Abstract
The ribulose monophosphate (RuMP) pathway is one of the metabolic pathways for the synthesis of compounds containing carbon-carbon bonds from one-carbon units and is found in many methane- and methanol-utilizing bacteria, which are known as methylotrophs. The characteristic enzymes of this pathway are 3-hexulose-6-phosphate synthase (HPS) and 6-phospho-3-hexuloisomerase (PHI), neither of which was thought to exist outside methylotrophs. However, the presumed yckG gene product (YckG) of Bacillus subtilis shows a primary structure similar to that of methylotroph HPS (F. Kunst et al., Nature 390:249–256, 1997). We have also investigated the sequence similarity between the yckF gene product (YckF) and methylotroph PHI (Y. Sakai, R. Mitsui, Y. Katayama, H. Yanase, and N. Kato, FEMS Microbiol. Lett. 176:125–130, 1999) and found that the yckG and yckF genes of B. subtilis express enzymatic activities of HPS and PHI, respectively. Both of these activities were concomitantly induced in B. subtilis by formaldehyde, with induction showing dependence on the yckH gene, but were not induced by methanol, formate, or methylamine. Disruption of either gene caused moderate sensitivity to formaldehyde, suggesting that these enzymes may act as a detoxification system for formaldehyde in B. subtilis. In conclusion, we found an active yckG (for HPS)-yckF (for PHI) gene structure (now named hxlA-hxlB) in a nonmethylotroph, B. subtilis, which inherently preserves the RuMP pathway.
The ribulose monophosphate (RuMP) pathway is involved in formaldehyde fixation in many methylotrophs (11, 15, 17, 22). Typically, the RuMP pathway has two characteristic enzymes, 3-hexulose-6-phosphate synthase (HPS) and 6-phospho-3-hexuloisomerase (PHI), and it also shares some enzymes with the pentose phosphate pathway and the glycolytic or Entner-Doudoroff pathway. PHI catalyzes isomerization between fructose 6-phosphate and hexulose 6-phosphate, which is synthesized from ribulose 5-phosphate (Ru5P) and formaldehyde by HPS. Although HPS and PHI have been detected in many methylotrophs (3, 12, 18–20, 23, 32, 34), the genes encoding these enzymes remain largely unknown. The hps gene, encoding HPS, was first cloned from an obligate methylotroph, Methylomonas aminofaciens 77a (44), and cloning of the gene (rmpB) encoding PHI from M. aminofaciens 77a (35) and a facultative methylotroph, Mycobacterium gastri MB19 (28), was recently reported. Neither enzyme was thought to exist outside methylotrophs, but sequence analysis of the Bacillus subtilis genome (24, 41) has suggested that yckG may be a homolog of the gene encoding HPS (44) in M. aminofaciens 77a. The yckG gene is located between yckF and yckH, at about 32° on the B. subtilis genome map (24) (Fig. 1a). Here, we present data showing that a nonmethylotroph, B. subtilis, contains genes (yckG and yckF) encoding two key enzymes of the RuMP pathway (HPS and PHI) and that the activities of both enzymes can be induced by formaldehyde with dependence on the yckH gene. These findings indicate the presence of characteristic enzymes from the RuMP pathway in a nonmethylotroph. The significance of detection of these enzymes in B. subtilis is discussed from the aspect of molecular evolution of the RuMP pathway.
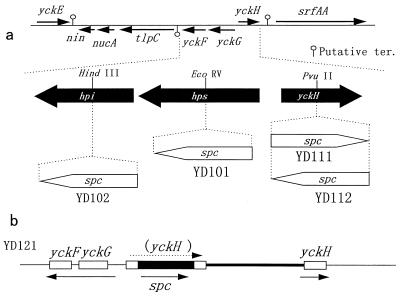
FIG. 1.
Diagram of the chromosomes of wild-type B. subtilis and its mutants. (a) Map of the yckF-yckG (phi-hps) and yckH loci showing the protein coding regions at 32° on the B. subtilis genome (24). Locations and orientations of the coding regions are indicated by thin black arrows, and potential transcription terminators, as proposed in the SubtiList data bank (41), are shown. The hps, phi, and yckH genes are expanded and depicted as thick black arrows below the map. Insertion of a spc cassette (thick white arrow) leading to disruption of hps in strain YD101, phi in YD102, or yckH in YD111 or YD112 is shown. (b) Chromosomal structure of strain YD121, which contains the disrupted and intact yckH genes. Heavy and thin lines depict vector and chromosomal DNA, respectively. Open boxes indicate the yckG, yckF, and yckH genes. The black box shows the spc gene, which disrupts the yckH gene. Thin arrows show the direction of transcription.
MATERIALS AND METHODS
Bacterial strains, oligonucleotide primers, and plasmids.
The wild-type strain B. subtilis 168 (1) was purchased from the American Type Culture Collection (Rockville, Md.), and Escherichia coli JM109 (45) was obtained from Takara Shuzo Co. (Kyoto, Japan). B. subtilis YD101 (yckG-deficient mutant) and YD102 (yckF-deficient mutant) were constructed, as described below, by inserting the spectinomycin resistance (spc) DNA fragment from pDG1726 (13) (Bacillus Genetic Stock Center, Columbus, Ohio) into the individual genes of the chromosome of strain 168. Strains YD111 (yckH::spc) and YD112 (yckH::spc) were also constructed from strain 168 in the same way (Fig. 1a). Strain YD121 (yckH yckH+) contained both the intact yckH gene and the defective one (Fig. 1b).
