Summary
The prevalence of sleep disorders has increased because of the fast-paced and stressful modern lifestyle, negatively impacting the quality of human life and work efficiency. It is crucial to address sleep problems. However, the current practice of diagnosing sleep disorders using polysomnography (PSG) has limitations such as complexity, large equipment, and low portability, hindering its practicality for daily use. To overcome these challenges, in this article an optical fiber sensor is proposed as a viable solution for sleep monitoring. This device offers benefits like low power consumption, non-invasiveness, absence of interference, and real-time health monitoring. We introduce the sensor with an optical fiber interferometer to capture ballistocardiography (BCG) and electrocardiogram (ECG) signals from the human body. Furthermore, a new machine learning method is proposed for sleep condition detection. Experimental results demonstrate the superior performance of this architecture and the proposed model in monitoring and assessing sleep quality.
Subject areas: Fiber optics, Health technology, Optics
Graphical abstract

Highlights
-
•
An optical fiber sensor based on fiber interferometer is proposed
-
•
A machine learning model is proposed to detect vital sleep conditions
-
•
The monitoring system has full potential in clinical applications
Fiber optics; Health technology; Optics
Introduction
As an essential part of life activities for mankind, sleep plays a vital role in the maintenance and regulation of a variety of biological functions in the molecular sense.1,2 Good sleep is beneficial for people to maintain not only physical and mental health but also normal brain function during the day.3,4,5,6 In the modern society with rapid developments, there are more and more people suffering from sleep disorders because of the increased stress of life or work, thus resulting in poor quality of sleep.7,8,9,10,11,12 For example, the occurrence of apnea during sleep causes sleep disruption at night, which affects the metabolism of other organs, thus causing a range of cardiovascular diseases, diabetes, and even mental issues. In particular, the problems caused by cardiovascular diseases become more serious with higher mortality rate in the elderly group than in the younger group.13,14 For this reason, more and more scholars and medical professionals have paid attention to the study of sleep, for better treatment of the diseases caused by sleep disorders through the analysis of various physiological indicators at each stage of the sleep process. With the change of sleep time, human brain state changes at different stages and various physiological parameters of the human body change accordingly.15,16,17,18,19,20 According to these changes, it can be observed that people experience several different stages during sleep, which means the sleep process can be operated by stages.21 The long-term monitoring of vital signs of the body during sleep can assist doctors in making diagnoses, further preventing serious health risks and promoting the development of a healthy lifestyle.22,23,24
Monitoring vital signs is essential in evaluating human states at the physiological level, with numerous applications, including smart home healthcare.25,26 One such vital sign is the ballistocardiography (BCG) signal, which is generated by the impact of blood flow on the blood vessels during a heartbeat.27,28,29 Flexible sensors that do not require physical contact can be employed to gather BCG signals, enabling the monitoring of health conditions without involving instruments.30 BCG is not only used for cardiac disease diagnosis but also as a means to assess sleep quality.31,32 Optical fiber is the technology associated with data transmission using light pulses traveling along a long fiber, which is usually made of plastic or glass; this has been widely used in optical communication or other fields. For example, the step-index plastic optical fiber is suitable for high-performance fiber links at short distances,33 the multimode optical fibers are widely used in smart healthcare, measurements,34,35 and so on. Various techniques have been proposed to detect BCG signals in recent times, with optical fiber sensors (OFS) receiving significant attention based on their advantageous features such as light weight, high flexibility, and their robust merit to interference by the electromagnet. OFS is created by embedding fiber optic cables beneath a sheet or mat,36,37 and they transmit vibrations when exposed to external pressure or disturbance. The vibration-related data are conveyed by the modulated signal in the fiber optic line, and by making an analysis on the fluctuation featured in the optical signals vibration signals on the outside can be monitored and evaluated to determine vital signs. OFS is ideal for the monitoring of BCG signals owing to its distributed measurement capabilities, low cost, high sensitivity, and invisibility, producing satisfactory outcomes.38
Physics-informed neural network (PINN) is a kind of neural network used to solve supervised learning tasks.39 It can not only learn the distribution rules of training data samples like traditional neural networks, but can also learn the physical laws described by mathematical equations. Compared with pure data-driven neural network learning, PINN imposes physical information constraints during the training process, enabling it to learn models with more generalization ability with fewer data samples. However, PINN is more difficult to handle with high-dimensional data because the required training data exponentially increases with the increase of equation dimensions, which greatly limits its ability to solve high-dimensional equations, and the PINN model runs slowly. In this article, a sleep condition monitoring and assessment sensor is designed according to optical fiber interferometer, which can be of good use to capture and record the signal of vital signs during sleep. Depending on the needs of high-precision scientific research and wearable sleep monitoring equipment as well as the signal characteristics of vital signs, an automatic sleep staging model is constructed through the support vector machine (SVM) algorithm and general regression neural network (GRNN), so as to monitor and classify the quality of sleep non-invasively by using optical fiber sensor. Different from PINN, GRNN is a type of radial basis function (RBF) neural network that exhibits good nonlinear mapping performance during the learning process, and has greater advantages than an RBF network. It will ultimately perform optimal regression when the sample size is aggregated, while also handling unstable sample data, and can still achieve good prediction results even when the sample data are small. GRNN does not have as high requirements for sample data as radial basis networks, which determines its great advantage in handling cases with poor data accuracy in actual sample data classification and model fitting processes. The ultimate goal of this study is to achieve the automatic staging and monitoring of sleep with high accuracy based on the characteristics of vital signs during sleep in different applications. On the one hand, the analysis of electrocardiogram (ECG) signals is purposed to enable the user to monitor the quality of sleep scientifically, conveniently, and with great comfort. On the other hand, the research and development of automatic sleep staging is intended to improve the work efficiency of sleep physicians and the consistency of sleep staging results. In this study, a mixed signal sleep staging algorithm based on SVM is implemented and its performance is evaluated to demonstrate its advantages of high accuracy and wide applicability. Therefore, it is applicable to domestic healthcare and clinical sleep staging diagnosis.
Results and discussions
The human heart has four chambers, which work together to receive and transmit blood from all parts of the body. As an organ capable of electrophysiological activities, conduction electrical stimulation in the heart causes the contraction of atrium and ventricle to complete the beating of the heart. The conduction system in the heart starts from the sinoatrial node. It sends a depolarization wave a short period of time to cause the depolarization of the heart and then the contraction of the heart. Then, the depolarization wave reaches the atrium to complete the contraction of the atrium and then continues to transmit to the nerve fibers of the atrioventricular bundle and the Purkinje fibers in the cardiac muscle. At this time, the ventricle is contracting, and after the contraction of the atrium and ventricle, it returns to the original state after a short period of time; this process is termed repolarization. During depolarization and repolarization, the heart will generate current that can be transmitted to the entire human body. At this time, if the electrode piece of the detected current is attached to the surface of the human body, the ECG waveform of the heart during the depolarization and repolarization activity will be described, that is to say, the ECG. Each depolarization and repolarization operation of the heart will cause a periodic change in the ECG, which includes the starting position, flow direction, amplitude, and duration of the current generated in the heart, as well as a large amount of physiological information in each activity of the heart.
