Abstract
Embryonic development is a complex process integrating cell fate decisions and morphogenesis in a spatiotemporally controlled manner. Previous studies with model organisms laid the foundation of our knowledge on post-implantation development; however, studying mammalian embryos at this stage is a difficult and laborious process. Early attempts to recapitulate mammalian development in vitro begun with embryoid bodies (EBs), in which aggregates of mouse embryonic stem cells (mESCs) were shown to differentiate into spatially arranged germ layers. A more revised version of EBs, gastruloids, improved the germ layer differentiation efficiency and demonstrated cell fate patterning on multiple axes. However, gastruloids lack anterior neural progenitors that give rise to brain tissues in the embryo. Here, we report a novel culture protocol to coax mESCs into post-implantation epiblast-like (EPI) aggregates in high throughput on bioengineered microwell arrays. We show that upon inhibition of the Wnt signaling pathway, EPI aggregates establish an extended axial patterning, leading to co-derivation of anterior neural progenitors and posterior tissues. Our approach is amenable to large-scale studies aimed at identifying novel regulators of gastrulation and anterior neural development that is currently out of reach with existing embryoid models. This work should contribute to the advancement of the nascent field of synthetic embryology, opening up exciting perspectives for various applications of pluripotent stem cells in disease modeling and tissue engineering.
Key features
A new gastruloid culture system to model post-implantation mouse embryonic development in vitro
High-throughput formation of epiblast-like aggregates on hydrogel microwells
Builds upon conventional gastruloid cultures and provides insight into the role of Wnt signaling for the formation of anterior neural tissues
Graphical overview
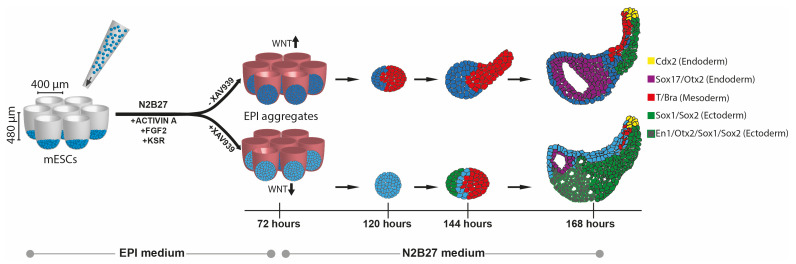
Keywords: Gastruloid, Organoid, Developmental biology, Embryoid, Synthetic embryos
Background
The first attempts to model mouse embryonic development in vitro begun with a coincidental observation. When teratocarcinomas from neonatal mouse testes were analyzed, scientists noticed a structural similarity to the developing mouse embryo (Stevens, 1959). Hence, they called these tumors “embryoid bodies” (Pierce et al., 1960). It was further shown that embryoid bodies retained the tumor-forming capacity to form cell types ranging from cartilage to neural tissue, a multipotency that was later attributed to the embryonal carcinoma (EC) cells (Kleinsmith and Pierce, 1964). The use of EC cells to mimic embryonic development was rapidly replaced by embryonic stem cells (ESCs) following their isolation in 1981 (Evans and Kaufman, 1981). Pioneering studies have demonstrated self-organization potential of embryoid bodies to recapitulate, to a limited extent, gastrulation-like events and antero-posterior axis determination (ten Berge et al., 2008). However, this was not accompanied by axial morphogenesis.
More recently, gastruloids have taken the extent of self-organization potential of mouse ESCs (mESCs) to demonstrate that embryoid bodies could undergo axial morphogenesis (van den Brink et al., 2014). In this model, when treated with Wnt agonist CHIR99021, small aggregates of mESCs (~300 cells) were shown to break symmetry and demonstrated polarized T/Bra expression in a reproducible way. When cultured further, gastruloids elongated and established patterning along antero-posterior, dorso-ventral, and medio-lateral axes (Beccari et al., 2018). Moreover, the multi-axial patterning was linked to spatiotemporal activation of Hox gene clusters, a phenomenon that is conserved across many species (Santini et al., 2003). Such self-organization potential of mESCs, in the absence of any extraembryonic tissue, was remarkably similar to the developing post-occipital region of the mouse embryo; however, tissues mapping to anterior brain regions were completely absent in gastruloids.
Mechanical forces and extracellular matrix composition have significant influence on the developing mouse embryo (Hiramatsu et al., 2013). When gastruloids are embedded in a basement membrane substitute, they remarkably organize into structures bearing somites and a central neural tube-like tissue (Veenvliet et al., 2020). Even under these conditions, gastruloids fail to form any anterior neural tissues corresponding to the brain regions.
The epiblast is the domain that forms the embryo proper. However, contribution from extraembryonic tissues is required since development is halted in their absence (Donnison et al., 2005; Rodriguez et al., 2005). Studies accounting for this necessity have therefore established hybrid embryoid models, combining embryonic stem cells with trophoblast stem cells (TSCs) (Harrison et al., 2017; Rivron et al., 2018) and/or extraembryonic endoderm cells (XENs) (Sozen et al., 2018). More recently, mESCs were co-cultured with transdifferentiated TSCs and XENs to generate structures that almost completely recapitulate E8.5 embryos, including brain tissues (Amadei et al., 2022; Tarazi et al., 2022). However, the complexity of the tri-culture system and necessity of special equipment to grow them until late stages remain as the major limitations in generating synthetic embryos.
