Abstract
Background
The small hive beetle (SHB), Aethina tumida, has emerged as a worldwide threat to honey bees in the past two decades. These beetles harvest nest resources, feed on larval bees, and ultimately spoil nest resources with gelatinous slime together with the fungal symbiont Kodamaea ohmeri.
Results
Here, we present the first chromosome-level genome assembly for the SHB. With a 99.1% representation of conserved (BUSCO) arthropod genes, this resource enables the study of chemosensory, digestive, and detoxification traits critical for SHB success and possible control. We use this annotated assembly to characterize features of SHB sex chromosomes and a female-skewed primary sex ratio. We also found chromosome fusion and a lower recombination rate in sex chromosomes than in autosomes.
Conclusions
Genome-enabled insights will clarify the traits that allowed this beetle to exploit hive resources successfully and will be critical for determining the causes of observed sex ratio asymmetries.
Keywords: pest, invasion, sex chromosome, genome assembly, recombination, sex ratio
Background
The small hive beetle, Aethina tumida (SHB, NCBI :txid116153), is a nest parasite of social bees. Outside its native range, SHB was first reported in the United States in 1996 and then further invaded Australia, Europe, and Asia [1–4]. This beetle is exceptionally damaging to managed honey bee colonies, accelerating colony decline and spoiling honey and other hive products [5]. The previous SHB draft genome identified genes involved in detoxification, physiological and chemosensory pathways, and supplemented mitochondrial markers used to track the ongoing diaspora of this pest species [6, 7]. As expected, the SHB movement largely follows international trade lines, and incipient populations fare well against SHB-naive hosts [8–12]. In Africa, worker honey bees mount a range of defenses against these beetles, attacking and isolating them, so they remain at low numbers. Naive honey bee populations seem to lack many of these defenses, consequently supporting substantially higher SHB populations [13]. When honey bee colony size decreases due to management, disease, or stress, SHB populations can rapidly take advantage, removing resources and eventually “sliming” the colony with a resinous substance. This slime, and indeed much of the biology of SHB, is linked with a commensal fungus, Kodamaea ohmeri [14–16]. Metabolites from this fungus are attractive to beetles, providing a bait to trap the SHB in the field [17]. However, the route to transfer this fungal symbiont to SHBs remains unclear, which is essential to understand the symbiosis.
In beehives, the observed SHB sex ratio is often female biased, a fact that was proposed to facilitate the global invasion [18, 19]. However, plausible mechanisms for such a skew remain unclear. In other insects, female-biased sex ratios have also been observed [20–22]. Selfish genetic elements and the symbiotic bacteria Wolbachia were found to act as sex ratio distorters, skewing ratios toward females [21, 23, 24]. In a previous metagenomic study, Wolbachia fragments were found in the small hive beetles [25]. It is challenging to explain the mechanism under the observed biased SHB sex ratio because it is impossible to determine the primary sex ratio. Therefore, a chromosomal-level SHB genome assembly and the identification of the sex chromosomes were urgently required.
Previously, we assembled a 234-Mbp SHB genome without context to the chromosomal structure. Here, we substantially improved the SHB genome and generated a 259-Mbp SHB genome assembly consisting of only 38 gapless contigs and scaffolded to 8 chromosomes. We identified and characterized the SHB sex chromosomes for the first time and established the egg sex ratios for this species. We have also used this complete assembly to estimate tandem repeats and recombination rates and produced a definitive gene set.
Analyses
Genome assembly statistics
The final assembly (GenBank accession: GCA_024364675.1) comprised 8 chromosomes and a mitochondrial genome (Table 1). These 9 genetic components were assembled from 38 contigs using Hi-C contacts and derived from A. tumida (Supplementary Table S1). The ancestral insect telomere motif (TTAGG)n was detected on the 5′ end of chromosomes 1, 2, 3, 4, 5, and 8 and the 3′ end of chromosomes 2, 3, 4, 5, and 6. The ancestral insect telomere motif was also detected at around 27.8 Mb and 37.6 Mb of chromosome 1, indicating recent chromosome fusion. The final assembly has a total length of 259 MB, which is about 10% larger than the genome size estimated by GenomeScope (Supplementary Fig. S1), which is likely due to highly repetitive regions in the assembly that did not contribute to the estimated size derived from k-mer analysis or an inflated assembly of the highly heterochromatic centromere regions. Corroborating the low proportion of artifact duplicates were the k-mer frequencies of the raw circular consensus sequencing (CCS) reads relative to the k-mers detected in the final assembly (Supplementary Fig. S2). Genomic completeness measured by the proportion of Endopterygota BUSCOs revealed a high level of completeness, with the genome containing 99.1% of expected genes (97.2% in a complete single copy and 1.9% complete but duplicated) and an annotated protein set containing 99.3% of expected genes (97.4% complete single copy and 1.9% complete but duplicated) (Supplementary Fig. S3). Additionally, 99.6% of genes were validated using transcriptomic data. We additionally analyzed 50 chromosome-level beetle genomes. On average, 13 ± 4 chromosomes were annotated in beetle genomes, and the genome size ranged from 132 to 2,533 Mbp. Compared with other beetle genomes, SHB showed a relatively compact genome size (one-sample t-test, P < 0.001) (Supplementary Fig. S4).
