Abstract
Hfq, an RNA chaperone, promotes the pairing of small RNAs (sRNAs) to target mRNAs, mediating post-transcriptional regulation of mRNA stability and translation. This regulation contributes to bacterial adaptation during stress and pathogenesis. Recent advances in sequencing techniques demonstrate the presence of sRNAs encoded not only in intergenic regions but also from the 3’ and 5’ UTRs of mRNAs, expanding sRNA regulatory networks. Additional layers of regulation by Hfq and its associated RNAs continue to be found. Newly identified RNA sponges modulate the activity of some sRNAs. A subset of sRNAs are proving to be bifunctional, able to pair with targets and also encoding small ORFs or binding other RNA binding proteins, such as CsrA. In addition, there are accumulating examples of Hfq inhibiting mRNA translation in the absence of sRNAs.
Keywords: Small RNAs, CsrA, Rho, Sponges
Bacteria are armed with various ways to respond to stress and adapt to changing growth conditions. Post-transcriptional regulation of mRNA translation and stability provides a rapid and efficient mechanism for responding to changing growth conditions and stresses. Here we review some general principles emerging from recent studies of the major RNA binding protein, Hfq, and the proteins and RNAs it interacts with.
Hfq, a homohexameric toroidal protein of the Sm/Lsm family, acts as an “RNA matchmaker” [1,2], bringing together regulatory small RNAs (sRNAs) and the target mRNAs they pair with. The specificity and timing of regulation depend, in most cases, on the pairing specificity and pattern of expression of the sRNA. Both sRNA and mRNA bind to Hfq, on different faces [2]. sRNA-mRNA pairing can lead to either negative regulation of the mRNA, by blocking ribosome entry and/or by increasing mRNA degradation [3,4], or positive regulation, by increasing accessibility to ribosome entry and translation or by preventing RNase E cleavage (reviewed in [5],[6], [7]) (Figure 1A). Hfq is found in 50% of bacterial species [8], and judging by the phenotypes of cells deleted for hfq, it has a strong effect on growth and virulence in many of them (reviewed in [9,10]). Initial studies in this field focused on sRNAs encoded in intergenic regions, suggested that all sRNAs might interact similarly with Hfq, and assumed that most sRNAs had only one primary function, to pair with mRNA targets. Over the last few years, new tools and expanding studies in multiple organisms have made it clear that each of these initial assumptions needs to be revisited.
Figure 1: Mechanisms of Hfq-dependent regulation.
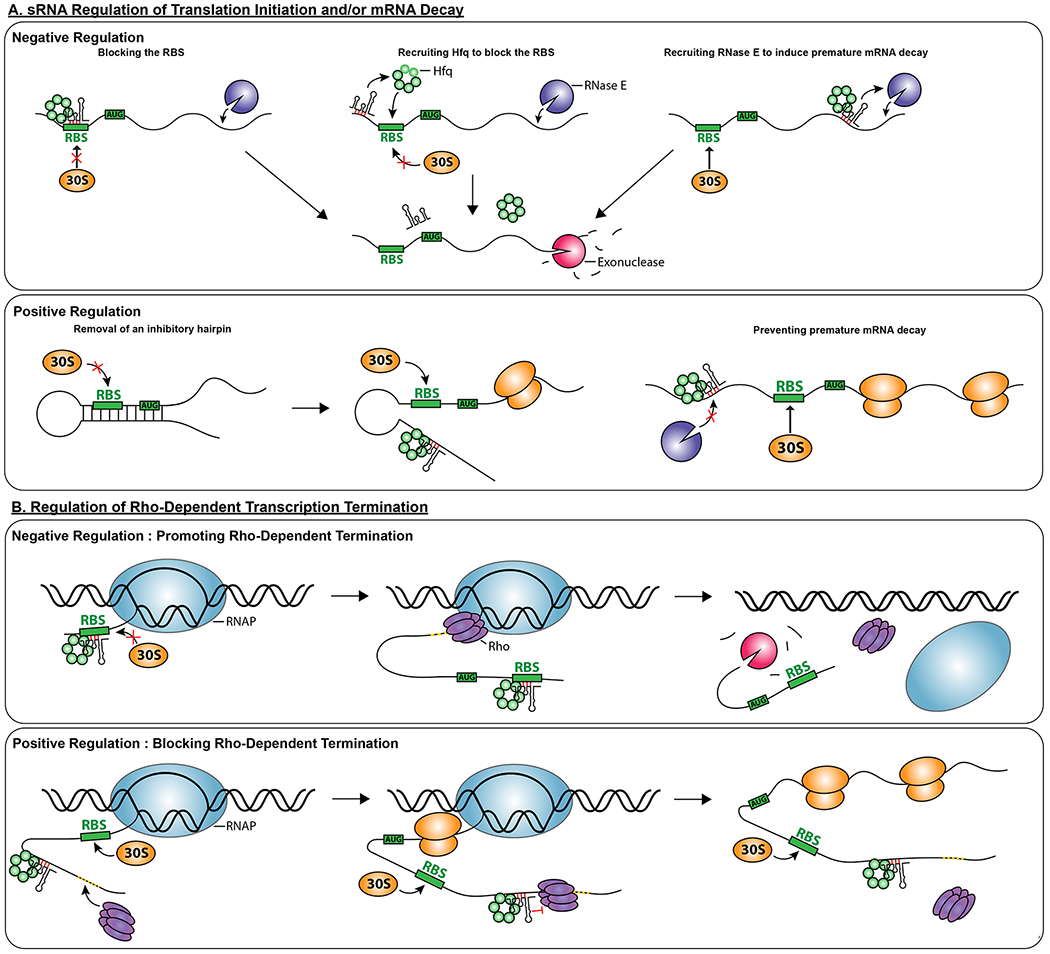
In general, binding of Hfq, with or without other partners (sRNAs in this figure) can change RNA folding and allow or block access of other RNA binding factors, therefore changing the fate of the mRNA.
