Abstract
Alzheimer's disease (AD) has attracted considerable attention from the public and scientific researchers, leading to a rapid growth in relevant research on this disorder in the last 10 years. The present study aimed to conduct a bibliometric analysis to elucidate the trends of global research on the role of apolipoprotein E in AD in the past decade. Three bibliometric software (CiteSpace, VOSviewer, and R Bibliometrix) were used to analyze the active journals, countries/regions, institutes, authors, co-cited references, and keywords in this field. The USA was the most influential country, and the University of California was the most productive institute. Zetterberg H contributed the highest number of publications, and Petersen RC was the most cited author in this field. On the basis of the co-cited reference analysis, knowledge base on biomarkers, risk factors, and mechanisms were updated in the past decade. Current research hotspots are shifting to tau-related mechanisms and identification of genetic risk factors. Our study provides insights into the developing knowledge base and trends related to research on apolipoprotein E in AD, which may provide new directions for further research in this field.
Keywords: Alzheimer's disease, Apolipoprotein E, Biomarker, Mechanism, Risk factor
Highlights
-
•
Research on apolipoprotein E in Alzheimer's disease has been burgeoning in the past decade.
-
•
The USA is the most influential country in this field.
-
•
Zetterberg H, Blennow K, and Petersen RC are the most influential authors.
-
•
Knowledge base has been updated for biomarkers, risk factors, and mechanisms.
-
•
Hotspots are shifting to tau-related mechanisms and genetic risk factor identification.
1. Introduction
Alzheimer's disease (AD), the leading cause of cognitive impairment and dementia in older individuals, is diagnosed in people with positive results for amyloid and tau biomarkers together with specific clinical phenotypes [1]. According to the US Alzheimer's Association, AD is ranked as the fifth leading cause of death among Americans aged 65 years and older and the seventh leading cause of death in the USA [2]. An estimated 6.5 million Americans aged 65 years and older are living with Alzheimer's dementia today, and this number could reach 13.8 million by 2060 [2]. The high prevalence, high fatality rate, high nursing cost, and increasing medical expenditure related to AD have led to increased public concern and scientific interest along with a rapid growth in research on neuroscience and AD in the past decades [3].
Human apolipoprotein E (APOE), the most abundant apolipoprotein in the brain, plays a crucial role in redistributing cholesterol and other lipids to neurons [4]. In the past decade, there has been sustained interest in understanding mechanisms and developing therapeutic approaches for targeting APOE in AD. The role of APOE in AD pathogenesis has broadened from amyloid-β (Aβ)-related mechanisms to tau neurofibrillary degeneration, microglial and astrocytic response, and disruption of the blood–brain barrier (BBB) [5]. APOE also influences AD pathogenesis through synaptic and mitochondrial functions, synaptogenesis, neuroinflammation, glucose metabolism, insulin signaling, and cerebrovascular integrity [See review [ [3,6,7]]]. Recently published studies have updated the knowledge base of AD. The enormous number of relevant published articles has, however, made it difficult to extract the key knowledge nodes and identify the research frontier in this field.
Bibliometrics is the analysis of published information (e.g., books, journal articles, datasets, and blogs) and its related metadata (e.g., abstracts, keywords, and citations) by using statistics to describe or show relationships between the published works [8]. Bibliometrics is based on the assumption that a research field's scholarly output can be captured in the published literature [9]. Bibliometric analysis uses comprehensive approaches such as mathematical methods, network analysis, and clustering algorithms to analyze the general profiles of published literatures and to show the current status and trends of various fields from a quantitative and objective perspective [10]. CiteSpace [11], VOSviewer [12], and R Bibliometrix [13] are the three most popular bibliometric software in recent years. Because of the free availability of these bibliometric tools and the exponential increase in published literature, bibliometric analysis has been used to analyze the development of many specific research fields in recent years [ [[14], [15], [16], [17]]].
To the best of our knowledge, there is currently a lack of systematic analysis on the development trend of research on APOE in AD in the past decade. Only one similar scientometric analysis on the role of APOE in the CNS was published by Gong et al. [18] in 2020, which suggested an outmoded research field of APOE in the CNS. However, in the era of AD, some significant changes have occurred with recent publication of pivotal studies, thereby providing additional data that revealed novel perspectives. Accordingly, we conducted a bibliometric analysis of publications on the role of APOE in AD to provide an overview of literature characteristics and explore the emerging trends in this domain.
2. Methods
We followed a systematic procedure [19] to analyze the literature on APOE in AD. Relevant papers were retrieved from the Web of Science Core Collection (WoSCC) database. A statistical descriptive analysis was performed using three bibliometric software [CiteSpace [11], VOSviewer [12], and R Bibliometrix [13]] with different dimensions and perspectives. Search strategy details and data analysis are shown in Methods in the supplementary material.
3. Results
3.1. General characteristics
A total of 9125 documents (7445 articles and 1680 reviews) related to APOE in AD were retrieved from the WoSCC database. Fig. 1 shows the number of documents by year and document type. There were 507 documents in 2011 and 756 documents in 2021, and the annual growth rate of documents was 4.08%. The 9125 documents of APOE in AD received 238,782 citations in total, with an average of 297 citations per document. The number and citations of relevant documents indicated a steady development of research in this domain in the past decade.
Fig. 1.
Distribution and growth trends of documents and citations on APOE in AD from 2011 to 2021.
The major funding agencies supporting the research in this domain included the National Institute on Aging, USA (3310 documents); the National Institutes of Health, USA (1445 documents); Alzheimer's Association, USA (759 documents); Medical Research Council, UK (687 documents); the National Institute of Neurological Disorders and Stroke, USA (608 documents); and the National Natural Science Foundation of China, China (534 documents).
3.2. Distribution of journals
A total of 1273 academic journals have published the abovementioned 9125 documents on APOE in AD. Table 1 lists the characteristics of 12 core journals that published 3021 articles, accounting for one-third of the total number of articles. The top three journals with most documents were Journal of Alzheimer's Disease (904 documents, IF2021 = 4.160), Neurobiology of Aging (387 documents, IF2021 = 5.133), and Alzheimer's & Dementia (302 documents, IF2021 = 16.655). The impact factor of JAMA Neurology (102 documents, IF2021 = 29.907), Alzheimer's & Dementia (302 documents, IF2021 = 16.655), and Brain (81 documents, IF2021 = 15.255) exceeded 10. Relevant documents published in Neurology, Neurobiology of Aging, Journal of Alzheimer's Disease, and Alzheimer's & Dementia received more than 10,000 local citations. Most of the top journals are specialized clinical neurology or neuroscience journals, and the majority are located in the USA and Europe.