Table 1:
Statistics of current and previous small hive beetle genome assemblies. Overall, the assembly statistics have been substantially improved compared with the previous version.
| icAetTumi1.1 GCA_024364675.1 (Current version) | Atum_1.0 GCA_001937115.1 (Previous version) | |
|---|---|---|
| Assembly level | Chromosome | Contig |
| Assembly size (Mbp) | 259.9 | 234.3 |
| Number of contigs | 38 | 3,063 |
| Contig N50 (kbp) | 11,742 | 298 |
| Number of gene | 14,581 | 14,076 |
| Number of mRNAs | 21,401 | 17,634 |
| BUSCO % | 99.1 | 97.5 |
XY sex determination in small hive beetles
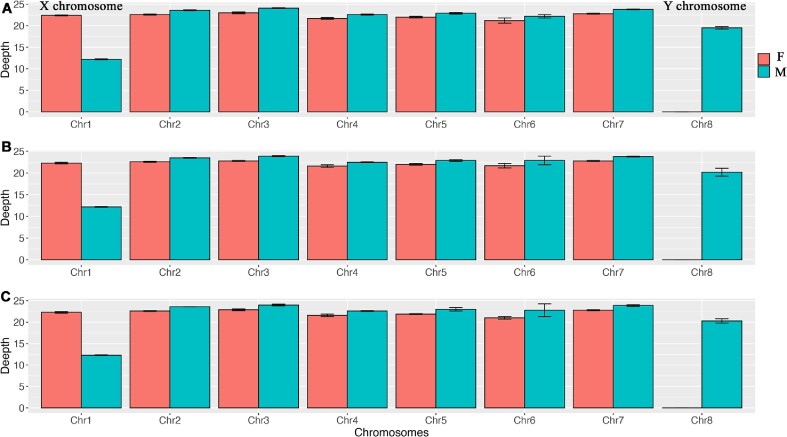
We established 3 beetle families, producing 49 offspring beetles with known sex (Fig. 1, Table 2). On average, 84 million reads (150-bp paired reads) were aligned per offspring. By plotting the alignment depth along the genome, we found that the shortest chromosome (Chr8) only exists in male beetles, defined as the Y chromosome. Comparatively, we did not identify any chromosome that only aligned in females. Additionally, the depth of the longest chromosome (Chr1) was twofold higher in female than male beetles, defined as the X chromosome (Fig. 2). In the remaining chromosomes (Chr2–Chr7), the depth ratio between males and females was approximately equal (paired t-test, P = 0.54), suggesting them to be autosomal.
Figure 1:
Life stages of small hive beetles. (A) Dorsal image of adult SHB. (B) ventral image of adult SHB. (C) Dorsal image of pupa. (D) Ventral image of pupa. (E) Dorsal image of larva. (F) Ventral image of larva.
Table 2:
Beetle families established to determine sex chromosomes. Several beetle pairs (a male and a female) were constructed, and 3 beetle families were successfully established, with both male and female offspring. Beetle family F17 first laid male and female offspring, followed by the family F31 and F8.
| Beetle family | Parental male | Parental female | Offspring male | Offspring female | Sex ratio |
|---|---|---|---|---|---|
| F8 | 1 | 1 | 5 | 10 | Chi-squared test, P = 0.76 |
| F17 | 1 | 1 | 8 | 8 | |
| F31 | 1 | 1 | 9 | 9 |
Figure 2:
The sequence alignment depth along each chromosome. To determine the sex chromosome, the alignment depth was calculated along the genome. Chromosome 1 (Chr1) is the longest, and the alignment depth was twofold higher in female than male beetles, suggesting that this is the X chromosome. Chromosome 8 (Chr8) is the shortest chromosome, explicitly associated with male beetles as the Y chromosome. This pattern was highly congruent in 3 independent beetle families, F8 (A), F17 (B), and F31 (C). Collectively, the data suggest an XY sex determination mechanism in small hive beetles. Red indicates female beetles; blue indicates male beetles; the error bar indicates standard error.
Chromosome fusion
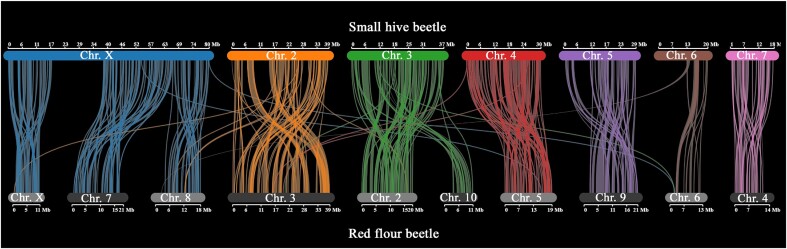
By aligning the protein-encoding sequences of the small hive beetle to that of the red flour beetle, the 2 beetle species shared 5,846 synteny blocks (>5 genes in a block). At the chromosome level, orthologous groups were well paired along the genome (Fig. 3). In the small hive beetle genome, the X chromosome was twice as large as the other autosomes and matched the X chromosome and 2 autosomes in the red flour beetle (Tribolium castaneum). The gene density of ChrY (139 kbp per gene) was over an order of magnitude lower than autosomes (11 kbp per gene). Even though the overall gene density in ChrX (13 kbp per gene) was similar to the autosomal density, the gene density at the 3′ end (11 kbp per gene) was twice higher than at the 5′ end (24 kbp per gene) in ChrX (Supplementary Fig. S5).
Figure 3:
Synteny between the small hive beetle and the red flour beetle genomes. The orthologs were generally well aligned in chromosomes. In the small hive beetle, the extra-long X chromosome seems analogous to a fusion of 2 autosomes and the X chromosome in the red flour beetle. Other chromosomes were generally well paired.
Reduced recombination rate in sex chromosome
Among the 3 beetle families, parental and offspring beetles shared 60,118 biallelic single-nucleotide variants (SNVs), generating 1,450 linkage groups. The X chromosome showed the lowest recombination rate (0.04 cM/Mbp), followed by the Y chromosome (0.06 cM/Mbp). Comparatively, chromosome 3 showed the highest recombination rate (2.3 cM/Mbp). By comparing the recombination rate between males and females, the variance was minor in autosomes (chi-squared test, P > 0.05). On average, the recombination rate was 30-fold higher in autosomes (1.5 cM/Mbp) than in sex chromosomes (0.05 cM/Mbp).