A. sRNA Regulation of Translation Initiation and/or mRNA Decay.
Negative Regulation: In most cases, negative regulation is via sRNA pairing with targets, as shown here. This can lead to inhibition of translation (left panel), recruitment of RNases (right panel), or both (center panel). Hfq helps to stabilize and recruit the sRNA, and may, in some cases, help to recruit ribonucleases.
Positive Regulation: sRNAs and Hfq binding can collaborate to change RNA folding, remodeling an inhibitory hairpin, for instance, to allow ribosome access (left and center panels), or blocking access of a ribonuclease such as RNase E (right panel), thus stabilizing an mRNA.
B. Regulation of Rho-Dependent Transcription Termination
Rho (purple hexamers) terminates transcription by first accessing naked RNA at a rut site (dotted yellow portion of mRNA), and then traveling along the elongating RNA to release the RNA from RNA polymerase (blue oval) (reviewed in [43]). Thus sRNAs that affect access to the rut site can regulate the ability of Rho to act.
Negative Regulation: Promoting Rho-Dependent Termination: In at least one case, sRNA pairing to a target blocks ribosome entry, allowing Rho to access the naked RNA and leading to Rho-dependent termination within the ORF, downstream of the pairing site [44].
Positive Regulation: Blocking Rho-Dependent Termination: sRNA binding may also block access of Rho to RNA, therefore allowing transcription of downstream genes. In the case examined, this positive regulation collaborates with remodeling of the 5’ UTR to allow both increased transcription and translation [45].
Hfq Regulatory Inputs: Where are Hfq-dependent sRNAs found?
The initial searches for Hfq-dependent sRNAs were focused on those encoded from intergenic regions, in part because it was easiest to define novel non-coding transcripts that were clearly distinct from mRNAs [11] [12] [13] (Figure 2A) . Recent advances in the global analysis of transcription, alone (RNA-seq) or combined with UV-crosslinking and immunoprecipitation (particularly CLIP-seq, RIL-seq and GRIL-seq [14–16]) have greatly simplified the identification of Hfq-dependent RNAs, where Hfq binds on these RNAs, and, for RIL-seq and GRIL-seq, provided evidence for the mRNA partners of the sRNAs. These studies led to the realization that sRNAs are not restricted to intergenic regions but may originate from many other genomic regions, including the 5’ UTR and 3’UTR of mRNAs [17–20] (Figure 2).
Figure 2: sRNA Synthesis.
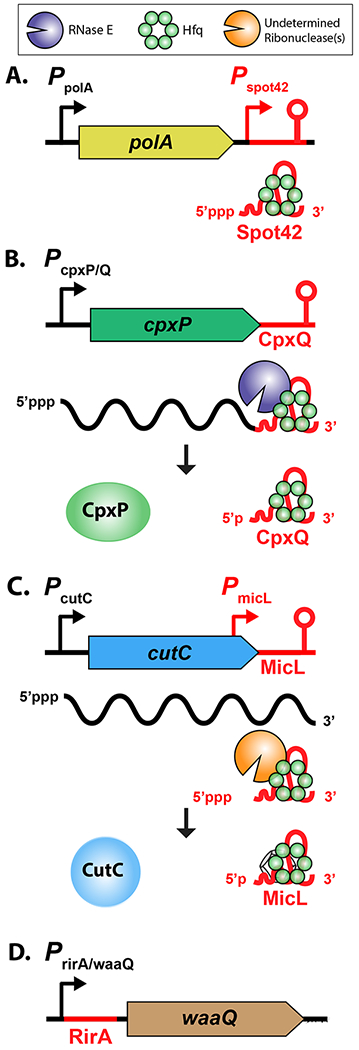
A. Many sRNAs are transcribed from intergenic regions as free-standing transcripts [11] [12] (Spot 42, reviewed in [55], is shown as an example); their abundance in the cell is primarily regulated at the level of transcription, and this is frequently highly regulated.
B. sRNAs can also be synthesized from processing of mRNAs (as for CpxP; [19]); in this case, regulatory signals for the mRNA promoter will also govern synthesis of the sRNA.
C. sRNA promoters may also be embedded within mRNA coding regions (as for MicL here; [18]). In this case, regulation of the upstream gene can be independent of transcription of the overlapping mRNA. The promoter for MicL is a sigma E-dependent promoter [18]; further processing takes place to create the final sRNA.
D. Evidence has also accumulated for sRNAs arising from the 5’ UTR of mRNAs [15]. In the case shown, it is not clear that RirA is an Hfq-dependent sRNA (RirA) [20].
