Flagella and motility represent two of the richest subjects in microbiology, involving not only bacterial genetics, molecular biology, and physiology but also bioenergetics, hydrodynamics, structural analysis, and macromolecular assembly.
Our knowledge that bacteria actively move goes as far back as the discovery of bacteria themselves (7). To quote from an article by Howard Berg in 1975 (4), written not long after the modern era of investigation of bacterial flagella, motility, and chemotaxis had begun:
When Antony van Leeuwenhoek looked through a single-lens microscope in 1676 and observed man’s first recorded glimpse of bacteria, it was their motion that most delighted him: “I must say, for my part, that no more pleasant sight has ever yet come before my eye than these many thousands of living creatures, seen all alive in a little drop of water, moving among one another, each several creature having its own proper motion.”
Leeuwenhoek goes on to say, in a charming phrase: “…I can make out no paws…[yet] I am persuaded that they too are furnished with paws withal.”
The bacterial “paw,” more commonly known as the flagellum, is a structure with a very long (ca. 10-μm), thin (ca. 20-nm-diameter) external filament. Besides its extreme thinness and length, the first thing that strikes one about the flagellar filament is its “waviness.” The active propagation of this wave during motility was evident from early high-speed movies, so there was no doubt that flagella were the organelles of bacterial motility. Cells typically displayed more than one type of movement: in some cases, simple forward and backward swimming and in other cases (e.g., Salmonella), swimming and tumbling.
Around 1970, the major questions about bacterial motility could be summarized as follows: (i) What is the shape of the wave, and is it intrinsic to the flagellar structure? (ii) How is the waveform propagated? (iii) What is the nature of the motor? (iv) What is responsible for the two types of motility (swimming and tumbling in the case of Salmonella)? (v) What is the energy source? In only a few years, the answers to these questions were obtained, at least in broad outline.
A HELICAL PROPELLOR
Kamiya and colleagues (13) carried out extensive in vitro studies of flagellar filaments. Their principal conclusions were that the waveform of a filament in aqueous suspension is a perfect helix, that the helicity is intrinsic (in vitro depolymerization and repolymerization occurs readily) and so is a cause rather than a consequence of motility, and that filaments exhibit polymorphism. At least two of the polymorphic forms are important to normal motility (23). The helicity of the filament is both remarkable and essential. Remarkable, because it is a consequence of a subtle breaking of symmetry in a polymer made (in many species) from identical subunits; normally, one would expect such a polymer to be straight. Essential, because without the helicity, propulsion would be impossible.
Structural studies of filament, hook, basal body, etc. by DeRosier, Namba, and others (mostly by analysis of electron microscopic images) have become ever more refined, so that the subunit shapes and their quaternary interactions are being seen in more and more detail, although the data still do not approach atomic resolution.
A REVERSIBLE ROTARY MOTOR
There was a general presumption until the early 1970s that the waveform was propagated as a conformational wave (much as one can drive a wave along a rope by wrist movement). There was no evidence in favor of such a model, and several good arguments against it. Yet the alternative—a rotational model—seemed to be unpalatable, even though it could accommodate many of the known observations (5). Then, in three back-to-back papers in 1974 from the laboratories of Simon, Adler, and Berg (3, 20, 32), the rotational model was established beyond doubt, mostly on the basis of experiments with tethered Escherichia coli cells, which whirled around merrily.
These experiments generated another, equally important, finding. Not only did the motor rotate, it rotated in both directions, clockwise (CW) and counterclockwise (CCW). It reversed stochastically and indefinitely in the absence of stimulation, rotated almost exclusively CCW upon addition of an attractant, and almost exclusively CW upon addition of a repellent. Thus, the basis for selective motion in response to chemical gradients, chemotaxis, was reduced to the simple notion of a binary switch whose CCW versus CW states had probabilities that were modulated by environmental signals.
A tethered cell presents an artificially high load to the motor, so the cell rotates relatively slowly (less than 500 rpm). At the much lower load of a freely rotating filament, the motor is capable of astonishing speeds, e.g., around 15,000 rpm in E. coli (21). The world record is for a Vibrio cell clocked at 100,000 rpm by laser microscopy (24)!