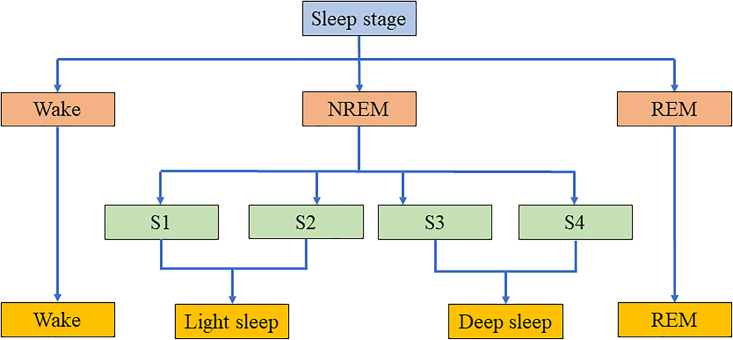
In the middle of the 20th century, N. Kleitman and Aserinsky proposed a method of dividing sleep phases;3 that is to say, according to the eye rotation during sleep, it is divided into two phases, non-rapid eye movement (NREM) and rapid eye movement (REM). On this basis, Kales et al. subdivided the REM phase and NREM phase according to the changes of the brain, eyeball, genial muscle, etc. during human sleep in the 1960s,5 that is to say, the NREM phase is divided into two stages, and REM and Wake constitute four stages of sleep. Until 1966, Rechtschaffen et al. put forward a staging rule of sleep stage based on previous studies.9 The rule summarized the sleep process into six stages: 0 (WAKE), 1 (S1), 2 (S2), 3 (S3), 4 (S4) and 5 (REM). REM can also be subdivided into Light Phase I and II and Deep Phase I and II. This rule is also known as the golden standard of R&K sleep staging. Figure 6 demonstrates the relationship among all sleep stages.
Figure 6.
The mechanism and illustration of sleep staging
Herein, the optimal SVM model based on the classification criteria of sleep stage is applied to stage the ECG signals collected from the sleep mattress based on piezoelectric perception. To begin with, ECG signals are extracted from the heartbeat interval sequence by using the adaptive heart rate calculation method which is based on Bayesian probability. The heartbeat interval sequences of every 5 min are taken as samples. After preprocessing, feature extraction, and feature dimensionality reduction, the proposed machine learning model is used to perform classification, with the classification results obtained to demonstrate the feasibility of the algorithm. Figures 8 and 9 show the sleep stage detection results based on the proposed model (LSVM) and the RBF model. Figure 10 shows the sleep stage detection and assessment results based on the GRNN model.
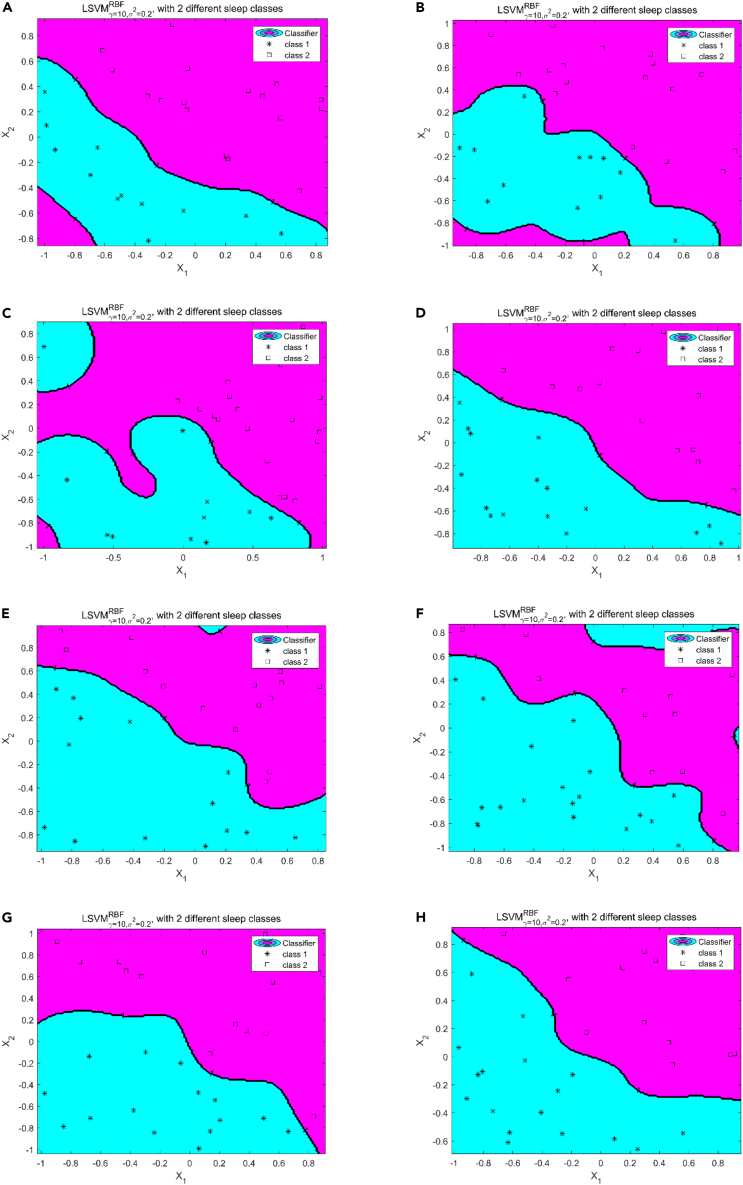
Figure 8.
Sleep stage detection and assessment results based on proposed LSVM model
8 h of raw ECG signal was obtained and used in experiments (class 1: deep sleep; class 2: light sleep).
(A), (B), (C), (D), (E), (F), (G), and (H) refer to the detection and assessment results of sleep stages for each hour within 8 h, respectively. In every figure, the blue-green area contains class 1, and the purplish red area contains class 2. The size of different colored areas represents the proportion of different sleep stages during that time period.
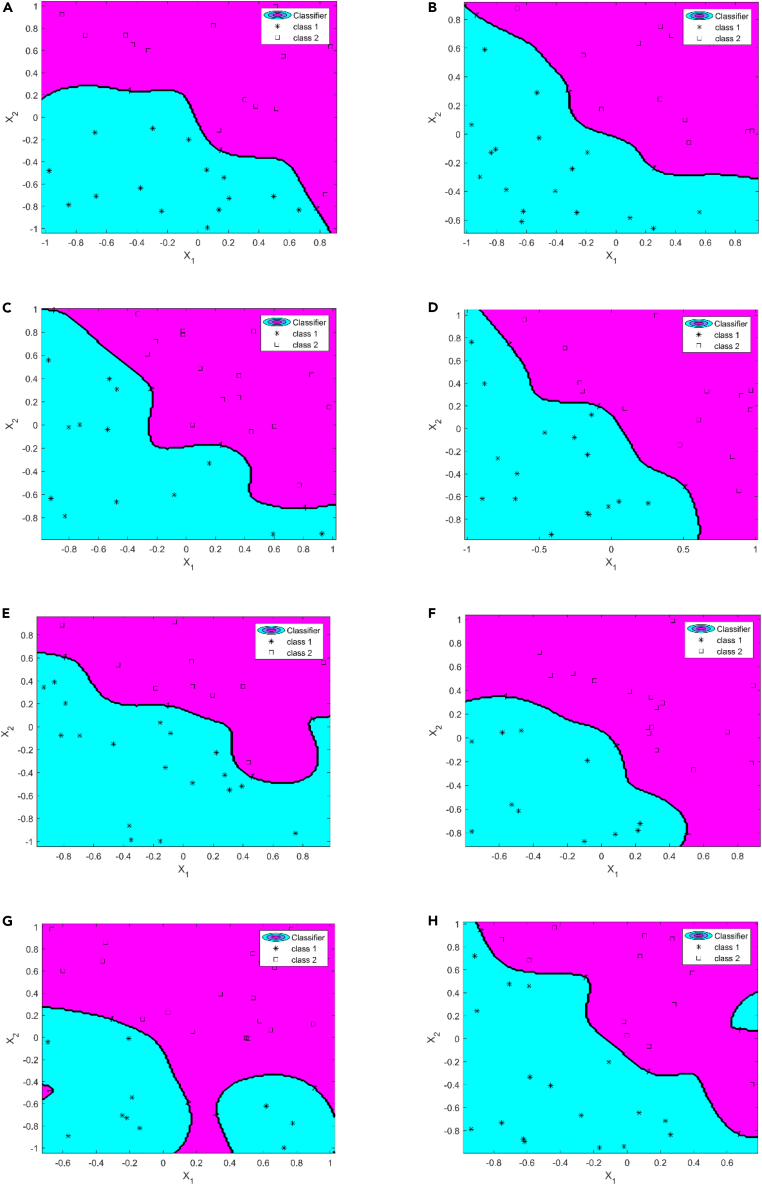
Figure 9.