These findings have significantly expanded the inventory of Hfq-binding sRNAs found in Escherichia coli and Salmonella as well the stress regulons that include sRNAs (reviewed in [21]). For instance, CpxQ, a conserved sRNA characterized in Salmonella, is processed from the 3’ end of the cpxP mRNA, and is the first sRNA effector of the Cpx regulon, controlling the response to inner membrane stress (Figure 2B) [19]. DapZ, a Salmonella-specific sRNA with its own promoter, is encoded at the 3’ end of dapB, and has targets similar to those of the broadly conserved GcvB sRNA [17]. MicL, an sRNA associated with the 3’ end of a mRNA, but transcribed from its own promoter, is a new member of the sigma E regulon (Figure 2C) [18]. It is now clear that new Hfq-associated sRNAs can lurk anywhere. Presumably the same is likely to be true for sRNAs not necessarily associated with Hfq, such as RirA, from the 5’ UTR of the waaQ gene (Figure 2D) [20]. Insertion sequences and repeated sequences are yet another possible source of regulatory RNAs that have not been fully explored. A recent report by Haniford and coworkers demonstrates a regulatory role for an RNA processed from the 5’ end of the mRNA encoding the transposase for IS200, a very widespread insertion sequence [22]. While it is not yet clear if this regulatory RNA makes use of Hfq (or another RNA binding protein), this work points to yet another source of RNA-based regulators.
The realization that sRNAs can be embedded in or can overlap with mRNAs leads to an additional issue – how can one define a functional (regulatory) RNA, and distinguish it from the mRNAs it may be processed from or may overlap with? If criteria for regulatory RNAs are developed, will bifunctional RNAs (for instance, sRNAs that also encode short ORFs or that bind multiple RNA chaperones) pass these tests? We consider these questions with respect to Hfq-binding sRNAs. An initial necessary criterion should be enrichment of the candidate sRNA relative to other RNAs by immunoprecipitation (IP) with Hfq or cross-linking to Hfq [23] [14]. However, Hfq binding does not distinguish regulatory sRNAs from targets (mRNAs). More information is provided by RIL-Seq [15], in which RNAs bound to Hfq are ligated to each other, providing chimeric RNAs composed of sRNA and mRNA targets. Two characteristics of these data can point to new sRNAs. First, the known regulatory sRNAs are primarily found as the second (3’) partner of these ligated RNA species, presumably reflecting the way in which these RNAs are bound to Hfq. Thus, other RNAs found primarily as the second partner in chimeras may also be sRNA-like in their Hfq association. Second, RNA species associated with multiple partners in chimeras are worth considering as sRNAs, since many sRNAs have multiple targets. CpxQ, mentioned above, from the 3’ UTR of cpxP [19], has more than 200 RNA chimeric partners, and is predominantly found as the second member of these chimeras [15]. The sucD 3’ UTR was previously identified from Hfq IP experiments and named RybD [23]; it has more than 30 RNA partners and is again found mostly as the 3’ member of these chimeras [15]. These observations are consistent with our results (De Mets et al., in preparation) demonstrating that RybD is a functional sRNA. Using these criteria, another 20 possible candidate sRNAs are suggested from the RIL-Seq data [15], but await further experimental confirmation.
A variety of evidence suggests another characteristic for evaluating Hfq-binding RNAs for whether they are regulatory sRNAs or targets is whether they contain a U-rich Rho-independent terminator. The need for this run of 3’ Us for sRNA stability and function has been demonstrated [24] [25], and recently the strength of the terminator has been shown to be an important and apparently regulated characteristic of whether an sRNA is made or whether, instead, transcription reads through [26]. The need for such a 3’ Rho-independent terminator suggests that, even for a 5’UTR-derived sRNA, such a structure likely needs to be present. U-rich sequences are known to bind to the proximal face of Hfq, while A rich sequences bind to the distal face of the ring (reviewed in [6] [2]). Mutation of a conserved Gln residue on the proximal face of Hfq (Gln8) dramatically reduces the stability of known sRNAs [27], and thus would be expected to have a similar effect on new candidate sRNAs.
When is a pairing sRNA more than a pairing sRNA?
In addition to the finding that many sRNAs are processed from what we can now call bifunctional mRNAs, it is becoming increasingly clear that a pairing sRNA need not only be a pairing sRNA. Bifunctional pairing sRNAs with short ORFs have been known for some time (reviewed in [28]). A growing list of sRNAs are now known to bind more than one RNA chaperone, leading to different and sometimes competing functions.
One well-studied example is McaS in E. coli. McaS is an Hfq-dependent sRNA, with pairing to some targets leading, for instance, to positive regulation of motility [29]. It also has been shown to activate biofilm by increasing expression of the pga operon, which encodes an outer membrane porin and the poly β-1,6-N-acetyl-D-glucosamine adhesin that it exports. This regulation, however, is not dependent on pairing. Instead, McaS has a second function, directly binding and thus titrating CsrA, a negative regulator of pga translation [30] (Figure 3A). The carbon storage regulator A (CsrA) is a small homodimeric protein that usually acts as a translational repressor by binding around the Shine-Dalgarno (SD) sequence of mRNA targets. Unlike Hfq, which generally requires sRNA binding partners to fulfill its function, CsrA regulates translation by direct sequence-specific binding to AUGGA motifs in mRNAs in a wide range of bacterial species [31] [14]. It is an important player in the regulation of physiology and virulence in many pathogenic and non-pathogenic bacterial species. The activity of CsrA is modulated, in large part, by the expression of sRNAs (CsrB and CsrC in E. coli) that carry repeats of the GGA binding motifs, and thus, when expressed, titrate the protein away from mRNA targets species (reviewed in [32]) (Figure 3A). McaS also contains two critical GGA binding motifs, and, judging by its role in promoting biofilm formation, is able to effectively remove CsrA from the pga RNA [30] (Figure 3A).
Figure 3: RNA binding proteins and regulation by titrating sRNAs.