STRUCTURE AND COMPOSITION OF THE FLAGELLAR MOTOR
What does this rotary motor look like? Electron micrographs of isolated flagella taken by Cohen-Bazire and London in 1967 (6) had revealed a basal structure containing four rings threaded by a rod. Subsequent work showed that two of them, the M and S rings, lay in the cytoplasmic membrane and just above it, respectively. Any rotary motor must have a rotor (the part that does external work) and a stator (the anchor), so it was natural to think that the M and S rings might fulfil such roles. The M and S rings, however, are a single, double-flanged ring made from subunits of just one protein (34). Also, studies of mutants showed that the MS ring does not contribute to torque generation. Other studies established that the stator consists of a series of membrane-imbedded studs or Mot complexes spaced around the MS ring (15). These studs contain two components, MotA and MotB, with the latter apparently binding to the peptidoglycan layer—about as good a cellular anchor as one can get. The rotor turned out to be an extensive structure projecting from the MS ring into the cytoplasm (10, 14, 16) and termed the C ring, a vital piece of the basal body that had escaped detection with the protocols used heretofore. The C ring contains three of the most interesting proteins in the flagellum, the motor/switch proteins. These work against the Mot complexes to generate torque, and they also have the ability to change their conformational state in a bimodal fashion, so that the motor direction can be switched from CCW to CW and vice versa. Despite much effort and the accumulation of much detailed structure-function information, the nature of the conformational change underlying motor switching remains elusive.
FLAGELLA ARE DRIVEN BY IONIC POTENTIALS, NOT ATP
Because of the large body of research into muscle and other biological structures whose function is to produce mechanical work (all of them driven by ATP hydrolysis), it was probably natural to suppose that ATP might drive the bacterial flagellar motor also. This notion was dispelled in 1974 by Larsen et al. (19), who showed that uncouplers of oxidative phosphorylation like carbonyl cyanide m-chlorophenylhydrazone, or mutations that uncouple the process, block motility even though ATP levels remain high. This was at a time when Peter Mitchell’s chemiosmotic hypothesis (27), while in general circulation, had not gained full understanding or acceptance. Thus, the Larsen et al. paper studiously avoids the statement that bacterial motility is driven by proton motive force. Only in 1977 was this term used explicitly, in a paper by Manson et al. (25) that included a staunch Mitchellian, Franklin Harold, as one of its authors. Many bacterial species, incidentally, use sodium motive force, arguing for an electrostatic mechanism and against a hydrogen-bonding one.
HOW DOES THE MOTOR WORK?
In the 25 years since the rotary mechanism and the ionic energy source were discovered, the bioenergetics of the motor have been studied in ever greater detail by the laboratories of Berg, Aizawa, and others, so that its empirical characteristics are well established. What is not well understood is the mechanism by which ionic energy is converted into mechanical work. Given the ubiquitous yet elusive character of the proton, this may not be too surprising.
Initially, a common assumption was that the proton (or sodium ion) would travel down its gradient via a series of binding sites contributed jointly by the stator (Mot complexes) and rotor (motor/switch complex), developing torque in the process. This notion has become less attractive as a result of recent mutational analyses by Zhou and coworkers (37), who have found that only one of the conserved protonatable residues in the five Mot and switch proteins is essential. They suggest that the proton conductance pathway may reside entirely within the Mot complexes and cause a conformationally strained structure which, interacting with the motor/switch complexes, relaxes to generate torque.
THE FLAGELLAR GENE SYSTEM IS COMPLEX
A structure such as the flagellum has, not surprisingly, a large genetic basis. Our current detailed understanding of the many genes, their products, and their transcriptional controls owes an enormous debt to the classical work that has been carried out over several decades since the 1950s in many laboratories (including, for E. coli and Salmonella alone, those of T. Iino, Komeda, K. Kutsukake, Parkinson, Simon, B. A. D. Stocker, and Yamaguchi). In the early 1950s, a distinction was made between mutants that lacked flagella (nonflagellate or fla mutants) and those that had flagella but could not move them (paralyzed or mot mutants) (33). Later, it was recognized that there were mutants that, though highly motile, were nonchemotactic (che mutants) (2). Their motility was unusual in that it consisted either of swimming with essentially no tumbling or tumbling with essentially no swimming (now recognized as being due to a high CCW bias or a high CW bias, respectively). There then followed an extended period of research in which mutants were divided into ever finer complementation groups, which now correspond to the genes that are known today by physical mapping and sequencing.
