOX challenge (β-Tubulin used as an internal control, n = 6 per group). (D) Relative mRNA levels of PHB2 in mouse heart lysates with and without DOX challenge quantified using RT-qPCR (GAPDH used as an internal control, n = 6 per group). (E) Representative immunoblotting of PHB2 in heart lysates from PHB2flox/flox and PHB2CKO mice (β-Tubulin used as an internal control). (F) Flow chart depicting experimental strategy for cardiac-specific PHB2 conditional knockout through tamoxifen administration in PHB2flox/flox mice in DOX cardiomyopathy. (G) Body weight and heart weight-to-tibial length ratio (n = 6–12 per group). (H) Serum levels of creatine kinase-muscle/brain (CK-MB) and lactic acid dehydrogenase (LDH) (n = 6 per group). (I) Representative images of hematoxylin and eosin (H&E) staining (upper), wheat germ agglutinin (WGA) staining (middle), and picrosirius red (PSR) staining (lower) of myocardial sections. (J) Quantification of cardiomyocyte area in WGA staining (n = 6 per group). (K) Quantitation of the fibrotic area in PSR staining (normalized to the entire field, n = 6 per group). (L and M) Representative echocardiographic images and echocardiographic measurements (n = 6–21 per group). IVSd, interventricular septum thickness at diastole; LVPWd, left ventricular posterior wall thickness at diastole; LVEDD, left ventricular end-diastolic dimension; LVESD, left ventricular end-systolic dimension; EF, ejection fraction; FS, fractional shortening; HR, heart rate; CO, cardiac output. Mean ± SEM. **p < 0.01, ****p < 0.0001, ns, no significance. For statistical analysis, unpaired student's t-test was used for C-D; two-way ANOVA with Tukey's test for multiple comparisons was used for G-H, J-K, and M. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Cardiac overexpression of PHB2 alleviated DOX-induced cardiomyopathy in mice
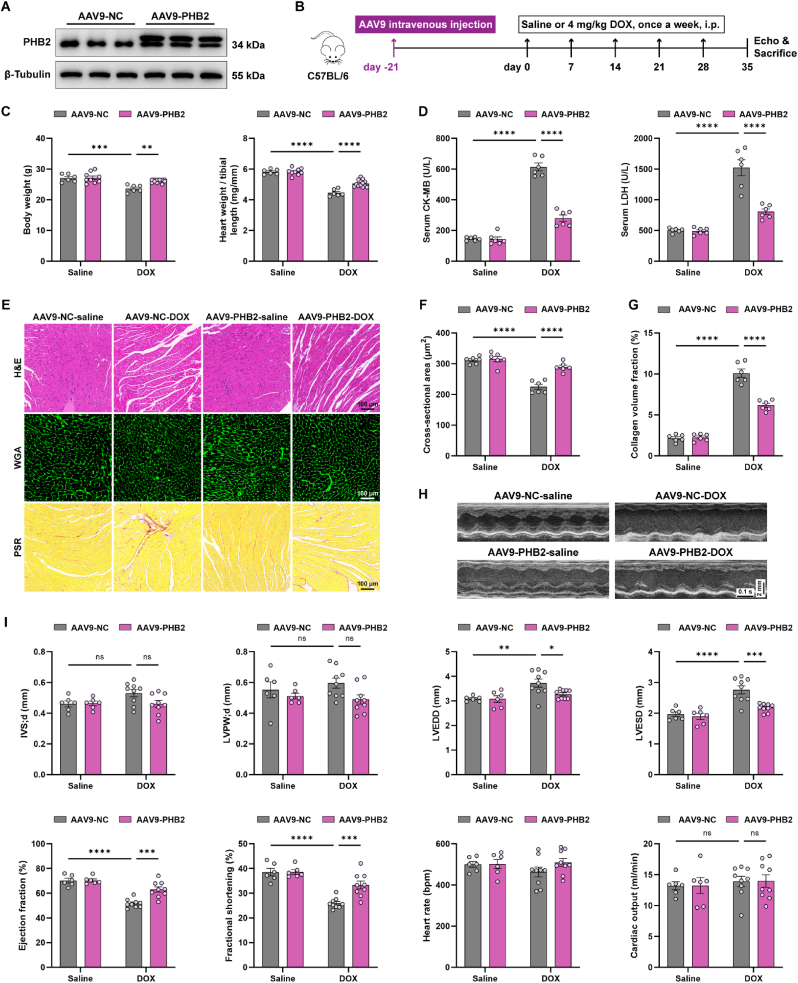
To test the gain-of-function for PHB2 in DOX cardiomyopathy, mice were transfected with PHB2 using adeno-associated virus serotype 9 (AAV9) [42]. The transfection efficacy was verified using mCherry fluorescence (Fig. S2A) and PHB2 was significantly overexpressed in mouse heart lysates (Fig. 2A). Three weeks following the AAV9 injection, mice were subjected to DOX challenge and were followed up as illustrated (Fig. 2B). Our data revealed that cardiac overexpression of PHB2 ameliorated DOX-induced body weight loss and cardiac atrophy as indicated by the heart weight-to-tibial length ratio (Fig. 2C). Moreover, DOX challenge evoked elevations in cardiac injury markers including CK-MB and LDH in mouse serum, the effects of which were partially attenuated by PHB2 overexpression (Fig. 2D). Besides, histological staining also displayed a reconciliation of DOX-induced cardiomyocyte atrophy and myocardial interstitial fibrosis in response to PHB2 overexpression (Fig. 2E-G). Echocardiographic data noted that PHB2 overexpression improved DOX-induced systolic dysfunction and cardiac remodeling, as evidenced by compromised ejection fraction and fractional shortening, as well as increased left ventricular dimension (Fig. 2H and I). Left ventricular wall thickness and cardiac output were unaffected by either DOX or PHB2 overexpression (Fig. 2I). Thus, cardiac overexpression of PHB2 using AAV9 alleviated DOX-induced cardiac dysfunction in mice.
Fig. 2.
Cardiac overexpression of PHB2 alleviated DOX-induced cardiomyopathy in mice.
(A) Representative immunoblotting images of PHB2 in mouse heart lysates to validate PHB2 overexpression (β-Tubulin used as an internal control). (B) Flow chart depicting experimental strategy for PHB2 overexpression using AAV9 delivery in DOX cardiomyopathy. (C) Body weight and heart weight-to-tibial length ratio (n = 6–12 per group). (D) Serum levels of CK-MB and LDH (n = 6 per group). (E) Representative images of H&E staining (upper), WGA staining (upper), and PSR staining (lower) of myocardial sections. (F) Quantification of cardiomyocyte area in WGA staining (n = 6 per group). (G) Quantitation of the fibrotic area in PSR staining (normalized to the entire field, n = 6 per group). (H and I) Representative echocardiography images and echocardiographic measurements of IVSd, LVPWd, LVEDD, LVESD, ejection fraction, fractional shortening, heart rate, and cardiac output of mice (n = 6–12 per group). Mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, no significance. For statistical analysis, two-way ANOVA with Tukey's test for multiple comparisons was used.