Sleep stage detection and assessment results based on RBF model
8 h of raw ECG signal was obtained and used in experiments (class 1: deep sleep; class 2: light sleep).
(A), (B), (C), (D), (E), (F), (G), and (H) refer to the detection and assessment results of sleep stages for each hour within 8 h, respectively. In every figure, the blue-green area contains class 1, and the purplish red area contains class 2. The size of different colored areas represents the proportion of different sleep stages during that time period.
Figure 10.
Sleep stage detection and assessment results based on GRNN model (0: WAKE; 1: S1; 2: S2; 3: S3; 4: S4; 5: REM)
(A) 4 different brainwaves extracted from raw vital signs signal; (B) Sleep stage detection and raw vital signs signal.
The experimental data are obtained from the sleep data collected by the sleep mattress integrated with the interferometric optical fiber sensor independently developed by the laboratory. The experimental object comprises the healthy experimental subjects in the laboratory. The mattress is used to record the ECG signal of the human body during sleep, with the heartbeat interval sequence extracted by calculating the adaptive heart rate based on the Bayesian probability. Owing to the impact of ectopic pulsation, errors tend to arise from the time and frequency domain analysis of heart rate variability (HRV), which makes it necessary to preprocess the interval sequence. To clarify it, the upper and lower boundaries are set to identify and rectify data that falls below 0.7 times the mean value of the R-R interval and above 1.3 times the mean value of the R-R interval. The correction method is to calculate the median value of 30 pieces of data adjacent to the outlier as the correction value of the outlier. After processing, the interval values fall within the normal threshold range.
After preprocessing of the collected heartbeat interval sequence, the data points within 5 min are taken as a sample, and the characteristics of the sample data in time and frequency domains are extracted. By using the mathematical statistics method and autoregressive (AR) model, the order of AR model is set to 16, so as to minimize the sum of forward and backward prediction error power. Levenson Durbin recursion is performed to solve the AR coefficient and estimate the power spectrum. Then, the analysis on the principal component is made to lower the dimensionality of the feature matrix, with fewer feature vectors used to represent the original data, so as to improve the efficiency of operation and extract the features of the samples.
With the extension of the acquisition time, that is to say, the sleep time, there is a gradual increase in the average value of the following sample data, indicating the start of transition from the awakening period to the shallow sleep period, the prolonged sequence time of heartbeat interval, and the decrease in heart rate. According to Table 1, which shows frequency domain characteristics, the energy of the collected sample data diminishes gradually with the increase of the acquisition time, indicating the entry into the sleep state and the start of an increase in the high-frequency component. As a result, the low-frequency constituent diminishes along with the ratio between the energy of the low-frequency range and the energy of the high-frequency range. It is indicated that with the deepening of sleep, the excitability of the parasympathetic nerve improves, but the excitability of the sympathetic nerve declines. Table 2 shows the comparison in average running time between the model proposed in this article and RBF model. It can be seen from the table that the proposed LSVM model significantly outperforms the others in running time, and it performs better in computational efficiency.
Table 1.
The decomposition nodes and corresponding frequencies contained in the rhythm wave
| Wave | Node | Frequency (Hz) |
|---|---|---|
| α | [31, 53] | 7.808–13.176 |
| β | [56, 122] | 13.908–30.012 |
| θ | [15, 28] | 3.904–7.076 |
| δ | [2, 11] | 0.488–2.928 |
Table 2.
Comparison of average running time with other method. All data are divided into 8 parts over 8 h
| Data No. | RBF | LSVM |
|---|---|---|
| #1 | 38s | 25s |
| #2 | 36s | 21s |
| #3 | 35s | 23s |
| #4 | 35s | 25s |
| #5 | 35s | 24s |
| #6 | 36s | 25s |
| #7 | 34s | 26s |
| #8 | 36s | 25s |
Compared with the existing works by Wang et al.14,28 and Chen et al.,23 it can be concluded that: (1) Loss-based sensors are easy to break during use, and their sensitivity is not as good as the optical fiber interferometer sensor; (2) The cost of grating-based sensors is high, and it is difficult to obtain practical applications; (3) The electrical based dual sensors have low sensitivity to weak vibration signals, and it is difficult to obtain high-quality signals in complex environments. The optical fiber interferometer sensor proposed in this article can effectively eliminate the background noise and improve the quality of the collected signal; the proposed monitoring system is simple, cost-effective, and non-invasive. As for vital sign signal processing part in the existing works, the fast Fourier transformation (FFT) and wavelet transformation (WT) used in previous studies are traditional algorithms. Traditional algorithms rely on design feature extraction methods to extract useful features, so the loss of some information is inevitable and the accuracy is relatively low (55%). As a crucial technology for monitoring the quality of sleep, an automatic sleep staging algorithm can assist in the research and auxiliary treatment of sleep disorders. Based on polysomnographic (PSG) signals, the traditional approach to sleep monitoring is reliant on artificial interpretation to score the quality of sleep. However, it has various disadvantages such as high cost, complexity, and heavy workload. With the development and application of artificial intelligence theory, researchers around the world have widely applied classification algorithms to study the automatic sleep staging models. However, the existing sleep staging algorithms have such shortcomings as low classification accuracy, various features to be extracted, and the necessity to collect multi-channel physiological signals. Traditional methods are labor-intensive, time-consuming, and relatively low accuracy because of the need for ECG or BCG recordings from several sensors attached to subjects over several nights, then various different features need to be extracted and processed. According to the American Academy of Sleep Medicine (AASM) sleep standard, different sleep stages usually have different salient waves in physiological signals. The existing method uses feature extraction to indirectly capture salient waves, the raw physiological signals are converted into time-frequency images as input to deep learning models for sleep staging.6 Although the above methods indirectly capture signal wave features, manually extracted features not only require prior knowledge but may also cause partial information loss. To address the main problems facing the current practice of monitoring the quality of sleep, in this article, we focus mainly on constructing the sleep staging model based on OFS to work out the design of a sleep monitoring assistant system that can be used to reduce the complexity of the system while improving both efficiency and accuracy.