The potential open reading frame (ORF) of the yckG or yckF gene was cloned by PCR amplification (29) using the oligonucleotide primer pair BsYck-G1 (5′-GAGTATCGATAAAATGGAATTACAGCTTGCATTAGACCTCGT-3′; the translation start codon is underlined, and the restriction enzyme site [ClaI] for ligation to the vector is italicized) plus BsYck-G6 (5′-AATTGTGGATCCCATTGAGAATTTCCGCTACGTATTCAGTCG-3′; the restriction enzyme site [BamHI] for ligation to the vector is italicized) or BsYck-F1 (5′-AAGCATCGATAAAATGAAAACGACTGAATACGTAGCGGAA-3′) plus BsYck-F2 (5′-ATCTTGGATCCGGTTGTGTGATGTTATTCAAGGTTTGCG-3′). The region containing the yckG and yckF genes was amplified by PCR using the primer pair BsYck-G1 plus BsYck-F2. PCR was performed with Takara LA Taq DNA polymerase (Takara Shuzo) for 28 cycles with purified B. subtilis genomic DNA. The PCR products were purified with a QIAquick gel extraction kit (Qiagen). The fragments thus obtained were ligated behind the E. coli trp promoter to mediate overexpression in E. coli (46). Each amplified DNA fragment was digested with restriction enzymes ClaI and BamHI, and the fragments were introduced into the vector to construct the expression vectors pT-Bsb-yckG6, pT-Bsb-yckF1, and pT-Bsb-yckGF1 for the overexpression of yckG, yckF, and yckGF, respectively. The fragment containing yckH was amplified by PCR using primers BsYck-H1 (5′-CAATGTTAACGTCAGGCTTTTGCTGGATCACTTCTGGCA-3′; the restriction enzyme site for cloning of the amplified DNA fragment is italicized) and BsYck-H2 (5′-GTGTACCGAATTCGTTTTTGTGCATCCGTTAAAGGGTA-3′). After digestion with HpaI and EcoRI, the yckH fragment was cloned into pACYC184 (Nippon Gene, Tokyo, Japan) treated with Bst1107I and EcoRI to construct pAY-Bsb-yckH.
For the purpose of gene disruption, the spc fragment was introduced into the cloned yckG and yckF genes to construct pT-Bsb-yckG-Sp5 and pT-Bsb-yckF-Sp6, respectively. The spc gene was also cloned into the PvuII site in the yckH gene to construct two plasmids, pAY-Bsb-yckH-SpA and pAY-Bsb-yckH-SpB, in which the direction of the spc gene was the same as and opposite, respectively, that of the yckG gene (Fig. 1a).
Construction of B. subtilis mutants.
B. subtilis mutants YD101, YD102, YD111, and YD112 were constructed by the introduction of pT-Bsb-yckG-Sp5, pT-Bsb-yckF-Sp6, pAY-Bsb-yckH-SpA, and pAY-Bsb-yckH-SpB, respectively, into the wild-type strain. YD121 was generated from the wild-type strain through homologous recombination with the yckH region by a Campbell-type event. Transformation of B. subtilis was performed by the competent-cell method (1), and spectinomycin (100 μg/ml) was used for selection of the transformants.
Preparation of cell extract from E. coli overexpressing the yckG and yckF genes.
E. coli JM109 was transformed with the expression plasmids (36). Transformants were grown at 37°C for 12 h in 5 ml of M9-Casamino Acids medium (36) supplemented with thiamine-HCl (2 μg/ml), indoleacrylic acid (25 μl/ml), and ampicillin (100 μl/ml). The cells were harvested by centrifugation and suspended in buffer A (50 mM potassium phosphate [pH 7.6], 1 mM MgCl2). After centrifugation, the washed cell pellet was resuspended in 1 ml of buffer B (50 mM potassium phosphate [pH 7.6], 5 mM MgCl2, 1 mM dithiothreitol). A cell extract was prepared by sonication with a disrupter (Bioruptor; Cosmobio, Tokyo, Japan) at maximum power with a standard oscillator. The sonicate was clarified by centrifugation at 15,000 × g for 20 min at 2°C, and the supernatant thus obtained was used as the cell extract.
SDS-PAGE analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (14% polyacrylamide) was performed by the method of Laemmli (25), using a ready-made gel system (TEFCO, Tokyo, Japan). Samples of cultured cells were collected by centrifugation, and the cell pellets were suspended in gel loading buffer and heated at 100°C for 5 min. Aliquots of 5 μl were loaded onto an SDS-polyacrylamide gel, which was stained with PhastGel Blue R (Coomassie blue R-350; Pharmacia AB). Molecular weight standards consisted of a marker mix consisting of seven recombinant proteins of 15, 25, 35, 50, 75, 100, and 150 kD (Novagen, Madison, Wis.).
Induction of expression of the yckG and yckF genes in B. subtilis.
After overnight growth in Luria-Bertani (LB) medium (36), cultures of wild-type and mutant B. subtilis were diluted to 1:20 in fresh medium. Cells were cultured at 37°C with shaking until the mid-log phase, and then formaldehyde or another reagent (methanol, formate, or methylamine) was added as specified in the text. Other cultures were exposed to the following stresses: heat shock, provided by culture at 48°C; salt stress, created by adding NaCl at a final concentration of 5% (wt/vol); and acid shock, provided by adding 0.5 N HCl to reduce the pH of the culture from 6.8 to 5.2. Incubation was continued for 90 min under these conditions. Next, the cells were harvested, washed, and resuspended in 50 mM potassium phosphate buffer (pH 7.5) containing 3 mM MgCl2 and 1 mM dithiothreitol. The cell extract was prepared for assay by sonication and centrifugation to remove debris.
Assay of HPS and PHI activities.
HPS activity was determined as described previously (3), with some modifications. The reaction mixture (0.95 ml) consisted of 50 mM potassium phosphate (pH 7.6), 5 mM MgCl2, 5 mM ribose-5-phosphate (Ri5P) (Nacalai Tesque, Inc., Kyoto, Japan), 2.5 mM NADP (Sigma, St. Louis, Mo.), 10 U of phosphoribose isomerase (PRI) (Sigma), 10 U of glucose-6-phosphate dehydrogenase (Boehringer Mannheim), 10 U of phosphoglucose isomerase (PGI) (Boehringer Mannheim), 50 U of recombinant PHI purified from E. coli overproducing M. aminofaciens 77a PHI (35), and the test sample of cell extract. The mixture was preincubated for 3 min at 37°C to achieve equilibrium of Ru5P and Ri5P (catalyzed by PRI), and then the reaction was started by adding formaldehyde at a final concentration of 5 mM. In the PHI assay, recombinant M. aminofaciens 77a HPS (35) was used instead of recombinant PHI, and both were eliminated from the reaction mixture when the serial reaction of HPS and PHI enzymes was assayed. Activity was assessed by monitoring the change in optical density (OD) at 340 nm with NADP as a cofactor, and correction was made for formaldehyde-independent activity. The protein concentration was determined by the method of Bradford (8) with a Bio-Rad protein assay kit (Bio-Rad Laboratories, Richmond, Calif.) and bovine serum albumin as the standard.