3.3. PHB2 deficiency aggravated DOX-induced mitochondrial fragmentation in mouse hearts
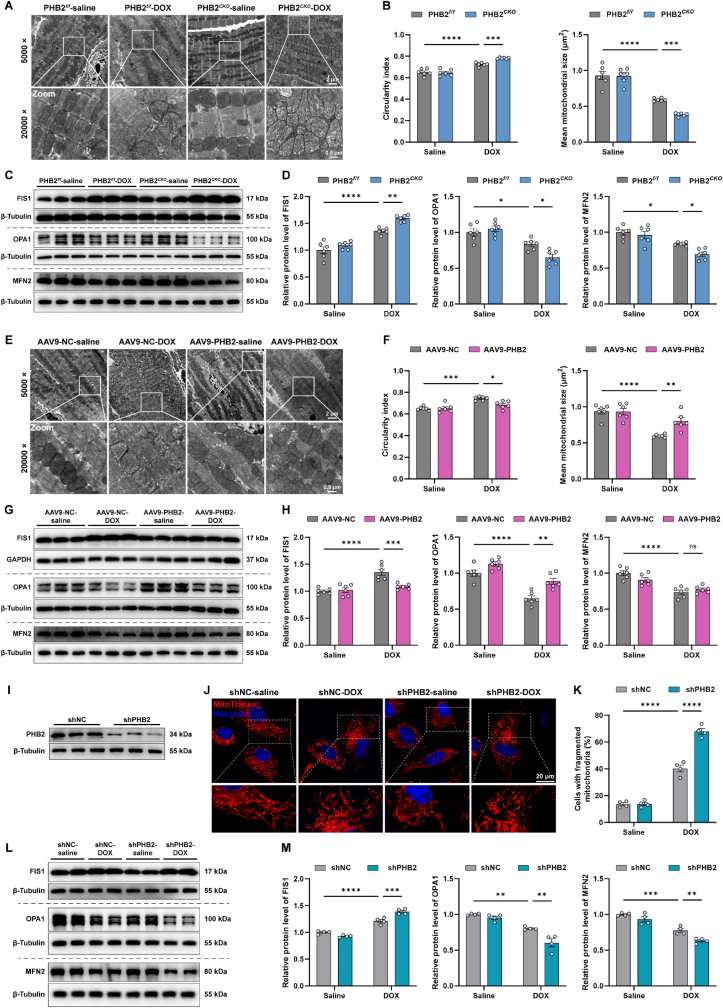
Previous studies showed that PHB2 regulated mitochondrial dynamics mainly through regulating mitochondrial fusion protein OPA1 processing and stabilization [25,43,44]. Therefore, additional studies were conducted to assess mitochondrial morphology and function in cardiac-specific PHB2 knockout mice. TEM results revealed that DOX resulted in disordered arrangements, abnormal density, and fragmentation of mitochondria with pronounced cristae disorganization, the effect of which was further exacerbated by PHB2 knockout (Fig. 3A). As a marker for mitochondrial swelling and injury, the circularity index (the minor axis-to-major axis length ratio) was elevated in DOX cardiomyopathy, the effect of which was accentuated by PHB2 deficiency (Fig. 3B). DOX-induced mitochondrial fragmentation was also exacerbated in PHB2CKO mice as indicated by mitochondrial size (Fig. 3B). In addition, mitochondrial fission protein FIS1 (mitochondrial fission protein 1) was upregulated while fusion proteins OPA1 (optic atrophy protein 1) and MFN2 (mitofusin 2) were downregulated in DOX-challenged mouse hearts, the effect of which was accentuated by PHB2 knockout (Fig. 3C and D). On the contrary, TEM findings noted ameliorated DOX-induced mitochondrial swelling and fragmentation with PHB2 overexpression using AAV9 delivery, as indicated by circularity index and mitochondrial size (Fig. 3E and F). DOX challenge led to increased levels of FIS1 and decreased levels of OPA1, the effect of which was reversed by PHB2 overexpression (Fig. 3G and H). Consistent with the TEM findings from mouse hearts, MitoTracker staining in primary neonatal cardiomyocytes (NRCMs) supported that DOX-induced mitochondrial fragmentation was accentuated by PHB2 silencing (Fig. 3I-K). Likewise, DOX-induced elevation in FIS1 and reduction in OPA1 and MFN2 were further aggravated by PHB2 knockdown (Fig. 3L and M). Altogether, these data indicated that PHB2 deficiency exacerbated DOX-induced mitochondrial fragmentation in vivo and in vitro.
Fig. 3.
PHB2 deficiency aggravated DOX-induced mitochondrial fragmentation in mouse hearts.
(A and B) Representative images of mitochondrial ultrastructure using TEM in myocardial sections from PHB2flox/flox and PHB2CKO mice, and quantification of circularity index and mitochondrial size based on TEM images (n = 6 per group). (C and D) Representative immunoblotting images and quantification of FIS1, OPA1, and MFN2 in heart lysates from PHB2flox/flox and PHB2CKO mice (β-Tubulin used as an internal control, n = 6 per group). (E and F) Representative images of mitochondrial ultrastructure using TEM in myocardial sections from AAV9-NC and AAV9-PHB2 mice, and quantification of circularity index and mitochondrial size based on TEM images (n = 6 per group). (G and H) Representative immunoblotting images and quantification of FIS1, OPA1, and MFN2 in heart lysates from AAV9-NC and AAV9-PHB2 mice (GAPDH and β-Tubulin used as internal controls, n = 6 per group). (I) Representative immunoblotting images of PHB2 knockdown using adenoviruses carrying shRNA in primary cardiomyocytes (β-Tubulin used as an internal control). (J and K) Representative images of MitoTracker staining in neonatal rat cardiomyocytes (NRCMs) with and without DOX challenge (0.1 μM) for 24 h after adenovirus transfection, and quantification of the percentage of cells with fragmented mitochondria (those with >50% fragmented mitochondria, n = 4 independent experiments per group and approximately 20 cells for each experiment). (L and M) Representative immunoblotting images and quantification of FIS1, OPA1, and MFN2 in primary cardiomyocytes with and without DOX challenge for 24 h after adenovirus transfection (β-Tubulin used as an internal control, n = 4 independent experiments per group). Mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, no significance. For statistical analysis, two-way ANOVA with Tukey's test for multiple comparisons was used.
3.4. PHB2 deficiency impaired mitochondrial OXPHOS in DOX-treated mouse hearts
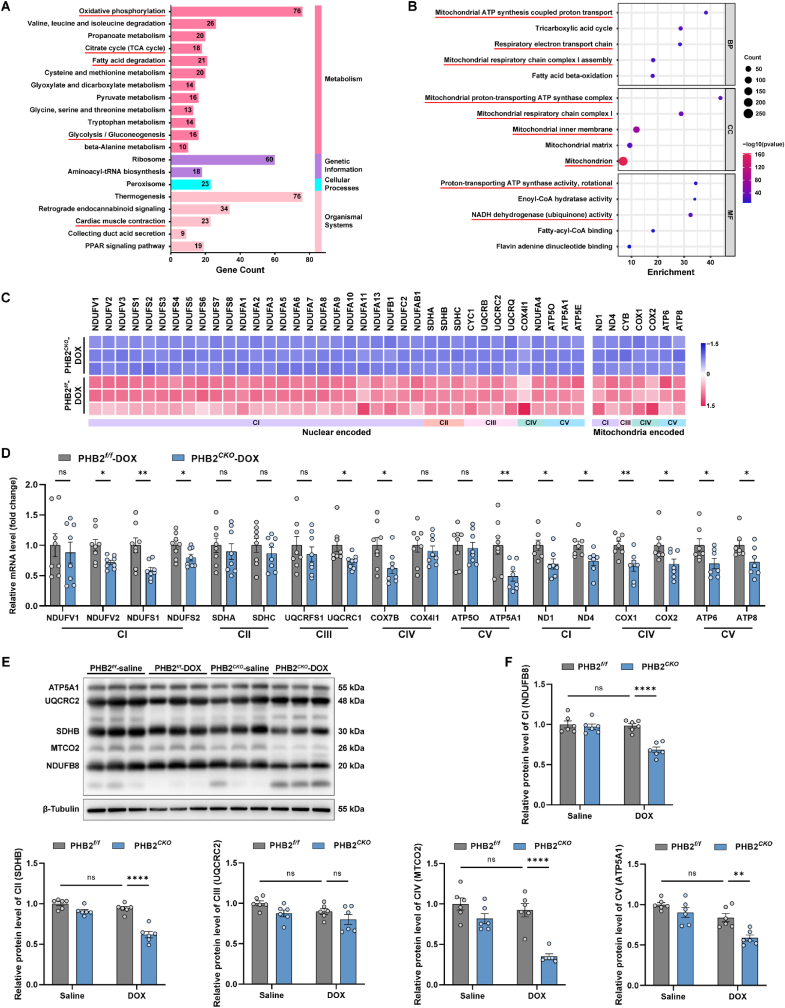
To gather insights into the underlying mechanisms through which PHB2 deficiency exacerbated DOX cardiomyopathy, TMT labeled quantitative proteomics was performed using heart lysates from PHB2flox/flox-DOX and PHB2CKO-DOX mice. A total of 1275 differentially expressed proteins were identified (FC > 1.5 and p < 0.05), among which 894 were upregulated and 381 were downregulated in PHB2CKO-DOX mice compared with PHB2flox/flox-DOX counterparts (Figs. S3A and S3B). KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis revealed that mitochondrial metabolic pathways including oxidative phosphorylation, citrate cycle, fatty acid degradation, and glycolysis were among the top enriched pathways (Fig. 4A). Similarly, Gene Set Enrichment Analysis (GSEA) showed aerobic respiration, oxidative phosphorylation, and respiratory electron transport chain as the top three enriched pathways, which were significantly inhibited by PHB2 knockout (Fig. S3C). Furthermore, GO analysis for biological process (BP) denoted that downregulated proteins were enriched in pathways including mitochondrial ATP synthesis coupled proton transport, tricarboxylic acid cycle, respiratory electron transport chain, mitochondrial respiratory chain complex I assembly, and fatty acid beta-oxidation (Fig. 4B). GO analysis for cellular component (CC) indicated enrichment for mitochondrial ATP synthase complex, mitochondrial respiratory chain complex I, and mitochondrial inner membrane, and GO analysis for molecular function (MF) indicated enrichment for ATP synthase activity and NADH dehydrogenase (ubiquinone) activity (Fig. 4B). Upregulated proteins were more likely to be associated with protein translation and folding (Fig. S3D).