Previously, it was shown that embryos lacking extraembryonic Wnt source could still break symmetry and initiate gastrulation, suggesting an autonomous developmental potential of epiblast cells (Yoon et al., 2015). Moreover, overactivation of Wnt signaling in the epiblast was shown to deplete anterior neural progenitors in the favor of mesoderm derivatives; this phenotype could be rescued by inhibition of Wnt signaling (Osteil et al., 2019). Therefore, we reasoned that a similar trade-off could be happening in conventional gastruloids, owing to overactivation of Wnt signaling by CHIR99021 treatment.
Here, we report a new model system based on aggregation of mESCs to derive post-implantation epiblast-like structures (EPI aggregates). We formulated a serum-free epiblast-induction medium comprising Activin-A (Tgf-β agonist), Fgf2 (Fgf agonist), and knockout serum replacement, which promoted the acquisition of epiblast identity, followed by their spontaneous symmetry breaking and subsequent morphogenesis without any external Wnt stimulation. Moreover, inhibition of Wnt signaling during early stages of EPI aggregate formation helped to maintain anterior neural progenitors, which then committed to generate forebrain-, midbrain-, and hindbrain-like tissues. This protocol expands the cell type repertoire that can be generated within gastruloids.
Materials and reagents
Mouse embryonic stem cell lines
SBr [SOX1-GFP; T/BRA-mCherry] mESC line (Deluz et al., 2016)
WNT [TLC-mCherry] mESC line (Ferrer-Vaquer et al., 2010; Faunes et al., 2013)
TGF-β [AR8-mCherry] mESC line (Serup et al., 2012)
Cell culture materials
Ultra-low attachment 96-well plate (Corning, catalog number: 7007)
6-well plate, tissue culture–treated (Falcon, catalog number: 38016)
6-well plate, non tissue culture–treated (Corning, catalog number: CLS3471)
PEG microwells (400 μm microwell diameter, 121 wells/array in 24-well format) (Brandenberg et al., 2020). Commercially available Gri3D® 96-well plate (400 μm microwell diameter, 121 wells/array) could be used alternatively.
10 cm plate, non tissue culture–treated (Corning, catalog number: CLS430591)
Pipettes (2, 5, 10, 25, and 50 mL)
Micropipettes (1–10, 2–20, 20–200, and 100–1,000 μL)
Pipette tips (10, 20, 200, and 1,000 μL)
Cell culture reagents
-
Base media and dissociation reagents
DMEM+Glutamax (Gibco, catalog number: 61965-026). Keep at 4 °C
DMEM/F12+GlutaMAX (Gibco, catalog number: 31331-028). Keep at 4 °C
Neurobasal (Gibco, catalog number: 21103-049). Keep at 4 °C
Accutase (Gibco, catalog number: A11105). Keep at 4 °C
PBS (Gibco, catalog number: 10010023). Keep at room temperature
-
Media supplements
ESC-qualified FBS (Gibco, catalog number: 16141-079). Keep stock at -80 °C, working aliquots at -20 °C
KnockOutTM serum replacement (Thermo Fisher Scientific, catalog number: 10828010). Keep stock at -80 °C, working aliquots at -20 °C
N2 supplement (Gibco, catalog number: 17502001). Keep at -20 °C
B27 supplement (Gibco, catalog number: 17504001). Keep at -20 °C
Sodium pyruvate (Gibco, catalog number: 11360-070). Keep at 4 °C
MEM non-essential amino acids (Gibco, catalog number: 11140-035). Keep at 4 °C
2-mercaptoethanol (Gibco, catalog number: 31350-010). Keep at 4 °C
Penicillin/Streptomycin (Gibco, catalog number: 15140-122). Keep working aliquots at -20 °C
GlutaMAX (Gibco, catalog number: 35050-038). Keep at room temperature
-
Growth factors and small molecule inhibitors
CHIR99021 (Tocris, catalog number: 4423). Keep stock at -80 °C, working aliquots at 4 °C
PD0325901 (Selleckchem, catalog number: S1036). Keep stock at -80 °C, working aliquots at 4 °C
LIF (in-house preparation). Keep stock at -80 °C, working aliquots at 4 °C
FGF2 (Gibco, catalog number: PMG0035). Keep stock at -80 °C, working aliquots at -20 °C. Use within three months after reconstitution
ACTIVIN A (R&D systems, catalog number: 338-AC). Keep stock at -80 °C, working aliquots at -20 °C. Use within three months after reconstitution
XAV939 (Tocris, catalog number: 3748). Keep stock at -80 °C, working aliquots at -20 °C
-
Immunostaining materials and reagents
4% PFA (Thermo Fisher, catalog number: J19943.K2)
DAPI (Sigma, catalog number: 9542)
Glass slides
Coverslips (1.5 thickness)
Mounting medium (Vectashield, catalog number: H-1000-10)
Nail polish
Triton X-100 (Sigma, catalog number: X100)
Mouse embryonic stem cell (mESC) maintenance medium (500 mL) (see Recipes)
EPI differentiation medium (500 mL) (see Recipes)
Primary and secondary antibodies (Table 1)
Table 1. List of primary and secondary antibodies.