Table 1.
Characters of core journals.
| Rank | Journal | Publications | Citations | Country/Region | IF2021 | Category and JCI Quartile |
|---|---|---|---|---|---|---|
| 1 | Journal of Alzheimer's Disease | 904 | 16,662 | Netherlands | 4.160 | Neurosciences, Q2 |
| 2 | Neurobiology of Aging | 387 | 17,846 | UK | 5.133 | Geriatrics & Gerontology, Q1; Neurosciences, Q1 |
| 3 | Alzheimer's & Dementia | 302 | 13,970 | USA | 16.655 | Clinical Neurology, Q1 |
| 4 | Plos One | 239 | 9202 | USA | 3.752 | Multidisciplinary Sciences, Q1 |
| 5 | Neurology | 225 | 28,963 | USA | 11.80 | Clinical Neurology, Q1 |
| 6 | Frontiers in Aging Neuroscience | 223 | 2412 | Switzerland | 5.702 | Geriatrics & Gerontology, Q2; Neurosciences, Q2 |
| 7 | Alzheimer's Research & Therapy | 189 | 2953 | UK | 8.835 | Clinical Neurology, Q1; Neurosciences, Q1 |
| 8 | Current Alzheimer Research | 148 | 2799 | U Arab Emirates | 3.040 | Clinical Neurology, Q3; Neurosciences, Q3 |
| 9 | Scientific Reports | 126 | 2721 | UK | 4.996 | Multidisciplinary Sciences, Q1 |
| 10 | JAMA Neurology | 102 | 4393 | USA | 29.907 | Clinical Neurology, Q1 |
| 11 | International Journal of Molecular Sciences | 95 | 1154 | Switzerland | 6.208 | Biochemistry & Molecular Biology, Q2; Chemistry, Multidisciplinary, Q2 |
| 12 | Brain | 81 | 7512 | UK | 15.255 | Clinical Neurology, Q1; Neurosciences, Q1 |
3.3. Distribution of countries/regions
A total of 101 countries/regions have published these 9125 relevant documents in this research field. Table 2 lists the characteristics of the top 10 most influential countries/regions. The USA was the most influential country, with the highest number of documents and citations (3392 documents and 160,697 citations), followed by the People's Republic of China (1178 documents and 22,360 citations), the UK (477 documents and 24,042 citations), Italy (341 documents and 8461 citations), and Australia (335 documents and 12,494 citations). The centralities of France (0.30), Germany (0.11), Canada (0.11), and Malaysia (0.11) were greater than 0.1, indicating that these countries might act as important intermediaries in this field. Most studies were completed in cooperation with researchers from multiple countries/regions. Fig. 2 shows the cooperation networks of the countries/regions. There were 1175 collaborating countries/regions worldwide. The top 3 of the most frequent cooperation networks were USA-UK, USA-China, and UK-Sweden.
Table 2.
Characters of the top 10 most productive countries/regions.
| Rank | Country | Documentsa | Citations |
|---|---|---|---|
| 1 | USA | 3392 | 160,697 |
| 2 | China | 1178 | 22,360 |
| 3 | UK | 477 | 24,042 |
| 4 | Italy | 341 | 8461 |
| 5 | Australia | 335 | 12,494 |
| 6 | Canada | 302 | 9300 |
| 7 | Germany | 297 | 9578 |
| 8 | Spain | 289 | 6593 |
| 9 | Sweden | 253 | 9011 |
| 10 | Netherlands | 250 | 9811 |
By corresponding author
Fig. 2.
Number of documents (A) and cooperation networks (B) of active countries/regions in this field.
3.4. Distribution of institutes
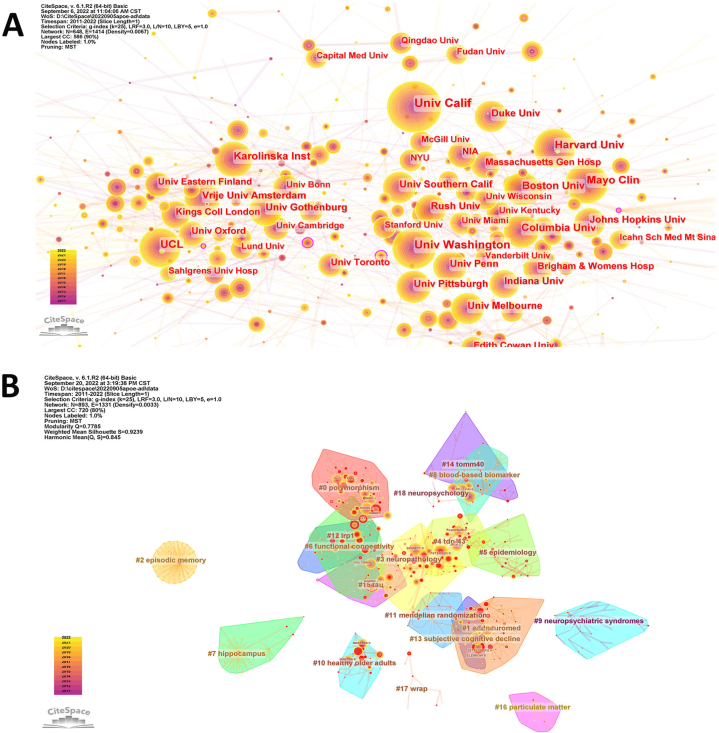
A total of 66,993 institutes contributed to this research field. Table 3 shows the top 10 institutes based on the number of documents and centrality. The University of California had the highest number of documents (3.76%), followed by Washington University (2.79%) and Boston University (1.65%). From the perspective of centrality, Lausanne University, Monash University, and Rhode Island Hospital showed the highest centrality (0.12). Most of the top institutes in this research field are located in the USA or Europe. A visualized co-occurrence network of major institutes is shown in Fig. 3A. These top organizations were grouped in accordance with their geographical locations on the whole.
Table 3.