Female-biased sex ratio in small hive beetle eggs
A pair of primers (SHB-Y) on the Y chromosome was designed to differentiate male and female eggs (Table 3). As the sex of adult beetles can be visually identified, we validated the primers in 15 adult male and female SHBs, respectively. The PCR product generated by the primer pair was approximately 569 bp (Supplementary Figs. S6, S7). The sensitivity and specificity were 100% in adult SHBs. A previously designed universal primer (SHB-universal) to detect SHB served as a positive control (Table 3). The eggs that amplified SHB-Y were defined as male eggs. The eggs that amplified SHB-universal, but not SHB-Y, were defined as female eggs. In total, 79 eggs were collected from the lab-reared SHBs, and 33 male and 46 female eggs were identified from pool-reared adults. Even though statistically insignificant, the egg sex ratio skewed toward females (chi-squared test, P = 0.37; Supplementary Table S2). In the adult beetles, slightly more females were pupated (27 females) than males (22 males) (chi-squared test, P = 0.76; Table 2).
Table 3:
Primer sequences to determine the egg sex ratio
| primers | F-sequence | B-sequence | Annealing temperature | Product size | Target region | Sensitivity | Specificity | Reference |
|---|---|---|---|---|---|---|---|---|
| SHB-Y | TGACAACTCATAACCTGTTGGAT | ACAGGATGGTTTCCCTGCTC | 60°C | 569 bp | Y chromosome | 100% | 100% | This study |
| Universal | GCTAAGTTAACTGAAGATCCACCAT | TAGTTCCACTAATACTAAGAGCCCC | 56°C | 190 bp | mitochondria | 100% | 52.63% | [76] |
Discussion
Coleoptera (beetles) make up 40% of all described insect species, including many agricultural parasites [26]. Emergent parasites can readily evade the defenses of their hosts, and the SHB is a perfect example of a parasite adept at exploiting naive host populations. Rarely seen in honey bee colonies in their historical range in Africa, SHB is now a notorious global parasite [5]. These beetles are remarkably fecund in weaker honey bee colonies, destroying food resources and feeding on developing bees. SHBs share many traits with invasive emergent pests, including high female fecundity, excellent dispersal and homing skills, broad diet preferences, and a female-biased adult sex ratio [20, 27–29]. Genomic resources can be used to compare SHB to other fully (chromosome-level assemblies) sequenced beetles and thereby help address beetle biological and evolutionary questions. Here we present a complete genome analysis of SHB and use this resource to characterize sex chromosome traits, develop a tool for genetically distinguishing male and female beetle eggs, and present global studies of chromosomal and gene traits.
Given the proposal that SHBs benefit from a sex-biased adult sex ratio as a part of their global dispersal [18, 19], our first goal was to characterize the genetic factors determining sex in these beetles. Sex determination is a fundamental biological character, substantially impacting organisms' effective population size and reproductive behavior. Across a subset of Coleoptera, 3,348 beetle species have an XY sex determination system, and 766 have an XO sex determination system [30]. In our study, bias in coverage across several chromosomes identified the sex chromosomes and suggested that SHB has an XY sex determination. In small hive beetles, female-biased sex ratios were observed in field and lab conditions [18, 19]. As a complete metamorphosis insect, the sex ratio can be biased in any stage of eggs, larvae, and pupation. In lab conditions, the mortality of pupation was not different between male and female SHBs, suggesting the biases originated from eggs or the competition of larvae [19]. In our study, the lab-reared eggs and pupated adults trended toward a female-biased sex ratio, although the skew was not statistically significant. In natural conditions, competition for mating and food resources may further skew secondary sex ratios even when the primary sex ratio is unbiased [31]. In this study, the identified sex chromosome and male-specific PCR primers allow future empirical testing of the sex ratio under different environmental conditions before apparent sexual traits are found in adults.
In synteny alignment, we surprisingly found that the X chromosome and 2 other autosomes of the red flour beetle aligned singularly to the X chromosome of SHB. The synteny analysis suggests that chromosome fusion occurred during SHB evolution. Additionally, 2 additional telomere motifs were detected in the SHB X chromosome, which supports chromosome fusion. Besides, the 0.5:1 coverage ratio between the X chromosome and autosomes in males further supports chromosome fusion. Otherwise, the coverage ratio should be 0.8:1. In other insects, chromosomal fusion has shown substantial impacts on speciation, genetic diversity, and genome size [32, 33]. SHB showed a relatively compact genome size compared with other beetles, which might be due to the parasitic life character [34, 35].
Recombination shuffles alleles to form novel genotypes, a fundamental advantage of sexual reproduction [36]. Constantly breaking linkages among genes is a central paradigm in coevolutionary biology, and parasite selection for host adaptation can promote increased host recombination frequency [37, 38]. In animals, an average recombination rate of 2.52 cM/Mb was observed; however, an exceptionally high recombination rate of 19 cM/Mb was found in social bees [39–41]. In our data, a recombination rate of 1.5 cM/Mb was observed, which is lower than Drosophila melanogaster at 2.05 cM/Mb [42] and slightly higher than the red flour beetle (T. castaneum) at 1.3 cM/Mb [43]. Recombination is a critical evolutionary trait in light of host–parasite interactions [44]. Increased recombination rates were observed in mosquitoes infected by microsporidian parasites [38]. The red flour beetle also showed benefits from recombination when infected by microsporidian parasites [37]. As an invasive pest, novel genotypes and broad food types facilitate population expansion.
Conclusions
This chromosome-level genome assembly allows for the identification of sex determination, recombination rate, and chromosomal fusion. The developed tool allows for deciphering mechanism under the female-biased sex ratio in future studies.