Fig. 4.
PHB2 deficiency impaired mitochondrial OXPHOS in DOX-treated mouse hearts.
(A) Top 20 KEGG pathways of differentially expressed proteins in mouse heart lysates between PHB2CKO-DOX and PHB2f/f-DOX groups. (B) Top 5 GO pathways of BP, CC, and MF analysis using downregulated proteins in PHB2CKO-DOX and PHB2f/f-DOX groups. (C) Relative protein levels of mitochondrial respiratory electron transport chain (ETC) subunits determined by proteomics. (D) Relative mRNA levels of nuclear-encoded OXPHOS subunits in mouse heart lysates quantified by RT-qPCR (18S used as an internal control, n = 8 per group). (E and F) Representative immunoblotting images and quantification of mitochondrial OXPHOS proteins in mouse heart lysates (antibodies against NDUFB8, SDHB, UQCRC2, MTCO2, and ATP5A1 used as representatives for mitochondrial complex I, II, III, IV, and V, and β-Tubulin used as an internal control, n = 6 per group). Mean ± SEM. *p < 0.05, **p < 0.01, ****p < 0.0001, ns, no significance. For statistical analysis, unpaired student's t-test was used for D; two-way ANOVA with Tukey's test for multiple comparisons was used for F.
Considering findings from the enrichment analysis, we focused on the mitochondrial respiratory electron transport chain (ETC) and oxidative phosphorylation. A robust decline was noted in protein levels of ETC subunits in PHB2CKO-DOX mouse hearts compared with those from PHB2flox/flox-DOX mice using proteomic profiling (Fig. 4C). To verify these results, RT-qPCR was employed and our results revealed declined mRNA levels in certain nuclear-encoded ETC subunits (particularly mitochondrial complex I subunits) and mitochondrial-encoded ETC subunits in PHB2CKO-DOX mouse hearts compared with PHB2flox/flox-DOX group (Fig. 4D). Moreover, protein levels of ETC complex subunits were determined using mouse heart lysates, which displayed an overt downregulation in complex I (NDUFB8) in PHB2CKO-DOX mice compared with PHB2flox/flox-DOX mice (Fig. 4E and F). These data indicated that PHB2 deficiency impaired protein expression and function of mitochondrial complex I in DOX-challenged mouse hearts.
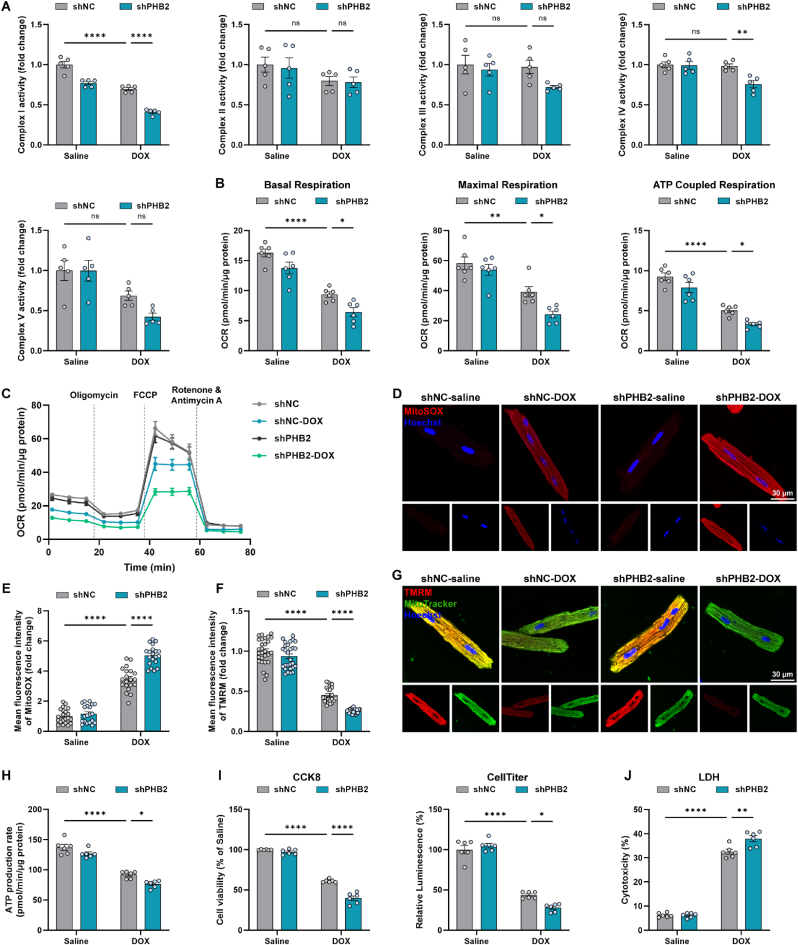
3.5. PHB2 deficiency aggravated DOX-induced mitochondrial dysfunction in cardiomyocytes
To examine the effects of PHB2 deficiency on mitochondrial function, PHB2 expression was manipulated in primary cardiomyocytes by infecting adenoviruses carrying shRNA targeting PHB2 (Fig. 3I). Enzymatic activity of mitochondrial respiratory complex I (NADH dehydrogenase) was overtly inhibited by PHB2 silencing in isolated mitochondria from DOX-challenged primary cardiomyocytes (Fig. 5A). Cell Mitochondrial Stress Test showed that basal respiration, maximal respiration, and ATP-coupled respiration were all decreased in DOX-challenged cardiomyocytes, the effect of which was exacerbated by PHB2 silencing (Fig. 5B and C). MitoSOX staining indicated elevated mitochondrial ROS (Reactive oxygen species) production following DOX challenge, the effect of which was aggravated by PHB2 deficiency (Fig. 5D and E). Tetramethylrhodamine methyl ester (TMRM) staining in AMCMs indicated that DOX challenge resulted in collapsed mitochondrial membrane potential, which was further declined by PHB2 knockdown (Fig. 5F and G). Meanwhile, DOX-induced impairment in ATP production was further exacerbated by shPHB2 (Fig. 5H). Consequently, CCK8 and CellTiter assay revealed that PHB2 silencing aggravated DOX-induced loss of cell viability (Fig. 5I). LDH release was also increased by PHB2 knockdown in DOX-challenged cardiomyocytes (Fig. 5J). These data favor that PHB2 deficiency aggravated DOX-induced mitochondrial dysfunction and cytotoxicity in vitro.
Fig. 5.
PHB2 deficiency aggravated DOX-induced mitochondrial dysfunction in cardiomyocytes.