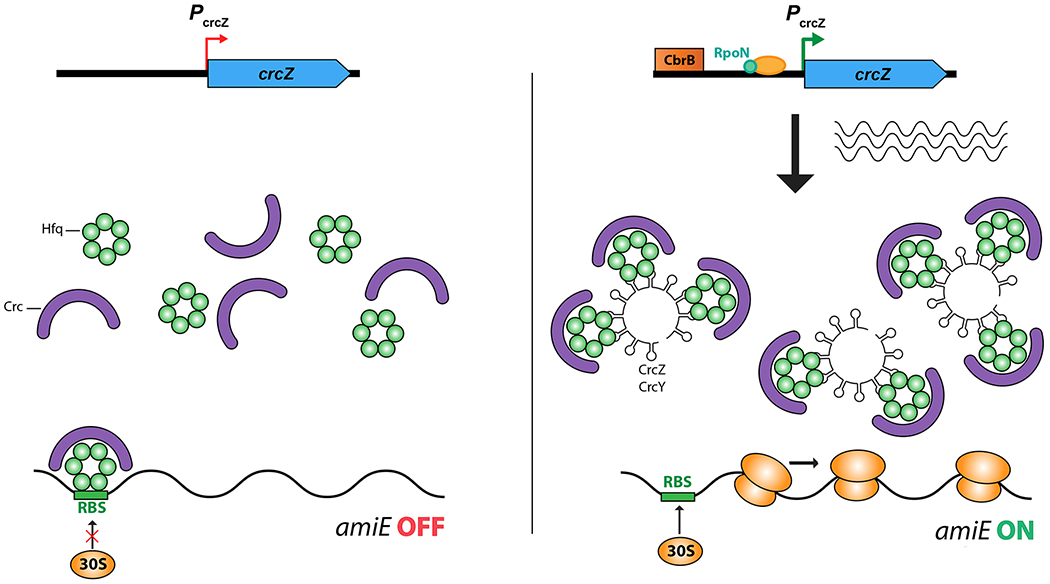
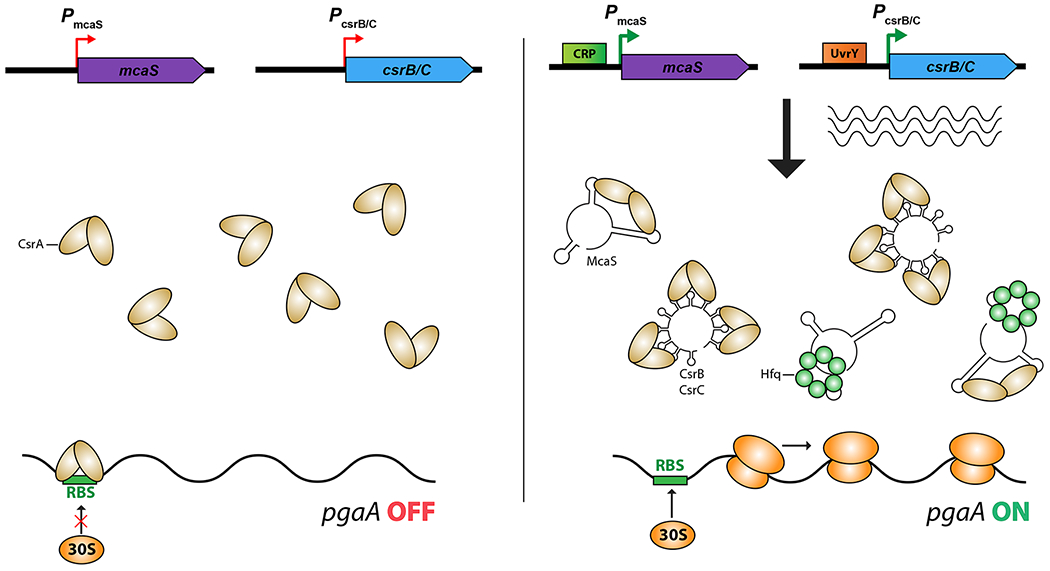
RNA binding proteins can be removed from their regulatory sites on target mRNAs by competing RNAs that also carry the regulatory sites. These titrating RNAs may be the major regulatory input affecting translational regulatory proteins.
A. Titration of CsrA: CsrA is a major regulatory protein that acts by binding to targets, frequently blocking ribosome access (reviewed in [32]). All bacteria that contain CsrA or its homologs also encode titrating sRNAs such as CsrB and CsrC, which each contain multiple CsrA binding sites. Recent work demonstrates that some Hfq-binding sRNAs that act by pairing, such as McaS, also contain CsrA binding sites and thus can titrate CsrA as well [30,33]. Different regulatory signals affect synthesis of different titrating sRNAs.
B. In Pseudomonads, Hfq acts in concert with another RNA binding protein, Crc, to carry out sRNA-independent catabolite repression at multiple sites. The CrcZ regulatory RNA, when synthesized, acts to remove Hfq and Crc from targets, allowing expression of genes such as amiE (shown here) [52]. The expression of CrcZ is dependent upon a response regulator, CbrB, which responds to the status of carbon sources to set the hierarchy of carbon utilization [51].