Conclusions
In this article, a sleep staging method based on SVM is proposed by us, which is developed and implemented for processing the ECG signals collected by OFS. In the algorithm, both mathematical statistics and the AR model are used to extract the characteristics of the heartbeat interval sequence extracted from the ECG signal. Besides, the analysis on the principal component is made to lower the dimensionality of the feature matrix, which not only reduces the redundancy and correlation of the data but also accelerates the operation. The optimal SVM model is applied to classify the heart rate variability characteristic matrix. The optimal SVM model is constructed by performing two sets of SVM experiments. The optimal model based on two sets of experiments is established after a series of operations, such as feature extraction, principal component extraction, SVM training, parameter optimization, the determination of the optimal model and optimal model testing. The four-classification model with high classification accuracy is treated as the classification model of this study. The optimal four-classification model is adopted to classify the sleep data collected from the mattress, and then the classification results are compared with the expert staging results of the ECG experiment. It is discovered that the accuracy of the algorithm in classifying the ECG signals collected from the mattress is 70.3%, which indicates the relatively high accuracy of the algorithm and its applicability to the monitoring of quality of sleep. Owing to the sleep condition is detected from the raw ECG or BCG signals obtained by the proposed optical fiber sensor, 60% is the lowest acceptable threshold.6 In some recent studies, some deep learning models pretrained by the public datasets have achieved better performance; it or its variant models can be applied to increase the accuracy. In the future work, we will use the deep learning models with graph convolutions for extracting spatial features and temporal convolutions for capturing the transition rules among sleep stages to increase the sleep condition detection performance better. In the present study, a sleep monitoring system is implemented as well, which is capable of sleep monitoring, sleep quality analysis, sleep statistics, and sleep knowledge display. For users, they can make use of it to view their real-time sleep status, sleep quality, historical sleep statistics, and the sleep knowledge that they want. In this way, the users can better understand their sleep status quantitatively, so as to improve problems such as poor quality of sleep. The system requires no physical contact with the user when the ECG signal is monitored and collected. Because the optical fiber sensor is embedded in the mattress, there is barely any disruption caused to sleep. In addition, it is easy to operate, affordable, and accurate in the staging results, which makes it ideal for domestic monitoring.
Limitations of the study
The sleep monitoring system mentioned herein can be of good use in monitoring the quality of sleep without interference, and yield more accurate sleep staging results. However, there remain some minor differences between the staging results and expert classification results. This may lead to the misjudgment on abnormal information about sleep, thus causing serious consequences. Therefore, it is still necessary to improve the algorithm proposed in this article, and continued optimization can be achieved for the parameters of the optimal SVM model constructed in this article. By selecting the sample data different from the experiment performed in this article, more characteristics of heart rate changes are extracted, with SVM training and parameter optimization carried out. In the algorithm, the heart rate interval sequence extracted from ECG signal is treated as the data of sleep stage. The correctness of heart rate interval sequence extraction has a direct effect on the classification results. For this reason, the preprocessing algorithm can be further improved to retain the characteristics of the processed signal, thus improving the accuracy of classification. In this algorithm, heartbeat interval sequence is used for sleep staging, and then multiple parameters are used to develop the sleep staging algorithm.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| MATLAB R2016a | MathWorks | https://www.mathworks.com/ |
| Python 3.8 | Python Software Foundation | https://www.python.org/ |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Changyuan Yu (Changyuan.yu@polyu.edu.hk).
Materials availability
This study did not generate new materials.
Experimental model and subject details
The proposed machine learning model is PSVM, and the RBF model is chosen as the baseline model. No subjects participated in the experiment.
Method details
Optical fiber interferometer
Optical fiber sensors are a versatile group of sensors that can be classified into several categories, depending on their intended use. These categories include temperature sensors, strain sensors, bending sensors, bending vector sensors, liquid level sensors, current sensors, refractive index sensors, pressure sensors, and others.40 Furthermore, the type of optical fiber sensor can also be categorized based on whether it can be continuously monitored with an increase in sensing distance, which includes point distributed optical fiber sensors, distributed optical fiber sensors, and quasi-distributed optical fiber sensors. Another factor to consider is whether the sensor is an interference or non-interference type. Compared to conventional metal wire sensors, optical fiber sensors offer numerous advantages such as compactness, lightweight, affordability, high safety performance and reliability, electromagnetic interference immunity, corrosion resistance, and the ability to measure diverse objects while causing minimal impact on the object being measured. As a result, optical fiber sensors have the potential for use in a broad range of fields, including agriculture, petroleum exploration, healthcare, aerospace, national defense, and security.
Optical fiber sensors are highly suitable for micro-manufacturing, which makes them particularly useful in the biomedical field for monitoring vital signs and developing minimally invasive surgical tools. In 2007, Takeuchi et al. conducted a study on pharyngeal manometry using a pharynx pressure sensor based on the Fabry-Perot interferometer.41 This sensor (FOP-MIV, FISO Technology) exhibited high consistency with the catheter-type reference sensor (p37-4109c05, zinatics). Its conduit structure had a small diameter of only 2.08mm, and it could measure pressures ranging from -30kPa to 30kPa at a sampling frequency of 250Hz. Moreover, optical fiber sensors are resistant to electromagnetic interference, allowing them to operate even in its presence. For instance, microbend optical fiber sensors can measure various vital signs such as heart rate (HR), respiratory rate (RR), and blood pressure (BP) in the presence of nuclear magnetic resonance.42,43
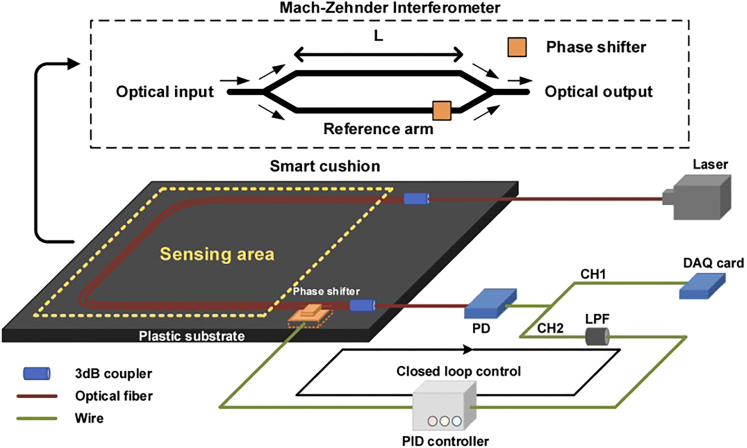
The present study uses a smart cushion as a non-invasive method for BCG monitoring. The monitoring system comprises a BCG monitor based on a Mach-Zehnder interferometer (MZI), a phase shifter, and a proportional-integral-derivative (PID) controller. The monitoring system comprises an MZI-based BCG monitor, a phase shifter, a DFB laser, and a low-speed photodetector (PD). The MZI, made up of two 3dB couplers functioning as an optical splitter and coupler, respectively, is mounted on a plastic substrate, which facilitates packaging as a smart cushion. The MZI and phase shifter are situated outside the sensing area and are fixed parallelly. The PD converts the BCG signal into an electrical signal, which is decomposed into two channels, CH1 and CH2. While CH1 contains the raw data, CH2 includes the data processed using a low-pass filter (LPF) as input for the proportional-integral-derivative (PID) controller. The controller compensates for phase drift to maintain the system operating at the quadrature point. The raw data from CH1 is captured by a data acquisition (DAQ) card (National Instrument, USB6001). Figure 1 illustrates the overall structure of the monitoring system based on the optical fiber sensor.