Assay of other enzymes.
NAD-dependent methanol dehydrogenase (MDH) and glutathione-dependent formaldehyde dehydrogenase (GS-FDH) were assayed by using the B. subtilis cell extract treated with formaldehyde. MDH activity was assayed as described by Arfman et al. (2); GS-FDH activity was assayed as described by Attwood. (6).
Effect of yckG and yckF mutations on cell growth in the presence of formaldehyde or NaCl.
B. subtilis 168 (wild type) and two isogenic mutant strains (YD101 and YD102) were cultured at 30°C with shaking in LB medium containing spectinomycin when necessary. Overnight cultures were inoculated into warmed fresh LB medium, or into medium containing formaldehyde or NaCl, and cell growth and lysis profiles were automatically recorded at an OD of 660 nm with a Bio-photorecorder (Advantec, Tokyo, Japan).
13C NMR analysis of the products obtained with [13C]formaldehyde as the substrate.
A reaction mixture containing 50 mM potassium phosphate (pH 7.6), 5 mM MgCl2, 5 mM Ri5P, 10 U of PRI, and 10 U of PGI was prepared. After preincubation at 37°C for 10 min to produce Ru5P from Ri5P, cell extract containing the yckG gene product (YckG) and yckF gene product (YckF) was added to the mixture. The serial YckG and YckF reaction was started by addition of [13C]formaldehyde as the substrate. After incubation at 37°C for 20 min, EDTA was added to the mixture to chelate Mg2+, which is required for HPS activity. The total activity in the mixture was assayed with 13C nuclear magnetic resonance (NMR) spectroscopy by monitoring the decrease of the resonance signal for the 13C atom of formaldehyde and the appearance of signals for the C atoms of fructose 6-phosphate and glucose 6-phosphate, which was yielded by addition of PGI. In the negative control sample, EDTA was added to the mixture immediately after addition of the cell extract.
NMR spectra were obtained at 100 MHz with a Fourier transform (FT) NMR spectrometer (JNM-A400; JEOL, Tokyo, Japan), using 0.6-ml samples containing 15% D2O in 5-mm-diameter tubes to provide a lock signal. A total of 256 transients were acquired by using 32k data points. Gated proton decoupling was used, and 45° pulses were applied, with a 2.5-s relaxation delay between pulses to give a repetition time of 3.9 s. During acquisition, the samples were kept at 25°C. [13C]formaldehyde (99% 13C atom) was purchased from Cambridge Isotope Laboratories (Andover, Mass.).
RNA analysis.
B. subtilis cells were grown to mid-log phase in LB medium at 37°C. After the addition of formaldehyde at final concentration of 0.5 mM, culture was continued for 45 min. Then RNA was extracted by using an ISOGEN kit (Nippongene, Toyama, Japan) and treated with RNase-free DNase (Takara Shuzo). About 1 μg of RNA was mixed with the YGF-R2 oligonucleotide DNA (5′-TTCAGGCCGGTTGTGTGATGC-3′), and cDNA was synthesized by using a reverse transcription (RT) kit (RNA LA PCR kit; Takara Shuzo). YGF-R2 corresponded to the minus strand of DNA just behind the yckF ORF. Then a 20-μl aliquot of the cDNA mixture was combined with DNA primers YGF-F1 (5′-ATGGAATTACAGCTTGCATTAGACC-3′) and YGF-R1 (5′-TGTGATGCTATTCAAGGTTTGC-3′), LA Taq DNA polymerase, and PCR buffer (Takara Shuzo) in a volume of 100 μl. RT-PCR was carried out according to the manufacturer’s instructions. YGF-F1 and YGF-R1 corresponded to the plus strand of DNA encoding the N-terminal region of YckG and to the minus strand of DNA encoding the C-terminal region of YckF, respectively. As a negative control, mixture without RT was used for PCR amplification.
Computer analysis.
Nucleotide and amino acid sequences were analyzed with the Genetyx-Mac computer program (Software Development Co., Tokyo, Japan). Alignment of deduced amino acid sequences was performed by comparison with entries in the SWISS-PROT database (release 35.0).
RESULTS
Overproduction and enzyme activity of the yckG and yckF gene products.
To identify the orthologs of the gene encoding PHI, FASTA searches (30, 31) of the microorganism protein sequence database were performed, using the amino acid sequence of the rmpB gene product (PHI) (35) of M. aminofaciens 77a as the query sequence. As a result, the primary sequence of the yckF gene product (YckF) in B. subtilis showed the highest structural similarity with that of PHI (Fig. 2). The yckF gene is situated five nucleotides downstream of the yckG gene in the B. subtilis genome (Fig. 1a), and the yckG gene product (YckG) is known to show structural similarity with HPS (24). Thus, both putative genes involved in one-carbon compound metabolism were situated side-by-side in the B. subtilis genome. Because B. subtilis cannot utilize methanol as its sole carbon source (i.e., it is a nonmethylotroph), the detection of such genes in this microorganism was interesting with respect to function and evolution.
FIG. 2.
Alignment of amino acid sequences of YckF of B. subtilis (24) and PHI of M. aminofaciens 77a (35). Identical amino acids and conservative replacements are indicated by asterisks and dots, respectively. Amino acids are numbered from the N terminus of each protein.