Characters of the top 10 institutes based on publications and centrality.
| Rank | Institute | Documents | Centrality | Location | Rank | Institute | Documents | Centrality | Location |
|---|---|---|---|---|---|---|---|---|---|
| 1 | University of California | 1905 | 0.03 | USA | 1 | Lausanne University | 51 | 0.12 | Switzerland |
| 2 | Washington University | 1413 | 0.01 | USA | 2 | Monash University | 51 | 0.12 | Australia |
| 3 | Boston University | 838 | 0.03 | USA | 3 | Rhode Island Hospital | 17 | 0.12 | USA |
| 4 | Columbia University | 741 | 0.03 | USA | 4 | University of Nottingham | 53 | 0.1 | UK |
| 5 | Harvard University | 733 | 0.01 | USA | 5 | Radboud University | 85 | 0.1 | Netherlands |
| 6 | Mayo Clinic | 703 | 0.02 | USA | 6 | University of Maryland | 52 | 0.09 | USA |
| 7 | Vrije University Amsterdam | 593 | 0.02 | Netherlands | 7 | Heidelberg University | 80 | 0.08 | German |
| 8 | University of Pittsburgh | 525 | 0.06 | USA | 8 | University of Iowa | 38 | 0.07 | USA |
| 9 | Karolinska Institute | 473 | 0.03 | Sweden | 9 | University of Geneva | 158 | 0.06 | Switzerland |
| 10 | Johns Hopkins University | 445 | 0.04 | USA | 10 | University of Pittsburgh | 525 | 0.06 | USA |
Fig. 3.
Co-citation networks of major institutes (A) and authors (B).
3.5. Distribution of authors
A total of 34,982 authors published these 9125 relevant documents in this research domain. The characteristics of the top 10 authors based on the number of documents are listed in Table 4. Zetterberg H from the University College London published the largest number of documents (164 documents), followed by Blennow K from the University of Gothenburg (159 documents) and Petersen RC from Mayo Clinic (136 documents). Petersen RC had the highest citations and citation index (15,837 citations, h-index: 56, g-index: 125), followed by Holtzman DM from Washington University (14,128 citations, h-index: 56, g-index: 98) and Bennett DA from Rush University (12,061 citations, h-index: 50, g-index: 109). Bartzokis G from the University of California had the highest centrality (centrality: 0.24), followed by Morris J from Washington University (centrality: 0.18), Mintun M from Washington University (centrality: 0.12), Bartha R from Western University (centrality: 0.12), and Graff-Radford N from Mayo Clinic (centrality: 0.11); these authors are considered an important bridge in different cooperation relationships.
Table 4.
Characters of the top 10 authors based on publications.
| Rank | Author | Documents | Citations | h-index | g-index | Centrality | Affiliation |
|---|---|---|---|---|---|---|---|
| 1 | Zetterberg H | 164 | 8355 | 46 | 87 | 0.05 | University College London |
| 2 | Blennow K | 159 | 10,205 | 47 | 98 | 0.01 | University of Gothenburg |
| 3 | Petersen RC | 136 | 15,837 | 56 | 125 | 0.03 | Mayo Clinic |
| 4 | Bennett DA | 127 | 12,061 | 50 | 109 | 0.02 | Rush University |
| 5 | Scheltens P | 126 | 8413 | 45 | 90 | 0.03 | Vrije University Amsterdam |
| 6 | Jack CR | 119 | 11,446 | 46 | 106 | 0.03 | Mayo Clinic |
| 7 | Knopman DS | 100 | 10,093 | 46 | 100 | 0.04 | Mayo Clinic |
| 8 | Holtzman DM | 98 | 14,128 | 56 | 98 | 0.05 | Washington University |
| 9 | Morris JC | 94 | 10,572 | 39 | 94 | 0.18 | Auburn University |
| 10 | Soininen H | 89 | 9279 | 39 | 89 | 0.05 | University of East Finland |
A visualized co-occurrence network of the major authors based on co-citation is shown in Fig. 3B. As illustrated in the network, various collaborations were built loosely in accordance with authors’ affiliations, countries, and research fields. Petersen RC, Jack CR, Knopman DS, and Graff-Radford N from Mayo Clinic and Bennett DA from Rush University are in a collaboration network interested in neuropathology of AD, which is partly overlapped with another collaboration network labeled as “tau” that includes Holtzman DM and Morris J from Washington University, where Morris J, Mintun M, and Graff-Radford N act as important bridges across the network. Zetterberg H from the University College London, Blennow K from the University of Gothenburg, and Scheltens P from Vrije University Amsterdam also loosely formed a collaboration network named as “AddNeuroMed.” Li Y from Washington University, Wang X from Mayo Clinic, Wang J from Beijing Normal University, and Wang Y from Chinese Academy of Sciences are in a loose collaboration network, and they have published relevant articles on polymorphisms. Martins R from Edith Cowan University and Masters C from the University of Melbourne formed another isolated collaboration network. Kim S from Indiana University and Lee J from Columbia University, who are interested in research on blood-based biomarkers, formed an isolated collaboration network.
3.6. Analysis of references
A total of 236,804 references were included in these 9125 documents on APOE in AD. When a reference was cited by multiple documents in this field, it was called a co-cited reference. Co-cited references constituted the knowledge base in a particular research field. The co-citation frequency of a literature indicated its degree of significance in this field. Table 5 shows the top 10 co-citation references (top 100 references, see Table S1 in the supplementary material). Five references had co-citation frequency of more than 1000. A report entitled “Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families” published by Corder EH et al. [20] in Science was the most co-cited paper (n = 1914), followed by a guideline entitled “Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease” published by McKhann G et al. [21] (n = 1204) and a meta-analysis entitled “Effects of Age, Sex, and Ethnicity on the Association Between Apolipoprotein E Genotype and Alzheimer Disease” published by Farrer LA et al. [22] (n = 1168).
Table 5.
Characters of the top 10 co-cited references.