Figure 1.
Overall structure of the experiment setup
The MZI design used in this study has two arms that are about 40cm in length, with a 5mm difference between them. The plastic substrate integrated cushion contains two bent arms that are arranged side by side in a semicircle shape and do not overlap. The body’s recoil from heartbeats when a subject sits on the cushion induces a phase difference in the interferometer. This phase difference can be used to detect the BCG signal through the intensity variation. The monitoring system utilizing the optical fiber sensor is illustrated in Figure 2. The average output light intensity of the optical fiber interferometer in this system is presented as Equation 1 in Figure 3, where D denotes the average output light intensity, I0 represents the peak intensity of interference fringes, k refers to the number of output light paths (k = 1, 2, 3), φ (t) indicates the phase difference signal of the sensor that is to say the measured signal, and ψ (t) denotes the phase difference caused by environmental changes.
| (Equation 1) |
Figure 2.
The proposed optical fiber sensor system
Figure 3.
Detail information about the proposed monitoring system
Sleep condition detection with machine learning
Calculation of arrangement entropy
The collected ECG signal can be used to record the change of potential difference at each point in time, showing continuity in the time domain to a certain extent. In the wavelet domain, there is usually a large modulus value of the wavelet coefficient as obtained through the decomposition of effective continuous signal.44 However, the noise made by the instrument itself is thermal noise in most cases, that is to say, Gaussian white noise, which lacks continuity in the time domain. Even after wavelet transform, the randomness of Gaussian white noise remains strong in the wavelet domain, where the coefficient corresponding to the effective continuous signal far exceeds the coefficient corresponding to the noise as a result. Therefore, the wavelet coefficient is processed by using the threshold function, and the processed signal is reconstructed by wavelet. In this way, the signal can be obtained after ECG denoising and smoothing. The design makes difference to the denoising and smoothing effect of ECG signals.
Butterworth filter can maximize the flatness of frequency response curve in the passband. The bandwidth of ECG signal ranges from 0 to 100Hz, while the effective signal falls into the range of 0∼30Hz. Therefore, the sampling frequency of Butterworth bandpass filter is set to 300 (>2 ∗ 100).45 For the purpose of wavelet filtering, soft threshold filtering is performed by default, Daubechies 8 wavelet base is adopted, the maximum level of wavelet decomposition is used, and wavelet reconstruction is performed after filtering. By comparing the outcome of filtering by the two filters on the same frame data, it can be found out that the wavelet threshold method leads to a significant improvement of the sharp noise signal and breakpoint after ECG denoising, and the peak value of ECG is relatively constant.
Wavelet packet decomposition (WPD) is performed on the data.46 In the context of wavelet transform, it is difficult to continue decomposing the high-frequency part of ECG. Unlike the wavelet decomposition, WPD is effective not only in decomposing and reconstructing the low-frequency part, but also in decomposing the approximate component and detail component again to analyze the high-frequency part of the signal. For the signals of different frequency bands, WPD can be carried out to adjust the frequency resolution by changing the number of decomposition layers and to identify the optimal basis function, which significantly enhances the advantages of local signal analysis.
The ECG signal is decomposed in n layers, and the minimum resolution is calculated according to the sampling law as follow
| (Equation 2) |
where fs represents the sampling frequency.
According to the frequency range of the four EEG rhythms (0∼30Hz), the minimum number of decomposition nodes contained in them is calculated respectively, the lower nodes in the calculation are detected, and then the corresponding frequency band filter is set according to the frequency of each rhythm wave. By using [i, j] to represent the i-th node to the j-th node of WPD, the frequency band interleaving in the node division of WPD is taken into consideration.
A total of four significant feature values with different and specific frequency can be extracted from the raw ECG signals during sleep, including Alpha wave (α), Beta wave (β), Delta wave (δ) and Theta wave (θ).
-
(1)
Delta Wave (δ): The frequency is between 0.5Hz and 4Hz, amplitude range is 20 μV∼200 μV. Delta Wave, as the lowest frequency signal wave spontaneously generated in the brain, can be detected during deep sleep, deep anesthesia hypoxia, or organic brain lesions.
-
(2)
Theta wave (θ): The frequency range is 4Hz∼8Hz, with an amplitude of approximately 10 μV∼50 μV. Theta wave has a lower frequency, and under normal circumstances, the brain does not spontaneously produce them when a person is awake, only when the central nervous system of a person is inhibited during fatigue and drowsiness, will there be Theta wave appear.
-
(3)
Alpha wave (α): The frequency range is 8Hz∼13Hz, and the signal amplitude is 20 μV∼100 μV. Alpha wave has a faster frequency and is the most rhythmic signal wave. Only when people relax or close their eyes, Alpha wave can be stably detected, and once the subject is moving, opening their eyes, or imagining movement, the wave will decrease or even disappear.
-
(4)
Beta wave (β): The frequency range is between 13Hz and 30Hz, and the signal amplitude range is 5 μV∼20 μV. According to different frequencies, Beta wave can be further divided into β1 wave and β2 wave. When this frequency band wave is detected, it indicates that the cerebral cortex is in a relatively excited state. At this time, people are more active in thinking, and may be thinking logically or experiencing great emotional fluctuations. Therefore, Beta wave can be considered as a sign of brain wakefulness.
The energy of each rhythm wave of ECG is denoted as Eα, Eβ, Eδ and Eθ, respectively. Given the variation in energy between different sleep periods, it can be taken as a characteristic parameter of sleep stages and can be extracted by MATLAB. The total energy of signal g (t) is expressed as Equation 3:
| (Equation 3) |
where j represents the rhythmic wave; Ej denotes the energy value after the reconstruction of the corresponding rhythmic wave signal g (t); i refers to the number of points of signal sampling (i = 0, 1, …, m); and xi indicates the corresponding amplitude of the sample point of the reconstructed signal.
To extract features from nonlinear data, it is crucial to extract not only energy features but also nonlinear features, such as calculating the arrangement entropy and sample entropy of each data frame.
-
(1)
The matrix is obtained by reconstructing the phase space of X as the time series and denoting the size of the phase space as m, where each row represents phase space length in the form of a sequence.
-
(2)
A sequence of symbols is obtained by recording the subscript order of the row way before sorting happens as the rearrangement of the matrix in each row is subject to an increasing order.
-
(3)
The count of subscript occurrences in each row/m! is calculated and used as the probability of the row. The arrangement entropy is then obtained as the sum of the information entropy of all rows in the time series.
By following the above calculation process, the entropy value of the arrangement is recorded as permutation entropy (PeEn) and reaches the maximum when the subscript symbol sequence probability of each line P=1/m!. At this time, the time series becomes more complex. There is no or limited repetition in the symbol sequence. Similarly, when the PeEn value decreases, the time series shows improved regularity.
Calculation of sample entropy
When the length of the original data time series is denoted as N, the sample entropy can be put as {u (i), 1 ≤ i ≤ N}, which is calculated in detail as follows.
-
(1)
The vector X (1), X (2), …, X (N-m+1) of the m-dimensional space is constructed, such that X (i) = {u (i), u (i+1), …, u (i+m – 1)} for i ranging from 1 to N-m+1.