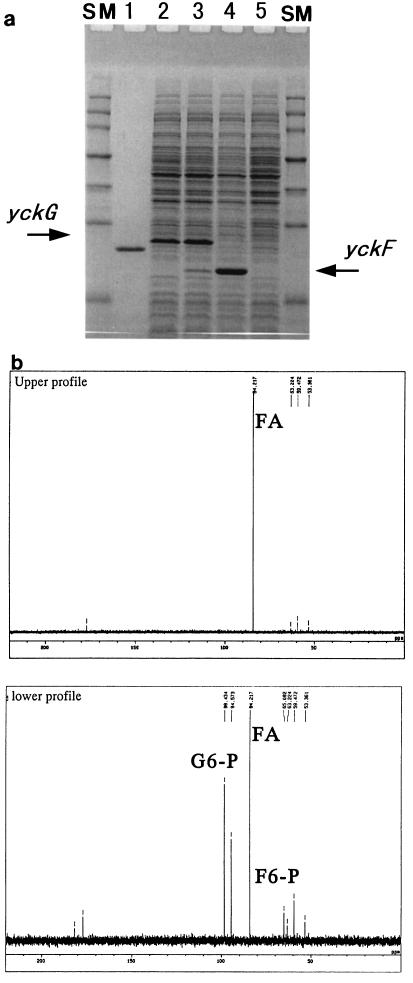
We first expressed the yckG- and/or yckF ORF in E. coli to evaluate whether the translated product of each ORF displayed HPS or PHI activity. After each corresponding DNA fragment with the postulated ORF was amplified by PCR, the fragments were inserted into the expression plasmid and overproduction was stimulated (Fig. 3a). As a result, the yckG gene yielded a dense protein band with the expected molecular mass of 23 kDa on SDS-PAGE, and the yckF gene yielded a protein with a molecular mass of 19 kDa. Furthermore, overexpression of a DNA fragment containing the yckG and yckF genes in the intact gene structure led to concomitant overproduction of both ORF products (Fig. 3a, lane 3), showing that termination of transcription did not occur between the yckG and yckF genes in E. coli.
FIG. 3.
Analysis of the products of yckG and yckF. (a) The yckG and/or yckF gene was overproduced in E. coli and analyzed by SDS-PAGE. Cells harboring the expression plasmid were cultured in M9 medium supplemented with Casamino Acids, thiamine, and metal ions. Indoleacrylic acid was added to induce the expression of each gene. Aliquots were taken from cells harboring pT-Bsb-yckG (lane 2), pT-Bsb-yckGF (lane 3), pT-Bsb-yckF (lane 4), and the control plasmid (lane 5). The single band in lane 1 indicates the position of purified HPS of M. aminofaciens 77a, and the positions of YckG and YckF are shown by arrows. The positions of migration of molecular mass standards (SM) are indicated on both sides of the gel (from top to bottom, 150, 100, 75, 50, 35, 25, and 15 kDa). (b) Formation of glucose-6-phosphate and fructose-6-phosphate by the condensation of 13C-labeled formaldehyde and Ru5P and the subsequent isomerization of sugar-phosphates was analyzed by 13C NMR spectrometry as described in the text. The upper profile exhibits the peak of formaldehyde (FA) in the test mixture, in which the reaction was immediately stopped after addition of cell extract containing YckG and YckF; the lower profile displays the carbon peaks in the reaction mixture after incubation for 20 min at 37°C. The C-1 (α,β anomers) positions of glucose-6-phosphate (G6-P) and fructose-6-phosphate (F6-P) are indicated.
The HPS and PHI activities of cell extracts were examined by the enzyme assay method. E. coli overexpressing yckG or yckF showed HPS or PHI activity, respectively (Table 1). To confirm that formaldehyde was incorporated into pentose phosphate, we used the 13C NMR technique with [13C]formaldehyde. Preincubation of the reaction mixture containing Ri5P, PRI and other chemicals yielded Ru5P, which is an acceptor of formaldehyde in the reaction with HPS. Addition of a cell extract containing YckG and YckF to the mixture caused the appearance of the corresponding resonance signals for [13C]fructose-6-phosphate and [13C]glucose-6-phosphate, along with a decrease of the [13C]formaldehyde signal (Fig. 3b). This showed that the serial reaction catalyzed by both gene products could progress, because YckG produced hexulose-6-phosphate from formaldehyde and Ru5P, while YckF and PGI then created fructose-6-phosphate and glucose-6-phosphate, respectively. Since the two enzymes were previously thought to be unique to methylotrophs (11, 15), it was surprising that B. subtilis contained nucleotide sequences that could express these enzyme activities.
TABLE 1.
HPS and PHI activity in E. coli bearing an expression plasmid
| Straina | Sp act (nmol of NADP reduced/min/mg of protein)b in cell extracts
|
||
|---|---|---|---|
| HPS | PHI | HPS + PHIc | |
| E. coli JM109/pT-Bsb-yckG6 | 3,160 | <1 | NDd |
| E. coli JM109/pT-Bsb-yckF1 | <1 | 25,900 | ND |
| E. coli JM109/pT-Bsb-yckGF1 | ND | ND | 3,970 |
| E. coli JM109/pT-Bs-ΔNcoe | <1 | <1 | ND |
Cells were grown in modified M9 Casamino Acids medium with indoleacrylic acid.
Mean value from three experiments.
Total activity for serial reaction of HPS and PHI.
ND, not determined.
Negative control.
Expression of yckG and yckF genes by B. subtilis.
To investigate whether the yckG and yckF genes intrinsically expressed any enzyme activity in B. subtilis, we assessed extracts of cells grown under various culture conditions. The extract of wild-type cells cultured in LB medium showed negligible HPS or PHI activity, but both activities were detected after culture in the presence of formaldehyde (Table 2). When various concentrations of formaldehyde were tested, significant HPS and PHI activity was found in the cell extract at formaldehyde concentrations of around 0.5 mM. At a concentration of over 1 mM, however, both activities almost disappeared.
TABLE 2.
HPS and PHI activities in B. subtilis 168 and deletion mutants under different growth conditions
| Strain | Growth conditionsa | Sp act (nmol of NADPH produced/ min/mg of protein)b in cell extracts
|
||
|---|---|---|---|---|
| HPS | PHI | HPS + PHI | ||
| 168 wild type | LB | <1 | <1 | NDc |
| LB + 0.5 mM FA | 690 | ND | 310 | |
| LB + 1.0 mM FA | 95 | ND | <1 | |
| LB + 2.0 mM FA | <1 | ND | <1 | |
| LB + 2% (vol/vol) MeOH | <1 | ND | <1 | |
| LB + 0.5 mM FM | <1 | ND | <1 | |
| YD101 (yckG) | LB + 0.5 mM FA | <1 | <1 | ND |
| YD102 (yckF) | LB + 0.5 mM FA | 290 | <1 | ND |
| YD111 (yckH) | LB + 0.5 mM FA | <1 | <1 | <1 |
| YD112 (yckH) | LB + 0.5 mM FA | <1 | <1 | <1 |
| YD121 (yckH yckH+) | LB + 0.5 mM FA | 1,050 | ND | 200 |
FA, formaldehyde; MeOH, methanol; FM, formate.