Recently, the RpoS-dependent GadY sRNA was also found to increase biofilm formation via pgaA activation. Like McaS, it contains GGA sites and thus may act at least in part by titrating CsrA [33]. Furthermore, a recent global study of RNA interactions with CsrA in Salmonella detected possible interactions of CsrA with yet other Hfq-dependent sRNAs that contain GGA binding sites [14]. As Hfq function is mainly dictated by the abundance of its sRNA binding partners, the shared sRNA could have regulatory effects of two types. First, the sRNA can regulate of CsrA availability, as has been shown for McaS and GadY. Secondly (but not yet demonstrated), CsrA could titrate these sRNAs from Hfq under some conditions, interfering with their ability to carry out Hfq-dependent regulation. Understanding when these alternative regulatory activities are most relevant will require further in vivo analysis of mutations that separate sRNA functions, as well as quantitative measurements of relative binding affinities for sRNAs and target mRNAs to Hfq and CsrA, understanding if these two proteins are mutually exclusive in their RNA binding, and understanding when sRNA levels are likely to be highest. Another RNA chaperone, ProQ, has also recently been described (reviewed in [34]); it may act more like Hfq in promoting RNA/RNA pairing, but in any case it, like CsrA, appears to bind some of the Hfq-binding sRNAs.
Hfq Regulatory Inputs: Sponges and decoys affect sRNA function and stability
Whether a given sRNA regulates its targets under a given growth condition will depend, in large part, on how abundant it is. The most important control of abundance will be the transcriptional regulation of synthesis, which is highly regulated for most sRNAs. The stability of the sRNA will, of course, also affect its availability. Recent work demonstrates that the mode of binding to Hfq can affect whether the sRNA is reused or decays after pairing [27] and thus how abundant it will be for pairing with less favored mRNA targets. Particularly (but not only) in the case in which an sRNA is degraded after it pairs, an abundant and preferred mRNA target may act to keep levels of the sRNA low. This interaction of different targets for a given sRNA has just begun to be explored [35,36] [37], but suggests that all mRNA targets may be considered part of the regulatory signaling for the sRNA that targets it. For an sRNA that can be reused multiple times, down-regulating its activity may require an active way to degrade it. This is the case for ChiX, an Hfq-binding sRNA that is destabilized when an inducible “decoy” mRNA pairs with it [38].
sRNA sponges are RNAs that can pair with and thus inactivate an sRNA, much like the decoy for ChiX mentioned above. Fragments of tRNA precursors, previously considered non-functional products of tRNA biogenesis, have now been shown to function as sponges for RyhB, an sRNA made when iron is limiting. This type of sponge, which is presumably constitutively expressed, may reduce the basal level of RyhB, avoiding any regulation by RyhB when it is not needed [39]. Another RNA sponge, SroC, was previously identified as an sRNA [40]. SroC is formed from decay of the gltIJKL mRNA and was found to trigger degradation of the sRNA GcvB. This is particularly intriguing since GcvB directly represses the glt locus, which encodes an amino acid transporter. Therefore, SroC participates in a feed-forward loop, relieving the GcvB-dependent repression of its parental mRNA. This cross-talk is particularly important for bacterial growth when peptides are the only souces of carbon and nitrogen [41]. A recent study in Salmonella suggests that SroC’s sponge activity may be broader; it can also pair to MgrR, modulating MgrR-mediated phenotypes [42].
Further RNA sponges may well be found among RNAs ligated to or associated with known regulatory sRNAs [15] [16,39], although in many cases it may not be easy to determine whether a ligated RNA is an mRNA target and/or a sponge or decoy.
Hfq Regulatory Outputs: sRNA roles in Rho-dependent anti-termination
Most of the described regulation by Hfq-binding sRNAs is post-transcriptional, affecting translation and/or stability of mRNAs. However, changes in RNA availability and folding, including those changes mediated by sRNAs or Hfq, can affect the ability of other proteins to access RNA. This includes regulating the ability of the transcriptional terminator Rho to act. Rho loads onto rut (Rho utilization) sites on RNA and moves along the RNA to cause termination (reviewed in [43]). ChiX sRNA pairs at the ribosome binding site of the chiPQ operon, blocking ribosome entry. This allows Rho entry and termination downstream, reinforcing negative regulation of the operon (Figure 1B) [44].
Recently, sRNAs were also found to have the opposite effect, reducing Rho termination by blocking Rho access to long 5’ UTRs [45]. Three sRNAs contribute to expression of the stationary phase sigma factor RpoS; they have been shown to positively regulate RpoS translation by pairing to the 5’ UTR, opening up an inhibitory hairpin to allow ribosome entry (reviewed in [46]). It now appears that in addition to this translational activation, the sRNAs also activate RpoS expression by interfering directly with Rho-dependent transcription termination [45] (Figure 1B). Global analysis of Rho termination suggests that sRNAs and Hfq may help to interfere with Rho-dependent termination on many genes with long 5’UTRS [45].
Hfq Regulatory Outputs: Hfq repression beyond sRNAs
While most of the regulation associated with Hfq can be attributed to the action of sRNAs, there are now a number of examples in which Hfq appears to act as a regulatory RNA binding protein, in the absence of sRNAs (Figure 4). This is most clearly demonstrated by the ability of the Hfq Q8A variant, mutated at a critical site on the proximal face needed for sRNA binding and stability, to still carry out regulation. This makes the assumption that all regulatory RNAs will bind to the proximal face and become unstable if binding is not possible. In E. coli, it was demonstrated that Hfq binding to the ribosome binding site (RBS) of Tn10 transposase mRNA leads to repression of IS10 transposase [47]. Recently, Hfq was also found to directly repress mutS mRNA, independently of the proximal sRNA binding face. The repression of mutS by Hfq contributes to increased stationary phase mutagenesis [48]. For repression of IS10 transposase, the Hfq binding site is near the RBS, directly leading to translation inhibition (Figure 4, upper example). However, in the case of mutS, Hfq binds to an (AAN)3 repeat 40 nt upstream of the RBS, rearranging mRNA secondary structure to inhibit translation (Figure 4, lower example). These examples suggest that more cases of Hfq acting alone to affect mRNA folding, translation and/or stability are likely to be found.