| Target | Species | Dilution | Catalogue number | Supplier |
|---|---|---|---|---|
| Anti-E-cadherin | Rabbit | 1:500 | 24E10 | Cell Signaling Technology |
| Anti-Sox2 | Rabbit | 1:400 | ab97959 | Abcam |
| Anti-Sox1 | Goat | 1:50 | af3369 | R&D Systems |
| Anti-Otx2 | Goat | 1:25 | af1979 | R&D Systems |
| Anti-Brachyury | Goat | 1:300 | sc-17745 (C-19) | Santa Cruz |
| Anti-Brachyury | Rabbit | 1:100 | ab209665 | Abcam |
| Anti-Oct4 | Mouse | 1:200 | sc-5270 (C-10) | Santa Cruz |
| Anti-Nanog | Rat | 1:300 | 14-5761-80 | Thermo Fisher |
| Anti-Cdx2 | Rabbit | 1:200 | ab76541 | Abcam |
| Anti- aPKC | Mouse | 1:100 | sc-17781 (H-1) | Santa Cruz |
| Anti-Sox17 | Goat | 1:200 | AF1924 | Abcam |
| Anti-Cdx2 | Rabbit | 1:200 | ab76541 | Abcam |
| Anti-mCherry | Rat | 1:400 | M11217 | Thermo Fisher |
| Phalloidin AF488 | 1:1,000 | A12379 | Thermo Fisher | |
| Phalloidin AF635 | 1:1,000 | A34054 | Thermo Fisher | |
| Anti-chicken Alexa Fluor 488 | Donkey | 1:500 | 703-545-155 | Jackson ImmunoResearch |
| Anti-rat Alexa Fluor 568 | Goat | 1:500 | A-21247 | Thermo Fisher |
| Anti-goat Alexa Fluor 568 | Donkey | 1:500 | A-11057 | Thermo Fisher |
| Anti-goat Alexa Fluor 647 | Donkey | 1:500 | A-21447 | Thermo Fisher |
| Anti-rabbit Alexa Fluor 488 | Donkey | 1:500 | A-21206 | Thermo Fisher |
| Anti-rabbit Alexa Fluor 568 | Donkey | 1:500 | A-10042 | Thermo Fisher |
| Anti-rabbit Alexa Fluor 647 | Donkey | 1:500 | A-31573 | Thermo Fisher |
| Anti-mouse Alexa Fluor 568 | Donkey | 1:50 | A-10037 | Thermo Fisher |
| Anti-mouse Alexa Fluor 647 | Donkey | 1:50 | A-31571 | Thermo Fisher |
Equipment
Incubator with regulated temperature and humidity
Centrifuge
Biological safety cabinet
4 °C fridge, -20 °C freezer, and -80 °C freezer
Eclipse inverted microscope (Nikon, catalog number: TS100)
Pipettes (single and multichannel)
Sterile tips
Hemocytometer and Countess 3 automated cell counter (catalog number: AMQAX2000)
Zeiss LSM700 inverted confocal microscope
Nikon Eclipse Ti inverted microscope
EC Plan-Neofluar 10×/0.30 and Plan-Apochromat 20×/0.80 air objectives
Software
Zen software (2009)
NIS elements (version 4.11.0)
Fiji ImageJ (version 2.0.0-rc-69/1.52n)
GraphPad Prism software (version 8.4.2)
RStudio (version 1.3.1056)
Procedure
-
Passaging of mouse embryonic stem cells
In a Falcon tube, prepare 4.5 mL of mESC maintenance medium (without CHIR99021, PD0325901, and LIF) to collect cells in. In a separate tube, prepare 3 mL of s2iL medium (see Recipe 1).
Check mESCs under microscope. The colonies should be round and not touching each other (Figure 1).
Remove medium from the mESCs and wash with 2 mL of PBS (without calcium and magnesium).
Add 550 μL of room-temperature accutase for dissociation. Incubate for 2-3 min at room temperature. Gently tap the plate to lift up colonies.
Pipette up and down 10–15 times to dissociate colonies into single cells. Make sure cells are single by checking under the microscope.
Place the cell suspension in previously prepared 4.5 mL of mESC maintenance medium.
Spin down at 200× g (1,000 rpm) for 5 min.
Remove supernatant and resuspend in 1 mL of s2iL medium.
Count cells with hemocytometer or automated cell counter.
Seed 50,000–60,000 cells per well of 6-well plate in s2iL medium. Distribute cells equally by shaking the plate back and forth and left and right.
Place the plate back in the incubator and culture for two or three days at 37 °C with 5% CO2 and 21% O2.
-
Preparing EPI aggregates on PEG microwells
Carefully remove PBS from PEG microwells (400 μm well diameter, 121 wells per array) without touching the hydrogel.
Add 50 μL of EPI differentiation medium on top of the array and place the microwells in the incubator for at least 30 min to equilibrate.
Meanwhile, remove medium from the mESCs and wash with 2 mL of PBS (without calcium and magnesium).
Add 550 μL of room-temperature accutase. Incubate for 2–3 min at room temperature. Gently tap the plate to lift up colonies.
Pipette up and down 10–15 times to dissociate colonies into single cells.
Place the cell suspension in previously prepared 4.5 mL of mESC maintenance medium.
Spin down at 200× g (1,000 rpm) for 5 min.
Remove supernatant and resuspend in 10 mL of PBS.
Spin down at 200× g (1,000 rpm) for 5 min.
Remove supernatant and resuspend in 10 mL of PBS.
Spin down at 200× g (1,000 rpm) for 5 min.
Resuspend in appropriate volume of EPI differentiation medium to obtain a cell suspension of 484,000 cells/mL.
Carefully remove the 50 μL of EPI differentiation medium from the microwell arrays and add 35 μL of cell suspension, to have 100–150 cells per each well of the microwell array.
Do not shake or swirl the plate as it will disturb the equal segregation of cells. Incubate at 37 °C for 30 min for cells to sediment.