Mean value from three experiments.
ND, not determined.
Formaldehyde is produced from methanol by methanol dehydrogenase and is also oxidized to formate by formaldehyde dehydrogenase in some methylotrophs (11). Therefore, we examined whether the synthesis of HPS and PHI could be induced by methanol or formate, but neither enzyme activity was detected after cells were grown in medium containing methanol or formate (Table 2). On the other hand, since formaldehyde is also synthesized by oxidation of methylamine in several methylotroph species (26), we looked for HPS or PHI activity in extracts of B. subtilis grown in medium containing methylamine hydrochloride (0.4 or 1.2% [wt/vol], final concentration) but did not observe the induction of these enzymes. Therefore, the nonmethylotroph B. subtilis possessed two key enzymes of the RuMP pathway, and the expression of both enzymes was induced by formaldehyde.
Analysis of yckG and/or yckF disruption mutants.
We next examined whether the yckG or yckF gene encoded the observed formaldehyde-inducible HPS or PHI activity. We constructed a yckG mutant (strain YD101) and a yckF mutant (strain YD102) by insertional mutagenesis (Fig. 1a) and analyzed the formaldehyde-inducible HPS or PHI activity of each strain. After induction with formaldehyde, we detected no significant enzyme activity in the YD101 strain and only HPS activity in the YD102 strain (Table 2). Thus, we identified YckG and YckF as the only formaldehyde-inducible HPS and PHI enzymes, respectively, and our results also suggested that a polar effect occurred between the yckG and yckF genes. To confirm the transcription unit of the two genes, total RNA was isolated from wild-type B. subtilis cells exposed to formaldehyde and analyzed by RT-PCR. The presence of mRNA species (of at least 1.2 kb) spanning the yckG ORF to the yckF ORF (data not shown) was revealed, suggesting that the two genes (yckG and yckF) were expressed as a polycistronic mRNA from an operon.
We also investigated the effect of formaldehyde on the phenotype of these mutants. In LB medium, the wild-type strain and the two mutant strains all showed typical growth and lysis curves (Fig. 4a). However, after addition of formaldehyde to the medium, the mutants showed a prolonged lag phase relative to the growth profile of the wild-type strain, which was dependent on the concentration of formaldehyde added (Fig. 4). These results indicated that the yckG-yckF system might endow B. subtilis with the ability to detoxify formaldehyde produced via endogenous metabolism or environmental changes.
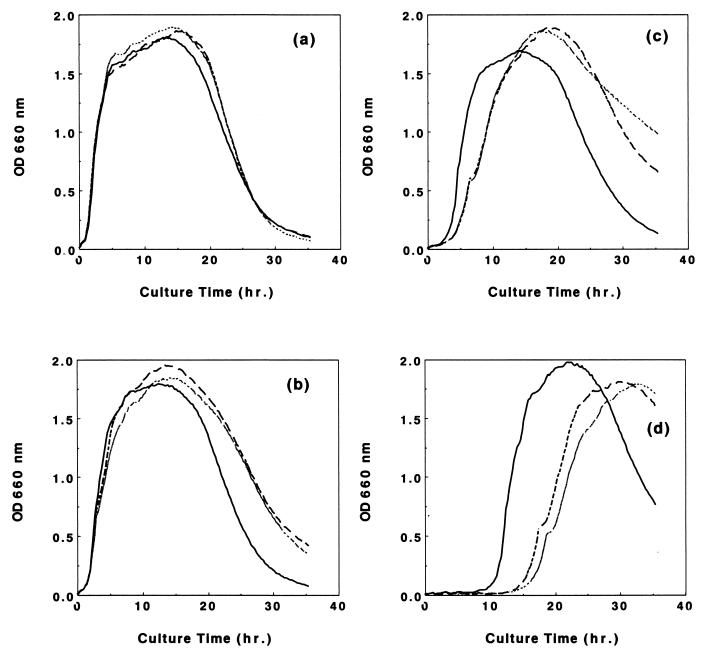
FIG. 4.
Effect of formaldehyde on the growth of B. subtilis. Overnight cultures of wild-type strain 168 (▄), YD101 (yckG) (•••••), and YD102 (yckF) (–––) in LB medium were inoculated into prewarmed LB medium containing formaldehyde at concentrations of 0.0 (a), 0.3 (b), 0.6 (c), and 0.9 (d) mM. Growth and lysis of cells at 37°C were monitored automatically at an OD of 660 nm.
It appeared that the cells were under stress when exposed to formaldehyde, since growth was arrested for a few hours in its presence (Fig. 4c and d). To examine whether the increase of HPS and PHI activities after exposure to formaldehyde was due to a general stress response rather than to formaldehyde itself, we assessed the levels of these enzymes in cells grown under heat (increase from 30 to 48°C), acid (decrease from pH 6.8 to 5.2), and salt (addition of 5% NaCl) stress. We also examined the growth profiles of wild-type and mutant cells under salt stress. Enzyme activities were never detected except after exposure to formaldehyde, but comparison of the growth profiles of cells exposed to salt stress (5% NaCl) revealed a slight delay in the growth of both mutants relative to the wild-type strain. This observation suggests that the yckG or yckF gene may be involved also in the salt stress response, although HPS or PHI activity was below the level of detection, as described above.
Effect of the yckH gene on expression of the yckG and yckF genes.