Methods
DNA extraction and genome sequencing
Genomic DNA was obtained from nucleic acid isolation of a single adult male A. tumida flash frozen in liquid nitrogen. High molecular weight DNA for sequencing was extracted using the fresh or frozen tissue protocol of the Qiagen MagAttract HMW DNA Kit (Qiagen, Hilden, Germany). Following isolation, genomic DNA was subjected to a 2.0× bead cleanup to improve sample purity and then quantified using the dsDNA Broad Range (BR) Qubit assay (Thermo Fisher Scientific, Waltham, MA, USA) and the fluorometer of a DS-11 Spectrophotometer and Fluorometer (DeNovix, Wilmington, DE, USA). Purity was determined using the UV-Vis spectrometer feature of the DS-11, which reports OD 260/230 (>2.0 and <2.2) and 260/280 ratios (>1.8 and <2.0). Following the first bead cleanup, the high molecular weight DNA sample was sheared to a mean size of 20 kb with the Megaruptor 2 (Diagenode, Denville, NJ, USA). Subsequent size distribution was assessed with the High Sensitivity (HS) Large fragment kit run on the Fragment Analyzer (Agilent Technologies, Santa Clara, CA, USA). A PacBio SMRTBell library was prepared using the sheared DNA and the SMRTBell Express Template Prep Kit 2.0 (Pacific Biosciences, Menlo Park, CA, USA). The prepared library was bound and sequenced at the USDA-ARS Genetics and Animal Breeding Research Unit in Clay Center, Nebraska, USA, on a Pacific Biosciences 8 M SMRT Cell on a Sequel IIe system (Pacific Biosciences) beginning with a 2-hour preextension followed by a 30-hour movie collection time. After sequencing, consensus sequences from the PacBio Sequel IIe subreads were obtained using the SMRTLink v8.0 software.
Concurrent to the PacBio HiFi library prep and sequencing, a Hi-C library was prepared from a second adult male A. tumida collected from the same Apis mellifera colony. The proximity-ligated sequencing library was prepared using the Arima Hi-C kit (Arima Genomics, San Diego, CA, USA) from crosslinked tissue prepared following the Arima Hi-C low-input protocol. Following proximity ligation, DNA was sheared using a Bioruptor Pico (Diagenode), and DNA fragments in the range of 200 to 600 bp were selected as the input for the Illumina library prep using the Swift Accel NGS 2S Plus kit (Integrated DNA Technologies, Coralville, IA, USA). Illumina 2 × 150-bp sequencing was performed on a NovaSeq 6000 (RRID:SCR_016387) at the Hudson Alpha Genome Sequencing Center (Huntsville, AL, USA), and adapter trimming after sequence collection was performed using BaseSpace software (Illumina, San Diego, CA, USA; RRID:SCR_011881).
Genome assembly
Prior to genome assembly, HiFi reads containing artifact adapter sequences were removed from the HiFi read pool using the program HiFiAdapterFilt v2.0 [45]. This filtered read set was assembled into a contig assembly using HiFiASM v0.16.1-r375 (RRID:SCR_021069) using the default parameters [46]. The output of HiFiASM was an assembly in .gfa format, which was converted to a .fasta format using any2fasta [47] The primary contig assembly was scaffolded following the Arima Genomics mapping pipeline and YaHS scaffolding software [48, 49]. The Arima Genomics mapping pipeline uses BWA-MEM2 (RRID:SCR_022192) to align the paired Illumina R1 and R2 reads separately to the reference contig assembly and applies the filtering script “filter_five_end.pl” to only retain reads that are mapped in the 5′ orientation [50]. Following filtering, the independently mapped R1 and R2 reads are paired using the script “two_read_bam_combiner.pl,” which results in a sorted and quality-filtered paired-end file in .bam format. The “MarkDuplicates” function of Picard Tools [51] was used to remove PCR duplicate artifacts from the mapped and paired .bam file, which, along with the reference contig assembly, served as the input files for the YaHS scaffolding software. The YaHS software was implemented using the “no contig error correcting” option, and YaHS outputs were converted using the “juicer_pre” function of YaHS to Juicebox-compatible files for the manual curation [52]. Following manual curation, edits were applied to the scaffold assembly using “juicebox_assembly_converter.py” from the Phase Genomics suite of juicebox_scripts [52]. To inform Hi-C scaffolding and identify contigs containing the ancestral Insecta telomere sequence motif (TTAGG)n, the software program Tandem Repeat Finder was run on the contig assembly using the recommended parameters (matching weight = 2, mismatching penalty = 7, indel penalty = 7, match probability = 80, indel probability = 10, minimum alignment score = 50, and maximum period size = 500) with an additional parameter to denote the longest allowable TR array (−l) of 17 million bp, which represented the longest contig in the assembly [53].
Assembly quality assessment
The Hi-C scaffold assembly and annotated protein set were assessed for completeness in terms of gene content with BUSCO (RRID:SCR_015008), using all relevant taxonomic databases for the genome (Eukaryota, Metazoa, Arthropoda, Insecta, and Endopterygota) and only the most derived database, Endopterygota, for the protein set. Ab initio annotations on the scaffold assembly were performed using Metaeuk v.4.a0f584d for the Eukaryota, Arthropoda, Insecta, and Endopterygota odb10 databases, and Augustus v3.4.0 was used to detect the Metazoa odb10 orthologs [54]. Designation of genes as a complete single copy, duplicated, fragmented, or missing was determined using BUSCO v5.2.2 in “genome” mode for the genome assembly and “protein” for the annotated protein set [55]. Identification of off-target (non–A. tumida) contigs in the assembly was performed by aligning all contigs to the NCBI nucleotide database (accessed 14 February 2022) using the “blastn” function (RRID:SCR_001598) of BLAST+ v2.5.9+ [56]. These contigs were secondarily aligned to the UniProt protein database (accessed March 2020) using Diamond (RRID:SCR_009457) [57]. Local alignments to the nucleotide and protein databases were then used to assign the A. tumida contigs to a taxon using the rule “bestsumorder” of blobtoolkit v.2.6.1, which assigns contigs to a taxon first based on alignments to the nucleotide database and then followed by alignments to the protein database if there were no hits to the nucleotide database [58]. Coverage per scaffold and contig record was calculated using minimap2 v2.2-r1101 (RRID:SCR_018550) [59]. Coverage, taxonomic assignment, and BUSCO results were aggregated using blobtoolkit and summarized using blobblurb v2.0 [45]. Expected genome size was estimated using GenomeScope v2.0 (RRID:SCR_017014), which uses k-mer frequency analysis of k-mer counts performed by KMC v3.2.1 (RRID:SCR_001245) [60, 61]. The level of duplicate artifacts in the assembly was assessed using BUSCO results for both the genome and the protein set and using k-mer abundance in the raw HiFi reads relative to their representation in the final assembly as determined by K-mer Analysis Toolkit v2.4.2 [62]. The gene features were annotated through the NCBI Eukaryotic Genome Annotation pipeline, and the RNA sequencing data (SRR1798556) of both males and females were used to support the annotation. The annotated features were displayed using the Rideogram package in R [63].