Just how extensive are these gene systems? In Salmonella, there are currently 44 known flagellar genes. Twenty-three of these encode structural components of the flagellum. Of these components, five (MotA, MotB, FliG, FliM, and FliN) are needed for torque generation and three of these five (FliG, FliM and FliN) are also needed for switching; these components are the heart of the motor. The principal remaining components are the filament (propellor), the hook (universal joint), and the basal body; the latter can be broken down into rod (transmission shaft), MS ring (motor mounting plate), and LP ring or outer cylinder (bushing).
Another five flagellar genes or so fulfil regulatory roles. There is a hierarchy of expression whose full complexity is just beginning to be realized. One regulatory feature involves a flagellum-specific sigma factor and its antagonist. The concept of a specialized sigma factor is not unusual these days. What is unusual is the mechanism by which the anti-sigma factor is inactivated at the appropriate point in flagellar assembly: it is exported from the cell by the same system that is responsible for assembling the flagellum itself (11, 18) (see below).
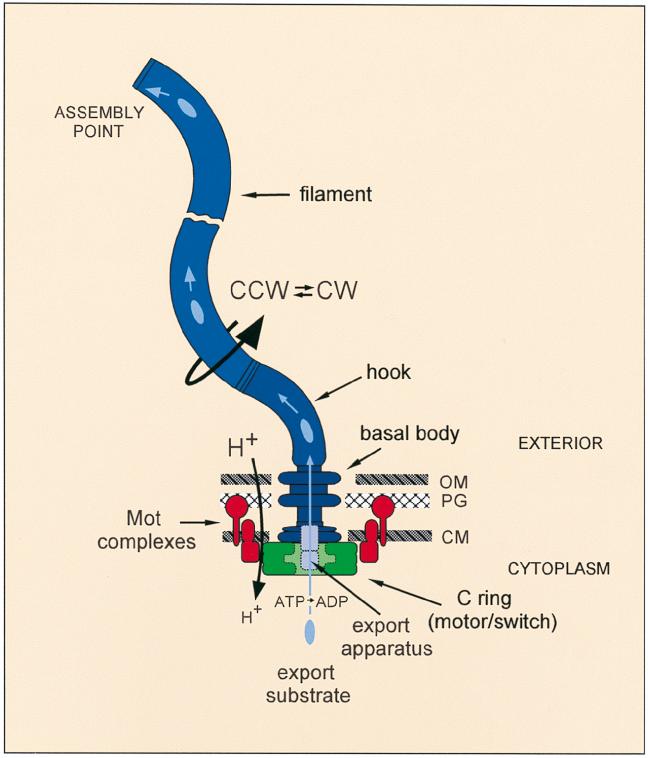
Almost all of the remaining genes (about 11 of them) appear to encode components that are responsible for flagellar assembly. This brings us to the point that—as the title to this Commentary indicates—the flagellum performs not just one function, but two (Fig. 1). Not only is it the organelle of propulsion for the bacterium but it also functions as a sophisticated export and self-assembly apparatus.
FIG. 1.
The flagellum is the motor organelle for bacterial propulsion. Driven by a transmembrane proton gradient (H+→h+), it rotates both CCW and CW; the filament is helical and so converts torque into thrust. The motor consists of stators or Mot complexes (red) and a rotor or C ring (green), which also serves as the CCW⇄CW switch. As well as being the organelle of motility, the flagellum is a specialized type III export apparatus (lilac), translocating subunits of its substrates (pale blue) in an ATP-dependent manner across the plane of the cytoplasmic membrane (CM) and delivering them into a central channel in the basal body-hook-filament structure where they eventually reach their assembly point at the distal end of the structure. PG, peptidoglycan layer; OM, outer membrane.
