ABSTRACT
Streptomyces bacteria have been studied for more than 80 years thanks to their ability to produce an incredible array of antibiotics and other specialized metabolites and their unusual fungal-like development. Their antibiotic production capabilities have ensured continual interest from both academic and industrial sectors, while their developmental life cycle has provided investigators with unique opportunities to address fundamental questions relating to bacterial multicellular growth. Much of our understanding of the biology and metabolism of these fascinating bacteria, and many of the tools we use to manipulate these organisms, have stemmed from investigations using the model species Streptomyces coelicolor and Streptomyces venezuelae. Here, we explore the pioneering work in S. coelicolor that established foundational genetic principles relating to specialized metabolism and development, alongside the genomic and cell biology developments that led to the emergence of S. venezuelae as a new model system. We highlight key discoveries that have stemmed from studies of these two systems and discuss opportunities for future investigations that leverage the power and understanding provided by S. coelicolor and S. venezuelae.
KEYWORDS: Streptomyces, Streptomyces coelicolor, Streptomyces venezuelae, differentiation, multicellular development, specialized metabolites, model species, antibiotic, regulation
HISTORY OF STREPTOMYCES
Streptomyces bacteria are estimated to have been inhabiting the Earth for ~400 million years (1). It has, however, only been within the last century that these remarkable organisms have been getting the attention they richly deserve. In the Journal of Bacteriology in 1943, the Streptomyces designation was bestowed by Selman Waksman and Arthur Henrici to describe these aerobic, filamentous, and spore-forming bacteria. This name derives from the Latin for “twisted” (Strepto) “fungus” (myces), but, despite the fungal reference, these microbes and their relatives have been recognized as bacteria since at least the 1940s (2). There are currently >1,100 named Streptomyces species (lpsn.dsmz.de), making it one of the largest bacterial genera.
Streptomyces bacteria are abundant in the environment and can be readily cultured. They are best known as soil bacteria; however, they have also been found in marine sediments (3) and freshwater ecosystems (4). While the vast majority of Streptomyces species studied to date are free living, these microbes are increasingly being found in association with other organisms, living as symbionts with insects and sponges and existing as plant endophytes. Notably, there are only a few pathogenic Streptomyces species that have been identified, which infect specific but diverse hosts spanning plants (e.g., Streptomyces scabies [5]) to humans (e.g., Streptomyces somaliensis [6]).
Beyond their ubiquitous distribution in the environment, there are a multitude of features that have made Streptomyces fascinating organisms to study. These include atypical genomic characteristics, an unusual fungal-like growth mode, and impressive metabolic capabilities, including the ability to produce diverse antibiotics and other bioactive natural products.
As befits many free-living environmental bacteria, Streptomyces species possess large chromosomes, ranging in size from ~6 Mbp to ~13 Mbp (7, 8) and averaging 8 to 9 Mbp. In addition to their notable size, Streptomyces chromosomes also have a highly skewed genomic composition, with greater than 70% of residues being G/C. Unusually for bacteria, they have linear, not circular, chromosomes (9). This raises unique challenges for chromosome replication, particularly in duplicating the chromosome ends (10, 11). The chromosome itself is intriguingly organized; most of the genes essential for viability are centered around the origin of replication within the “core” region of the chromosome (12). Flanking this core region are the chromosome “arms,” within which many species-specific genes are located, including the majority of natural product biosynthetic gene clusters. The distinctive genomic characteristics of the streptomycetes also extend to their transcriptional units, where greater than 20% of genes are transcribed as leaderless messages (i.e., lacking a conventional ribosome binding site) (13).
Within the 5,000 to 10,000 proteins encoded by any given Streptomyces species are an extraordinary number of regulatory factors. For example, the Streptomyces coelicolor chromosome encodes 66 sigma factors, the majority of which fall into the compact “extracytoplasmic function” (ECF) family of transcription initiators. In comparison, Bacillus subtilis encodes 7 ECF sigma factors (and 18 sigma factors in total), while Escherichia coli has 2 ECF sigma factors (and 7 in total). S. coelicolor is also a rich source of transcription factors and encodes an abundance of two-component regulatory systems, including 69 kinase/regulator pairs, plus additional orphaned (unpaired) kinases and response regulators (14), 34 serine/threonine kinases (15), and many so-called one-component regulators, including 153 TetR-family regulators alone (16). This strong emphasis on regulation is likely due to many factors, including the need to rapidly sense and respond to dynamic environmental conditions, the sheer number of genes that require control in these organisms, and the complex developmental and metabolic programs that need to be coordinated.