Slowly add 1 mL of EPI differentiation medium or EPI+XAV differentiation medium (see Recipe 2) from the notch. Do not dispense liquid directly on the microwell array, as it will cause cells to lift up.
Place the plate back in the incubator and culture for at least 72 h at 37 °C with 5% CO2 and 21% O2. Cells should sediment at the bottom of microwells and form clumps within an hour of seeding (Figure 2).
-
Transferring EPI aggregates into 96-well plates
At approximately 75 h of culture, carefully remove the EPI differentiation medium from the microwells. Dispense 1 mL of fresh N2B27 directly on the microwell array at different locations to lift EPI aggregates up.
Collect EPI aggregates in non-tissue culture–treated 10 cm dishes. Repeat the collection with 1 mL of N2B27 five times. Add 5 mL of N2B27 directly to the 10 cm dish to add up to 10 mL.
Observe EPI aggregates under the microscope. It is advised to do the picking under sterile conditions, under the cell culture hood.
Pick EPI aggregates under the microscope in 10 μL and dispense directly in a well of ultra-low attachment 96-well plate filled with 180 μL of N2B27. Make sure that transferred EPI aggregates are not damaged and maintain smooth edges.
Repeat this step until the 96-well plate is completely filled. It usually takes 30 min to 1 h to fill a full plate.
Place the plate in the incubator.
Every 24 h until 168 h, change 150 μL of medium with fresh N2B27 using a multichannel pipette (Figure 3).
-
Preparing gastruloids
Follow steps B1–B11.
Resuspend in appropriate volume of N2B27 medium to obtain a cell suspension of 7,500 cells/mL. For example, 37,500 cells should be added in 5 mL of N2B27 to prepare a full 96-well plate of gastruloids.
Add 40 μL of the cell suspension per well of a 96-well plate using a multichannel pipette to target 300 cells/well. Place the plate in the incubator for 48 h at 37 °C with 5% CO2 and 21% O2.
At 48 h, prepare 15 mL of N2B27 with 3 μM of CHIR99021. Add 150 μL per well. Note that the final CHIR99021 concentration is 2.36 μM. Place the plate back in the incubator.
At 72 h, gently flush gastruloids to lift up shed cells. Wait 1 min for the main aggregate to sediment. Carefully remove 150 μL of medium and add exact volume of fresh N2B27 medium.
Repeat step D5 every 24 h until 168 h (Figure 4).
-
Immunostaining and imaging
Collect EPI aggregates and gastruloids from 96-well plates with a cut 1,000 μL pipette tip and transfer to low attachment 6-well plates in 3 mL of PBS/well. Incubate for 10 min at room temperature.
Transfer into a new well containing 3 mL of 4% PFA and fix for 2 h at 4 °C.
After fixation, transfer into new wells to do three serial PBS washes (3 mL) of 20 min at room temperature. It is very important to coat the pipette tips with coating solution (PBS + 10% FBS) before transferring fixed aggregates, to prevent sticking on the walls of the tip.
Transfer into a new well containing 3 mL of blocking solution (PBS + 10% FBS + 0.3% Triton X-100) and incubate for 1 h at room temperature.
Transfer to low attachment 24-well plates in 300 μL of blocking solution containing primary antibodies and DAPI. List of primary antibodies used can be found in Table 1. Incubate for at least 24 h at 4 °C on a shaker. Cover the plate with aluminum foil to preserve fluorescence intensity of the reporters.
Next day, transfer back to low attachment 6-well plate and wash away primary antibodies by three serial PBS washes (3 mL) of 20 min at room temperature.
Transfer to low attachment 24-well plates in 300 μL of blocking solution containing secondary antibodies (Table 1) and DAPI. Incubate for at least 24 h at 4 °C on a shaker. Cover the plate with aluminum foil to preserve fluorescence intensity of the reporters.
Next day, transfer back to low attachment 6-well plate and wash away secondary antibodies by three serial PBS washes (3 mL) of 20 min at room temperature.
Carefully aspirate aggregates in 100 μL and place them on glass slide. Remove excess PBS around without touching the aggregates. Add 20–30 μL of mounting medium dropwise. Place the coverslip on top and seal with nail polish. Keep the mounted samples in the dark until imaging.
Image with Zeiss LSM700 inverted confocal microscope with EC Plan-Neofluar 10×/0.30 or Plan-Apochromat 20×/0.80 air objectives (Figure 5).
-
Preparation of EPI aggregates for bulk RNA sequencing
EPI aggregates were lysed with 200 µL TRIzol, followed by addition of 70 µL of chloroform to trigger phase separation. Then, the aqueous phase was collected.
The extraction process was repeated a second time, and an equal volume of isopropanol was added to precipitate the RNA, which was collected by centrifugation at 20,000× g for 30 min.
The pellet was washed with 15 mM sodium acetate in aqueous 70% ethanol, followed by salt-free 70% ethanol, before picking up in RNase-free water.
Figure 1. Representative immunofluorescence image of mouse embryonic stem cells (mESCs) grown in s2iL medium stained for SOX2 (green) and F-Actin (red) (left panel), and KLF4 (green) and TFCP2L1 (red) (right panel).