(A) Enzymatic activities of OXPHOS complex I–V in isolated mitochondria from primary cardiomyocytes with and without DOX challenge (0.1 μM) for 24 h after adenovirus transfection (n = 5 independent experiments per group). (B and C) OCR curves and quantification of basal respiration, maximal respiration, and ATP-coupled respiration in primary cardiomyocytes under various treatment settings (n = 6 independent experiments per group). (D and E) Representative images and quantification of fluorescence intensity of MitoSOX staining in adult mouse cardiomyocytes (AMCMs) under various treatment settings (n = 20 independent experiments per group). (F and G) Representative images and quantification of fluorescence intensity of TMRM staining in AMCMs under various treatment settings (n = 20–30 independent experiments per group). (H) ATP production in primary cardiomyocytes under various treatment settings (n = 6 independent experiments per group). (I) Measurement of cell viability using CCK-8 and CellTiter-Glo luminescent assay in primary cardiomyocytes under various treatment settings (n = 6 independent experiments per group). (J) LDH leakage in supernatants of primary cardiomyocytes under various treatment settings (n = 6 independent experiments per group). Mean ± SEM. *p < 0.05, **p < 0.01, ****p < 0.0001, ns, no significance. For statistical analysis, two-way ANOVA with Tukey's test for multiple comparisons was used.
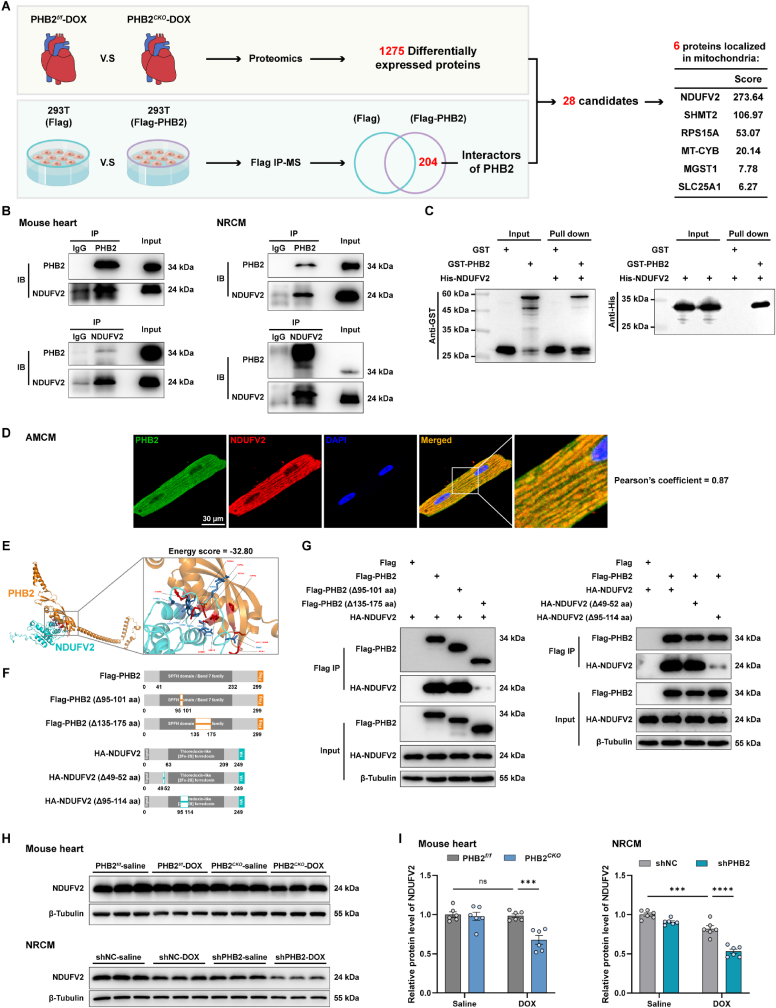
3.6. PHB2 interacted with NDUFV2 to regulate its expression level
Given the pivotal role of PHB2 in mitochondrial function, we explored downstream signaling mechanisms through which PHB2 deficiency modulated mitochondrial bioenergetics in the face of DOX cardiomyopathy. As aforementioned, TMT-labeled quantitative proteomic profiling using heart lysates detected 1275 differentially expressed proteins in PHB2CKO-DOX mice compared with PHB2flox/flox-DOX mice (Figs. S3A and S3B). In addition, 204 potential interactors of PHB2 were revealed using immunoprecipitation-mass spectrometry analysis after the exclusion of non-specific interactors (Fig. 6A). An effort to identify overlaps between differentially expressed proteins and PHB2 interactors yielded 28 candidate proteins as listed in Table S6, six of which were localized in mitochondria, including NDUFV2 (NADH-ubiquinone oxidoreductase core subunit V2), SHMT2 (Serine hydroxymethyltransferase, mitochondrial), PRS15A (Glutathione S-transferase D1), MT-CYB (Cytochrome b), MGST1 (Microsomal glutathione S-transferase 1) and SLC25A1 (Tricarboxylate transport protein, mitochondrial). NDUFV2 ranked first based on the Score Sequest HT in the protein-protein interaction analysis (Fig. 6A).
Fig. 6.
PHB2 interacted with NDUFV2 to regulate its expression level.
(A) Illustrations depicting the experimental approach to screen proteins downstream of PHB2 using proteomic profiling in conjunction with immunoprecipitation-mass spectrometry analysis. Quantitative proteomics using mouse heart lysates revealed 1275 differentially expressed proteins between PHB2CKO-DOX and PHB2f/f-DOX groups (n = 3 per group, differentially expressed proteins were identified using FC > 1.5 and p < 0.05). Meanwhile, immunoprecipitation-mass spectrometry (IP-MS) analysis identified 204 potential interactors of PHB2 after the exclusion of non-specific ones. Overlap between the 1275 differentially expressed proteins and 204 PHB2 interactors yielded 28 candidate proteins (listed in Table S6), and then these candidates were narrowed down to 6 proteins localized in mitochondria, among which NDUFV2 ranked the first based on the Score Sequest HT in the protein-protein interaction analysis. (B) Representative immunoblotting images following co-immunoprecipitation (co-IP) using anti-PHB2 and anti-NDUFV2 antibodies, respectively, in mouse heart lysates and isolated NRCMs. (C) GST-pulldown assay indicating physical interaction between PHB2 and NDUFV2 proteins. (D) Representative immunofluorescence images depicting colocalization between PHB2 and NDUFV2 in AMCMs. (E) Structure-based protein interaction interface analysis between PHB2 and NDUFV2 proteins with interactive residues predicted. (F and G) Construction of wild-type and truncated mutants of PHB2 and NDUFV2 vectors, and representative immunoblotting images of co-IP assays in transfected HEK293T cells. (H and I) Representative immunoblotting images and quantification of NDUFV2 in mouse heart lysates and NRCMs (β-Tubulin used as an internal control, n = 6 per group). NRCMs were treated with DOX challenge (0.1 μM) for 24 h after adenovirus transfection. Mean ± SEM. ***p < 0.001, ****p < 0.0001, ns, no significance. For statistical analysis, two-way ANOVA with Tukey's test for multiple comparisons was used.
Co-immunoprecipitation assays identified an interaction between PHB2 and NDUFV2 in mouse hearts and primary cardiomyocytes (Fig. 6B). Next, in vitro pull-down assay also supported the presence of physical interaction between these two proteins (Fig. 6C). Immunofluorescence staining in primary cardiomyocytes also consolidated an interaction between PHB2 and NDUFV2, with a Pearson's correlation coefficient of 0.87 (Fig. 6D). To determine the molecular domains responsible for protein interaction, molecular docking was carried out to predict possible interacting sites (Fig. 6E). Accordingly, truncated mutants for PHB2 and NDUFV2 were constructed for co-immunoprecipitation assays using the HEK293T cell line (Fig. 6F). Results showed that NDUFV2 interacted with full-length PHB2 and PHB2 Δ95-101 aa mutant, but not with the PHB2 Δ135-175 aa mutant when co-expressed in HEK293T cells, indicating an obligatory role for 135–175 aa domain of PHB2 for interaction with NDUFV2 (Fig. 6G). On the other hand, PHB2 interacted with full-length NDUFV2 and NDUFV2 Δ49-52 aa mutant, but not NDUFV2 Δ95-114 aa mutant, suggesting a role for 95–114 aa domain of NDUFV2 for interaction with PHB2 (Fig. 6G).
To verify results from proteomic profiling, Western blot was performed to examine protein levels in heart lysates. A significant decrease in NDUFV2 was observed in PHB2CKO-DOX mice compared with PHB2flox/flox-DOX mice (Fig. 6H and I), consistent with the protein level of mitochondrial complex I maker NDUFB8 (Fig. 4E and F). Moreover, DOX-induced decline in NDUFV2 was exacerbated by PHB2 knockdown in primary cardiomyocytes (Fig. 6H and I). Taken together, these results favor that PHB2 interacted with NDUFV2, and PHB2 deficiency instigated the downregulation of NDUFV2.