Figure 4: Hfq Regulation in the absence of sRNAs.
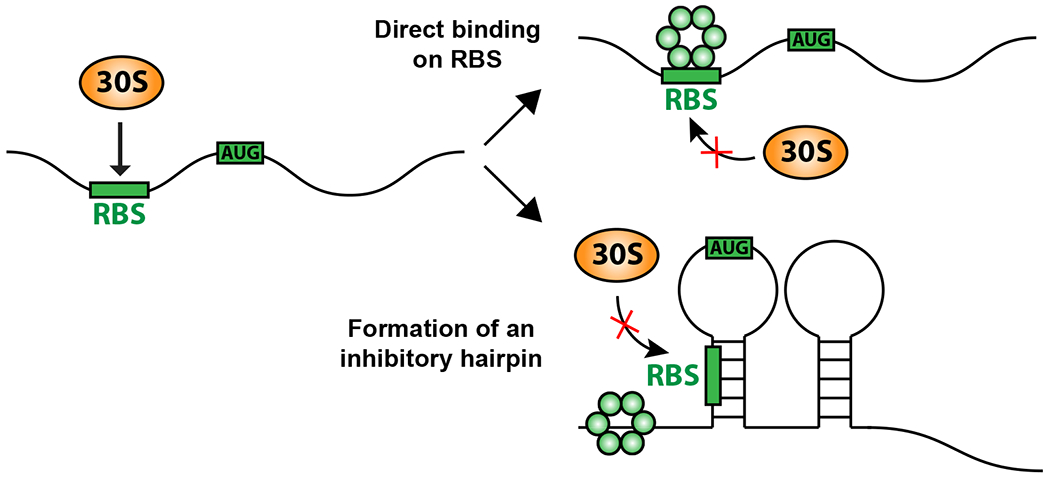
Hfq is able to bind RNA to repress mRNA translation. In some cases, this binding is close to or at the ribosome binding site, directly blocking ribosome access [47]. In at least one other case, binding is upstream of the ribosome binding site but leads to remodeling of the RNA to block ribosome access [48].
How RNA binding by Hfq in the absence of sRNAs might be regulated remains to be explored, as does understanding when Hfq can act by itself and when it must be recruited by an sRNA, as has been shown for Hfq repression of sdhC [49]. If Hfq is acting without an sRNA, what are the regulatory inputs? Do Hfq levels change, or do other (competing or sponge) RNAs or other protein partners change the availability of Hfq? Possibly in some of the organisms that contain Hfq but in which it appears not to be important for most sRNA-based regulation (for instance, in S. aureus [50]), Hfq may act as an mRNA binding protein rather than as a chaperone for sRNA pairing.
Recent work in Pseudomonads on Hfq-dependent catabolite repression provides a very clear example of how Hfq translational repression, in the absence of pairing sRNAs, can be regulated (Figure 3B). In E. coli and many other bacteria, transcriptional regulation mediated by cyclic AMP and CRP sets a hierarchy for use of carbon sources. However, in Pseudomonads, the hierarchy appears to be set via translational repression that is dependent on the small Crc protein [51]. A-rich sequences, called CA (for catabolite activity) motifs, near the ribosome binding site of catabolite repressed genes are critical for repression [51]. Recent studies demonstrate that Crc itself does not bind to RNA. Instead, Hfq binds to the CA motifs, using its distal face and Crc stabilizes that binding, in a way that is not currently fully understood [52]. When catabolite repression is not needed (i.e. when only a poorly metabolized carbon source is available), Crc/Hfq repression is relieved by expression of a non-coding RNA, CrcZ or its homologs, that carries multiple copies of the CA motif, and thus can sequester Hfq (Figure 3B) [52,53]. The expression of CrcZ is regulated at the transcriptional level, by the availability of carbon sources [54] [51]. Thus, in these organisms, Hfq, with the help of Crc, bypasses the need for a pairing RNA and acts as a sequence-specific translational repressor, but is regulated by a titrating sRNA, all reminiscent of how CsrA acts and is regulated.
Conclusions/Future Prospects:
Finding regulatory RNAs and their targets has been made significantly easier by recent advances in technology. However, this onslaught of information still leaves many challenges – understanding the hierarchy of sRNA use, dissecting how various regulons are intertwined, and evaluating the conditions under which various levels of regulation are most important. In addition to sRNAs that act primarily by pairing to target mRNAs, it is becoming increasingly clear that other regulatory RNAs (sponges, decoys, and titrating RNAs) may act to modulate the availability and stability of the primary regulators. Both sRNAs and RNA binding proteins have the potential to restructure RNA, changing accessibility to ribosomes, other RNA binding proteins, nucleases and termination factors, and it should be expected that investigations will continue to uncover variations on these possibilities. Finally, as more investigations are done both in E. coli and in other organisms, striking parallels for different modes of mRNA regulation are found, and are likely to be continue to be found. In E. coli, for instance, Hfq is thought to primarily act with sRNA partners, with regulatory input via transcription of the sRNAs, while CsrA directly represses multiple genes, with regulatory input primarily from synthesis of titrating sRNAs. However, in Pseudomonads, Hfq, with the help of a second protein, Crc, acts in a “CsrA-like” fashion on many but not all its targets, regulated by a titrating sRNA. Thus the major caution for the future is to keep our minds open to new ways in which the translation of genetic information may be regulated.
Highlights.