| Rank | First author | Title | Type | Journal | Publication year | Citations | DOI |
|---|---|---|---|---|---|---|---|
| 1 | Corder EH | Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families | report | Science | 1993 | 1914 | 10.1126/SCIENCE.8,346,443 |
| 2 | Mckhann G | Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease | guideline | Neurology | 1984 | 1204 | 10.1212/WNL.34.7.939 |
| 3 | Farrer LA | Effects of Age, Sex, and Ethnicity on the Association Between Apolipoprotein E Genotype and Alzheimer Disease | meta-analysis | JAMA | 1997 | 1168 | 10.1001/JAMA.278.16.1349 |
| 4 | Folstein MF | “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician | article | J Psychiat Res | 1975 | 1108 | 10.1016/0022–3956 (75)90,026-6 |
| 5 | Liu CC | Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy | review | Nat Rev Neurol | 2013 | 1014 | 10.1038/NRNEUROL.2012.263 |
| 6 | Mckhann GM | The diagnosis of dementia due to Alzheimer's disease: Recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease | guideline | Alzheimers Dement | 2011 | 774 | 10.1016/J.JALZ.2011.03.005 |
| 7 | Harold D | Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease | article | Nat Genet | 2009 | 707 | 10.1038/NG.440 |
| 8 | Strittmatter WJ | Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. | article | P Natl Acad Sci Usa | 1993 | 702 | 10.1073/PNAS.May 90, 1977 |
| 9 | Braak H | Neuropathological stageing of Alzheimer-related changes | review | Acta Neuropathol | 1991 | 669 | 10.1007/BF00308809 |
| 10 | Lambert JC | Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease | meta-analysis | Nat Genet | 2013 | 669 | 10.1038/NG.2802 |
CiteSpace software was used to calculate the centrality of references, determine the burst detection of references (Fig. 4A) and construct the cluster and timeline maps of references (Fig. 4B and C). The article entitled “Clinical and Biomarker Changes in Dominantly Inherited Alzheimer's Disease” published in New England Journal of Medicine had the largest centrality (0.32), indicating that it was the most acknowledged article and had a significant influence on the work of other researchers. The top 25 references with the strongest citation bursts are shown in Fig. 4A. As illustrated in the figure, 9 references showed citation bursts till 2022 [ [7,[23], [24], [25], [26], [27], [28], [29], [30]]].
Fig. 4.
Top 25 references with the strongest citation burst (A). Top 12 co-citation clusters of references with the highest connected components and their landmark references in cluster view (B) and timeline view (C).
Forty clusters were identified in the network of co-citation references for these 9125 relevant documents. The top 12 co-citation clusters with the highest connected components and their landmark references are shown in Fig. 4B and C. The modularity Q and silhouette scores (Q = 0.7281, S = 0.924) indicated a significant delineation structure and convincing clustering results, respectively. In the timeline view (Fig. 4C), lines and labels from red to yellow indicate more recent references. The top 12 clusters of co-cited references were as follows: #0 cerebrospinal fluid (CSF) biomarker, #1 mouse brain, #2 targeting apolipoprotein, #3 Han Chinese, #4 polygenic risk score, #5 triggering receptor, #6 AD biomarker, #7 genome-wide association study, #8 sex difference, #9 emerging role, #10 middle-aged adult, #11 single-subject probabilistic prediction, and #12 temporal neocortex. The identification of biomarkers and risk factors and the exploration of pathological mechanisms are the core components of knowledge base in these publications.
3.7. Analysis of keywords
A total of 11,203 author's keywords were included in these 9125 documents. The top 10 keywords with the highest frequency were APOE, Alzheimer, dementia, risk, mild cognitive impairment, association, brain, Aβ, genome-wide association, and APOE genotype. The keywords in the studies suggested the current focus and trends of research in this domain. Combined networks including network, overlay, and density maps of keywords plus in the WoSCC database are shown in Fig. 5A–C. Keywords were clustered in eight research categories and shown with different colors in the network map (Fig. 5A). Keywords (occurrence >300) in the major clusters in the network map were as follows: red clusters included the keywords APOE, Alzheimer, Aβ, brain, mouse model, CSF, amyloid precursor protein (APP), expression, protein, CNS, and oxidative stress; yellow clusters included the keywords risk, association, genome-wide association, age, allele, identifies variants, meta-analysis, and onset; blue clusters included the keywords dementia, APOE genotype, decline, impairment, cognitive decline, and population; light blue clusters included the keywords national institute, diagnosis, disease, biomarkers, diagnostic guidelines, and pathology; and purple clusters included the keywords risk-factors, cognitive impairment, and prevalence. These popular keywords mainly referred to related mechanisms (Aβ, brain, mouse model, CNS, CSF, APP, expression, and oxidative stress), risk factors (risk, risk-factors, association, genome-wide association, age, allele, identifies variants, and onset), cognitive impairment (dementia, decline, impairment, cognitive decline, and cognitive impairment), and diagnostic approach (diagnosis, biomarkers, and diagnostic guidelines). The average publication years of individual keywords are shown in different colors in the overlay map (Fig. 5B), with purple to yellow indicating more recent publication years. Tau-mediated neurodegeneration and neurofilament light were the two most recent keywords that emerged from 2020 (average publication year), following other keywords such as polygenic risk, risk loci, insights, TDP-43 pathology, and TREM2 deficiency that emerged from 2019. Findings from the overlay map indicated mechanism exploration was the current hotspot in the era of APOE in AD. The density map shows the co-occurrence and co-cited frequencies of keywords, wherein the size of the word is positively related to the co-occurrence frequency and the opacity of yellow is related to the co-cited frequency (Fig. 5C). In the retrieved 9125 documents, APOE, Alzheimer, and dementia were the top 3 keywords with the highest co-occurrence, while nursing-home residents, CSF Aβ(42), and imputation were the top 3 keywords with the highest co-cited frequencies in the past decade.
Fig. 5.
Cluster maps of keywords in three visualizations: network (A), overlay (B), and density (C). Top 25 keywords with the strongest citation burst (D).
We screened the top 25 keywords with the strongest citation bursts (Fig. 5D). As shown in Fig. 5D, the hotspots have been shifting from Aβ-related mechanisms, vascular dementia, and gene variation to pathological changes in the BBB and cerebral amyloid angiopathy, transgenic animal models, polymorphisms, and cognitive function and then to tau-related mechanisms and meta-analysis. Keywords such as disease, neurodegeneration, tau, meta-analysis, and pathology burst from 2020 until 2022.
4. Discussion
4.1. General trends
In the present article, we used several innovative bibliometric tools, including CiteSpace, VOSviewer, and R Bibliometrix, to analyze and visualize the characteristics of research studies on the role of APOE in AD published in the past decade. The contributions of journals, countries/regions, institutes, and authors to this research domain were analyzed. Knowledge base and hot topics in the coming years were also identified.
A total of 9125 papers related to APOE in AD were published in 1273 journals with 236,804 cited references and 34,982 authors from 101 countries/regions. The steady increase in the number of documents and citations suggested sustained interest in this domain in the past decade.
4.2. Influential countries/regions, institutes, and authors
The USA is the most influential country in this research domain, with the highest number of documents and citations; the strongest collaboration network worldwide, and the majority of top funding agencies, top institutes, and top authors. The UK has a comparable number of core journal agencies to that in the USA, the second largest number of citations and collaborations, and the third largest funding agency, and it is supposed to be the second leading country in this field. China is the third leading country with the number of documents second only to the USA, the third largest number of citations and collaborations, and the fifth largest funding agency. China, however, lacks influential journal agencies and top institutes in this field. In contrast, Europe as a whole has the number of documents, citations, top institutes, and authors second only to the USA.