Small hive beetle rearing and genome resequencing
Adult beetles were captured from collapsed honey bee (Apis cerana) hives in Hainan, China, and reared in the lab according to the standard method for small hive beetle research [64]. The pupae were preserved in plastic cups individually. After hatching, individual male and female beetles were paired and kept in a plastic container until the first batch of larvae was pupated in the soil. The parental and offspring beetles were then collected and preserved in liquid nitrogen until DNA extraction. DNA was extracted from each beetle using the Magnetic Universal Genomic DNA kit (TianGen, Beijing, China). Next, DNA for each beetle was used to prepare libraries using the NEB Next Ultra DNA Library Pre Kit (BioLabs, ipswich, massachusetts, USA). The beetles were individually sequenced on an Illumina Novaseq 6000 machine. The DNA sequencing reads were filtered through Fastp (version 0.20.1; RRID:SCR_016962) with default parameters [65] and then were aligned to the small hive beetle genome assembly (GCF_024364675.1) using Bowtie (version 0.7.17-r1188; RRID:SCR_005476) with default parameters [50]. To calculate the alignment depth, the number of reads aligned to the assembly was calculated on a 5-kbp sliding widow using Jvarkit bioalcidae [66]. The numbers of aligned reads for each 5-kbp window were normalized using count per million reads for each library [67].
Eggs collection and DNA extraction
The offspring were from paired male and female beetles. The sex ratio was determined by counting the emerged adult beetles. Additionally, we developed a primer based on the Y chromosome, which allowed us to distinguish the eggs developed to males. We amplified the intergenic region to avoid nonspecific amplification. In addition, the universal primers that amplify both males and females were used as a positive control for DNA quality. The eggs were collected from pooled SHBs in the lab. The DNA was extracted using DNA isolation Kit (Omega, Norcross, Georgia, USA).
Sex chromosome identification and synteny analysis
After normalization, there were regions with an extremely high or low number of aligned reads, which might have been alignment artifacts on repetitive regions. To exclude this bias, the second quartile was used to represent the alignment depth. First, we examined the existence of the Y or W chromosome, which is associated with either male or female beetles. Then we examined the ratio of alignment depth between male and female beetles. The small hive beetle protein sequences were aligned to T. castaneum (GCA_000002335.3) to infer the synteny using MCScanX (RRID:SCR_022067) with default parameters [68, 69]. SynVisio was used to view the synteny along the genome [70].
Recombination rate analysis
SNVs were identified from individual beetles using the GATK pipeline (RRID:SCR_001876) with default parameters [71]. In each beetle family, the parents' genotypes allow inference of crossovers in the offspring based on linkage equilibrium. We assume that any SNVs found in offspring should also be found in the parents. Therefore, only the SNVs identified in both parents and offspring were kept for further analysis. The package Lep-MAP3, supporting the integration of parents and offspring to determine recombination events using genome-wide SNVs, was used to determine the recombination events in the offspring beetles [72].
Supplementary Material
Christopher Cunningham -- 3/13/2023 Reviewed
Andrew Gordus, Ph.D. -- 4/16/2023 Reviewed
Andrew Gordus, Ph.D. -- 5/22/2023 Reviewed
Acknowledgement
We appreciate Prof. Nancy A. Moran for her comments on sex chromosome identification, Dawn Lopez for lab support, and members of the USDA-ARS Ag100Pest Team for sequencing and analysis support.
Contributor Information
Qiang Huang, Honeybee Research Institute, Jiangxi Agricultural University, 330045, Nanchang, China; Department of Integrative Biology, The University of Texas at Austin, Austin, TX 78712, USA.
Sheina B Sim, Daniel K. Inouye US Pacific Basin Agricultural Research Center Tropical Pest Genetics and Molecular Biology Research, USDA, Agricultural Research Service, Hilo, HI 96720, USA.
Scott M Geib, Daniel K. Inouye US Pacific Basin Agricultural Research Center Tropical Pest Genetics and Molecular Biology Research, USDA, Agricultural Research Service, Hilo, HI 96720, USA.
Anna Childers, Beltsville Agricultural Research Center, Bee Research Laboratory, USDA, Agricultural Research Service, Beltsville, MD 20705, USA.
Junfeng Liu, Periodicals Agency, Jiangxi Agricultural University, 330045, Nanchang, China.
Xiuxiu Wei, Honeybee Research Institute, Jiangxi Agricultural University, 330045, Nanchang, China.
Wensu Han, Environment and Plant Protection Institute, Chinese Academy of Tropical Agricultural Sciences, Haikou, 571101, China.
Francisco Posada-Florez, Beltsville Agricultural Research Center, Bee Research Laboratory, USDA, Agricultural Research Service, Beltsville, MD 20705, USA.
Allen Z Xue, Department of Integrative Biology, The University of Texas at Austin, Austin, TX 78712, USA.
Zheng Li, Department of Integrative Biology, The University of Texas at Austin, Austin, TX 78712, USA.