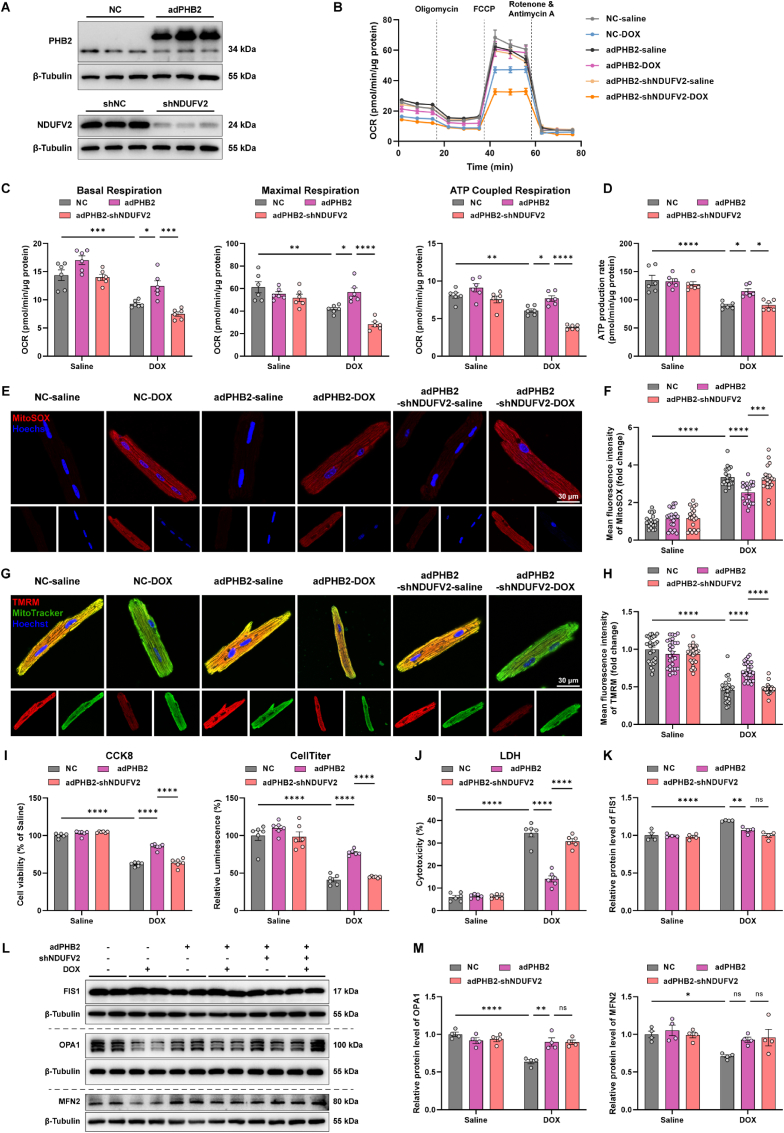
3.7. PHB2 overexpression alleviated DOX-induced mitochondrial dysfunction through NDUFV2 in cardiomyocytes
To explore the role of PHB2-NDUFV2 interaction in DOX cardiotoxicity, expression of PHB2 and NDUFV2 were manipulated in cardiomyocytes using adenoviruses carrying vectors encoding PHB2 or shRNA targeting NDUFV2 (Fig. 7A). PHB2 overexpression reversed the DOX-induced decrease in mitochondrial basal respiration, maximal respiration, and ATP-coupled respiration in murine cardiomyocytes, the effect of which was diminished by NDUFV2 silencing (Fig. 7B and C). In addition, PHB2 overexpression improved DOX-reduced ATP production, the effect of which was diminished by NDUFV2 knockdown (Fig. 7D). MitoSOX probes displayed a significant increase in mitochondrial oxidative stress and ROS production following DOX treatment, whereas such a rise in mitochondrial oxidative stress was partially removed by PHB2 overexpression. NDUFV2 silencing abrogated the protective effect of PHB2 in cardiomyocytes (Fig. 7E and F). TMRM finding denoted that DOX-induced collapse in mitochondrial membrane potential was rescued by PHB2 overexpression, the effect of which was ablated by shNDUFV2 (Fig. 7G and H). Consequently, PHB2 overexpression reversed DOX-induced loss of cell viability and increase in LDH release, the effect of which was diminished by NDUFV2 knockdown (Fig. 7I and J). These findings suggested that PHB2 overexpression alleviated DOX-induced mitochondrial dysfunction and cardiotoxicity through the governance of NDUFV2 in cardiomyocytes. Besides, DOX-induced elevation in FIS1 and decline in OPA1 were reversed by PHB2 overexpression in primary cardiomyocytes, although NDUFV2 silencing failed to affect these protein levels (Fig. 7K–M).
Fig. 7.
PHB2 overexpression alleviated DOX-induced mitochondrial dysfunction through NDUFV2 in cardiomyocytes.
(A) Representative immunoblotting images of PHB2 overexpression and NDUFV2 knockdown using adenoviruses in primary cardiomyocytes (β-Tubulin used as an internal control). (B and C) OCR curves and quantification of basal respiration, maximal respiration, and ATP-coupled respiration in primary cardiomyocytes with and without DOX challenge (0.1 μM) for 24 h after adenovirus transfection (n = 6 independent experiments per group). (D) ATP production rate in primary cardiomyocytes under various treatment settings (n = 6 independent experiments per group). (E and F) Representative images and quantification of fluorescence intensity of MitoSOX staining in AMCMs under various treatment settings (n = 20 independent experiments per group). (G and H) Representative images and quantification of fluorescence intensity of TMRM staining in AMCMs under various treatment settings (n = 20–30 independent experiments per group). (I) Measurement of cell viability using CCK-8 and CellTiter-Glo luminescent assay in primary cardiomyocytes under various treatment settings (n = 6 independent experiments per group). (J) LDH leakage in supernatants of primary cardiomyocytes under various treatment settings (n = 6 independent experiments per group). (K, L, and M) Representative immunoblotting images and quantification of FIS1, OPA1, and MFN2 in primary cardiomyocytes under various treatment settings (β-Tubulin used as an internal control, n = 4 independent experiments per group). Mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, no significance. For statistical analysis, two-way ANOVA with Tukey's test for multiple comparisons was used.
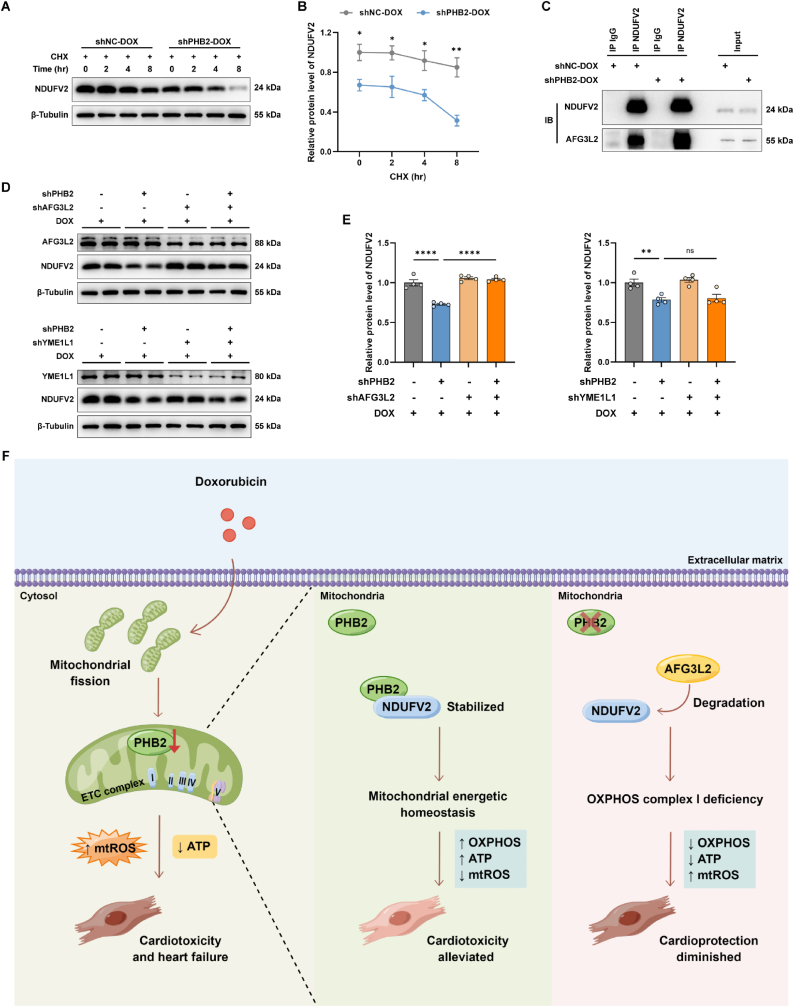
3.8. PHB2 regulated NDUFV2 expression by facilitating its stabilization
To examine the mechanisms underscoring how PHB2 deficiency affected NDFUV2 expression, a cycloheximide (CHX) chase assay was conducted in primary cardiomyocytes, which evaluates the degradation of endogenous proteins by inhibition of protein translation. PHB2 silencing significantly accelerated the degradation of NDUFV2 in DOX-challenged cardiomyocytes, indicating the possible role of PHB2 in NDUFV2 stabilization (Fig. 8A and B).