University of California, which includes 10 campuses, 6 academic centers, and 3 laboratories, is the most productive institute in this research domain. Bartzokis G from the University of California has the highest value of centrality, thus indicating a pivotal role in the field of APOE and AD. However, University of California as a whole did not show high centrality, while some institutes with much less production played an important role in the collaboration network.
Among the 34,982 authors who published relevant documents in this domain, approximately 10% have contributed at least five documents. This authorship distribution almost followed Lotka's law [31], in which the number of authors with n papers tended to be inversely proportional to n2. Zetterberg H from the University College London contributed to most documents, with h-index, g-index, and citations ranked 7th, 10th, and 19th, respectively. The most cited document by Zetterberg H was “Defeating Alzheimer's disease and other dementias: a priority for European science and society” published in Lancet Neurology [32]. Zetterberg H focused on exploring biomarkers for AD, according to his recent documents. Blennow K from the University of Gothenburg is the second largest contributor to documents in this domain. His most cited document was “Two phase 3 trials of bapineuzumab in mild-to-moderate Alzheimer's disease” published in New England Journal of Medicine [33]. Both Zetterberg H and Blennow K are in a collaboration network named as “AddNeuroMed,” which is the European collaboration for the discovery of novel biomarkers for AD. Petersen RC from Mayo Clinic has the most citations and index as well as the third largest number of relevant documents. The most cited document by Petersen RC was “Tracking pathophysiological processes in Alzheimer's disease: an updated hypothetical model of dynamic biomarkers” published in Lancet Neurology [34]. Recent documents by Petersen RC were related to the neuropathologic process, risk factor identification, and scoring of AD.
4.3. Hotspots and trending
Our keyword co-occurrence and reference co-citation network maps indicated that APOE continued to attract considerable interest in research on AD pathogenesis, expanding from Aβ mechanisms to tau degeneration, microglial and astrocytic responses, and BBB disruption. Potential biomarkers and auxiliary diagnostic approach have also been developed. Many genome-wide association studies and associated secondary analyses have also been conducted to identify potential risk factors and blood biomarkers for the timely and accurate diagnosis of AD.
4.3.1. Amyloid-β
The amyloid cascade hypothesis is the most prominent hypothesis for AD. It is assumed that Aβ is the primary causative agent in AD pathology, and brain amyloid deposits cause downstream events that finally lead to cognitive impairment and dementia [35]. The accumulation of the Aβ peptide is believed to be an initial event in the pathological process of AD. APOE partially contributes to AD risk by differentially regulating overall Aβ clearance through the BBB, cellular uptake and subsequent degradation, enzymatic degradation, clearance through interstitial fluid bulk flow, and CSF absorption into the circulatory and lymphatic systems in a dose- and isoform-dependent manner (ε4 > ε3 > ε2) [7]. In the past decade, a series of trials on antiamyloid therapies for AD, such as amyloid antibodies, active vaccine, and β-secretase inhibitors, have been conducted; however, most of them failed to show clinical benefits in phase II or phase III trials [ [36,37]]. To the best of our knowledge, only aducanumab, an antiamyloid monoclonal antibody (AAMA), has been approved using the accelerated approval pathway by FDA on the basis of its amyloid-lowering property [38]. However, the lack of clinical benefits of aducanumab has raised serious concerns and controversies. The Centers for Medicare and Medicaid Services finally adopted a coverage of aducanumab and other AAMAs with the evidence development approach for the treatment of AD recently [39]. Beyond the therapeutic effect, Aβ has been investigated as a biomarker for AD. Nakamura et al. [40] reported a validated method of high-performance mass spectrometry coupled with immunoprecipitation to detect APP669–711/AβAβ1–42 and Aβ1–40/Aβ1–42 ratios and their composites in patients with AD. These biomarkers showed high performance when predicting brain Aβ burden, which is confirmed by Aβ-PET imaging, thus demonstrating the potential clinical utility of Aβ as plasma biomarkers [40].
4.3.2. Tau
The pathological aggregation of tau, a microtubule-associated protein, also strongly correlates with the patterns of neurodegeneration and clinical manifestations in AD. Shi et al. [29] generated P301S tau transgenic mice with different APOE genotypes and cultured P301S tau-expressing neurons with ε4-expressing mixed glia to determine the role of APOE in tau-related neuropathology. The results demonstrated that APOE affects tau pathogenesis, neuroinflammation, and tau-mediated neurodegeneration independent of Aβ pathology [29]. APOE ε4 aggravates neurodegeneration, whereas the absence of APOE exerts a strong neuroprotective effect [29]. Owing to its inherently higher innate immune reactivity, APOE may aggravate neuroinflammation and then lead to neurodegeneration by directly targeting injured neurons and by activating toxic A1 astrocytes [29]. The latest study by Lee et al. [41] showed that regional interactions between Aβ and tau promoted the spread of tau pathology in AD. However, similar to aducanumab, the monoclonal anti-tau antibody semorinemab failed to slow down clinical AD progression as compared to placebo [42]. Blood phospho-tau is also a promising diagnostic and prognostic biomarker for AD. Barthélemy et al. [43] used nano liquid chromatography coupled with tandem mass spectrometry to determine plasma p-tau isoforms and investigated the relationship between the CSF and plasma tau isoforms. Plasma levels of p-tau-217 and p-tau-181 were correlated with those in the CSF and were highly specific for amyloid plaque pathology [43].
4.3.3. Blood-brain barrier
BBB breakdown is an early biomarker of human cognitive dysfunction. APOE accelerates BBB breakdown and the degeneration of brain capillary pericytes responsible for BBB integrity [44]. Montagne et al. [44] found that BBB breakdown in the hippocampus and medial temporal lobe in APOE ε4 carriers is different from that in noncarriers. The level of soluble PDGFRβ in the CSF as a BBB pericyte injury biomarker adequately predicted future cognitive decline in APOE ε4 carriers but not in noncarriers, even after controlling for Aβ and tau status [44]. APOE ε4 activated the cyclophilin A-matrix metalloproteinase-9 pathway in the CSF, which may accelerate BBB degradation and thereby cause neuronal and synaptic dysfunction [44].