Jay D Evans, Beltsville Agricultural Research Center, Bee Research Laboratory, USDA, Agricultural Research Service, Beltsville, MD 20705, USA.
Data Availability
Raw WGS HiFi and Hi-C Illumina sequence data were deposited at DDBJ/ENA/GenBank within BioProject PRJNA825637, under the Sequence Read Archive accessions SRX14827166 and SRX14828569, respectively. The annotated primary assembly version icAetTumi1.1 accession GCA_024364675.1 (Annotation Release 101, BioProject PRJNA825637) and icAetTumi1.1 alternate haplotype assembly version accession GCA_024364635.1 (BioProject PRJNA825646) were described in this article. Both assemblies are under the Ag100Pest umbrella project, BioProject PRJNA555319. The primary assembly and annotations are also available at the i5k Workspace@NAL [73]. The genome assembly and gene annotation are available at NCBI to download [74]. The beetle family genome resequencing reads were deposited to BioProject PRJNA776042. All supporting data and materials are available in the GigaScience GigaDB database [75].
Abbreviations
bp: base pairs; BUSCO: Benchmark of Universal Single-Copy Orthologs; kb: kilobase; kbp: kilobase pair; Mb: megabase; Mbp: megabase pair; NCBI: The National Center for Biotechnology Information; SHB: small hive beetle; SNV: single-nucleotide variant.
Competing Interests
The authors declare that they have no competing interests.
Funding
This work was supported by the US Department of Agriculture, Agricultural Research Service (USDA-ARS; S.B.S., S.M.G., A.C., F.P.-F., J.D.E.), the initiation package of Jiangxi Agricultural University 050014/923230722 (Q.H.), and the Hainan Province Science and Technology special fund ZDYF2021XDNY282 (W.H.). A.Z.X. is supported by the US Army Research Office MURI award W911NF-20–1-0195. The genome assembly was generated as part of the USDA-ARS Ag100Pest Initiative. This research used resources provided by the SCINet project of the USDA Agricultural Research Service, project number 0500–00093-001–00-D. This project is a component of the USDA-ARS AgPest100 Genome Project (USDA-ARS). The US Department of Agriculture, Agricultural Research Service is an equal opportunity/affirmative action employer, and all agency services are available without discrimination. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. NSF Postdoctoral Research Fellowships in Biology Program, DBI-2109306 (Z.L.).
Authors’ Contributions
Q.H. and J.D.E. planned and managed the project. Q.H., S.B.S., and J.D.E. wrote the manuscript. S.B.S., A.C., and S.M.G. assembled the genome and analyzed repetitive regions. J.L. and F.P.-F. reared and collected beetles. X.W. and W.H. analyzed the egg sex ratio. A.Z.X. and Z.L. analyzed the synteny.
Additional Files
Supplementary Table S1. Statistics of the assembled chromosomes.
Supplementary Table S2. Sex ratio of SHB eggs.
Supplementary Table S3. Quality metric for the HIFI reads.
Supplementary Figure S1. Estimated genome size from GenomeScope.
Supplementary Figure S2. K-mer frequency reveals low levels of artifact duplicates.
Supplementary Figure S3. The genome and assembly statistics of the small hive beetle.
Supplementary Figure S4. Scatter plot of genome size and chromosome number in beetles.
Supplementary Figure S5. Rideogram of the small hive beetle genome assembly.
Supplementary Figure S6. primers to distinguish male (M) and female (F) small hive beetles.
Supplementary Figure S7. Amplified region in ChrY.
References
- 1. Hood WM. Overview of the small hive beetle, aethina tumida, in North America. Bee World. 2000;81:129–37. [Google Scholar]
- 2. Gillespie P, Staples J, King C, et al. Small hive beetle aethina tumida (Murray) (Coleoptera: nitidulidae) in New South Wales. Gen Appl Entomol. 2022;32:5–72. [Google Scholar]
- 3. Mohamadzade Namin S, Koh Y, Osabutey AF et al. Invasion pathway of the honeybee pest, small hive beetle, aethina tumida (Coleoptera: nitidulidae) in the Republic of Korea inferred by mitochondrial DNA sequence analysis. J Asia Pac Entomol. 2019;22:963–8. [Google Scholar]
- 4. Palmeri V, Scirtò G, Malacrinò A, et al. A scientific note on a new pest for European honeybees: first report of small hive beetle aethina tumida (Coleoptera: nitidulidae) in Italy. Apidologie. 2014;46:527–9. [Google Scholar]
- 5. Neumann P, Pettis JS, Schäfer MO. Quo vadis aethina tumida? Biology and control of small hive beetles. Apidologie. 2016;47:427–66. [Google Scholar]
- 6. Evans JD, Pettis JS, Hood WM et al. Tracking an invasive honey bee pest: mitochondrial DNA variation in North American small hive beetles. Apidologie. 2003;34:103–9. [Google Scholar]
- 7. Evans JD, McKenna D, Scully E et al. Genome of the small hive beetle (Aethina tumida, Coleoptera: nitidulidae), a worldwide parasite of social bee colonies, provides insights into detoxification and herbivory. Gigascience. 2018;7:giy138. 10.1093/gigascience/giy138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Y, Henkel J, Beaurepaire A, et al. Comparative genomics suggests local adaptations in the invasive small hive beetle. Ecol Evol. 2021;11:15780–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu Y, Han W, Gao J et al. Out of Africa: novel source of small hive beetles infesting Eastern and Western honey bee colonies in China. J Apic Res. 2021;60:108–10. [Google Scholar]
- 10. Idrissou FO, Huang Q, Yañez O, et al. International beeswax trade facilitates small hive beetle invasions. Sci Rep. 2019;9:10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bai WF, Liu J, Liu Y et al. Phylogenetic analysis of small hive beetles from native to introduced populations. Front Genet. 2022;13:900795. 10.3389/fgene.2022.900795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neumann P, Ellis JD. The small hive beetle (Aethina tumida Murray, Coleoptera: nitidulidae): distribution, biology and control of an invasive species. J Apic Res. 2008;47:181–3. [Google Scholar]
- 13. Ouessou Idrissou F, Straub L, Neumann P. Keeping a low profile: small hive beetle reproduction in African honeybee colonies. Agric For Entomol. 2019;21:136–8. [Google Scholar]
- 14. Benda ND, Boucias D, Torto B et al. Detection and characterization of Kodamaea ohmeri associated with small hive beetle aethina tumida infesting honey bee hives. J Apic Res. 2008;47:194–201. [Google Scholar]