FLAGELLAR PROTEINS TRAVEL THROUGH THEIR OWN STRUCTURE
The story of flagellar protein export begins with two reports published by Iino in 1969 (12) and by Emerson et al. in 1970 (8); both reports presented convincing evidence that new flagellin monomers are added to growing filaments at their distal end. It was known from structural studies in the early 1960s (e.g., reference 22) that flagellar filaments were hollow tubes, and we now know that this is true of the basal-body rod and the hook also. Iino speculated on roles that the flagellar basal structure might play: flagellin synthesis (where he was wrong [there is no flagellar ribosome]), flagellar motion, and initiation of flagellar assembly. Presciently, he suggested that “…accumulation of flagellin molecules in the basal structure might cause the efficient diffusion or pushing of the molecules through the hole to the tip of the flagellum.”
AN ANTI-SIGMA FACTOR, A MURAMIDASE, AND A HOOK-LENGTH PROTEIN?
Most of the flagellar proteins that are exported are structural components, but there are three interesting exceptions: One is the anti-sigma factor that was alluded to earlier—when you no longer need it, get rid of it! Another is a muramidase (28). Why would the cell want a flagellum-specific muramidase, and why would it want to export this enzyme? The answer, for which we already have some experimental support, is almost certainly to punch a hole in the peptidoglycan layer in the early stage of flagellar morphogenesis, in order to let the nascent rod penetrate it. The third example is a fascinating protein that is implicated in controlling the length of the flagellar hook; it has only recently been shown that it is exported, and how it functions in the process of length control is not well understood.
THE FLAGELLAR EXPORT APPARATUS
After the papers reporting distal growth, there followed a long hiatus in the investigation of flagellar protein export, perhaps because everyone was too busy examining flagellar structure, composition, function, genetics, etc. But as the function of more and more flagellar genes was established, it became evident that there was a residuum with no known function. This triggered (at least in my mind) the realization that there was also a function, flagellar protein export, with no known genes. Maybe there was a match? In 1991, we performed a very simple experiment that yielded the first tentative identification of a few export component genes (36); one of these components is an ATPase that, inexplicably, is related to the catalytic subunit of the F0F1 ATPase (1, 36). Subsequently, the total number of components of the flagellar export apparatus has risen to at least 8 (26) and perhaps as high as 13 if one includes components that may have specialized functions such as chaperones.
AN EXPORT APPARATUS WITHIN THE BASAL-BODY MS RING?
Six of the export components are integral membrane proteins, three of which we have already shown to be associated with the basal body (9, 26). Given that their substrates have to be delivered into the hollow channel in the rod-hook-filament structure, it seems difficult to avoid the conclusion that the export apparatus resides in a patch of membrane within the central pore of the MS ring. Soluble components, such as the ATPase, presumably interact in a dynamic fashion with this membrane complex. Efforts to build up evidence in support of this model represent an active area of current research.
FLAGELLAR PROTEINS AND MANY VIRULENCE FACTORS ARE EXPORTED BY RELATED PATHWAYS
Bacteria export or secrete proteins by several different pathways (of which perhaps the best known is the type II Sec-dependent general secretory pathway or GSP (30), which entails signal peptide cleavage during translocation of the protein across the cytoplasmic membrane). In the field of bacterial pathogenesis, genes needed for export of virulence factors by the so-called type III pathway (35), whose characteristics include a lack of signal peptide cleavage, were rapidly being discovered during the 1990s, and as their sequences and the sequences of putative flagellar export genes became available, there was an almost overnight realization by many laboratories that the flagellar export pathway and type III export pathways for virulence factors are closely related (see, e.g., references 29 and 31). I consider, in fact, that the flagellar pathway is a type III pathway, differing only in the nature of its export substrates and in the fact that it operates via a working organelle of propulsion.
If the flagellar export apparatus resides within the basal body, is there a corresponding structure for the virulence factor export apparatus? The answer appears to be yes. Kubori et al. (17) have recently described the existence in Salmonella of a “needle complex” that is constructed from components of the export pathway and closely resembles a flagellar basal body with an elongated rod (but of course no hook or filament!). Ironically, S.-I. Aizawa (an author on the paper by Kubori et al.) and I saw these structures in 1984 but dismissed them as being, perhaps, virus-related—a clear illustration that (to misquote Pasteur) chance disfavors the unprepared mind.