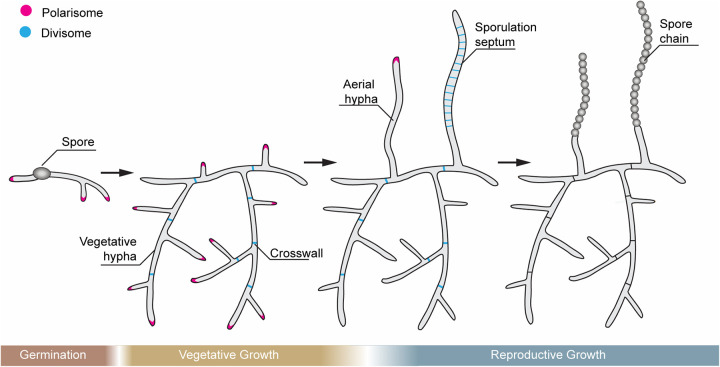
Streptomyces species have a multitude of strategies that enable them to survive—and thrive—in their environmental niches. Their classical life cycle (Fig. 1), which closely resembles that of the filamentous fungi and appears to have arisen by convergent evolution (17), is defined by a period of filamentous hyphal growth, followed by reproductive sporulation. Hyphal growth occurs at the tips of the filamentous cells where it is directed by a biosynthetic complex known as the polarisome (Fig. 1) (18). Hyphal branching is also frequently observed in vegetative cells. By contrast, cell division leading to crosswall formation within the hyphae (where a septum is laid down to subdivide hyphal compartments but where cell separation does not occur) is far less common, and, consequently, the resulting vegetative hyphal compartments contain multiple chromosomes. Reproductive growth initiates with the emergence of spatially and physically distinct nonbranching hyphae that extend into the air and are encased within a coat of hydrophobic proteins (19–22). Within these aerial filaments, the chromosomes are segregated at regular intervals, and the hyphae undergo a synchronous round of cell division to generate chains of unigenomic prespore compartments. These cellular compartments ultimately mature to form dormant spores that effectively resist many environmental stresses.
FIG 1.
Classic Streptomyces life cycle. Spore germination involves swelling and the emergence of one or two germ tubes that grow by a combination of tip growth and branching into a dense vegetative mycelial network that penetrates the surrounding growth substrate. During vegetative growth, cell division occurs occasionally, leading to the synthesis of crosswalls that segment the growing hyphae into multigenomic compartments. To reproduce, streptomycetes raise aerial (reproductive) hyphae that escape the vegetative colony surface and grow into the air. The aerial hyphae undergo a synchronous cell division event leading to the synthesis of dozens of unigenomic prespore compartments. These further mature to give thick-walled, pigmented spores, which are then released into the environment to restart the life cycle. Two major biosynthetic protein complexes are highlighted: The polarisome (magenta), which drives the polar tip growth of vegetative and aerial hyphae, and the divisome (blue), which leads to crosswalls and sporulation septa.
Streptomyces spp. lack conventional motility organelles (e.g., flagella, pili, and focal adhesion complexes), and to move within their environment they have historically relied on the release and dispersal of individual spores via water, wind, or other organisms. A fascinating strategy to encourage dispersal involves the coupling of sporulation with the release of the earthy-scented volatile compound geosmin (and its counterpart 2-methylisoborneol). Geosmin is a potent attractant for arthropods, with these animal vectors serving as effective spore dispersal agents (23).
Geosmin is one of the few specialized metabolites that is produced by virtually all streptomycetes. Indeed, most streptomycetes have unique metabolic repertoires, with any one species encoding the genes specific for the production of 20 to 50 distinct bioactive natural products. These molecules are collectively known as secondary or specialized metabolites, and many of these have been coopted for use in medicine and agriculture thanks to their antimicrobial, antifungal, antiparasitic, anticancer, and immunosuppressive properties. As is the case with geosmin, the production of many of these compounds coincides with the onset of reproductive growth, although their production is usually spatially segregated from the sporulating compartments (24).