4.3.4. Microglia
A prominent feature of AD is the proliferation and activation of microglia concentrated around amyloid plaques. A recent study reported that microglia in the brain can highly and selectively express many risk genes for AD [45]. Krasemann et al. [25] identified the triggering receptor expressed on myeloid cells 2 (TREM2)-APOE pathway as the major regulator of microglial functional phenotype, which switches the cellular state from homeostasis to the neurodegenerative state. These authors proposed that the activation of the TREM2-APOE pathway reduces the ability of MGnD microglia to prevent neuronal loss and delivery of tolerogenic signals to T cells [25]. The modulation targeting the TREM2-APOE pathway may serve as an approach to restore the homeostasis of microglia and treat neurodegenerative disorders [25].
4.3.5. Genetic risk factors
Accumulating evidence suggests that APOE is an important genetic risk factor for AD. There are three major allelic variants of APOE, namely APOE ε2, APOE ε3, and APOE ε4, which are defined by two single nucleotide polymorphisms, i.e., rs429358 and rs7412 [5]. Among these variants, APOE ε4 is associated with an increased risk of late-onset AD (LOAD) [46], whereas APOE ε2 homozygote shows an exceptionally low likelihood to promote the development of AD-related dementia [ [7,47]]. The magnitude of the APOE ε4-associated risk might also be related to ethnicity [7]. In addition to the classic amyloid cascade hypothesis, Frisoni et al. [48] proposed an alternative model in which the penetrance of the amyloid cascade is directly proportional to the penetrance of genetic risk factors. These authors identified the stochastic factors of three variants in the probabilistic model that played an increasingly important role: autosomal dominant AD, APOE ε4-related sporadic AD, and APOE ε4-unrelated sporadic AD [48].
Multiple innovative omics tools have been used in the risk assessment of AD. Mathys et al. [49] analyzed single-nucleus transcriptomes of individuals with AD and detected strongest disease-associated changes at early stages in specific cell types, while genes involved in global stress response were upregulated at late stages across different cell types. Sex difference was observed in transcriptional responses in several cell types [49]. The authors also suggested that myelination plays an important role in AD pathophysiology [49]. Liu et al. [50] integrated hippocampal gene expression and genome-wide association data to identify genes that are significantly expressed in the hippocampus of patients with AD. They prioritized 24 AD-related genes, including APOE and two novel genes (PTPN9 and PCDHA4) [50]. These novel genes were associated with several functions, including Aβ formation, phosphorylation/dephosphorylation, neuronal apoptosis, neurogenesis, and telomerase-related processes, and the expression levels of these genes were correlated with the hippocampal volume [50]. Bellenguez et al. [51] performed a two-stage genome-wide association study and identified 75 risk loci, including 42 new loci. Pathway enrichment analyses confirmed the involvement of amyloid/tau and microglia and the new genetically associated processes that included the tumor necrosis factor-α (TNF-α) pathway through the linear ubiquitin chain assembly complex [51]. A new genetic risk score beyond the effects of age and APOE ε4 was also obtained to improve the prediction ability of AD [51]. Ming et al. [52] identified consensus copy number variations (CNVs) from the whole-genome sequencing data and found 3012 rare AD-specific CNVs. Their residing genes are enriched for cellular glucuronidation and neuron projection pathways, and their mRNA expression is involved in major histocompatibility complex class II receptor activity, thus indicating novel pathways and targets for treating AD [52].
Meta-analysis as a statistical method has been used to integrate findings from independent studies investigating the genetic risk factors of AD. Neu et al. [53] conducted a meta-analysis to determine how sex and APOE genotype affect the risk for developing mild cognitive impairment and AD. The authors found that although men and women with the APOE ε3/ε4 genotype have comparable odds of developing AD across the age span of 55–85 years, women are at a higher risk during their younger age than men [53]. Several genome-wide association meta-analyses have also been conducted to identify other risk loci for AD. Jansen et al. [24] performed a large genome-wide association study of clinically diagnosed AD and AD-by-proxy and identified 29 risk loci, thus implicating 215 potential causative genes that are strongly expressed in immune-related tissues and cell types. Lipid-related processes for APP degradation are the underlying biological mechanisms responsible for AD [24]. Kunkle et al. [26] performed a large genome-wide association meta-analysis of patients with clinically diagnosed LOAD and confirmed 20 previous and 5 new risk loci for LOAD. The locus HLA-DR15 was a risk factor for LOAD [26]. Immunity, lipid metabolism, tau-binding proteins, and APP metabolism were potential mechanisms implicated in patients with LOAD [26].
4.4. Limitations
Our present study had several limitations. First, literature retrieval was conducted from the WoSCC database alone and was restricted to articles and reviews published in journals; this could have limited the generalization of the findings. The retrieval of relevant publication had a cutoff publication date of Aug 31, 2022. Some of the later published articles are already available online but were not included in this work; hence, this work does not fully reflect the actual research conducted in 2022. Second, the importance of articles was assessed on the basis of citations and burst strength, which could be significantly influenced by the publication date. Studies published recently may not be efficiently cited although they have a high academic value. Moreover, citation bias was not evaluated in this study. A reference can be cited not only for its quality or relevance but also for the purpose of self-citation, the authority of authors, and the impact factor of journals. Third, the analyses might have been affected by the presupposed criteria of selection. For example, in the keyword analysis, the threshold of keyword occurrence was five, and keywords with occurrence lesser than five were not included in the subsequent analysis. Therefore, latest research trends pertaining to the related keywords might have been excluded. Fourth, the bibliometric method is based on natural language processing, which may be biased. For example, keywords in different situations may have diferent meanings, or different words may represent the same terms.
5. Conclusion
The present study showed that research on the role of APOE in AD has been burgeoning in the past decade. The influencial countries, institutes, and authors; the development of knowledge base; and the emerging trends in this era were identified from an objective and quantitative point of view. On the basis of pivotal publications related to biomarkers, risk factors, pathogenetic mechanisms, and prediction models, the knowledge base in this domain has been greatly updated in the past decade. The precise mechanism through which APOE is involved in AD pathogenesis is not yet fully understood; however, our analysis suggests a comprehensive interpretation of APOE-related effects, with current research trends focusing on the associated mechanisms and biomarkers targeting Aβ, tau, microglia, and BBB disruption. Many studies have attempted to investigate antiamyloid and anti-tau antibodies as treatment regimens for AD; however, clinical benefits of these therapies are required to be validated in the future. Potential risk factors such as PTPN9, PCDHA4, TNF-α, and HLA-DR15 have been identifed in recent studies, thus shedding light on the complex and multifactorial nature of AD and indicating novel targets for AD. Our study provided insights into the developing knowledege base and trends of APOE in AD, which may show new directions for conducting further research in this domain.