Upstream of the yckG-yckF genes, the yckH gene is an ORF transcribed in the reverse direction from the operon (Fig. 1a). This gene arrangement led us to speculate that yckH might be a regulator of yckG-yckF expression, by analogy with the l-arabinose (ara) operon structure (araC and araBAD) (39) in E. coli. It has been reported that yckH encodes a protein of 120 amino acids (41), but its function remains unclear. Although the overall amino acid sequence does not share any significant homology with DNA-binding proteins, a helix-turn-helix motif that is characteristic of nucleic acid-binding proteins (16) was indicated at the carboxyl terminus by our prediction of the secondary structure (data not shown). To examine the effect of the yckH gene on expression of yckG-yckF, the yckH locus was inactivated by insertion of an spc gene. We constructed two mutant strains, YD111 and YD112, in which the directions of the spc gene were the same as and opposite, respectively, that of to the yckH gene (Fig. 1a). Although read-through of transcription of the spc gene could proceed into the yckG-yckF region of the chromosome in YD112, neither of the yckH-disrupted mutants (strains YD111 and YD112) could express detectable HPS and PHI activity when exposed to formaldehyde (Table 2). We thought that in these mutants, the promoter element for the yckG-yckF genes might have been destroyed by spc gene insertion, since the location of the promoter region was not identified. Therefore, we constructed a new strain, YD121 (yckH yckH+) (Fig. 1b), in which the DNA region just adjacent to the yckG-yckF locus remained disrupted as in YD111 but the intact yckH gene was located away from the yckG-yckF region past the intervening sequence of the integrated plasmid. We examined YD121 for formaldehyde-induced HPS or PHI activity and again detected the induction of both activities. This indicated that the yckH gene is required for the expression of HPS and PHI activities.
DISCUSSION
Recently, bioinformatics analysis has suggested that hps gene homologs may be widespread in bacteria (33). We also found a structural similarity between PHI from a methylotroph (35) and YckF from B. subtilis and noticed that the hps homolog (yckG) and phi homolog (yckF) were adjacent in the B. subtilis genome. In E. coli, it was reported that the SgaH (ORF o216) and SgbH (ORF o220) proteins exhibit sequence similarity to the HPS of M. aminofaciens (33). Therefore, we assessed the enzymatic activity of the sgaH and sgbH gene products from strain W3110 by overexpression of the corresponding genes but could detect no HPS activity. We also examined HPS activity in extracts of E. coli cells cultured under several stresses, including formaldehyde exposure, but the activity was below the level of detection (data not shown). Although HPS activity in E. coli might be induced by a specific effector, it appears from our results that E. coli does not possess the enzyme. Our homology search indicated that the E. coli genome does not contain a homolog of the rmpB (phi) gene (data not shown). These findings suggested that the E. coli hps homologs may possess a function other than hexulose phosphate synthesis or that during its evolution the bacterium has accumulated mutations which impede the expression of HPS activity. According to the evolutionary history of prokaryotes based on phylogenetic analysis (43), B. subtilis is more proximal than E. coli to the branch point of the three kingdoms, Archaea, Bacteria, and Eucarya (7). Therefore, it appears that the presence of a set of hps and phi genes in the genome is critical, and the existence of both genes in the B. subtilis genome suggests that the gene products were of significant value for this microorganism in its original environment.
Expression of the yckG and yckF genes was induced in B. subtilis by formaldehyde, similar to what has been found for two gram-positive methylotrophs, Arthrobacter P1 (27) and Bacillus methanolicus (4). Formaldehyde is produced from methanol by MDH in the first stage of the utilization of carbon and energy sources by methylotrophs. In B. methanolicus, the synthesis of HPS is also enhanced by culture with methanol (2), but methanol could not induce HPS or PHI activity in B. subtilis. Having found that HPS and PHI enzymes exist in B. subtilis, we examined the activity of MDH in this microbe. The structure of the cytoplasmic NAD-dependent MDH of B. methanolicus C1 was described by De Vries et al. (9). Using their sequence information, we performed a homology search for the mdh gene in the B. subtilis genome database and found no significant homologs. In addition, we could not detect NAD-dependent MDH activity in cell extracts of B. subtilis cultured under various conditions (data not shown). Thus, it appears that B. subtilis does not possess an NAD-dependent MDH homolog derived from B. methanolicus. However, it is interesting that the possible yeaC gene product of B. subtilis (24, 41) displays similarity to the product of mxaR (moxR) required for activity of the pyrrolo-quinoline quinone (PQQ)-linked MDH in a gram-negative methylotroph (42). Besides the methanol oxidation pathway, formaldehyde is also synthesized from methylamine in several species of methylotrophs which are able to utilize methylamine as the sole source of carbon and energy (21, 26, 27). The yckG-yckF system might be involved in methylamine metabolism, since some heterotrophs are able to use methylamine as a nitrogen source, and methylamine, which is more stable than formaldehyde, is widely distributed in marine environment (21). However, no detectable HPS or PHI activity was found in the extract of B. subtilis cells cultured with methylamine, and methylamine could not support the growth of B. subtilis as a nitrogen source (data not shown).
This study also indicated a possible role of YckG and YckF in the detoxification of formaldehyde. As noted by Attwood and Quayle (5), it seems possible that the HPS and PHI system is very efficient for trapping free formaldehyde. However, B. subtilis may also equip other detoxification systems for formaldehyde, since our yckG or yckF mutants did not show marked formaldehyde sensitivity. In E. coli, GS-FDH is the primary enzyme that detoxifies formaldehyde (14). Therefore, it is possible that B. subtilis also possesses a reliable detoxification system employing the corresponding enzyme. Indeed, our homology search showed that the adhB gene product in B. subtilis is similar to GS-FDH in Methylobacter marinus (40), although we detected no of GS-FDH activity in cell extracts from B. subtilis exposed to formaldehyde, as was the case for the methanotroph. In addition, we found that both mutants (yckG and yckF) were slightly more sensitive to salt (NaCl) stress than the wild-type strain, although the HPS or PHI activity in cells stressed by salt was too low to be measured. This implies that the promoter responsible for the salt stress response might exist in front of both genes or that salt stress could induce the accumulation of formaldehyde during metabolism. The relationship between the salt stress response and the function of yckG and yckF remains to be determined.
The results presented here demonstrate that a nonmethylotroph, B. subtilis, can synthesize two key enzymes (HPS and PHI) that were previously believed to be specific to methylotrophs employing the RuMP pathway. In B. subtilis, the pentose phosphate pathway has been identified by metabolic flux analysis (37, 38). Therefore, we suggest that B. subtilis preserves the RuMP pathway. Although De Wulf assumed the presence of this pathway in B. subtilis on the basis of a preliminary physiological experiment (10), his conclusion that formic acid in the culture medium was effectively utilized by B. subtilis via the RuMP pathway is not compatible with our results, because formic acid could not induce HPS and PHI activities in the present study. This discrepancy might be related to differences in the B. subtilis strains used. Either way, the existence of two key enzymes was substantially established by our research.