-
(2)
The maximum difference between the corresponding elements of vectors X (i) and X (j) is used to define the distance d [X (i), X (j)], which is denoted as max |u (i+k) - u (j+k)|.
-
(3)
For each value of {i, 1 ≤ i ≤ N - m+1}, the number of d [X (i), X (j)] < r is counted and represented as Nm (i) when the allowable deviation is denoted as r. The ratio of Nm (i) to the total distance between vectors is calculated as Cmi (r).
-
(4)
The value of i in average is calculated as φm(r) through Equation 4:
| (Equation 4) |
-
(5)
After the dimension is increased to m+1, the above procedures (1) to (4) are repeated for the reconstructed sequence vector of m+1 dimension to obtain Cm+1(r), with φm+1(r) expressed as Equations 5 and 6.
| (Equation 5) |
| (Equation 6) |
-
(6)
If a finite value goes to N, the sample entropy of this sequence is expressed as Equation 7.
| (Equation 7) |
In this study, LSVM represents a new machine learning method. Since the statistical theory adopts the structural risk minimization (SRM) criterion, the structural risk can also be minimized when the minimum sample points are obtained, thus improving the adaptability and generalization ability of the model. Besides, there is no limit imposed by the data dimension. In the process of linear classification, the classification surface is taken at a place distant from the two types of samples. By transforming the problem into a high-dimensional space, LSVM converts the nonlinear classification problem into a linear classification problem. Nonlinear mapping to high-dimensional space is necessary for LSVM in theory. However, instead of nonlinear mapping, LSVM relies on the inner product kernel function. The key idea of LSVM is to find the optimal hyperplane for feature space division by maximizing the classification margin. LSVM possesses the following advantages.
-
(1)
The decision in LSVM classification heavily relies on support vectors, which are the training outcome. Hence, the model must possess strong algorithm robustness while requiring minimal storage space;
-
(2)
There are neither preconditions nor probability measures involved.
Neural network
GRNN is a type of radial basis function neural network, which achieves an excellent nonlinear mapping performance in the learning process, showing greater advantages over RBF network.47,48 It performs optimal regression when the sample size is aggregated, and can be used to process unstable sample data. In the case of less sample data available, it remains capable to produce better prediction results. Since GRNN does not require as much sample data as RBF network, it is significantly advantageous in dealing with low data accuracy in the process of actual sample data classification and model fitting. As for the construction of the generalized regression neural network prediction model, it involves three steps: data preprocessing, model parameter optimization, and model implement.
Firstly, a large amount of data is required by the training, evaluation and testing of artificial neural networks. Data preprocessing plays an important role in data mining and calculation, involving various processes such as cleaning, integration, transformation, and specification before the collected data is classified or grouped.49 Data preprocessing is essential for the construction of network models, and it can often determine the outcome of training. At present, there are three commonly used data preprocessing methods: zero mean; normalization; and principal component analysis and whitening. The zero mean value is used to subtract the average value of each one-dimensional data from the original data for replacing the original data with the result. Normalization is purposed to normalize the original data to the same scale. There are two main ways of normalization. One is to carry out the zero mean of the original data, and then the data of each dimension is divided by the standard deviation of each dimension. The other is to normalize the data of different dimensions to the same numerical range. This approach to normalization is applicable given the same importance attached to the data of different dimensions. Principal component analysis and whitening are performed to reduce the dimensionality and variance of the data, thus reducing the input parameters of the model, improving the calculation efficiency and accelerating the learning process.
Secondly, the model parameters are optimized. Cross validation is a widely used method of precision test for the construction of artificial neural network models, which is also known as cyclic estimation. To achieve the cycle verification of cross validation, most of the samples are used for modeling in the given modeling samples, the remaining samples are tested with the established model, and the sum of the squares of the test errors of these small samples is calculated. This process continues to cycle until all the samples are tested. Finally, the model parameters that can reach the error limit and have the minimum error are selected. Two cross-validation methods are widely used: 10-fold and 4-fold cross-validation. To enhance the prediction accuracy, we employ the 10-fold one during our research. To begin with, the input data is randomly divided into 10 samples, of which the first 9 samples are taken for training each time, with the last one verified until all samples are tested. Then, the optimal parameters are obtained.
In the third and last one, the dataset pretreated in the first part as the input and output parameters are inputted into the model with the optimal parameter value as obtained through the second part of cross validation for processing. Also, the experimental value is used to conduct validation analysis and error analysis.
GRNN shows similarity to RBF in structure, representing an improved version of RBF. The difference between GRNN and RBF lies in the introduction of a summation layer into GRNN and the removal of weight connection between the output layer and the hidden layer, which means the least squares superposition of Gaussian weights. As shown in Figures 4 and 7, the corresponding neural network topology is composed of both input and output layers, a mode layer, and a summation layer, with the corresponding network input of X = [x1, x2, …, xn]T and the output of Y = [y1, y2, …, yn]T.
Figure 4.
Basic structure of the GRNN model
Figure 7.
Flow chart of GRNN building for sleep staging and assessment
Input layer
The input layer is intended mainly to determine the input dimension of the data in the model. Then, the inputted data is transferred directly and linearly to the next layer for preliminary processing.
Mode layer
The model layer neuron is a nonlinear function, whose neuron is a radial basis function neuron. The structural model is illustrated in Figure 4. To begin with, RBF neurons receive the output vector of the input layer Xn = [xn1, xn2, …, xnM]T, that is to say, the input vector of the neuron (where n represents the training sample). Then, the Euclidean distance between the center vector and the input vector dist is calculated as cj = [cj1, cj2, …, cjM], where j represents the j-th neuron.
| (Equation 8) |
Finally, the distance between the weight value dist and the input variable is taken as the independent variable. After being multiplied by the threshold value denoted as b, it is transferred to the activation function of the neuron, that is to say, the transfer function. Radial basis function refers to the activation function of radial basis function neurons. The activation function is expressed as follow
| (Equation 9) |
The threshold b is applied to adjust the sensitivity of neurons. Figure 5 shows the basic structure of RBF model. However, Gaussian kernel function is the commonly used RBF function in RBF neurons, so that the activation function of RBF neurons can be expressed as follow
| (Equation 10) |
where Xn represents the n input sample; cj denotes the center of the j radial basal neuron; σ refers to the variance of Gaussian kernel function as the smoothing factor. Then, the output value of the first neuron input from the first training sample is Pnj = R (Xn - cj).
Figure 5.
Basic structure of the RBF model
In GRNN, the rules of setting some parameters in the model layer neurons are specified. Initially, the quantity of neurons in this layer is equivalent to that of training samples (N) and σ is set manually when the network is established. The output vector of the mode layer is expressed as follow
| (Equation 11) |
where n stands for the quantity of training samples.
Summation layer
Two different types of neurons are involved in the summation layer, and one of them is the arithmetic summation in terms of the output for all neurons in the mode layer. Its output is expressed as follow
| (Equation 12) |
The other type of neuron is the weighted summation of the outputs of all mode-layer neurons. We denote the connection weight of this type of neuron as yjk, which represents the k element for the j output vector within the training sample. Its output is expressed as follow
| (Equation 13) |
where n represents the n training sample, k refers to the k-molecular summation neuron (n = 1, 2, ..., N and k = 1, 2, ..., K).
Output layer
The output layer has neurons, with its number equivalent to a dimension of the output vector in the learning sample K. The output of each neuron in the output layer expresses itself as follow
| (Equation 14) |
where k represents the k-output neuron (n = 1, 2, ···, N and k = 1, 2, ···, K).