Funding
This work was supported by the Natural Science Foundation of Guangdong Province (grant number 2021A1515011325), Science and Technology Plan Project of Guangzhou (grant number 202102080030, 202201010736), Science and Technology Plan Project of Guangdong Province (2019B0303160), and Guangzhou Municipal Key Discipline in Medicine (2021–2023).
Author contribution statement
Zhanzhang Wang: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Xiuqing Zhu: Analyzed and interpreted the data; Wrote the paper.
Yuguan Wen; Dewei Shang: Conceived and designed the experiments.
Data availability statement
Data will be made available on request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to express our gratitude to CM Chen, M Aria, C Cuccurullo, and Leiden University's Centre for Science and Technology Studies, who invented CiteSpace, R-bibliometrix, and WOSviewer, which is free to use. We also thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17987.
Contributor Information
Yuguan Wen, Email: wenyuguande@163.com.
Dewei Shang, Email: shang_dewei@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Dubois B., Villain N., Frisoni G.B., Rabinovici G.D., Sabbagh M., Cappa S., et al. Clinical diagnosis of Alzheimer's disease: recommendations of the international working Group. Lancet Neurol. 2021;20(6):484–496. doi: 10.1016/s1474-4422(21)00066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzheimer's Association Alzheimer's disease facts and figures. Alzheimers Dement. 2022;18(4):700–789. doi: 10.1002/alz.12638. [DOI] [PubMed] [Google Scholar]
- 3.Serrano-Pozo A., Aldridge G.M., Zhang Q. Four decades of research in Alzheimer's disease (1975-2014): a bibliometric and scientometric analysis. J. Alzheimers Dis. 2017;59(2):763–783. doi: 10.3233/jad-170184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hasel P., Liddelow S.A. Isoform-dependent APOE secretion modulates neuroinflammation. Nat. Rev. Neurol. 2021;17(5):265–266. doi: 10.1038/s41582-021-00483-y. [DOI] [PubMed] [Google Scholar]
- 5.Serrano-Pozo A., Das S., Hyman B.T. APOE and Alzheimer's disease: advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 2021;20(1):68–80. doi: 10.1016/s1474-4422(20)30412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lozupone M., Imbimbo B.P., Balducci C., Lo Vecchio F., Bisceglia P., Latino R.R., et al. Does the imbalance in the apolipoprotein E isoforms underlie the pathophysiological process of sporadic Alzheimer's disease? Alzheimers Dement. 2022;19(1):353–368. doi: 10.1002/alz.12728. [DOI] [PubMed] [Google Scholar]
- 7.Yamazaki Y., Zhao N., Caulfield T.R., Liu C.C., Bu G. Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat. Rev. Neurol. 2019;15(9):501–518. doi: 10.1038/s41582-019-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broadus R.N. Toward a definition of “bibliometrics”. Scientometrics. 1987;12(5):373–379. doi: 10.1007/BF02016680. [DOI] [Google Scholar]
- 9.Lewison G., Devey M.E. Bibliometric methods for the evaluation of arthritis research. Rheumatology. 1999;38(1):13–20. doi: 10.1093/rheumatology/38.1.13. [DOI] [PubMed] [Google Scholar]
- 10.Liu C., Li W., Xu J., Zhou H., Li C., Wang W. Global trends and characteristics of ecological security research in the early 21st century: a literature review and bibliometric analysis. Ecol. Indicat. 2022;137 doi: 10.1016/j.ecolind.2022.108734. [DOI] [Google Scholar]
- 11.Chen C. CiteSpace II: detecting and visualizing emerging trends and transient patterns in scientific literature. J. Am. Soc. Inf. Sci. Technol. 2006;57(3):359–377. doi: 10.1002/asi.20317. [DOI] [Google Scholar]
- 12.van Eck N.J., Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. 2010;84(2):523–538. doi: 10.1007/s11192-009-0146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aria M., Cuccurullo C. bibliometrix: an R-tool for comprehensive science mapping analysis. J. Informetr. 2017;11(4):959–975. doi: 10.1016/j.joi.2017.08.007. [DOI] [Google Scholar]
- 14.Ahmad P., Slots J. A bibliometric analysis of periodontology. Periodontol. 2000;85(1):237–240. doi: 10.1111/prd.12376. [DOI] [PubMed] [Google Scholar]
- 15.Hassan W., Zafar M., Duarte A.E., Kamdem J.P., Teixeira da Rocha J.B., Pharmacological Research A bibliometric analysis from 1989 to 2019. Pharmacol. Res. 2021;169 doi: 10.1016/j.phrs.2021.105645. [DOI] [PubMed] [Google Scholar]
- 16.Wilson M., Sampson M., Barrowman N., Doja A. Bibliometric analysis of neurology articles published in general medicine journals. JAMA Netw. Open. 2021;4(4) doi: 10.1001/jamanetworkopen.2021.5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun H.L., Bai W., Li X.H., Huang H., Cui X.L., Cheung T., et al. Schizophrenia and inflammation research: a bibliometric analysis. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.907851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong K., Chen Y., Liu W., Wang Z. Global research trends of Apolipoprotein E in central nervous system: a scientometric analysis. Int. Immunopharm. 2021;98 doi: 10.1016/j.intimp.2021.107919. [DOI] [PubMed] [Google Scholar]
- 19.Berta A., Miguel Ángel C., Clara G.-S., Rubén H. A bibliometric analysis of 10 years of research on symptom networks in psychopathology and mental health. Psychiatr. Res. 2022;308 doi: 10.1016/j.psychres.2021.114380. [DOI] [PubMed] [Google Scholar]
- 20.Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 21.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA work Group under the auspices of department of health and human Services Task Force on Alzheimer's disease. Neurology. 1984;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 22.Farrer L.A., Cupples L.A., Haines J.L., Hyman B., Kukull W.A., Mayeux R., et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278(16):1349–1356. [PubMed] [Google Scholar]
- 23.Jack C.R., Jr., Bennett D.A., Blennow K., Carrillo M.C., Dunn B., Haeberlein S.B., et al. NIA-AA Research Framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14(4):535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen I.E., Savage J.E., Watanabe K., Bryois J., Williams D.M., Steinberg S., et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk. Nat. Genet. 2019;51(3):404–413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krasemann S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017;47(3):566–581.e569. doi: 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunkle B.W., Grenier-Boley B., Sims R., Bis J.C., Damotte V., Naj A.C., et al. Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019;51(3):414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livingston G., Sommerlad A., Orgeta V., Costafreda S.G., Huntley J., Ames D., et al. Dementia prevention, intervention, and care. Lancet. 2017;390(10113):2673–2734. doi: 10.1016/s0140-6736(17)31363-6. [DOI] [PubMed] [Google Scholar]