MODEL SYSTEMS: STREPTOMYCES COELICOLOR AND STREPTOMYCES VENEZUELAE
Initial genetic and molecular studies in the streptomycetes centered on multiple genera, including Streptomyces avermitilis (producer of avermectin, a molecule that led to a Nobel Prize for Satoshi Ōmura) and Streptomyces griseus (led by Sueharu Horinouchi). While beautiful work has continued in these (and other) streptomycetes, much of our understanding of Streptomyces biology stems from work done with two different model systems: S. coelicolor and S. venezuelae. Here, we highlight how studying S. coelicolor and S. venezuelae has contributed to our knowledge of the biosynthesis and regulation of specialized metabolites and bacterial multicellular development.
STREPTOMYCES COELICOLOR GENETICS OPENED THE DOOR TO ENGINEERING NATURE’S MEDICINES AND UNDERSTANDING COMPLEX BACTERIAL LIFE CYCLES
The discovery of streptomycin, neomycin, and tetracycline moved members of the genus Streptomyces into the spotlight as producers of medically important antibiotics. At that time, however, virtually nothing was known about the genetics of these bacteria. This limited the discovery of new and more potent antibiotics and at the same time spurred the need for establishing tools and methodologies to genetically manipulate Streptomyces.
The founding member of the Streptomyces genetics era and the best-known representative of the genus is S. coelicolor A3(2). S. coelicolor A3(2) has a single linear 8.6-Mbp chromosome (72.1% GC content), encoding ~7,800 genes (12). In addition, S. coelicolor A3(2) harbors two episomes, the linear plasmid SCP1 (356 kb; 69.1% GC content; 351 coding sequences) (25) and the circular plasmid SCP2 (31 kb; 72.1% GC content; 34 coding sequences) (26). Across its genome, S. coelicolor A3(2) has at least 27 biosynthetic gene clusters encoding specialized metabolites.
There are two common plasmid-free derivatives of S. coelicolor A3(2) that were isolated independently: strains M145 and M600 (27, 28). It is worth noting that M145 and M600 display phenotypic differences, likely due to their lineage history (29, 30). Importantly, M145 was used to generate the annotated genome sequence of S. coelicolor A3(2) (12) and shows an advantageous antibiotic production and growth profile compared to M600. These characteristics have led to M145 being the more widely used laboratory “wild-type” strain (referred to as S. coelicolor henceforth).
The first milestone in the history of Streptomyces genetics was the discovery of genetic recombination in S. coelicolor (31–34), which enabled the systematic mapping of genes on the chromosome (35). The genetic analysis of Streptomyces was further supported by a series of ground-breaking technical advances that were driven in part by the discovery of conjugative plasmids (36), the characterization of several bacteriophage attachment sites for the site-specific integration of donor DNA (37, 38), the development of an efficient conjugation system (39), and the preparation of an ordered library of large S. coelicolor chromosomal inserts in an E. coli cosmid vector (40), which provided the material for sequencing the entire genome (12). These technical advances facilitated everything from cloning the first Streptomyces gene to heterologously expressing entire biosynthetic pathways in S. coelicolor (41, 42).
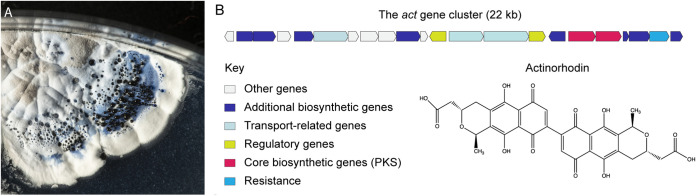
One of the reasons that S. coelicolor was initially such an attractive model system is that it produces pigments that make outstanding genetic markers. The species name “coelicolor,” which means “heavenly colored” or “sky colored” in Latin, was inspired by the observation that S. coelicolor produces a distinctive blue-pigmented molecule called actinorhodin, which accumulates in the culture medium and on the surface of S. coelicolor colonies (Fig. 2A) (43). In addition to actinorhodin, S. coelicolor hyphae also produce red (undecylprodigiosin) and yellow (coelimycin) specialized metabolites alongside a gray pigment that is associated with mature spores (44–46). Mutants blocked at different steps in the pigment biosynthesis pathway can be easily identified by visually screening for S. coelicolor colonies with altered colony coloration.
FIG 2.
Streptomyces coelicolor. (A) Image of an S. coelicolor colony with aerial mycelium and gray-pigmented spores. The dark halo around the colony and the droplets on top of the colony are secreted actinorhodin. (B) Schematic showing the actinorhodin biosynthetic gene cluster (SCO5071 to SCO5092) and its chemical structure.