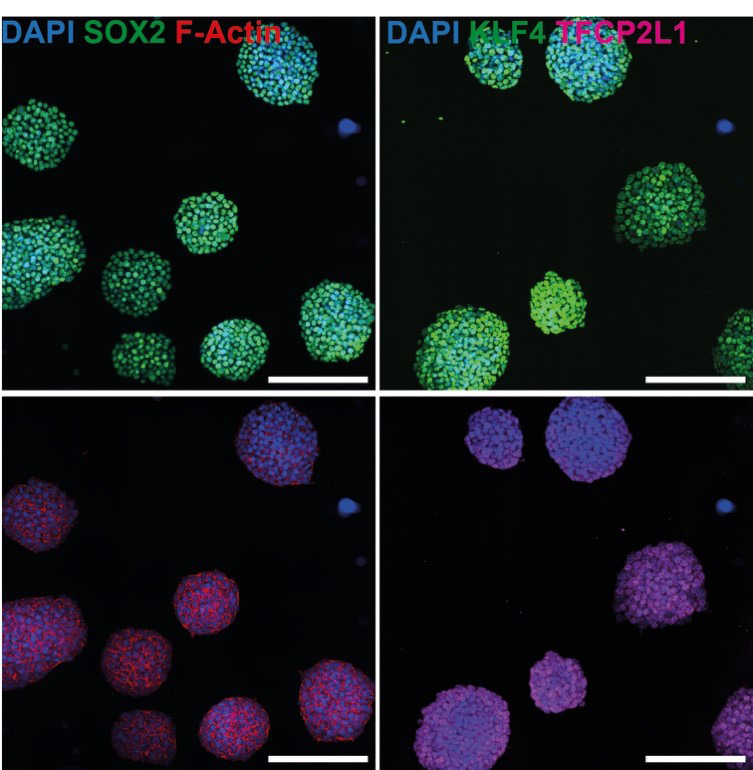
Scale bars = 200 µm.
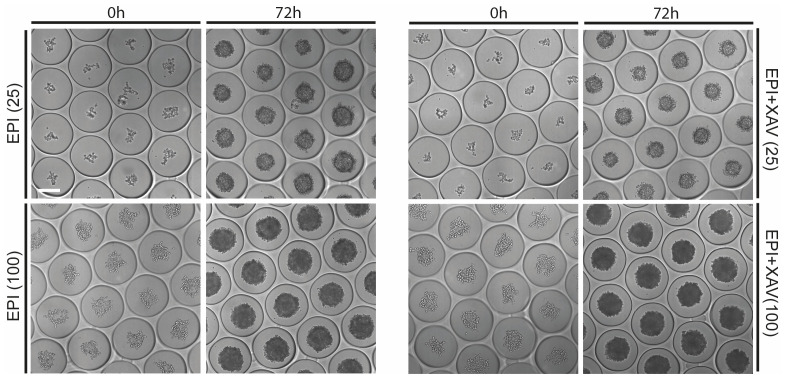
Figure 2. Representative images showing mouse embryonic stem cells (mESCs) seeded at indicated numbers at 0 h and resulting in EPI or EPI+XAV aggregates at 72 h.
Scale bar = 200 μm.
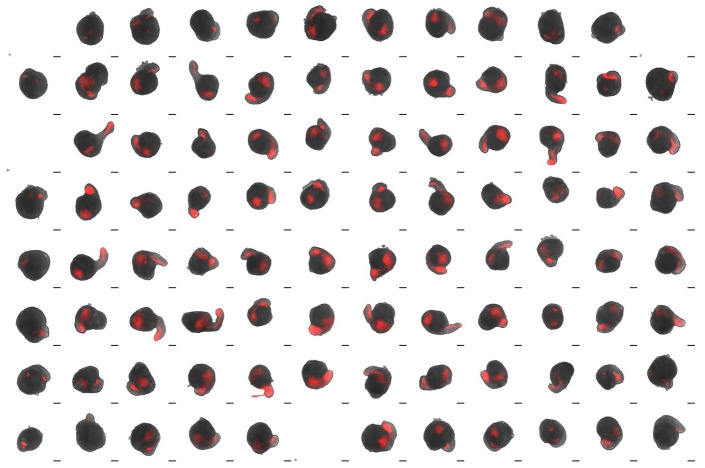
Figure 3. Montage of EPI or EPI+XAV aggregates formed from 100 cells/well shown at 144 h and 168 h after segmentation.
T/Bra expression is shown in red. Sox1 expression is shown in green. Note the Sox1-positive domain localized on the opposite end of T/Bra-positive domain in EPI+XAV aggregates.
Figure 4. Montage of a full 96-well plate of gastruloids at 168 h after segmentation.
T/Bra expression is shown in red. Note the autofluorescent signal over the darker, anterior regions. Scale bars = 200 μm.
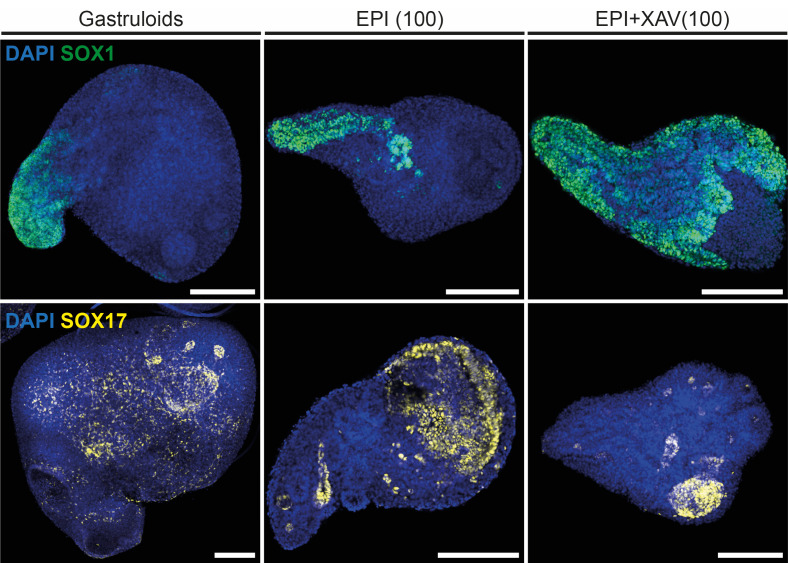
Figure 5. Representative immunofluorescence images of gastruloids, EPI, and EPI+XAV aggregates at 168h stained for SOX1 (top) or SOX17 (bottom).