The organization of divergent transcription between the two transcription terminators in the B. subtilis genome suggested that yckH may be a regulator gene (24). The putative gene product of yckH is estimated to be a polypeptide composed of 120 amino acids (41). However, our disruption experiment indicated that the yckH gene is required for the expression of HPS and PHI activities, and it appears that yckH (now named hxlR) may positively regulate yckG-yckF gene expression through DNA binding either directly or indirectly after stimulation by formaldehyde. We also suggest that yckG and yckF (now named hxlA and hxlB, respectively) are organized into an operon structure, based on the following data: (i) the existence of mRNA species covering the hxlA and hxlB ORFs, (ii) the polar effect between hxlA and hxlB genes, (iii) the simultaneous expression of the two genes in response to an inducer, (iv) the lack of hxlB expression concomitant with abolition of hxlA expression in an hxlR (yckH)-deficient mutant, and (v) the gene organization of hxlA-hxlB followed by a typical transcriptional terminator. Recently, it was shown that M. gastri MB19 contains a phi (rmpB)-hps (rmpA) operon (28), although the hps (rmpA) and phi (rmpB) genes of M. aminofaciens 77a are separated by an insertion element (35). We found that the hps (hxlA)-phi (hxlB) gene organization was conserved as a set of both genes even in a nonmethylotroph. Curiously, the genes of B. subtilis and M. gastri were arranged in reverse order. Therefore, more detailed bioinformatic analysis of the conserved features and differences in gene organization among three types of microbes (facultative methylotrophs, obligate methylotrophs, and nonmethylotrophs) may provide insight into the evolution of the RuMP pathway.
ACKNOWLEDGMENTS
We gratefully acknowledge N. Kato, Y. Sakai, and R. Mitsui (Kyoto University) for the generous gift of the enzyme samples (HPS and PHI from M. aminofaciens 77a) and for helpful discussion; E. Suzuki and N. Ootu for NMR analysis; and K. Sato, M. Ooba, and K. Kobayashi for technical support.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirement for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arfman N, Watling E M, Clement W, van Oosterwijk R J, de Vries G E, Harder W, Attwood M M, Dijkhuizen L. Methanol metabolism in thermotolerant methylotrophic Bacillus strains involving a novel catabolic NAD-dependent methanol dehydrogenase as a key enzyme. Arch Microbiol. 1989;152:280–288. doi: 10.1007/BF00409664. [DOI] [PubMed] [Google Scholar]
- 3.Arfman N, Bystrykh L, Govorukhina N I, Dijkhuizen L. 3-Hexulose-6-phosphate synthase from thermotolerant methylotrophic Bacillus C1. Methods Enzymol. 1990;188:391–397. doi: 10.1016/0076-6879(90)88062-f. [DOI] [PubMed] [Google Scholar]
- 4.Arfman N, de Vries K J, Moezelaar H R, Attwood M M, Robinson G K, van Geel M, Dijkhuizen L. Environmental regulation of alcohol metabolism in thermotolerant methylotrophic Bacillus strains. Arch Microbiol. 1992;157:272–278. doi: 10.1007/BF00245161. [DOI] [PubMed] [Google Scholar]
- 5.Attwood M M, Quayle J R. Formaldehyde as a central intermediary metabolite of methylotrophic metabolism. In: Crawford R L, Hanson R S, editors. Proceedings of the 4th International Symposium on Microbial Growth on C1 Compounds. Washington, D.C.: American Society for Microbiology; 1984. pp. 315–323. [Google Scholar]
- 6.Attwood M M. Formaldehyde dehydrogenases from methylotrophs. Methods Enzymol. 1990;188:314–327. [Google Scholar]
- 7.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–251. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.De Vries G E, Arfman N, Terpstra P, Dijkhuizen L. Cloning, expression, and sequence analysis of the Bacillus methanolicus C1 methanol dehydrogenase gene. J Bacteriol. 1992;174:5346–5353. doi: 10.1128/jb.174.16.5346-5353.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Wulf P. Presence of the ribulose monophosphate pathway in Bacillus subtilis. Microbiology. 1998;144:596–597. doi: 10.1099/00221287-144-3-596. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 11.Dijkhuizen L, Levering P R, de Vries G E. The physiology and biochemistry of aerobic methanol-utilizing Gram-negative and Gram-positive bacteria. In: Murrell J C, Dalton H, editors. Methane and methanol utilizers. New York, N.Y: Plenum Press; 1992. pp. 149–181. [Google Scholar]
- 12.Ferenci T, Strøm T, Quayle J R. Purification and properties of 3-hexulose phosphate synthase and phospho-3-hexuloisomerase from Methylococcus capsulatus. Biochem J. 1974;144:477–486. doi: 10.1042/bj1440477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guérout-Fleury A-M, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 14.Gutheil W G, Holmquist B, Vallee B L. Purification, characterization, and partial sequence of the glutathione-dependent formaldehyde dehydrogenase from Escherichia coli: a class III alcohol dehydrogenase. Biochemistry. 1992;31:475–481. doi: 10.1021/bi00117a025. [DOI] [PubMed] [Google Scholar]
- 15.Hanson R S, Hanson T E. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honikoff S, Haughn G W, Calvo J M, Wallance J C. A large family of bacterial activator proteins. Proc Natl Acad Sci USA. 1988;85:6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson P A, Quayle J R. Microbial growth on C1 compounds. Synthesis of cell constituents by methane- and methanol-grown Pseudomonas methanica. Biochem J. 1965;95:859–867. doi: 10.1042/bj0950859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato N, Ohashi H, Hori T, Tani Y, Ogata K. Properties of 3-hexulose phosphate synthase and phospho-3-hexuloisomerase of a methanol-utilizing bacterium, 77a. Agric Biol Chem. 1977;41:1133–1140. [Google Scholar]