- 28.Selkoe D.J., Hardy J. The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol. Med. 2016;8(6):595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi Y., Yamada K., Liddelow S.A., Smith S.T., Zhao L., Luo W., et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature. 2017;549(7673):523–527. doi: 10.1038/nature24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theurich S., Tsaousidou E., Hanssen R., Lempradl A.M., Mauer J., Timper K., et al. IL-6/Stat3-Dependent induction of a distinct, obesity-associated NK cell subpopulation deteriorates energy and glucose homeostasis. Cell Metabol. 2017;26(1):171–184.e176. doi: 10.1016/j.cmet.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 31.Lotka A.J. The frequency distribution of scientific productivity. J. Architect. Plann. Res. 1926;16:317–323. [Google Scholar]
- 32.Winblad B., Amouyel P., Andrieu S., Ballard C., Brayne C., Brodaty H., et al. Defeating Alzheimer's disease and other dementias: a priority for European science and society. Lancet Neurol. 2016;15(5):455–532. doi: 10.1016/s1474-4422(16)00062-4. [DOI] [PubMed] [Google Scholar]
- 33.Salloway S., Sperling R., Fox N.C., Blennow K., Klunk W., Raskind M., et al. Two phase 3 trials of bapineuzumab in mild-to-moderate Alzheimer's disease. N. Engl. J. Med. 2014;370(4):322–333. doi: 10.1056/NEJMoa1304839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jack C.R., Jr., Knopman D.S., Jagust W.J., Petersen R.C., Weiner M.W., Aisen P.S., et al. Tracking pathophysiological processes in Alzheimer's disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12(2):207–216. doi: 10.1016/s1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardy J.A., Higgins G.A. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256(5054):184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 36.Kim C.K., Lee Y.R., Ong L., Gold M., Kalali A., Sarkar J. Alzheimer's disease: key insights from two decades of clinical trial failures. J. Alzheimers Dis. 2022;87:83–100. doi: 10.3233/JAD-215699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubin R. Much anticipated alzheimer disease prevention trial finds No clinical benefit from drug targeting amyloid; highlights need to consider other approaches. JAMA. 2022;328(10):907–910. doi: 10.1001/jama.2022.11490. [DOI] [PubMed] [Google Scholar]
- 38.U.S. Food and Drug Administration . 2021. Grants Accelerated Approval for Alzheimer's Drug. [Google Scholar]
- 39.Centers for Medicare & Medicaid Services . 2022. Monoclonal Antibodies Directed against Amyloid for the Treatment of Alzheimer's Disease.https://www.cms.gov/medicare-coverage-database/view/ncacal-decision-memo.aspx?proposed=Y&NCAId=305 [Google Scholar]
- 40.Nakamura A., Kaneko N., Villemagne V.L., Kato T., Doecke J., Doré V., et al. High performance plasma amyloid-β biomarkers for Alzheimer's disease. Nature. 2018;554(7691):249–254. doi: 10.1038/nature25456. [DOI] [PubMed] [Google Scholar]
- 41.Lee W.J., Brown J.A., Kim H.R., La Joie R., Cho H., Lyoo C.H., et al. Regional Aβ-tau interactions promote onset and acceleration of Alzheimer's disease tau spreading. Neuron. 2022;110(12):1932–1943.e1935. doi: 10.1016/j.neuron.2022.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teng E., Manser P.T., Pickthorn K., Brunstein F., Blendstrup M., Sanabria Bohorquez S., et al. Safety and efficacy of semorinemab in individuals with prodromal to mild alzheimer disease: a randomized clinical trial. JAMA Neurol. 2022;79(8):758–767. doi: 10.1001/jamaneurol.2022.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barthélemy N.R., Horie K., Sato C., Bateman R.J. Blood plasma phosphorylated-tau isoforms track CNS change in Alzheimer's disease. J. Exp. Med. 2020;217(11) doi: 10.1084/jem.20200861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montagne A., Nation D.A., Sagare A.P., Barisano G., Sweeney M.D., Chakhoyan A., et al. APOE4 leads to blood-brain barrier dysfunction predicting cognitive decline. Nature. 2020;581(7806):71–76. doi: 10.1038/s41586-020-2247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang X., Wu H., Colt M., Guo X., Pluimer B., Zeng J., et al. Microglia and its genetics in Alzheimer's disease. Curr. Alzheimer Res. 2021;18(9):676–688. doi: 10.2174/1567205018666211105140732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strittmatter W.J., Saunders A.M., Schmechel D., Pericak-Vance M., Enghild J., Salvesen G.S., et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reiman E.M., Arboleda-Velasquez J.F., Quiroz Y.T., Huentelman M.J., Beach T.G., Caselli R.J., et al. Exceptionally low likelihood of Alzheimer's dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat. Commun. 2020;11(1):667. doi: 10.1038/s41467-019-14279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frisoni G.B., Altomare D., Thal D.R., Ribaldi F., van der Kant R., Ossenkoppele R., et al. The probabilistic model of Alzheimer disease: the amyloid hypothesis revised. Nat. Rev. Neurosci. 2022;23(1):53–66. doi: 10.1038/s41583-021-00533-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathys H., Davila-Velderrain J., Peng Z., Gao F., Mohammadi S., Young J.Z., et al. Single-cell transcriptomic analysis of Alzheimer's disease. Nature. 2019;570(7761):332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu N., Xu J., Liu H., Zhang S., Li M., Zhou Y., et al. Hippocampal transcriptome-wide association study and neurobiological pathway analysis for Alzheimer's disease. PLoS Genet. 2021;17(2) doi: 10.1371/journal.pgen.1009363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bellenguez C., Küçükali F., Jansen I.E., Kleineidam L., Moreno-Grau S., Amin N., et al. New insights into the genetic etiology of Alzheimer's disease and related dementias. Nat. Genet. 2022;54(4):412–436. doi: 10.1038/s41588-022-01024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ming C., Wang M., Wang Q., Neff R., Wang E., Shen Q., et al. Whole genome sequencing–based copy number variations reveal novel pathways and targets in Alzheimer's disease. Alzheimers Dement. 2021;18(10):1846–1867. doi: 10.1002/alz.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neu S.C., Pa J., Kukull W., Beekly D., Kuzma A., Gangadharan P., et al. Apolipoprotein E genotype and sex risk factors for alzheimer disease: a meta-analysis. JAMA Neurol. 2017;74(10):1178–1189. doi: 10.1001/jamaneurol.2017.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.