Fig. 8.
PHB2 regulated NDUFV2 expression by facilitating its stabilization.
(A and B) Representative immunoblot images and quantification of NDUFV2 in cycloheximide (CHX) chase assay in primary cardiomyocytes from shNC-DOX and shPHB2-DOX groups (n = 3 per group). Cells were treated with DOX (0.1 μM) for 24 h before the addition of CHX. (C) Representative immunoblot images following co-IP with anti-NDUFV2 antibody in primary cardiomyocytes. (D and E) Representative immunoblot images and quantification of NDUFV2 under knockdown of PHB2, AFG3L2, or YME1L1 in primary cardiomyocytes with DOX challenge (0.1 μM) for 24 h (β-Tubulin used as an internal control, n = 4 per group). (F) Schematic diagram depicting the purposed role of PHB2 in DOX cardiomyopathy. PHB2 protects against DOX-induced cardiomyopathy through interacting with OXPHOS complex I core subunit NDUFV2 to preserve mitochondrial energetic homeostasis. PHB2 deficiency leads to the degradation of NDUFV2 and impairment of OXPHOS, thus inhibiting mitochondrial bioenergetics and exacerbating DOX-induced cardiotoxicity and heart failure. Mean ± SEM. *p < 0.05, **p < 0.01, ****p < 0.0001, ns, no significance. For statistical analysis, unpaired student's t-test was used for B; one-way ANOVA with Tukey's test for multiple comparisons was used for E.
PHB1 and PHB2 form a multimeric ring-like structure in the inner mitochondrial membrane (IMM) to partner with the m-AAA protease AFG3L2 to suppress its proteolytic activity [22,45]. AFG3L2 is an evolutionarily conserved m-AAA proteolytic component responsible for protein homeostasis in IMM [36,46]. Given that PHB2 deficiency dampened NDUFV2 stabilization, we next asked if AFG3L2 served as a proteolytic unit for NDUFV2 degradation. Immunoprecipitation assays exhibited an enhanced interaction between AFG3L2 and NDUFV2 by PHB2 silencing in DOX-challenged cardiomyocytes, suggesting that PHB2 deficiency enabled the AFG3L2 access to NDUFV2 (Fig. 8C). Next, we knocked down the m-AAA protease AFG3L2 to test whether it reversed the loss of NDUFV2 by PHB2 deficiency. Notably, AFG3L2 depletion rescued the decrease in the NDUFV2 protein level in cardiomyocytes with PHB2 deficiency, indicating that AFG3L2 was required for the degradation of NDUFV2 (Fig. 8D and E). In contrast, depletion of the i-AAA protease YME1L1 failed to rescue NDUFV2 protein levels in cardiomyocytes with PHB2 knockdown (Fig. 8D and E). These results indicated that PHB2 stabilized NDUFV2 through suppression of the proteolytic activity of AFG3L2 to restrict its access to the substrate NDUFV2.
4. Discussion
The salient findings from our current study revealed a protective role of mitochondrial protein PHB2 against DOX cardiomyopathy. Our results noted that PHB2 deficiency exacerbated cardiac contractile dysfunction and mitochondrial injury in DOX-challenged mice and cardiomyocytes both in vivo and in vitro. Mechanistically, PHB2 directly interacted with the mitochondrial respiratory complex I core subunit NDUFV2 to preserve its expression, thus promoting mitochondrial oxidative phosphorylation and energy production. On the other side of the coin, overexpression of PHB2 effectively alleviated DOX cardiotoxicity and improved mitochondrial function. Taken together, these findings suggest that elevating PHB2 levels may represent a novel therapeutic avenue in the combat against DOX cardiomyopathy (Fig. 8F).
In our hands, cardiac overexpression of PHB2 alleviated cardiac atrophy, myocardial fibrosis, systolic dysfunction, and cardiac injury evoked by DOX challenge. On the other side of the coin, cardiac-specific PHB2 conditional knockout aggravated DOX-provoked cardiac dysfunction and mitochondrial damage in vivo and in vitro. To-date, the cardioprotective property of PHB2 has been indicated in pathological settings including cardiac remodeling, heart failure, ischemia-reperfusion injury, and diabetic cardiomyopathy [27,47,48]. Here in our work, we generated cardiac-specific PHB2 knockout mice for the first time by crossing PHB2flox/flox mice with αMyHC-Cre mice, which turned out to exhibit cardiac enlargement and severe heart failure at an early age (data not shown), consistent with an earlier report [25]. To this end, tamoxifen-induced conditional cardiac-specific PHB2 knockout mice were created instead by crossing PHB2flox/flox mice with Myh6-Cre-ERT2 mice. Tamoxifen was delivered to mice at 8 weeks of age, and DOX was administrated 2 weeks later. The efficacy of PHB2 knockout was verified using Western blot and immunohistochemistry staining. PHB2flox/floxMyh6-CreERT2 mice exhibited normal cardiac morphology, contractile function, and survival in the absence of stress, at least three months following tamoxifen administration. An earlier study employed cardiac-specific PHB2 knockout mice by crossing PHB2flox/flox mice with Mlc2v-Cre mice [27]. These PHB2 knockout mice exhibited severe heart failure at an early age (8 weeks old) as evidenced by dilated left ventricles, systolic dysfunction, and interstitial fibrosis, and died at an age of 10 weeks (50% mortality by day 70), accompanied with lipid accumulation and mitochondrial dysfunction in mouse hearts. The authors speculated PHB2 as a pivotal regulator for fatty acid oxidation, while PHB2 deficiency evoked mitochondrial dysfunction due to the impairment of fatty acid metabolism, accumulation of lipid droplets, and lack of energy provision [27]. The apparent discrepancies in phenotypes following PHB2 knockout between our study and this earlier report may be attributed to the timing of gene ablation (embryonic deletion versus conditional removal at adulthood). To the best of our knowledge, this represents the first study concerning the role of PHB2 as a mitochondrial regulator in the face of DOX cardiomyopathy. Our findings have provided compelling evidence for the possible role of PHB2 as a key modulator of mitochondrial OXPHOS, bioenergetics, mitochondrial dynamics, and morphology.