Which is the original pathway, the one for flagellar proteins or the one for virulence factors? Flagella are very ancient organelles, predating by far the targets for bacterial pathogenesis—plants, mammals, etc. So, it seems to me that the rest of the type III pathways must have evolved from the flagellar one.
FINAL COMMENTS
I leave the reader to contemplate the flagellum in all of its wonderful complexity. It is an organelle that receives sensory information from the cytoplasm, yet extends far beyond the cell itself; it rotates at high speed, and switches rotation in a controlled fashion; at the same time, it exports its own component proteins through itself and assembles them at its distant tip; and together with its cognate sensory transduction system, it generates a behavior, chemotaxis, that is critical for the cell’s survival. Although we have come a long way in our understanding of the flagellum, much remains to be done.
ACKNOWLEDGMENTS
Work in my laboratory is supported in part by USPHS grants AI12202 and GM40335.
Footnotes
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Albertini A M, Caramori T, Crabb W D, Scoffone F, Galizzi A. The flaA locus of Bacillus subtilis is part of a large operon coding for flagellar structures, motility functions, and an ATPase-like polypeptide. J Bacteriol. 1991;173:3573–3579. doi: 10.1128/jb.173.11.3573-3579.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong J B, Adler J, Dahl M M. Nonchemotactic mutants of Escherichia coli. J Bacteriol. 1967;93:390–398. doi: 10.1128/jb.93.1.390-398.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg H C. Dynamic properties of bacterial flagellar motors. Nature (London) 1974;249:77–79. doi: 10.1038/249077a0. [DOI] [PubMed] [Google Scholar]
- 4.Berg H C. How bacteria swim. Sci Am. 1975;233:36–44. [PubMed] [Google Scholar]
- 5.Berg H C, Anderson R A. Bacteria swim by rotating their flagellar filaments. Nature (London) 1973;245:380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- 6.Cohen-Bazire G, London J. Basal organelles of bacterial flagella. J Bacteriol. 1967;94:458–465. doi: 10.1128/jb.94.2.458-465.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobell C. Antony van Leeuwenhoek and his “little animals.” Dover. 1960. New York, N.Y. [Google Scholar]
- 8.Emerson S U, Tokuyasu K, Simon M I. Bacterial flagella: polarity of elongation. Science. 1970;169:190–192. doi: 10.1126/science.169.3941.190. [DOI] [PubMed] [Google Scholar]
- 9.Fan F, Ohnishi K, Francis N R, Macnab R M. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol Microbiol. 1997;26:1035–1046. doi: 10.1046/j.1365-2958.1997.6412010.x. [DOI] [PubMed] [Google Scholar]
- 10.Francis N R, Sosinsky G E, Thomas D, DeRosier D J. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 11.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 12.Iino T. Polarity of flagellar growth in Salmonella. J Gen Microbiol. 1969;56:227–239. doi: 10.1099/00221287-56-2-227. [DOI] [PubMed] [Google Scholar]
- 13.Kamiya R, Hotani H, Asakura S. Polymorphic transition in bacterial flagella. In: Amos W B, Duckett J G, editors. Prokaryotic and eukaryotic flagella. Cambridge, United Kingdom: Cambridge University Press; 1982. pp. 53–76. [PubMed] [Google Scholar]
- 14.Katayama E, Shiraishi T, Oosawa K, Baba N, Aizawa S-I. Geometry of the flagellar motor in the cytoplasmic membrane of Salmonella typhimurium as determined by stereo-photogrammetry of quick-freeze deep-etch replica images. J Mol Biol. 1996;255:458–475. doi: 10.1006/jmbi.1996.0038. [DOI] [PubMed] [Google Scholar]
- 15.Khan S, Dapice M, Reese T S. Effects of mot gene expression on the structure of the flagellar motor. J Mol Biol. 1988;202:575–584. doi: 10.1016/0022-2836(88)90287-2. [DOI] [PubMed] [Google Scholar]