Novel cross-linking and sequencing methods uncover new sRNAs and sRNA networks.
Many sRNAs are encoded within the 3’ and 5’ UTRs of mRNA as well as elsewhere.
sRNA availability can be regulated post-transcriptionally by decoy and sponge RNAs.
Hfq can regulate mRNA translation without sRNAs, alone and with other proteins.
Some sRNAs that use Hfq also bind and regulate translational repressor CsrA.
Acknowledgements/Funding:
The writing of this review was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. F. de Mets is a PhD candidate supported by a fellowship from the Fund for Research Training in Industry and Agriculture (FRIA). We thank G. Storz, J. Chen, T. Updegrove, I. Hamdallah and K. Borden for their constructive comments on the review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Rajkowitsch L, Schroeder R: Dissecting RNA chaperone activity. RNA 2007, 13:2053–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Updegrove TB, Zhang A, Storz G: Hfq: the flexible RNA matchmaker. Curr Opin Microbiol 2016, 30:133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G: The Escherichia coli oxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J 1998, 17:6069–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prévost K, Desnoyers G, Jacques J-F, Lavoie F, Massé E: Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev 2011, 25:385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papenfort K, Vanderpool CK: Target activation by regulatory RNAs in bacteria. FEMS Microbiol Rev 2015, 39:362–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogel J, Luisi BF: Hfq and its constellation of RNA. Nat Rev Microbiol 2011, 9:578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Lay N, Schu DJ, Gottesman S: Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem 2013, 288:7996–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X, Zhulin I, Wartell RM: Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res 2002, 30:3663–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valentin-Hansen P, Eriksen M, Udesen C: The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol 2004, 51:1525–1533. [DOI] [PubMed] [Google Scholar]
- 10.Chao Y, Vogel J: The role of Hfq in bacterial pathogens. Curr Opin Microbiol 2010, 13:24–33. [DOI] [PubMed] [Google Scholar]
- 11.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S: Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 2001, 11:941–950. [DOI] [PubMed] [Google Scholar]
- 12.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S: Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 2001, 15:1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rivas E, Klein RJ, Jones TA, Eddy SR: Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr Biol 2001, 11:1369–1373. [DOI] [PubMed] [Google Scholar]
- 14. Holmqvist E, Wright PR, Li L, Bischler T, Barquist L, Reinhardt R, Backofen R, Vogel J: Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J 2016, 35:991–1011. • Global analysis of Hfq and CsrA binding sites in the transcriptome of Salmonella by UV crosslinking better defines their binding sites and reveals overlaps between these regulators.
- 15. Melamed S, Peer A, Faigenbaum0Rmm R, Gatt YE, Reiss N, Bar A, Altuvia Y, Argaman L, Margalit H: Global mapping of small RNA-target interactions in bacteria. Mol Cell 2016, 63:884–897. •• The authors developed a new method (RIL-Seq) that identifies mRNA targets of sRNAs by ligating them on Hfq. This allows a global view of sRNA-based regulatory networks and highlights additional candidate sRNAs.
- 16. Han K, Tjaden B, Lory S: GRIL-seq provides a method for identifying direct targets of bacterial small regulatory RNA by in vivo proximity ligation. Nat Microbiol 2016, 2:16239. • In vivo ligation of sRNA and mRNA (GRIL-Seq) provides an overview of sRNA-based networks in Pseudomonas aeruginosa.
- 17.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J: An atlas of Hfq-bound transcripts reveals 3’ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J 2012, 31:4005–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo MS, Updegrove TB, Gogol EB, Shabalina SA, Gross CA, Storz G: MicL, a new sE-dependent sRNA, combat envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev 2014, 28:1620–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chao Y, Vogel J: A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol Cell 2016, 61:352–363. • The investigation of CpxQ, an sRNA processed from the 3’ UTR of the cpxP gene in Salmonella, provides evidence for the first sRNA of the Cpx membrane stress response. This study demonstrates the potential for 3’ UTR-derived sRNAs to lead to sRNA members of many regulons.
- 20.Klein G, Stupak A, Biernacka D, Wojtkiewica P, Lindner B, Raina S: Multiple transcriptional factors regulate transcription of the rpoE gene in Escherichia coli under different growth conditions and when the lipopolysaccharide biosynthesis is defective. J Biol Chem 2016, 291:22999–23019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyakoshi M, Chao Y, Vogel J: Regulatory small RNAs from the 3’ regions of bacterial mRNAs. Curr Opin Microbiol 2015, 24:132–139. [DOI] [PubMed] [Google Scholar]
- 22.Ellis MJ, Trussler RS, Charles O, Haniford DB: A transposon-derived small RNA regulates gene expression in Salmonella Typhimurium. Nucleic Acids Res 2017, 45:5470–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S: Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 2003, 50:1111–1124. [DOI] [PubMed] [Google Scholar]
- 24.Otaka H, Ishikawa H, Morita T, Aiba H: PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci U S A 2011, 108:13059–13064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauer E, Weichenrieder O: Structural basis for RNA 3′-end recognition by Hfq. Proc Natl Acad Sci USA 2011, 108:13065–13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morita T, Nishino R, Aiba H: Role of the terminator hairpin in the biogenesis of functional Hfq-binding sRNAs. RNA 2017, 23:1419–1431. • This group has previously demonstrated the necessity for rho-independent terminators in the stability and function of sRNAs, via Hfq binding. Here they find that these terminators are optimized in strength, and that, under stress conditions, readthrough can change the availability of the sRNA.