The production of actinorhodin is among the first and probably most thoroughly investigated pathways in S. coelicolor. Actinorhodin is not only a blue-pigmented molecule but also an antibiotic (47). Screening for Streptomyces mutants that had either lost or regained this blue color enabled the isolation and mapping of the actinorhodin biosynthetic pathway genes in vivo. Early research on actinorhodin production revealed several important features about antibiotic synthesis, including the fact that genes encoding the biosynthetic enzymes are arranged together in so-called biosynthetic gene clusters (BGCs) (Fig. 2B). These BGCs include all the genes required for metabolite production, export, and self-resistance. Unusually, given that BGCs are generally considered to be nonessential genes, they are found predominantly on the chromosome rather than on plasmids (48, 49).
Actinorhodin belongs to the class of polyketide antibiotics and is produced by an aromatic type II polyketide synthase (PKS) (50), a family of large multidomain enzymes comprising separate modules with dedicated enzymatic activities. Through investigations into the genetics of polyketide production, the actinorhodin biosynthetic pathway became the first target for genetic engineering resulting in a unique “hybrid” antibiotic (51). This discovery led to the concept of combinatorial biosynthesis of small molecules by the rational engineering of the core modular biosynthetic enzymes (52).
In addition to directing the production of actinorhodin, sequencing of the S. coelicolor genome revealed that this strain has the capacity to produce a wide array of structurally distinct specialized metabolites beyond just polyketide antibiotics. These compounds include peptides derived from nonribosomal peptide synthetases (NRPS), lanthipeptides, bacteriocins, terpenoids, and aminoglycosides, in addition to many other natural products (53). Such structural diversity in the specialized metabolic output of S. coelicolor is now known to be a hallmark of all streptomycetes. The building blocks used to assemble these specialized metabolites are often derived from primary metabolism (54, 55). Intriguingly, these linkages to primary metabolism extend beyond metabolite assembly to include transcriptional control, with many regulators of central metabolic pathways (e.g., AfsQ1 [nitrogen assimilation] and PhoP [phosphate utilization]) directly or indirectly controlling the expression of BGCs (56–58).
While the pigmented antibiotics produced by S. coelicolor represented an outstanding genetic marker for studies into specialized metabolism, its visually distinguishable developmental stages proved equally attractive for those interested in Streptomyces differentiation. The first seminal insight into the regulatory networks governing this multicellular developmental program came from isolating two groups of S. coelicolor mutants that were defective in aerial mycelium formation and sporulation. The mutations associated with these morphological defects were subsequently mapped to genomic loci that defined the two major classes of developmental regulators in Streptomyces: the Bld (bald) and Whi (white) regulators (59, 60). Bld regulators control the formation of aerial (reproductive) hyphae, and mutations in bld loci result in a so-called “bald” phenotype, with colonies that have waxy surfaces because they lack the hair-like layer of aerial hyphae. Whi regulators are required to differentiate the aerial hyphae into spore-bearing structures, and whi mutants fail to synthesize the colored pigment associated with mature spores (e.g., gray for S. coelicolor and green for S. venezuelae). Thus, the resulting colonies have a characteristic light/white color.
Although using S. coelicolor as a model system has provided the foundational understanding of Streptomyces development, the effective application of global omics technologies and cell biological tools and techniques has been limited by its complex lifestyle. Like many other Streptomyces species, S. coelicolor only sporulates on solid medium in an asynchronous manner, and it forms large mycelial clumps in liquid medium that can impact reproducible cultivation. To overcome these technical challenges and to advance our understanding of the signals and regulatory pathways controlling the progression through the Streptomyces life cycle, Streptomyces venezuelae was adopted as an experimental model organism for developmental studies.
STREPTOMYCES VENEZUELAE PROVIDES NEW AVENUES TO STUDY THE CELL BIOLOGY UNDERPINNING THE GROWTH AND DEVELOPMENT IN STREPTOMYCETES
S. venezuelae, as its name suggests, was first isolated in Venezuela from a soil sample and was initially described as a chloramphenicol producer (61, 62). The common laboratory strain is S. venezuelae NRRL B-65442, which has an 8.2-Mbp, GC-rich (72.5%) linear chromosome, encoding ~7,100 proteins, and a 158-kb circular plasmid, pSVJI1 (70.1% GC content and 163 coding sequences). Genome sequencing further indicated the presence of at least 34 BGCs, the majority of which are not expressed under standard laboratory conditions (63). Comparative sequence analysis revealed that ~85% of the protein-coding genes on the S. venezuelae chromosome have orthologues in S. coelicolor A3(2), with an average protein sequence identity of more than 64%.