- 19.Kato N, Ohashi H, Tani Y, Ogata K. 3-Hexulosephosphate synthase from Methylomonas aminofaciens 77a: purification, properties and kinetics. Biochim Biophys Acta. 1978;523:236–244. doi: 10.1016/0005-2744(78)90026-8. [DOI] [PubMed] [Google Scholar]
- 20.Kato N, Miyamoto N, Shimao M, Sakazawa C. 3-Hexulose phosphate synthase from a new facultative methylotroph, Mycobacterium gastri MB19. Agric Biol Chem. 1988;52:2659–2661. [Google Scholar]
- 21.Kelly D P, Malin G, Wood A P. Microbial transformations and biogeochemical cycling of one-carbon substrates containing sulphur, nitrogen or halogens. In: Murrell J C, Kelly D P, editors. Microbial growth on C1 compounds. Andover, Mass: Intercept Ltd.; 1993. pp. 47–64. [Google Scholar]
- 22.Kemp M B, Quayle J R. Microbial growth on C1 compounds. Uptake of (14C)formaldehyde and (14C)formate by methane-grown Pseudomonas methanica and determination of the hexose labelling pattern after brief incubation with (14C)methanol. Biochem J. 1967;102:94–102. doi: 10.1042/bj1020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemp M B. Hexulose phosphate synthase from Methylococcus capsulatus makes d-arabino-3-hexulose phosphate. Biochem J. 1974;139:129–134. doi: 10.1042/bj1390129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunst F, Ogasawara N, Moszer I, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Large P, Green J. Oxidation of mono-, di-, and trimethylamine by methanotrophic yeasts: properties of the microsomal and peroxisomal enzymes involved and comparison with bacterial enzyme systems. In: Crawford R L, Hanson R S, editors. Microbial growth on C1 compounds. Washington, D.C.: American Society for Microbiology; 1984. pp. 155–164. [Google Scholar]
- 27.Levering P R, Croes L M, Dijkhuizen L. Regulation of methylamine and formaldehyde metabolism in Arthrobacter P1. Formaldehyde is the inducing signal for the synthesis of the RuMP cycle enzyme hexulose phosphate synthase. Arch Microbiol. 1985;144:272–278. [Google Scholar]
- 28.Mitsui R. Organization and regulation of the genes involved in the ribulose monophosphate pathway in methylotrophic bacteria. Ph.D. thesis. Kyoto, Japan: Kyoto University; 1998. [Google Scholar]
- 29.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 30.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Person W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 32.Quayle J R. 3-Hexulose-6-phosphate synthase from Methylomonas (Methylococcus) capsulatus. Methods Enzymol. 1982;90:314–319. doi: 10.1016/s0076-6879(82)90147-1. [DOI] [PubMed] [Google Scholar]
- 33.Reizer J, Reizer A, Saier M H., Jr Is the ribulose monophosphate pathway widely distributed in bacteria? Microbiology. 1997;143:2519–2520. doi: 10.1099/00221287-143-8-2519. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 34.Sahm H, Schütte H, Kula M-R. 3-Hexulose-phosphate synthase from Methylomonas M15. Methods Enzymol. 1982;90:319–323. doi: 10.1016/s0076-6879(82)90148-3. [DOI] [PubMed] [Google Scholar]
- 35.Sakai Y, Mitsui R, Katayama Y, Yanase H, Kato N. Organization of the genes involved in the ribulose monophosphate pathway in an obligate methylotrophic bacterium, Methylomonas aminofaciens 77a. FEMS Microbiol Lett. 1999;176:125–130. doi: 10.1111/j.1574-6968.1999.tb13652.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Sauer U, Hatzimanikatis V, Hohmann H-P, Manneberg M, van Loon A P G M, Bailey J E. Physiology and metabolic fluxes of wild-type and riboflavin-producing Bacillus subtilis. Appl Environ Microbiol. 1996;62:3687–3696. doi: 10.1128/aem.62.10.3687-3696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauer U, Hatzimanikatis V, Bailey J E, Hochuli M, Szyperski T, Wüthrich K. Metabolic fluxes in riboflabin-producing Bacillus subtilis. Nature Biotechnol. 1997;15:448–452. doi: 10.1038/nbt0597-448. [DOI] [PubMed] [Google Scholar]
- 39.Schleif R. Regulation of the l-arabinose catabolic operon araBAD. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 643–665. [Google Scholar]
- 40.Speer B S, Chistoserdova L, Lidstrom M E. Sequence of the gene for a NAD(P)-dependent formaldehyde dehydrogenase (class III alcohol dehydrogenase) from a marine methanotroph Methylobacter marinus A45. FEMS Microbiol Lett. 1994;121:349–356. doi: 10.1111/j.1574-6968.1994.tb07125.x. [DOI] [PubMed] [Google Scholar]
- 41.SubtiList . Data release R14.2. In: Mozer I, Danchin A, editors. http://www.pasteur.fr./Bio/SubtiList.html.[Online.] Paris, France: Institut Pasteur; 1997. [Google Scholar]
- 42.Van Spanning R J M, Wansell C W, De Boer T, Hazelaar M J, Anazawa H, Harms N, Oltmann L F, Stouthamer A H. Isolation and characterization of the moxJ, moxG, moxI, and moxR genes of Paracoccus denitrificans: inactivation of moxJ, moxG, and moxR and the resultant effect on methylotrophic growth. J Bacteriol. 1991;173:6948–6961. doi: 10.1128/jb.173.21.6948-6961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yanase H, Ikeyama K, Mitsui R, Ra S, Kita K, Sakai Y, Kato N. Cloning and sequence analysis of the gene encoding 3-hexulose-6-phosphate synthase from the methylotrophic bacterium, Methylomonas aminofaciens 77a, and its expression in Escherichia coli. FEMS Microbiol Lett. 1996;135:201–205. doi: 10.1111/j.1574-6968.1996.tb07990.x. [DOI] [PubMed] [Google Scholar]
- 45.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 46.Yasueda H, Nakanishi K, Kumazawa Y, Nagase K, Motoki M, Matsui H. Tissue-type transglutaminase from red sea bream (Pagrus major): sequence analysis of the cDNA and functional expression in Escherichia coli. Eur J Biochem. 1995;232:411–419. doi: 10.1111/j.1432-1033.1995.tb20826.x. [DOI] [PubMed] [Google Scholar]