Given the contribution of PHB2 deficiency in the exacerbated cardiac dysfunction, downstream signaling cascade of PHB2 was evaluated with a particular focus on the mitochondrial complex I subunit NDUFV2. Previous studies revealed a fundamental role of mitochondrial bioenergetics in various forms of cardiovascular diseases such as heart failure and aortic aneurysm development [9,38]. Mitochondrial respiratory complexes, especially complex I, are becoming critical modulators for cardiac function. Cardiac expression of NDUFS1 (NADH-ubiquinone oxidoreductase core subunit S1) was low in heart failure patients and post-myocardial infarction mouse models. Cardiac-specific overexpression of NDUFS1 alleviated cardiac dysfunction following myocardial infarction by ameliorating mitochondrial respiratory function and reducing apoptosis [49]. Another core subunit of mitochondrial complex I NDUFV2 has been reported to modulate mitochondrial function in adipose tissues and metabolic syndrome in a sex-specific manner, by promoting oxidative phosphorylation, supercomplex assembly, mitochondrial biogenesis, and regulating mtDNA content [50]. However, the precise role of NDUFV2 in DOX cardiomyopathy remains elusive. Our study demonstrated that cardiac-specific PHB2 deficiency accentuated DOX-evoked mitochondrial metabolic dysfunction likely through downregulation of NDUFV2 expression, with a major role in mitochondrial oxidative phosphorylation and energy production. Nevertheless, the DOX-induced mitochondrial fission was reversed by PHB2 overexpression while NDUFV2 silencing failed to affect these fission molecules. These findings indicated the possible contribution of other pathways through which PHB2 regulated mitochondrial dynamics, given the role of PHB2 as a multi-functional protein in the mitochondrial inner membrane [29,43].
Experimental limitations: Several drawbacks may exist in our current study. First, considering the importance of mitochondrial proteins in energy metabolism and free radical production under DOX cardiomyopathy, possible contribution from other IMM proteins deserves further scrutiny. In addition, complicated mechanisms may participate in DOX-induced cardiomyopathy including DNA injury, oxidative stress, and cell death, other than PHB2-governed mitochondrial function [51,52]. Second, AAV9 was employed to overexpress PHB2 in mouse hearts through intravenous injection, and this maneuver is widely used for cardiac overexpression [32,53,54]. Although cardiac-specific promoter cTNT was utilized, AAV9 may also cause off-target forced expression in non-cardiac organs including liver, lung, muscle and nervous system [55]. Therefore, transgenic mice would be more appropriate for the gain-of-function study for PHB2 in DOX cardiomyopathy. Third, different half-lives of proteins were not taken into consideration in our screening of PHB2 downstream targets. Besides, the role of NDUFV2 in cardiac mitochondrial metabolism should be further tested both in vivo and in vitro. A cardiac-specific NDUFV2 overexpression mouse model would be required to delineate its unique role in DOX cardiomyopathy. Last but not the least, given the interrelationship between PHB1 and PHB2, possible involvement of PHB1 in DOX cardiomyopathy should not be ignored at this point. In particular, FL3 was identified as the synthetic ligand for PHBs to promote their mitochondrial translocation. FL3 induced heterodimerization of PHB1 with STAT3 and thereby STAT3 phosphorylation, which protected mice from DOX cardiotoxicity [56]. Although this earlier study mainly focused on the role of FL3 through mediating PHB1 [56], more studies are warranted to delineate the possible interaction of PHB2 with FL3.
DOX is an antitumor agent widely used in the clinical treatment of solid tumors and hematologic tumors, whereas its usage is partially restricted by its cardiotoxicity. In conclusion, our current study highlights PHB2 overexpression as a potential therapeutic target in DOX cardiomyopathy by stabilization of NDUFV2 to promote mitochondrial bioenergetic metabolism. Our findings are in line with previous notions for the role of PHB2 in cardiovascular diseases [27], and offer further support for PHB2 as an essential regulator of mitochondrial homeostasis. Taken together, cardiac overexpression of PHB2 may serve as an effective therapeutic target in DOX cardiotoxicity, and more clinical research is still warranted to consolidate the benefit of PHB2 in DOX cardiomyopathy among cancer patients.
Author contributions
MY, MA, XW, YZhang, and JR were responsible for the design, data collection, data analysis and interpretation, and manuscript preparation. YZhou performed molecular docking and provided helpful comments and necessary resources for this study. Yingmei Zhang and Jun Ren provided financial support and overall guidance for this study and served as guarantors for this work.
Declaration of competing interest
None of the authors declare any potential conflict of interest.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (82130011 and 82270267) and the Program of Shanghai Academic/Technology Research Leader 20XD1420900. In addition, we would appreciate the generous assistance from Dr. Yu Kong and Xu Wang (Electron Microscopy Facilities of Center for Excellence in Brain Science and Technology, Chinese Academy of Science) in TEM sample preparation and image analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102812.
Contributor Information
Yingmei Zhang, Email: zhang.yingmei@zs-hospital.sh.cn.
Jun Ren, Email: ren.jun@zs-hospital.sh.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Gabizon A.A., Patil Y., La-Beck N.M. New insights and evolving role of pegylated liposomal doxorubicin in cancer therapy. Drug Resist. Updates. 2016;29:90–106. doi: 10.1016/j.drup.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Cardinale D., et al. Early detection of anthracycline cardiotoxicity and improvement with heart failure therapy. Circulation. 2015;131(22):1981–1988. doi: 10.1161/CIRCULATIONAHA.114.013777. [DOI] [PubMed] [Google Scholar]
- 3.Yeh E.T., Bickford C.L. Cardiovascular complications of cancer therapy: incidence, pathogenesis, diagnosis, and management. J. Am. Coll. Cardiol. 2009;53(24):2231–2247. doi: 10.1016/j.jacc.2009.02.050. [DOI] [PubMed] [Google Scholar]
- 4.Chang H.M., et al. Cardiovascular complications of cancer therapy: best practices in diagnosis, prevention, and management: Part 1. J. Am. Coll. Cardiol. 2017;70(20):2536–2551. doi: 10.1016/j.jacc.2017.09.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang H.M., et al. Cardiovascular complications of cancer therapy: best practices in diagnosis, prevention, and management: Part 2. J. Am. Coll. Cardiol. 2017;70(20):2552–2565. doi: 10.1016/j.jacc.2017.09.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan G.C., et al. Heat shock protein 20 interacting with phosphorylated Akt reduces doxorubicin-triggered oxidative stress and cardiotoxicity. Circ. Res. 2008;103(11):1270–1279. doi: 10.1161/CIRCRESAHA.108.182832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S., et al. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat Med. 2012;18(11):1639–1642. doi: 10.1038/nm.2919. [DOI] [PubMed] [Google Scholar]
- 8.Lopaschuk G.D., et al. Cardiac energy metabolism in heart failure. Circ. Res. 2021;128(10):1487–1513. doi: 10.1161/CIRCRESAHA.121.318241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhuang L., et al. DYRK1B-STAT3 drives cardiac hypertrophy and heart failure by impairing mitochondrial bioenergetics. Circulation. 2022;145(11):829–846. doi: 10.1161/CIRCULATIONAHA.121.055727. [DOI] [PubMed] [Google Scholar]