Importantly, unlike S. coelicolor, S. venezuelae grows rapidly in a highly dispersed manner and sporulates comprehensively and almost synchronously in liquid culture (64). Although some regulatory details may differ between both species, all known regulatory mutations that block sporulation on solid medium also prevent sporulation in liquid-grown S. venezuelae cells. In addition to there being a generalized transducing phage available for use in S. venezuelae (65), most shuttle vectors, DNA transfer, and recombinant DNA techniques developed for S. coelicolor can be seamlessly used to clone and manipulate genes in S. venezuelae.
Consequently, the introduction of S. venezuelae as a new model system has greatly facilitated global functional analyses and provided a detailed understanding of the regulatory networks underpinning the complex multicellular developmental program from germination to sporulation (66, 67). Here, we highlight a few recent studies demonstrating the benefits of using S. venezuelae as a model species for developmental and cell biological studies.
The first pioneering study in S. venezuelae that combined chromatin immunoprecipitation sequencing (ChIP-seq) and global transcriptional profiling came from Bibb et al. (68), who set out to determine the regulon of the developmental regulator BldN. BldN is an ECF sigma factor that directs the transcription of genes required to synthesize the major components of the hydrophobic sheath (comprising the chaplin and rodlin proteins, discussed below) encasing the aerial hyphae. This study laid the foundation for many others that have followed and, collectively, provided comprehensive insight into the developmental regulatory networks governing Streptomyces differentiation.
To complete the life cycle and transform aerial hyphae into chains of spores, the function of multiple Whi regulators is required. Two transcription factors WhiA and WhiB sit at the top of this regulatory cascade and are essential for coordinating the cessation of aerial growth and the initiation of sporulation septation. Work by Bush et al. revealed that together WhiA and WhiB control the expression of about 240 genes, including those that encode proteins with roles in polar growth, cell division, and spore maturation as well as many proteins of unknown function (69, 70).
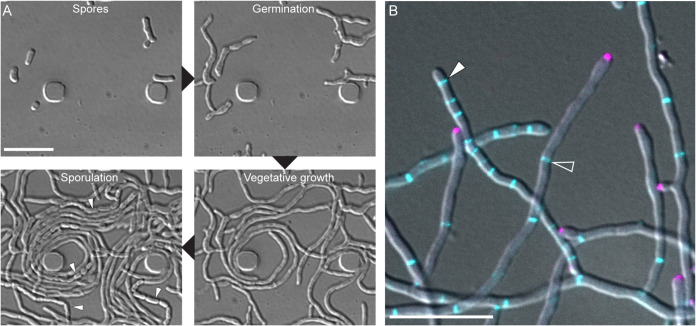
The fact that S. venezuelae completes its life cycle in liquid has also proven to be a compelling characteristic to study the spatiotemporal control of proteins that drive central developmental processes at the cellular level. In addition, the establishment of microfluidic-based time-lapse fluorescence imaging techniques has allowed the entire spore-to-spore life cycle to be captured (Fig. 3A) (71). This has opened up the field of Streptomyces cell biology and led to a number of important advances in our understanding of how critical cellular events, such as polar growth, chromosome arrangement and cell division, are organized in space and time (Fig. 3B) (72–78). For example, a recent study identified an essential component for crosswall formation (Fig. 1). The nature and significance of crosswalls, compared to the functionally distinct division septa formed during sporulation, had been elusive for decades. Using a combination of live-cell imaging and molecular genetics, Bush et al. (74) demonstrated that hyphal compartmentalization via crosswalls is crucial for the ordered progression through the developmental life cycle. This hyphal compartmentalization appears to ensure efficient sporulation and may also impact other cellular processes like specialized metabolite production.
FIG 3.
Streptomyces venezuelae. (A) Time-lapse images of wild-type S. venezuelae grown in liquid using a microfluidic system showing the cellular development during the Streptomyces life cycle, including germination, vegetative growth, and sporulation (white arrowheads). (B) Composite light microscopy image of S. venezuelae producing fluorescently tagged DivIVA (magenta) and FtsZ (blue) to visualize growing hyphae and sites of cell division, respectively. Open arrowheads indicate crosswalls, and filled arrowheads point to hyphae undergoing sporulation septation. Scale bars, 10 μm.