Quantification and statistical analysis
All machine learning methods were implemented via MATLAB R2016a and Python 3.8 with NVIDIA GeForce GTX1080Ti graphics card and Intel i7-8700 CPU. The RBF algorithm and analysis of average running time were realized with the assistance of the “Radial Basis Function” toolbox in MATLAB R2016a.
Additional resources
We have no relevant resources.
Acknowledgments
The authors gratefully acknowledge the financial supports of the Non-wearable and non-invasive photonic sleep monitoring system based on optical fiber sensor with machine learning (HK PolyU 1-WZ01) and the Shenzhen-Hong Kong-Macao Science and Technology Plan C SGDX (2020110309520303 K-ZGCQ).
Author contributions
Q.W.: Writing–original draft, software, and experiment; W.L. and J.Z.: Analysis and validation; C.Y.: Supervision and funding acquisition.
Declaration of interests
The authors declare no conflicts of interest.
Published: June 30, 2023
Data and code availability
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
References
- 1.Zamora A.N., Watkins D.J., Peterson K.E., Téllez-Rojo M.M., Hu H., Meeker J.D., Cantoral A., Mercado-García A., Jansen E.C. Prenatal maternal pesticide exposure in relation to sleep health of offspring during adolescence. Environ. Res. 2022;204 doi: 10.1016/j.envres.2021.111977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ziporyn T.D., et al. Adolescent sleep health and school start times: setting the research agenda for California and beyond: a research summit summary: a research summit summary. Sleep Health. 2022:661. doi: 10.1016/j.sleh.2021.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Aserinsky E., Kleitman N. A motility cycle in sleeping infants as manifested by ocular and gross bodily activity. J. Appl. Physiol. 1955;8:11–18. doi: 10.1152/jappl.1955.8.1.11. [DOI] [PubMed] [Google Scholar]
- 4.Lane J.M., Qian J., Mignot E., Redline S., Scheer F.A.J.L., Saxena R. Genetics of circadian rhythms and sleep in human health and disease. Nat. Rev. Genet. 2023;24:4–20. doi: 10.1038/s41576-022-00519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kales A., Hoedemaker F.S., Jacobson A., Kales J.D., Paulson M.J., Wilson T.E. Mentation during sleep: REM and NREM recall reports. Percept. Mot. Skills. 1967;24:555–560. doi: 10.2466/pms.1967.24.2.555. [DOI] [PubMed] [Google Scholar]
- 6.Phan H., Andreotti F., Cooray N., Chen O.Y., De Vos M. SeqSleepNet: end-to-end hierarchical recurrent neural network for sequence-to-sequence automatic sleep staging. IEEE Trans. Neural Syst. Rehabil. Eng. 2019;27:400–410. doi: 10.1109/TNSRE.2019.2896659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Sahakian B.J., Kang J., Langley C., Zhang W., Xie C., Xiang S., Yu J., Cheng W., Feng J. The brain structure and genetic mechanisms underlying the nonlinear association between sleep duration, cognition and mental health. Nat. Aging. 2022;2:425–437. doi: 10.1038/s43587-022-00210-2. [DOI] [PubMed] [Google Scholar]
- 8.Chaput J.-P., McHill A.W., Cox R.C., Broussard J.L., Dutil C., da Costa B.G.G., Sampasa-Kanyinga H., Wright K.P., Jr. The role of insufficient sleep and circadian misalignment in obesity. Nat. Rev. Endocrinol. 2023;19:82–97. doi: 10.1038/s41574-022-00747-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rechtschaffen A., Hauri P., Zeitlin M. Auditory awakening thresholds in REM and NREM sleep stages. Percept. Mot. Skills. 1966;22:927–942. doi: 10.2466/pms.1966.22.3.927. [DOI] [PubMed] [Google Scholar]
- 10.Harris J.J., Kollo M., Erskine A., Schaefer A., Burdakov D. Natural VTA activity during NREM sleep influences future exploratory behavior. iScience. 2022;25 doi: 10.1016/j.isci.2022.104396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albrecht J.N., Werner H., Rieger N., Widmer N., Janisch D., Huber R., Jenni O.G. Association between homeschooling and adolescent sleep duration and health during COVID-19 pandemic high school closures. JAMA Netw. Open. 2022;5:e2142100. doi: 10.1001/jamanetworkopen.2021.42100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong L., Xie Y., Zou X. Association between sleep duration and depression in US adults: A cross-sectional study. J. Affect. Disord. 2022;296:183–188. doi: 10.1016/j.jad.2021.09.075. [DOI] [PubMed] [Google Scholar]
- 13.Townsend N., Kazakiewicz D., Lucy Wright F., Timmis A., Huculeci R., Torbica A., Gale C.P., Achenbach S., Weidinger F., Vardas P. Epidemiology of cardiovascular disease in Europe. Nat. Rev. Cardiol. 2022;19:133–143. doi: 10.1038/s41569-021-00607-3. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q., Zhang Y., Chen G., Chen Z., Hee H.I. Assessment of heart rate and respiratory rate for perioperative infants based on ELC model. IEEE Sens. J. 2021;21:13685–13694. [Google Scholar]
- 15.Siddiqi S.H., Kording K.P., Parvizi J., Fox M.D. Causal mapping of human brain function. Nat. Rev. Neurosci. 2022;23:361–375. doi: 10.1038/s41583-022-00583-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang A.C., Vest R.T., Kern F., Lee D.P., Agam M., Maat C.A., Losada P.M., Chen M.B., Schaum N., Khoury N., et al. A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature. 2022;603:885–892. doi: 10.1038/s41586-021-04369-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joutsa J., Moussawi K., Siddiqi S.H., Abdolahi A., Drew W., Cohen A.L., Ross T.J., Deshpande H.U., Wang H.Z., Bruss J., et al. Brain lesions disrupting addiction map to a common human brain circuit. Nat. Med. 2022;28:1249–1255. doi: 10.1038/s41591-022-01834-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winkler E.A., Kim C.N., Ross J.M., Garcia J.H., Gil E., Oh I., Chen L.Q., Wu D., Catapano J.S., Raygor K., et al. A single-cell atlas of the normal and malformed human brain vasculature. Science. 2022;375 doi: 10.1126/science.abi7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Na S., Russin J.J., Lin L., Yuan X., Hu P., Jann K.B., Yan L., Maslov K., Shi J., Wang D.J., et al. Massively parallel functional photoacoustic computed tomography of the human brain. Nat. Biomed. Eng. 2022;6:584–592. doi: 10.1038/s41551-021-00735-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez H., Mei W., Robles I., Hagerling C., Allen B.M., Hauge Okholm T.L., Nanjaraj A., Verbeek T., Kalavacherla S., van Gogh M., et al. Cellular architecture of human brain metastases. Cell. 2022;185:729–745.e20. doi: 10.1016/j.cell.2021.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahrami M., Forouzanfar M. Sleep apnea detection from single-lead ECG: A comprehensive analysis of machine learning and deep learning algorithms. IEEE Trans. Instrum. Meas. 2022;71:1–11. [Google Scholar]