Scale bars: 200 μm.
Data analysis
-
Image analysis
Live imaging of EPI aggregates was performed with a Nikon Eclipse Ti inverted microscope, objective 10×, 0.3 N.A., using an Andor/iXon DU-888 camera (pixel size 1.2265 μm), equipped with an incubation chamber at 37 °C, 5% CO2. The images were analyzed with ImageJ using custom ActionBar (Mutterer, 2017) and BIOP basics (BIOP Basics ActionBar, c4science) plugins.
For measuring reporter activity in EPI aggregates and gastruloids, brightfield, GFP (for SOX1), and mCherry (for T/BRA-mCherry, TLC-mCherry, AR8-mCherry) channels were acquired. Thresholding and segmentation were performed sequentially for each channel by the custom script (Guiet et al., 2022; DOI:10.5281/zenodo.7409423). The coverage index was calculated by dividing the area of the object identified in mCherry channel to the brightfield area.
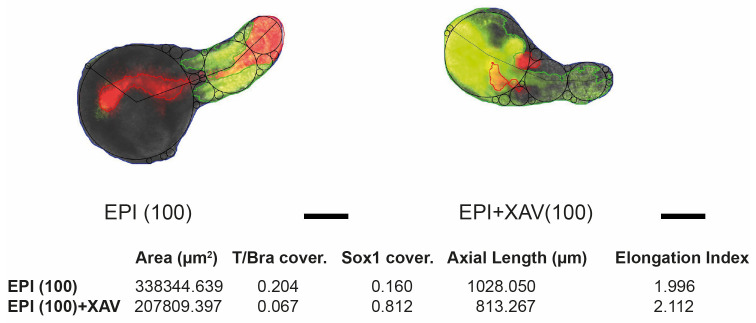
For morphology measurements, a custom script was used (Guiet et al., 2021; doi:10.5281/zenodo.4544370). Brightfield images were thresholded and segmented. Maximum inscribed circle function was used to fit circles in the identified object. Axial length was determined by connecting centers of the fit circles. Elongation index was calculated by dividing axial length to the diameter of the maximum inscribed circle (Figure 6).
GraphPad prism was used to analyze the data and calculate significance. In all experiments, the data were collected from three independent experiments, and at least 24 Epi aggregates or gastruloids per experiment were analyzed. Number of data points and statistical tests performed can be found on the legends of the figures in the original paper (Girgin et al., 2021).
-
Bulk RNA sequencing analysis
RNA quantity and quality were assessed on NanoDrop, qubit, and Agilent TapeStation 4200 profiling, and showed absorbance ratios 260/280 of 1.85 ± 0.12 and RNA integrity numbers of 9.9 ± 0.2 (average ± SD), supporting good purity and absence of degradation. TruSeq stranded mRNA LT libraries were prepared according to Illumina protocol 15031047 Rev. E, starting from 300 ng of RNA, quantified by qubit DNA HS, profiled on TapeStation 4200, and sequenced on an Illumina HiSeq 4000 at a targeted depth of 36 Mreads/sample and paired-end read length of 81,8i,8i,81. The reads were trimmed for adapters with bcl2fastq v2.20.0, aligned to the mouse genome mm10 with STAR 2.7.0e, and a count matrix was assembled using the cellranger v4.0 curation of ENSEMBL annotations. In the manuscript, “gene expression” refers to natural logarithm of counts per million for bulk RNA-seq data. The data was collected from four independent experiments. Cell type signatures were allocated based on previous reports (Pijuan-Sala et al, 2019). All RNA-seq datasets produced in this study are publicly available in the Gene Expression Omnibus (GEO) database under accession code GEO: GSE171210.
Figure 6. Representative post-analysis images of EPI and EPI+XAV aggregates at 168 h showing T/Bra (red) and Sox1 (green) expression domains and calculated coverage indices.
Black lines and circles indicate axial length and inscribed circles, respectively. Elongation index is calculated by dividing the axial length with the diameter of largest inscribed circle. Scale bars = 200 μm.
Notes
The passage number of mESCs should be taken into consideration when generating EPI aggregates or gastruloids. Mouse ESCs kept in culture for more than 15–20 passages might lead to inefficient generation of EPI aggregates or gastruloids.
The quality of mESC culture is critical to successfully generate EPI aggregates. Different cell lines might need different seeding densities and passaging frequency. Make sure that mESC cultures are not confluent and colonies do not fuse. In such cases, passage them at lower density and delay EPI aggregate preparation.
When seeding on PEG microwells, an overestimation of 1.5–2× of the desired cell number per well is recommended. During the seeding process, some cells will sediment into grooves and gaps between the microwells and will therefore not contribute to the aggregate.