- 15. Amos BA, Leemon DL, Hayes RA, et al. Associations between the small hive beetle and the yeast kodamaea ohmeri throughout the host life cycle. J Econ Entomol. 2018;111:1501–8. [DOI] [PubMed] [Google Scholar]
- 16. Amos BA, Hayes RA, Leemon DM et al. Small hive beetle (Coleoptera: nitidulidae) and the yeast, kodamaea ohmeri: a facultative relationship under laboratory conditions. J Econ Entomol. 2019;112:515–24. [DOI] [PubMed] [Google Scholar]
- 17. Stuhl CJ. The development of an attract-and-kill bait for controlling the small hive beetle (Coleoptera: nitidulidae). Apidologie. 2020;51:428–35. [Google Scholar]
- 18. Spiewok S, Neumann P. Sex ratio and dispersal of small hive beetles. J Apic Res. 2012;51:216–7. [Google Scholar]
- 19. Papach A, Gonthier J, Williams GR et al. Sex ratio of small hive beetles: the role of pupation and adult longevity. Insects. 2019;10:133. 10.3390/insects10050133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo J-W, Yang F, Li P, et al. Female bias in an immigratory population of Cnaphalocrocis medinalis moths based on field surveys and laboratory tests. Sci Rep. 2019;9:18388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dyson EA, Hurst GDD. Persistence of an extreme sex-ratio bias in a natural population. Proc Natl Acad Sci USA. 2004;101:6520–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kobayashi K, Hasegawa E. A female-biased sex ratio reduces the twofold cost of sex. Sci Rep. 2016;6:23982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fukui T, Kawamoto M, Shoji K et al. The endosymbiotic bacterium Wolbachia selectively kills male hosts by targeting the masculinizing gene. PLoS Pathog. 2015;11:e1005048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leclercq S, Thézé J, Chebbi MA et al. Birth of a W sex chromosome by horizontal transfer of Wolbachia bacterial symbiont genome. Proc Natl Acad Sci USA. 2016;113:15036–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang Q, Lopez D, Evans JD. Shared and unique microbes between small hive beetles (Aethina tumida) and their honey bee hosts. Microbiologyopen. 2019;8:e899. 10.1002/mbo3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bouchard P, Bousquet Y, Davies A et al. Family-group names in Coleoptera (Insecta). Zookeys. 2011;88:1–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Courant J, Vogt S, Marques R et al. Are invasive populations characterized by a broader diet than native populations?. PeerJ. 2017;5:e3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paini DR, Sheppard AW, Cook DC et al. Global threat to agriculture from invasive species. Proc Natl Acad Sci USA. 2016;113:7575–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nime MF, Fachinetti R, Pedemonte L et al. Potential fecundity, larval development, and survival of two invasive species of Arhopalus (Coleoptera: cerambycidae) coexisting in southern South America. Caldasia. 2019;41:268–77. [Google Scholar]
- 30. Bachtrog D, Mank JE, Peichel CL, et al. Sex determination: why so many ways of doing it?. PLoS Biol. 2014;12:e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Silk JB, Brown GR. Local resource competition and local resource enhancement shape primate birth sex ratios. Proc R Soc B. 2008;275:1761–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cicconardi F, Lewis JJ, Martin SH et al. Chromosome fusion affects genetic diversity and evolutionary turnover of functional loci but consistently depends on chromosome size. Mol Biol Evol. 2021;38:4449–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Vos JM, Augustijnen H, Bätscher L et al. Speciation through chromosomal fusion and fission in Lepidoptera. Philos Trans R Soc B Biol Sci. 2020;375:20190539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schrader L, Pan H, Bollazzi M et al. Relaxed selection underlies genome erosion in socially parasitic ant species. Nat Commun. 2021;12:2918. 10.1038/s41467-021-23178-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pombert J-F, Blouin NA, Lane C et al. A lack of parasitic reduction in the obligate parasitic green alga helicosporidium. PLoS Genet. 2014;10:e10043552014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barton NH, Charlesworth B. Why sex and recombination?. Science. 1998;281:1986–90. [PubMed] [Google Scholar]
- 37. Fischer O, Schmid-Hempel P. Selection by parasites may increase host recombination frequency. Biol Lett. 2005;1:193–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zilio G, Moesch L, Bovet N et al. The effect of parasite infection on the recombination rate of the mosquito Aedes aegypti. PLoS One. 2018;13:e0203481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beye M, Gattermeier I, Hasselmann M, et al. Exceptionally high levels of recombination across the honey bee genome. Genome Res. 2006;16:1339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wilfert L, Gadau J, Schmid-Hempel P. Variation in genomic recombination rates among animal taxa and the case of social insects. Heredity. 2007;98:189–97. [DOI] [PubMed] [Google Scholar]
- 41. Stapley J, Feulner PGD, Johnston SE, et al. Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Phil Trans R Soc B. 2017;372:20160455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Comeron JM, Ratnappan R, Bailin S. The many landscapes of recombination in drosophila melanogaster. PLoS Genet. 2012;8:e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lorenzen MD, Doyungan Z, Savard J et al. Genetic linkage maps of the red flour beetle, tribolium castaneum, based on bacterial artificial chromosomes and expressed sequence tags. Genetics. 2005;170:741–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Salathé M, Kouyos RD, Regoes RR et al. Rapid parasite adaptation drives selection for high recombination rates. Evolution. 2008;62:295–300. [DOI] [PubMed] [Google Scholar]
- 45. Sim SB, Corpuz RL, Simmonds TJ et al. HiFiAdapterFilt, a memory efficient read processing pipeline, prevents occurrence of adapter sequence in PacBio HiFi reads and their negative impacts on genome assembly. BMC Genomics. 2022;23:157. 10.1186/s12864-022-08375-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cheng H, Concepcion GT, Feng X et al. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 2021;18:170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Seemann T: any2fasta. Github. 2019. https://github.com/tseemann/any2fasta.