- 22.Yu C., et al. IEEE; 2017. Non-invasive Smart Health Monitoring System Based on Optical Fiber interferometers.” 2017 16th International Conference on Optical Communications and Networks (ICOCN) [Google Scholar]
- 23.Chen Z., Hu J., Yu C. IEEE; 2013. Fiber Senor for Long-Range and Biomedical measurements.” 2013 12th International Conference on Optical Communications and Networks (ICOCN) [Google Scholar]
- 24.Lin W., Lai S., Lu D., Zhang Q., Lin X., Lin J., Wang J., Huang Z. An acousto-assisted liquid-marble-based microreactor for quantitative SERS detection of alkaline phosphatase. Sensor. Actuator. B Chem. 2022;356 [Google Scholar]
- 25.Lyu W., Xu W., Yang F., Chen S., Tan F., Yu C. Non-invasive measurement for cardiac variations using a fiber optic sensor. IEEE Photon. Technol. Lett. 2021;33:990–993. [Google Scholar]
- 26.Chen S., Tan F., Lyu W., Yu C. Ballistocardiography monitoring system based on optical fiber interferometer aided with heartbeat segmentation algorithm. Biomed. Opt Express. 2020;11:5458–5469. doi: 10.1364/BOE.403086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mai Y., Chen Z., Yu B., Li Y., Pang Z., Han Z. Non-Contact Heartbeat Detection Based on Ballistocardiogram Using UNet and Bidirectional Long Short-Term Memory. IEEE J. Biomed. Health Inform. 2022;26:3720–3730. doi: 10.1109/JBHI.2022.3162396. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., You M., Zhang Y., Wu S., Zhang Y., Yang H., Zheng T., Chen X., Chen Z., Xie X., Zheng X. Noninvasive measurement of the vital signs of cancer patients with a dual-path microbend fiber sensor. Biomed. Opt Express. 2022;13:982–994. doi: 10.1364/BOE.450258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y., Chen Z., Hee H.I. Noninvasive measurement of heart rate and respiratory rate for perioperative infants. J. Lightwave Technol. 2019;37:2807–2814. [Google Scholar]
- 30.Wang Q., Lyu W., Cheng Z., Yu C. Noninvasive Measurement of Vital Signs with the Optical Fiber Sensor Based on Deep Learning. J. Lightwave Technol. 2023:1–11. [Google Scholar]
- 31.Li J., Liu B., Liu J., Shi J.L., He X.D., Yuan J., Wu Q. Low-cost wearable device based D-shaped single mode fiber curvature sensor for vital signs monitoring. Sensor Actuator Phys. 2022;337 [Google Scholar]
- 32.Zhao C., Liu D., Xu G., Zhou J., Zhang X., Liao C., Wang Y. Recent advances in fiber optic sensors for respiratory monitoring. Opt. Fiber Technol. 2022;72 [Google Scholar]
- 33.Savović S., Djordjevich A., Savović I. Theoretical investigation of bending loss in step-index plastic optical fibers. Optics. 2020;475 [Google Scholar]
- 34.Warren-Smith S.C., Kilpatrick A.D., Wisal K., Nguyen L.V. Multimode optical fiber specklegram smart bed sensor array. J. Biomed. Opt. 2022;27:067002. doi: 10.1117/1.JBO.27.6.067002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brientin A., et al. Optics & Laser Technology143; 2021. Numerical and Experimental Study of a Multimode Optical Fiber Sensor Based on Fresnel Reflection at the Fiber Tip for Refractive Index Measurement. [Google Scholar]
- 36.Zhao J., Zhang S., Sun Y., Zhou N., Yu H., Zhang H., Jia D. Wearable optical sensing in the medical internet of things (MIoT) for pervasive medicine: Opportunities and challenges. ACS Photonics. 2022;9:2579–2599. [Google Scholar]
- 37.Han P., Li L., Zhang H., Guan L., Marques C., Savović S., Ortega B., Min R., Li X. Low-cost plastic optical fiber sensor embedded in mattress for sleep performance monitoring. Opt. Fiber Technol. 2021;64 [Google Scholar]
- 38.Yang F., Xu W., Lyu W., Tan F., Yu C., Dong B. High Fidelity MZI-BCG Sensor With Homodyne Demodulation for Unobtrusive HR and BP Monitoring. IEEE Sens. J. 2022;22:7798–7807. [Google Scholar]
- 39.Zhang E., Dao M., Karniadakis G.E., Suresh S. Analyses of internal structures and defects in materials using physics-informed neural networks. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abk0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu W., Lin X., Chen X., Wang Q., Wang X., Yang B., Cai N., Chen R., Chen G., Lin Y. Vision-Based Estimation of MDS-UPDRS Scores for Quantifying Parkinson’s Disease Tremor Severity. Med. Image Anal. 2023;85 doi: 10.1016/j.media.2023.102754. [DOI] [PubMed] [Google Scholar]
- 41.Takeuchi S., et al. Biomedical Microdevices9; 2007. An Optic Pharyngeal Manometric Sensor for Deglutition Analysis; pp. 893–899. [DOI] [PubMed] [Google Scholar]
- 42.Wang Q., Liu W., Chen X., Wang X., Chen G., Zhu X. Quantification of scar collagen texture and prediction of scar development via second harmonic generation images and a generative adversarial network. Biomed. Opt Express. 2021;12:5305–5319. doi: 10.1364/BOE.431096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feitosa F.G.A.M., Feitosa A.D.M., Paiva A.M.G., Mota-Gomes M.A., Barroso W.S., Miranda R.D., Barbosa E.C.D., Brandão A.A., Lima-Filho J.L., Sposito A.C., et al. Impact of the COVID-19 pandemic on blood pressure control: a nationwide home blood pressure monitoring study. Hypertens. Res. 2022;45:364–368. doi: 10.1038/s41440-021-00784-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q., Liu W., Wang X., Chen X., Chen G., Wu Q. A Spatial-Temporal Graph Model for Pronunciation Feature Prediction of Chinese Poetry. IEEE Transact. Neural Networks Learn. Syst. 2022:1–15. doi: 10.1109/TNNLS.2022.3165554. [DOI] [PubMed] [Google Scholar]
- 45.Jahmunah V., Ng E.Y.K., Tan R.S., Oh S.L., Acharya U.R. Uncertainty quantification in DenseNet model using myocardial infarction ECG signals. Comput. Methods Progr. Biomed. 2023;229 doi: 10.1016/j.cmpb.2022.107308. [DOI] [PubMed] [Google Scholar]
- 46.Liu X., Han J., Xu H., Xiao X., Wen Z., Liang S. An indirect method for rail corrugation measurement based on numerical models and WPD. Measurement. 2022;191 [Google Scholar]
- 47.Mohanta H.K., Mohanta H.K., Pani A.K. Adaptive non-linear soft sensor for quality monitoring in refineries using Just-in-Time Learning—Generalized regression neural network approach. Appl. Soft Comput. 2022;119 [Google Scholar]
- 48.Chandrakar R., Raja R., Miri R., Sinha U., Kumar Singh Kushwaha A., Raja H. Enhanced the moving object detection and object tracking for traffic surveillance using RBF-FDLNN and CBF algorithm. Expert Syst. Appl. 2022;191 [Google Scholar]
- 49.Lai S., Liu Y., Fang S., Wu Q., Fan M., Lin D., Lin J., Feng S. Ultrasensitive detection of SARS-CoV-2 antigen using surface-enhanced Raman spectroscopy-based lateral flow immunosensor. J. Biophot. 2023 doi: 10.1002/jbio.202300004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.