For making EPI aggregates and gastruloids, different cell lines might need titration of starting cell numbers. When testing a new cell line, a range of 100–300 cells and 150–600 cells should be tested for EPI aggregates and gastruloids, respectively. For gastruloids, testing of final CHIR99021 concentrations (2–5 μM) is important to achieve the most efficient elongation.
Do not use FGF2 and ACTIVIN-A proteins reconstituted more than three months ago.
When transferring EPI aggregates into a 96-well plate, make sure the aggregates do not spend more than 1 h outside of the incubator. If transfer is taking too long, have 15 min incubation periods in between.
Make sure that transferred aggregates have smooth edges and a size of 200–220 μm diameter. Too small or too big aggregates will not elongate properly.
During daily medium exchanges, gently flush the EPI aggregates and gastruloids to lift off shed cells and remove them. Accumulation of shed cells have a negative impact on optimal development.
The quality of N2B27 medium is crucial for proper differentiation and axial elongation. Make sure to use fresh N2 and B27 supplements and do not use complete N2B27 medium older than three weeks.
Recipes
-
Mouse embryonic stem cell (mESC) maintenance medium (500 mL)
434 mL of DMEM+Glutamax
50 mL of ESC-qualified FBS. Final concentration: 10%
5 mL of sodium pyruvate. Final concentration: 1 mM
5 mL of MEM non-essential amino acids. Final concentration: 1×
1 mL of 2-mercaptoethanol. Final concentration: 0.1 mM
5 mL of penicillin/streptomycin. Final concentration: 50 U/mL
CHIR99021. Final concentration: 3 μM
PD0325901. Final concentration: 1 μM
LIF (in-house preparation). Final concentration: 0.1 mg/mL
Prepare base medium (steps a–f) and use within a month. Add CHIR99021, PD0325901, and LIF fresh on the day of culture to complete s2iL medium.
-
EPI differentiation medium (500 mL)
237 mL of DMEM/F12+ GlutaMAX
237 mL of neurobasal
2.5 mL of N2 supplement. Final concentration: 0.5×
5 mL of B27 supplement. Final concentration: 0.5×
2.5 mL of GlutaMAX. Final concentration: 0.5×
5 mL of sodium pyruvate. Final concentration: 1 mM
5 mL of MEM non-essential amino acids. Final concentration: 1×
1 mL of 2-mercaptoethanol. Final concentration: 0.1 mM
5 mL of penicillin/streptomycin. Final concentration: 50 U/mL
FGF2. Final concentration: 12 ng/mL
ACTIVIN A. Final concentration: 20 ng/mL
KnockOutTM serum replacement. Final concentration: 1%
Prepare N2B27 medium (steps a–i) and use within three weeks. Add FGF2, ACTIVIN A, and KnockOut serum replacement fresh on the day of culture to complete EPI differentiation medium. To make EPI+XAV medium, add 10 μM XAV939.
Acknowledgments
We thank Giuliana Rossi and Alfonso Martinez-Arias for useful feedback on the original manuscript. We thank Sylke Hoehnel and Nathalie Brandenberg (SUN biosciences) for providing PEG microwell plates and technical support. We thank members of the Lutolf laboratory for discussions and sharing materials. We thank Romain Guiet and Olivier Burri for providing the image analysis codes, and Arne Seitz and other members of Bioimaging and Optics Facility (EPFL) for microscopy support. We thank all personnel of the Histology Core Facility for their technical support. This work was funded by EPFL. This protocol was adapted from previous work (Girgin et al., 2021). This work was funded by a Sinergia grant (CRSII5_189956) from the Swiss National Science Foundation, the National Center of Competence in Research (NCCR) Bio-Inspired Materials and EPFL.
Competing interests
The authors declare no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1.Amadei G., Handford C. E., Qiu C., De Jonghe J., Greenfeld H., Tran M., Martin B. K., Chen D. Y., Aguilera-Castrejon A., Hanna J. H., et al.(2022). Embryo model completes gastrulation to neurulation and organogenesis. Nature 610(7930): 143-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beccari L., Moris N., Girgin M., Turner D. A., Baillie-Johnson P., Cossy A. C., Lutolf M. P., Duboule D. and Arias A. M.(2018). Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 562(7726): 272-276. [DOI] [PubMed] [Google Scholar]
- 3.Brandenberg N., Hoehnel S., Kuttler F., Homicsko K., Ceroni C., Ringel T., Gjorevski N., Schwank G., Coukos G., Turcatti G., et al.(2020). High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays. Nat Biomed Eng 4(9): 863-874. [DOI] [PubMed] [Google Scholar]
- 4.Deluz C., Friman E. T., Strebinger D., Benke A., Raccaud M., Callegari A., Leleu M., Manley S. and Suter D. M.(2016). A role for mitotic bookmarking of SOX2 in pluripotency and differentiation. Genes Dev 30(22): 2538-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnison M., Beaton A., Davey H. W., Broadhurst R., P. L’Huillier and Pfeffer P. L.(2005). Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development 132(10): 2299-2308. [DOI] [PubMed] [Google Scholar]
- 6.Evans M. J. and Kaufman M. H.(1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292(5819): 154-156. [DOI] [PubMed] [Google Scholar]
