Abstract
MicroRNAs (miRNAs) are approximately 22 nucleotide-long non-coding RNAs that are encoded in the genome. miRNAs form base pairs with target mRNAs in the RNA-induced silencing complex and repress their expression through a mechanism called RNA silencing. Expression profiles of miRNAs differ between cells and tissues. In this study, we performed cytosine β-D-arabinofuranoside (AraC)-induced neuron-like differentiation of human NTERA2/D1 (NT2) cells and quantified endogenous miRNA levels using quantitative RT-PCR. In conclusion, pre-mir-106b and pre-mir-19b levels were decreased after AraC-induced neuron-like differentiation of NT2 cells, indicating the functional relevance of miRNAs in the differentiation of mammalian cells.
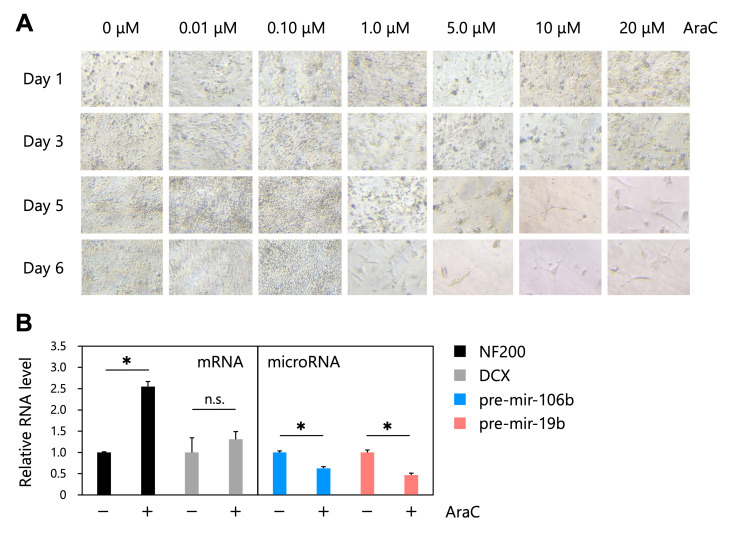
Figure 1. AraC-induced neuron-like differentiation of human NTERA2/D1 (NT2) cells and quantification of endogenous RNA levels .
( A ) Microscopic images of AraC-induced neuron-like differentiation of human NTERA2/D1 (NT2) cells. NT2 cells were treated with various concentrations of AraC (0, 0.01, 0.1, 1, 5, 10, and 20 μM) and cultured for up to 6 days. ( B ) Relative RNA levels of NF200, DCX, pre-mir-106b, and pre-mir-19b. Cells that were treated with 0 or 20 μM AraC for 6 days were collected and were performed qRT-PCR using specific primers for NF200, DCX, pre-mir-106b, and pre-mir-19b (n=3).
Description
MicroRNAs (miRNAs) are approximately 22 nucleotide-long non-coding RNAs that are encoded in the genome (Bartel et al. 2004; Wilson et al. 2013) . miRNAs form base pairs with target mRNAs in the RNA-induced silencing complex (RISC) and repress their expression through a mechanism called RNA silencing. According to the miRNA database miRBase, the human genome encodes 1,917 miRNA precursors (pre-miRNAs) (Kozomara et al. 2019) , and the expression profiles of miRNAs differ between cells and tissues (Ludwig et al. 2016; de Rie et al. 2017; Moore et al. 2020) . In this study, we performed cytosine β-D-arabinofuranoside (AraC)-induced neuron-like differentiation of human NTERA2/D1 (NT2) cells (González-Burguera et al. 2016) and quantified endogenous miRNA levels using quantitative RT-PCR (qRT-PCR).
NT2 cells derived from human testicular embryonic carcinoma cells (Andrews 1984) can be differentiated into neuron-like cells by retinoic acid (RA) treatment (Pleasure et al. 1992) ; however, AraC treatment can induce neuron-like differentiation of NT cells more efficiently than RA treatment in a short period of time (González-Burguera et al. 2016). AraC-induced neuron-like NT2 cells display glutamatergic and cholinergic neurotransmitter phenotypes, which differ from those of RA-induced neuron-like NT2 cells (González-Burguera et al. 2016).
NT2 cells were treated with various concentrations of AraC (0, 0.01, 0.1, 1, 5, 10, and 20 μM) and cultured for up to 6 days ( Figure 1A ). On day 6 after AraC treatment, the cells that were treated with 1, 5, 10, or 20 μM AraC showed neuron-like morphological changes, whereas cells that were treated with 0, 0.01, or 0.1 μM AraC showed no morphological changes and continued to proliferate ( Figure 1A ). To measure the expression of genes encoding neuronal phenotype markers, we used cells that were treated with 0 or 20 μM AraC for 6 days and performed qRT-PCR using specific primers for neuron-specific cytoskeletal proteins neurofilament 200 kDa (NF200) and doublecortin (DCX) ( Figure 1B ). The results showed that the expression of NF200 was significantly increased by AraC treatment for 6 days, indicating that 20 μM AraC treatment for 6 days induced neuron-like differentiation of NT2 cells. Similarly, the expression of DCX was increased by AraC treatment, but this difference was not statistically significant. We then quantified the endogenous miRNAs pre-mir-106b and pre-mir-19b using specific primers ( Figure 1B ). The quantified pre-miRNA levels were normalized by the mRNA level of Tubulin. The results showed that the levels of pre-mir-106b and pre-mir-19b were decreased by AraC treatment.
In conclusion, we found that the pre-mir-106b and pre-mir-19b levels were decreased after AraC-induced neuron-like differentiation of NT2 cells, indicating the functional relevance of miRNAs in the differentiation of mammalian cells.