- 27.Schu DJ, Zhang A, Gottesman S, Storz G: Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J 2015, 34:2557–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanderpool CK, Balasubramanian D, Lloyd CR: Dual-function RNA regulators in bacteria. Biochimie 2011, 93:1943–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomason MK, Fontaine F, De Lay N, Storz G: A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol 2012, 84:1365–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jorgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G: Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev 2013, 27:1132–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubey AK, Baker CS, Romeo T, Babitzke P: RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 2005, 11:1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vakulskas CA, Potts AH, Babitzke P, Ahmer BM, Romeo T: Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev 2015, 79:193–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker A, Cureoglu S, De Lay N, Majdalani N, Gottesman S: Alternative pathways for Escherichia coli biofilm formation revealed by sRNA overproduction. Mol Microbiol 2017, 105:309–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olejniczak M, Storz G: ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers? Mol Microbiol 2017, 104:905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jost D, Nowojewski A, Levine E: Regulating the many to benefit the few: Role of weak small RNA targets. Biophys J 2013, 104:1773–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fei J, SIngh D, Zhang Q, Park S, Balsubramanian D, GOlding I, Vanderpool CK, Ha T: Determination of in vivo target search kinetics of regulatory noncoding RNA. Science 2015, 347:1371–1374. •• Super-resolution imaging of sRNA SgrS pairing with its targets in the cell allows quantitative analysis of the parameters that affect pairing.
- 37.Bossi L, Figueroa-Bossi N: Competing endogenous RNAs: a target-centric view of small RNA regulation in bacteria. Nat Rev Microbiol 2016, 14:775–784. [DOI] [PubMed] [Google Scholar]
- 38.Figueroa-Bossi N, Valentini M, Malleret L, Bossi L: Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev 2009, 23:1981–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lalaouna D, Carrier MC, Semsey S, Brouard JS, Wang J, Wade JT, Masse E: A 3’ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol Cell 2015, 58:393–405. •• Using a novel method to identify RNAs associated with sRNAs of interest (MAPS), the authors identified products of tRNA processing as sponges for sRNAs, reducing the basal level of sRNA function from transcriptional noise.
- 40.Vogel J, Bartels V, Tang HH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EGH: RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res 2003, 31:6435–6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyakoshi M, Chao Y, Vogel J: Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J 2015, 34:1478–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Acuna LG, Barros MJ, Penaloza D, Rodas PI, Paredes-Sabja D, Fuentes JA, Gil F, Calderon IL: A feed-forward loop between SroC and MgrR small RNAs modulates the expression of eptB and the susceptibility to polymyxin B in Salmonella Typhimurium. Microbiol 2016, 162:1996–2004. [DOI] [PubMed] [Google Scholar]
- 43.Mitra P, Ghosh G, Hafeezunnisa M, Sen R: Rho Protein: Roles and Mechanisms. Annu Rev Microbiol 2017, 71:687–709. [DOI] [PubMed] [Google Scholar]
- 44.Bossi L, Schwartz A, Guillermardet B, Boudvillain M, Figueroa-Bossi N: A role for rho-dependent polarity in gene regulation by a noncoding small RNA. Genes Dev 2012, 26:1864–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sedlyarova N, Shamovsky I, Bharati BK, Epshtein V, Chen J, Gottesman S, Schroeder R, Nudler E: sRNA-mediated control of transcription termination in E. coli. Cell 2016, 167:111–121. • These authors find that sRNAs pairing with long 5’ UTRs can block Rho loading and termination within the 5’ UTR, providing a new activating role for sRNAs.
- 46.Battesti A, Majdalani N, Gottesman S: The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 2011, 65:189–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ellis MJ, Trussler RS, Hanford DB: Hfq binds directly to the ribosome-binding site of IS10 transposase mRNA to inhibit translation. Mol Microbiol 2015, 96:633–650. • sRNAs derived from the 5’ UTR of the IS200 transposase gene have a wide-spread regulatory effect in Salmonella, and provide evidence for unexpected roles for transposon transcripts.
- 48. Chen J, Gottesman S: Hfq links translation repression to stress-induced mutagenesis in E. coli. Genes Dev 2017, 31:1382–1395. •• Hfq was found to limit expression of the MutS mismatch repair protein, leading to increased mutagenesis in stationary phase. Hfq represses both via sRNAs and independently of sRNAs, by remodeling the mutS 5’ UTR.
- 49.Desnoyers G, Masse E: Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev 2012, 26:726–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bohn C, Rigoulay C, Bouloc P: No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol 2007, 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sonnleitner E, Abdou L, Haas D: Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 2009, 106:21866–21871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sonnleitner E, Blasi U: Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa carbon catabolite repression. PLoS Genet 2014, 10:e1004440. •• This work shows that, in contrast to previous work, Hfq binding to RNA is necessary for catabolite repression by Crc in Pseudomonads. The complex of Crc and Hfq repress many genes; repression is relieved by the CrcZ sRNA, which competes for Hfq binding to mRNAs..
- 53.Sonnleitner E, Prindl K, Blasi U: The Pseudomonas aeruginosa CrcZ RNA interferes with Hfq-mediated riboregulation. PLoS One 2017, 12:e0180887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hernandez-Arranz S, Sanchez-Hevia D, Rojo F, Moreno R: Effect of Crc and Hfq proteins on the transcription, processing, and stability of the Pseudomonas putida CrcZ sRNA. RNA 2016, 22:1902–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baekkedal C, Haugen P: The Spot 42 RNA: A regulatory small RNA with roles in the central metabolism. RNA Biol 2015, 12:1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]