- 7.Faunes F., Hayward P., Descalzo S. M., Chatterjee S. S., Balayo T., Trott J., Christoforou A., Ferrer-Vaquer A., Hadjantonakis A. K., Dasgupta R., et al.(2013). A membrane-associated beta-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development 140(6): 1171-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrer-Vaquer A., Piliszek A., Tian G., Aho R. J., Dufort D. and Hadjantonakis A. K.(2010). A sensitive and bright single-cell resolution live imaging reporter of Wnt/ß-catenin signaling in the mouse. BMC Dev Biol 10: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girgin M. U., Broguiere N., Mattolini L. and Lutolf M. P.(2021). Gastruloids generated without exogenous Wnt activation develop anterior neural tissues. Stem Cell Rep 16(5): 1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guiet R., Burri O., Girgin M. and Lutolf M.(2022). Measuring reporter activity domain in EPI aggregates and Gastruloids.ijm. Zenodo. 10.5281/zenodo.7409423. [DOI] [Google Scholar]
- 11.Guiet R., Burri O., Girgin M. U. and Lutolf M.(2021). Elongation Index(Fiji script)(Version v0). Zenodo. 10.5281/zenodo.4544370. [DOI] [Google Scholar]
- 12.Harrison S. E., Sozen B., Christodoulou N., Kyprianou C. and Zernicka-Goetz M.(2017). Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science 356(6334): eaal1810.. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu R., Matsuoka T., Kimura-Yoshida C., Han S. W., Mochida K., Adachi T., Takayama S. and Matsuo I.(2013). External mechanical cues trigger the establishment of the anterior-posterior axis in early mouse embryos. Dev Cell 27(2): 131-144. [DOI] [PubMed] [Google Scholar]
- 14.Kleinsmith L. J. and Pierce G. B., Jr (1964). Multipotentiality Of Single Embryonal Carcinoma Cells. Cancer Res 24: 1544-1551. [PubMed] [Google Scholar]
- 15.Mutterer J.(2017). Custom toolbars and mini applications with Action Bar. Figshare. [Google Scholar]
- 16.Osteil P., Studdert J. B., Goh H. N., Wilkie E. E., Fan X., Khoo P. L., Peng G., Salehin N., Knowles H., Han J. J., et al.(2019). Dynamics of Wnt activity on the acquisition of ectoderm potency in epiblast stem cells. Development 146(7): dev172858. [DOI] [PubMed] [Google Scholar]
- 17.Pierce G. B. Jr. Dixon F. J. Jr.and Verney E. L.(1960). Teratocarcinogenic and tissue-forming potentials of the cell types comprising neoplastic embryoid bodies. Lab Invest 9: 583-602. [PubMed] [Google Scholar]
- 18.Pijuan-Sala B., Griffiths J. A., Guibentif C., Hiscock T. W., Jawaid W., Calero-Nieto F. J., Mulas C., Ibarra-Soria X., Tyser R. C. V., Ho D. L. L., et al.(2019). A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566(7745): 490-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivron N. C., Frias-Aldeguer J., Vrij E. J., Boisset J. C., Korving J., Vivie J., Truckenmuller R. K., van Oudenaarden A., van Blitterswijk C. A. and Geijsen N.(2018). Blastocyst-like structures generated solely from stem cells. Nature 557(7703): 106-111. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez T. A., Srinivas S., Clements M. P., Smith J. C. and Beddington R. S.(2005). Induction and migration of the anterior visceral endoderm is regulated by the extra-embryonic ectoderm. Development 132(11): 2513-2520. [DOI] [PubMed] [Google Scholar]
- 21.Santini S., Boore J. L. and Meyer A.(2003). Evolutionary conservation of regulatory elements in vertebrate Hox gene clusters. Genome Res 13(6A): 1111-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serup P., Gustavsen C., Klein T., Potter L. A., Lin R., Mullapudi N., Wandzioch E., Hines A., Davis A., Bruun C., et al.(2012). Partial promoter substitutions generating transcriptional sentinels of diverse signaling pathways in embryonic stem cells and mice. Dis Model Mech 5(6): 956-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sozen B., Amadei G., Cox A., Wang R., Na E., Czukiewska S., Chappell L., Voet T., Michel G., Jing N., et al.(2018). Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nature cell biology 20(8): 979-989. [DOI] [PubMed] [Google Scholar]
- 24.Stevens L. C.(1959). Embryology of testicular teratomas in strain 129 mice. J Natl Cancer Inst 23: 1249-1295. [PubMed] [Google Scholar]
- 25.Tarazi S., Aguilera-Castrejon A., Joubran C., Ghanem N., Ashouokhi S., Roncato F., Wildschutz E., Haddad M., Oldak B., Gomez-Cesar E., et al.(2022). Post-gastrulation synthetic embryos generated ex utero from mouse naive ESCs. Cell 185(18): 3290-3306 e3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D. ten Berge, Koole W., Fuerer C., Fish M., Eroglu E. and Nusse R.(2008). Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell 3(5): 508-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van den Brink S. C., Baillie-Johnson P., Balayo T., Hadjantonakis A. K., Nowotschin S., Turner D. A. and Martinez Arias A.(2014). Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development 141(22): 4231-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veenvliet J. V., Bolondi A., Kretzmer H., Haut L., Scholze-Wittler M., Schifferl D., Koch F., Guignard L., Kumar A. S., Pustet M., et al.(2020). Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. Science 370(6522): eaba4937. [DOI] [PubMed] [Google Scholar]
- 29.Yoon Y., Huang T., Tortelote G. G., Wakamiya M., Hadjantonakis A. K., Behringer R. R. and Rivera-Perez J. A.(2015). Extra-embryonic Wnt3 regulates the establishment of the primitive streak in mice. Dev Biol 403(1): 80-88. [DOI] [PMC free article] [PubMed] [Google Scholar]