Methods
Cell culture
NT2 cells were obtained from the ATCC (#CRL-1973) and were cultured in Dulbecco’s Modified Eagle’s medium (Wako) containing 10% fetal bovine serum (NICHIREI) and antibiotics (100 U/ml of penicillin and 100 µg/ml of streptomycin, Wako) at 37°C in an atmosphere containing 5% CO 2 .
AraC-induced neuron-like differentiation
NT2 cells were treated with AraC as described previously (González-Burguera et al. 2016). Briefly, NT2 cells were seeded in a 12-well plate. Cells at 80–90% confluence were treated with 0, 0.01, 0.1, 1, 5, 10, and 20 μM of AraC (Sigma Aldrich, #C1768). The medium containing AraC was replaced with fresh medium every 2 days.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted using a FastGene RNA Premium Kit with FastGene miRNA enhancer (Nippon Genetics). The extracted RNA was used for complementary DNA (cDNA) synthesis using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qRT-PCR was performed using the KAPA SYBR Fast qPCR Master Mix ABI Prism Kit (Kapa Biosystems) and the QuantStudio Real-Time PCR system (Thermo Fisher Scientific). Primer sequences used in this study are listed in Reagents.
Reagents
PCR primers used in this study for qRT-PCR
|
Name |
Sequence (5’ to 3’) |
|
NF200-F |
TAACTGAGTACCGGCGTCAGC |
|
NF200-R |
TGCTGAATGGCTTCCTGGTAGG |
|
DCX-F |
TGTGGGCATGTGTGAGGAAAC |
|
DCX-R |
TGGTGGAACCTCAGAGACTGAC |
|
Tubulin-F |
CTGGCACCATGGACTCTG |
|
Tubulin-R |
TCGGCTCCCTCTGTGTAG |
|
pre-mir-106b-F |
GCTGACAGTGCAGATAGTGGTC |
|
pre-mir-106b-R |
GCAGCAAGTACCCACAGTGC |
|
pre-mir-19b-F |
AGTTTTGCAGGTTTGCATCCAG |
|
pre-mir-19b-R |
TTGCATGGATTTGCACAGCA |
Funding Statement
This work was supported by grants provided by the Secom Foundation and the Takeda Science Foundation to T.T.
References
- Andrews PW. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev Biol. 1984 Jun 1;103(2):285–293. doi: 10.1016/0012-1606(84)90316-6. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004 Jan 23;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- de Rie D, Abugessaisa I, Alam T, Arner E, Arner P, Ashoor H, Åström G, Babina M, Bertin N, Burroughs AM, Carlisle AJ, Daub CO, Detmar M, Deviatiiarov R, Fort A, Gebhard C, Goldowitz D, Guhl S, Ha TJ, Harshbarger J, Hasegawa A, Hashimoto K, Herlyn M, Heutink P, Hitchens KJ, Hon CC, Huang E, Ishizu Y, Kai C, Kasukawa T, Klinken P, Lassmann T, Lecellier CH, Lee W, Lizio M, Makeev V, Mathelier A, Medvedeva YA, Mejhert N, Mungall CJ, Noma S, Ohshima M, Okada-Hatakeyama M, Persson H, Rizzu P, Roudnicky F, Sætrom P, Sato H, Severin J, Shin JW, Swoboda RK, Tarui H, Toyoda H, Vitting-Seerup K, Winteringham L, Yamaguchi Y, Yasuzawa K, Yoneda M, Yumoto N, Zabierowski S, Zhang PG, Wells CA, Summers KM, Kawaji H, Sandelin A, Rehli M, FANTOM Consortium. Hayashizaki Y, Carninci P, Forrest ARR, de Hoon MJL. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat Biotechnol. 2017 Aug 21;35(9):872–878. doi: 10.1038/nbt.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A, Kaul R, Halow J, Van Nostrand EL, Freese P, Gorkin DU, Shen Y, He Y, Mackiewicz M, Pauli-Behn F, Williams BA, Mortazavi A, Keller CA, Zhang XO, Elhajjajy SI, Huey J, Dickel DE, Snetkova V, Wei X, Wang X, Rivera-Mulia JC, Rozowsky J, Zhang J, Chhetri SB, Zhang J, Victorsen A, White KP, Visel A, Yeo GW, Burge CB, Lécuyer E, Gilbert DM, Dekker J, Rinn J, Mendenhall EM, Ecker JR, Kellis M, Klein RJ, Noble WS, Kundaje A, Guigó R, Farnham PJ, Cherry JM, Myers RM, Ren B, Graveley BR, Gerstein MB, Pennacchio LA, Snyder MP, Bernstein BE, Wold B, Hardison RC, Gingeras TR, Stamatoyannopoulos JA, Weng Z. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020 Jul 29;583(7818):699–710. doi: 10.1038/s41586-020-2493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Burguera I, Ricobaraza A, Aretxabala X, Barrondo S, García del Caño G, López de Jesús M, Sallés J. Highly efficient generation of glutamatergic/cholinergic NT2-derived postmitotic human neurons by short-term treatment with the nucleoside analogue cytosine β-D-arabinofuranoside. Stem Cell Res. 2016 Mar 3;16(2):541–551. doi: 10.1016/j.scr.2016.02.038. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019 Jan 8;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E, Keller A. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016 Feb 25;44(8):3865–3877. doi: 10.1093/nar/gkw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasure SJ, Page C, Lee VM. Pure, postmitotic, polarized human neurons derived from NTera 2 cells provide a system for expressing exogenous proteins in terminally differentiated neurons. J Neurosci. 1992 May 1;12(5):1802–1815. doi: 10.1523/JNEUROSCI.12-05-01802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]