- 10.Ren J., et al. Mitochondrial biogenesis in the metabolic syndrome and cardiovascular disease. J. Mol. Med. (Berl.) 2010;88(10):993–1001. doi: 10.1007/s00109-010-0663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung K.G., et al. Sirtuin-3 (SIRT3) protein attenuates doxorubicin-induced oxidative stress and improves mitochondrial respiration in H9c2 cardiomyocytes. J. Biol. Chem. 2015;290(17):10981–10993. doi: 10.1074/jbc.M114.607960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marques-Aleixo I., et al. Physical exercise prior and during treatment reduces sub-chronic doxorubicin-induced mitochondrial toxicity and oxidative stress. Mitochondrion. 2015;20:22–33. doi: 10.1016/j.mito.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Osataphan N., et al. Effects of doxorubicin-induced cardiotoxicity on cardiac mitochondrial dynamics and mitochondrial function: insights for future interventions. J. Cell Mol. Med. 2020;24(12):6534–6557. doi: 10.1111/jcmm.15305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010;11(12):872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt K., et al. Circadian control of DRP1 activity regulates mitochondrial dynamics and bioenergetics. Cell Metabol. 2018;27(3):657–666.e5. doi: 10.1016/j.cmet.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 16.Xia Y., et al. LCZ696 improves cardiac function via alleviating Drp1-mediated mitochondrial dysfunction in mice with doxorubicin-induced dilated cardiomyopathy. J. Mol. Cell. Cardiol. 2017;108:138–148. doi: 10.1016/j.yjmcc.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Galán-Arriola C., et al. Remote ischaemic preconditioning ameliorates anthracycline-induced cardiotoxicity and preserves mitochondrial integrity. Cardiovasc. Res. 2021;117(4):1132–1143. doi: 10.1093/cvr/cvaa181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P., et al. SESN2 protects against doxorubicin-induced cardiomyopathy via rescuing mitophagy and improving mitochondrial function. J. Mol. Cell. Cardiol. 2019;133:125–137. doi: 10.1016/j.yjmcc.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Wei Y., et al. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2017;168(1–2):224–238.e10. doi: 10.1016/j.cell.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernando-Rodríguez B., Artal-Sanz M. Mitochondrial quality control mechanisms and the PHB (prohibitin) complex. Cells. 2018;7(12) doi: 10.3390/cells7120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Signorile A., et al. Prohibitins: a critical role in mitochondrial functions and implication in diseases. Cells. 2019;8(1) doi: 10.3390/cells8010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merkwirth C., Langer T. Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochim. Biophys. Acta. 2009;1793(1):27–32. doi: 10.1016/j.bbamcr.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Bavelloni A., et al. Prohibitin 2: at a communications crossroads. IUBMB Life. 2015;67(4):239–254. doi: 10.1002/iub.1366. [DOI] [PubMed] [Google Scholar]
- 24.Jian C., et al. Deficiency of PHB complex impairs respiratory supercomplex formation and activates mitochondrial flashes. J. Cell Sci. 2017;130(15):2620–2630. doi: 10.1242/jcs.198523. [DOI] [PubMed] [Google Scholar]
- 25.Anderson C.J., et al. Prohibitin levels regulate OMA1 activity and turnover in neurons. Cell Death Differ. 2020;27(6):1896–1906. doi: 10.1038/s41418-019-0469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan C., et al. PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy. 2020;16(3):419–434. doi: 10.1080/15548627.2019.1628520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu D., et al. Prohibitin 2 deficiency impairs cardiac fatty acid oxidation and causes heart failure. Cell Death Dis. 2020;11(3):181. doi: 10.1038/s41419-020-2374-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia Y., et al. Circ Res; 2022. PHB2 Maintains the Contractile Phenotype of VSMCs by Counteracting PKM2 Splicing. 101161circresaha122321005. [DOI] [PubMed] [Google Scholar]
- 29.Li L., et al. In vivo stabilization of OPA1 in hepatocytes potentiates mitochondrial respiration and gluconeogenesis in a prohibitin-dependent way. J. Biol. Chem. 2019;294(34):12581–12598. doi: 10.1074/jbc.RA119.007601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sohal D.S., et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 2001;89(1):20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 31.Madisen L., et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han D., et al. The tumor-suppressive human circular RNA CircITCH sponges miR-330-5p to ameliorate doxorubicin-induced cardiotoxicity through upregulating SIRT6, survivin, and SERCA2a. Circ. Res. 2020;127(4):e108–e125. doi: 10.1161/CIRCRESAHA.119.316061. [DOI] [PubMed] [Google Scholar]
- 33.Yang M., et al. Deletion of the E3 ubiquitin ligase, Parkin, exacerbates chronic alcohol intake-induced cardiomyopathy through an Ambra1-dependent mechanism. Br. J. Pharmacol. 2021;178(4):964–982. doi: 10.1111/bph.15340. [DOI] [PubMed] [Google Scholar]
- 34.Maurya S.K., et al. Western Diet Causes Heart Failure With Reduced Ejection Fraction and Metabolic Shifts After Diastolic Dysfunction and Novel Cardiac Lipid Derangements. 2023;8(4):422–435. doi: 10.1016/j.jacbts.2022.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ackers-Johnson M., et al. A simplified, langendorff-free method for concomitant isolation of viable cardiac myocytes and nonmyocytes from the adult mouse heart. Circ. Res. 2016;119(8):909–920. doi: 10.1161/CIRCRESAHA.116.309202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T., et al. C9orf72 regulates energy homeostasis by stabilizing mitochondrial complex I assembly. Cell Metabol. 2021;33(3):531–546.e9. doi: 10.1016/j.cmet.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalkhoran S.B., et al. Hydralazine protects the heart against acute ischaemia/reperfusion injury by inhibiting Drp1-mediated mitochondrial fission. Cardiovasc. Res. 2022;118(1):282–294. doi: 10.1093/cvr/cvaa343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun L.Y., et al. Circulation; 2022. Nuclear Receptor NR1D1 Regulates Abdominal Aortic Aneurysm Development by Targeting the Mitochondrial Tricarboxylic Acid Cycle Enzyme Aconitase-2. 101161circulationaha121057623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tunyasuvunakool K., et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596(7873):590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baspinar A., et al. PRISM: a web server and repository for prediction of protein-protein interactions and modeling their 3D complexes. Nucleic Acids Res. 2014;42:W285–W289. doi: 10.1093/nar/gku397. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S.L., et al. A GYS2/p53 negative feedback loop restricts tumor growth in HBV-related hepatocellular carcinoma. Cancer Res. 2019;79(3):534–545. doi: 10.1158/0008-5472.CAN-18-2357. [DOI] [PubMed] [Google Scholar]
- 42.Daya S., Berns K.I. Gene therapy using adeno-associated virus vectors. Clin. Microbiol. Rev. 2008;21(4):583–593. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang T., et al. Mitoquinone attenuates blood-brain barrier disruption through Nrf2/PHB2/OPA1 pathway after subarachnoid hemorrhage in rats. Exp. Neurol. 2019;317:1–9. doi: 10.1016/j.expneurol.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 44.De Rasmo D., et al. PBMC of multiple sclerosis patients show deregulation of OPA1 processing associated with increased ROS and PHB2 protein levels. Biomedicines. 2020;8(4) doi: 10.3390/biomedicines8040085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osman C., Merkwirth C., Langer T. Prohibitins and the functional compartmentalization of mitochondrial membranes. J. Cell Sci. 2009;122(Pt 21):3823–3830. doi: 10.1242/jcs.037655. [DOI] [PubMed] [Google Scholar]
- 46.Patron M., Sprenger H.G., Langer T. m-AAA proteases, mitochondrial calcium homeostasis and neurodegeneration. Cell Res. 2018;28(3):296–306. doi: 10.1038/cr.2018.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K., et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat. Commun. 2014;5:3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 48.Dong W.Q., et al. Prohibitin overexpression improves myocardial function in diabetic cardiomyopathy. Oncotarget. 2016;7(1):66–80. doi: 10.18632/oncotarget.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qi B., et al. Cardiac-specific overexpression of Ndufs1 ameliorates cardiac dysfunction after myocardial infarction by alleviating mitochondrial dysfunction and apoptosis. Exp. Mol. Med. 2022;54(7):946–960. doi: 10.1038/s12276-022-00800-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chella Krishnan K., et al. Sex-specific genetic regulation of adipose mitochondria and metabolic syndrome by Ndufv2. Nat Metab. 2021;3(11):1552–1568. doi: 10.1038/s42255-021-00481-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H., et al. Self-maintenance of cardiac resident reparative macrophages attenuates doxorubicin-induced cardiomyopathy through the SR-A1-c-myc Axis. Circ. Res. 2020;127(5):610–627. doi: 10.1161/CIRCRESAHA.119.316428. [DOI] [PubMed] [Google Scholar]
- 52.Heidt T., et al. Differential contribution of monocytes to heart macrophages in steady-state and after myocardial infarction. Circ. Res. 2014;115(2):284–295. doi: 10.1161/CIRCRESAHA.115.303567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanhoutte D., et al. Thbs1 induces lethal cardiac atrophy through PERK-ATF4 regulated autophagy. Nat. Commun. 2021;12(1):3928. doi: 10.1038/s41467-021-24215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y., et al. HINT1 (histidine triad nucleotide-binding protein 1) attenuates cardiac hypertrophy via suppressing HOXA5 (homeobox A5) expression. Circulation. 2021;144(8):638–654. doi: 10.1161/CIRCULATIONAHA.120.051094. [DOI] [PubMed] [Google Scholar]
- 55.Zincarelli C., et al. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16(6):1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 56.Qureshi R., et al. FL3, a synthetic flavagline and ligand of prohibitins, protects cardiomyocytes via STAT3 from doxorubicin toxicity. PLoS One. 2015;10(11) doi: 10.1371/journal.pone.0141826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.