- 48. Juric I, Yu M, Abnousi A, et al. MAPS: model-based analysis of long-range chromatin interactions from PLAC-seq and HiChIP experiments. PLoS Comput Biol. 2019;15:e1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou C, McCarthy SA, Durbin R. YaHS: yet another Hi-C scaffolding tool. Bioinformatics. 2023;39:btac808. 10.1093/bioinformatics/btac808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Broad Institute . Picard Toolkit. Github. 2023. https://github.com/broadinstitute/picard
- 52. Durand NC, Robinson JT, Shamim MS et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016;3:99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Levy Karin E, Mirdita M, Söding J. MetaEuk-sensitive, high-throughput gene discovery, and annotation for large-scale eukaryotic metagenomics. Microbiome. 2020;8:48. 10.1186/s40168-020-00808-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Manni M, Berkeley MR, Seppey M et al. BUSCO: assessing genomic data quality and beyond.Curr Protoc J. 2021;1:e323. 10.1002/cpz1.323. [DOI] [PubMed] [Google Scholar]
- 56. Camacho C, Coulouris G, Avagyan V et al. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Buchfink B, Reuter K, Drost H-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 2021;18:366–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Challis R, Richards E, Rajan J, et al. BlobToolKit—interactive quality assessment of genome assemblies. G3. 2020;10:1361–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kokot M, Długosz M, Deorowicz S. KMC 3: counting and manipulating k-mer statistics. Bioinformatics. 2017;33:2759–61. [DOI] [PubMed] [Google Scholar]
- 61. Ranallo-Benavidez TR, Jaron KS, Schatz MC. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 2020;11:1432. 10.1038/s41467-020-14998-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mapleson D, Garcia Accinelli G, Kettleborough G, et al. KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics. 2017;33:574–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hao Z, Lv D, Ge Y et al. RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput Sci. 2020;6:e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Neumann P, Evans JD, Pettis JS, et al. Standard methods for small hive beetle research. J Apic Res. 2013;52:1–32. [Google Scholar]
- 65. Chen S, Zhou Y, Chen Y et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lindenbaum P, Redon R. bioalcidae, samjs and vcffilterjs: object-oriented formatters and filters for bioinformatics files. Bioinformatics. 2018;34:1224–5. [DOI] [PubMed] [Google Scholar]
- 67. R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. http://www.R-project.org/2013. [Google Scholar]
- 68. Richards S, Gibbs RA, Weinstock GM et al. The genome of the model beetle and pest tribolium castaneum. Nature. 2008;452:949–55. [DOI] [PubMed] [Google Scholar]
- 69. Wang Y, Tang H, Debarry JD et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bandi V, Gutwin C. Interactive exploration of genomic conservation. In: Proceedings of the 46th Graphics Interface Conference on Proceedings of Graphics Interface 2020 (GI’20). 2020. https://github.com/kiranbandi/synvisio.
- 71. Van der Auwera GA, Carneiro MO, Hartl C et al. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;43:1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rastas P. Lep-MAP3: robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics. 2017;33:3726–32. [DOI] [PubMed] [Google Scholar]
- 73. Poelchau M, Childers C, Moore G, et al. The i5k Workspace@NAL–enabling genomic data access, visualization and curation of arthropod genomes. Nucleic Acids Res. 2015;43:D714–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Small hive beetle consortum: Small hive beetle genome assembly. 2022. https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/024/364/675/GCF_024364675.1_icAetTumi1.1.
- 75. Huang Q, Sim SB, Geib SM et al. Supporting data for “Identification of Sex Chromosomes and Primary Sex Ratio in the Small Hive Beetle, a Worldwide Parasite of Honey Bees.” GigaScience Database. 2023. http://gigadb.org/dataset/102405. [DOI] [PMC free article] [PubMed]
- 76. Idrissou FO, Huang Q, Yañez O et al. PCR diagnosis of small hive beetles. Insects. 2018;9:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Huang Q, Sim SB, Geib SM et al. Supporting data for “Identification of Sex Chromosomes and Primary Sex Ratio in the Small Hive Beetle, a Worldwide Parasite of Honey Bees.” GigaScience Database. 2023. http://gigadb.org/dataset/102405. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Christopher Cunningham -- 3/13/2023 Reviewed
Andrew Gordus, Ph.D. -- 4/16/2023 Reviewed
Andrew Gordus, Ph.D. -- 5/22/2023 Reviewed
Data Availability Statement
Raw WGS HiFi and Hi-C Illumina sequence data were deposited at DDBJ/ENA/GenBank within BioProject PRJNA825637, under the Sequence Read Archive accessions SRX14827166 and SRX14828569, respectively. The annotated primary assembly version icAetTumi1.1 accession GCA_024364675.1 (Annotation Release 101, BioProject PRJNA825637) and icAetTumi1.1 alternate haplotype assembly version accession GCA_024364635.1 (BioProject PRJNA825646) were described in this article. Both assemblies are under the Ag100Pest umbrella project, BioProject PRJNA555319. The primary assembly and annotations are also available at the i5k Workspace@NAL [73]. The genome assembly and gene annotation are available at NCBI to download [74]. The beetle family genome resequencing reads were deposited to BioProject PRJNA776042. All supporting data and materials are available in the GigaScience GigaDB database [75].