- 16.Khan S, Khan I H, Reese T S. New structural features of the flagellar base in Salmonella typhimurium revealed by rapid-freeze electron microscopy. J Bacteriol. 1991;173:2888–2896. doi: 10.1128/jb.173.9.2888-2896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán J E, Aizawa S-I. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 18.Kutsukake K. Excretion of the anti-sigma factor through a flagellar structure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet. 1994;243:605–612. doi: 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- 19.Larsen S H, Adler J, Gargus J J, Hogg R W. Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc Natl Acad Sci USA. 1974;71:1239–1243. doi: 10.1073/pnas.71.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen S H, Reader R W, Kort E N, Tso W-W, Adler J. Change in direction of flagellar rotation is the basis of the chemotactic response in Escherichia coli. Nature (London) 1974;249:74–77. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- 21.Lowe G, Meister M, Berg H C. Rapid rotation of flagellar bundles in swimming bacteria. Nature (London) 1987;325:637–640. [Google Scholar]
- 22.Lowy J, Hanson J. Electron microscope studies of bacterial flagella. J Mol Biol. 1965;11:293–313. doi: 10.1016/s0022-2836(65)80059-6. [DOI] [PubMed] [Google Scholar]
- 23.Macnab R M, Ornston M K. Normal-to-curly flagellar transitions and their role in bacterial tumbling. Stabilization of an alternative quaternary structure by mechanical force. J Mol Biol. 1977;112:1–30. doi: 10.1016/s0022-2836(77)80153-8. [DOI] [PubMed] [Google Scholar]
- 24.Magariyama Y, Sugiyama S, Muramoto K, Maekawa Y, Kawagishi I, Imae Y. Very fast flagellar rotation. Nature (London) 1994;371:752. doi: 10.1038/371752b0. [DOI] [PubMed] [Google Scholar]
- 25.Manson M D, Tedesco P, Berg H C, Harold F M, van der Drift C. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci USA. 1977;74:3060–3064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minamino T, Macnab R M. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol. 1999;181:1388–1394. doi: 10.1128/jb.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell P. Chemiosmotic coupling and energy transduction. Theor Exp Biophys. 1969;2:159–216. [Google Scholar]
- 28.Nambu T, Minamino T, Macnab R M, Kutsukake K. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J Bacteriol. 1999;181:1555–1561. doi: 10.1128/jb.181.5.1555-1561.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plano G V, Barve S S, Straley S C. LcrD, a membrane-bound regulator of the Yersinia pestis low-calcium response. J Bacteriol. 1991;173:7293–7303. doi: 10.1128/jb.173.22.7293-7303.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramakrishnan G, Zhao J-L, Newton A. The cell cycle-regulated flagellar gene flbF of Caulobacter crescentus is homologous to a virulence locus (lcrD) of Yersinia pestis. J Bacteriol. 1991;173:7283–7292. doi: 10.1128/jb.173.22.7283-7292.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverman M, Simon M. Flagellar rotation and the mechanism of bacterial motility. Nature (London) 1974;249:73–74. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- 33.Stocker B A D, Zinder N D, Lederberg J. Transduction of flagellar characters in Salmonella. J Gen Microbiol. 1953;9:410–433. doi: 10.1099/00221287-9-3-410. [DOI] [PubMed] [Google Scholar]
- 34.Ueno T, Oosawa K, Aizawa S-I. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J Mol Biol. 1992;227:672–677. doi: 10.1016/0022-2836(92)90216-7. [DOI] [PubMed] [Google Scholar]
- 35.Van Gijsegem F, Genin S, Boucher C. Conservation of secretion pathways for pathogenicity determinants of plant and animal bacteria. Trends Microbiol. 1993;1:175–180. doi: 10.1016/0966-842x(93)90087-8. [DOI] [PubMed] [Google Scholar]
- 36.Vogler A P, Homma M, Irikura V M, Macnab R M. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J Bacteriol. 1991;173:3564–3572. doi: 10.1128/jb.173.11.3564-3572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J, Sharp L L, Tang H L, Lloyd S A, Billings S, Braun T F, Blair D F. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol. 1998;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]