RESOURCES AND TOOLS TO STUDY STREPTOMYCES
The go-to resource for many Streptomyces researchers over the past decades has been the book “Practical Streptomyces Genetics” (79), which contains a comprehensive collection of laboratory protocols, media recipes, plasmid details, and other useful information on how to grow, manipulate, and evaluate Streptomyces species. Although this handbook was last updated in 2000 and thus lacks information on more recent developments in the field, it remains an invaluable and freely available resource that can be downloaded from StrepDB (http://strepdb.streptomyces.org.uk). StrepDB is the Streptomyces genomes annotation browser that hosts the genome and plasmid sequences of many of the best studied Streptomyces species, including S. coelicolor and S. venezuelae.
The most common methods of genetically manipulating S. coelicolor and S. venezuelae involve using the ReDirect technique, which is a λ RED-based gene disruption protocol (80, 81), and CRISPR-Cas9-mediated genome editing (82, 83). The design of single guide RNA sequences for CRISPR-based genome engineering has been further supported by the interactive online tool CRISPy-web (https://crispy.secondarymetabolites.org/#/input) (84).
Further advances in Streptomyces genetics that have facilitated the cloning, expression, and manipulation of genes and biosynthetic pathways in nonmodel Streptomyces species (see also reference 85) include the development of inducible promoters to control the conditional expression of genes (86, 87) and synthetic promoter systems for targeted gene expression studies (88–90).
A multitude of databases and computational approaches that have been developed to mine the ever increasing number of Streptomyces genomes for novel BGCs complement these genetic tools. The bioinformatic pipeline antiSMASH (https://antismash.secondarymetabolites.org/#!/start) is a widely used online tool that predicts BGCs in streptomycetes and other microbial genomes (91). The relative ease of cluster identification has in turn fueled an interest in cloning and heterologously expressing BGCs of interest. Heterologous expression has been greatly facilitated by the generation of a suite of S. coelicolor “superhosts” (92). These strains lack multiple endogenous BGCs, including those for actinorhodin and prodiginine, and have been further genetically modified to increase specialized metabolite production.
Given the wealth of genetic and genomic tools that have been developed up to this point, there is considerable interest in bringing the field together to both collate existing techniques and resources and develop additional community-wide tools and resources aimed at advancing and exploiting Streptomyces biology. One such initiative is ActinoBase (http://actinobase.org/index.php/Main_Page), a recently established online platform that aims to integrate freely accessible scientific resources, including experimental protocols covering more recent technical advancements (93). Considering the long history of Streptomyces genetics (94), the development of a comprehensive Wiki-based platform represents a valuable resource and one with considerable scope for future growth and utility for the growing Streptomyces community.
NOTABLE DISCOVERIES
The experimental and technical foundations laid by decades of work in S. coelicolor and S. venezuelae systems have enabled a multitude of pioneering discoveries, spanning everything from unique genetic regulatory mechanisms to novel developmental trajectories and community-based survival strategies.
Regulatory innovations.
From genetic and genomic perspectives, the streptomycetes have proven to be unrivaled in their regulatory innovations. Early evidence of this came in the 1980s, when it was discovered that a key developmental and metabolic determinant known as bldA (95) corresponded to a leucyl tRNA needed to translate the rare TTA/UUA codon (96). The fact that a tRNA was not essential for bacterial viability was surprising, and the phenotypic effects associated with its mutation suggested that it had been coopted for the translational control of development and antibiotic production. Analysis of the S. coelicolor genome sequence revealed that fewer than 2% of all genes possessed a TTA codon. Included among these was the developmental regulator-encoding adpA/bldH gene, which is now known to be solely responsible for the developmental block observed in the bldA mutant (97, 98). Additional TTA codons were present in cluster-situated regulators of multiple BGCs, including the actII-ORF4 (actinorhodin) and redZ (undecylprodigiosin) regulators in S. coelicolor. Indeed, expression of bldA has been effectively exploited to stimulate cryptic antibiotic production in other streptomycetes (e.g., Kalan et al. [99]).
Beyond translational control, Streptomyces are also masters of transcriptional regulation, as evidenced by their abundance of sigma factors and transcription factors. In the early (pregenomic) 1990s, comparative analysis of the S. coelicolor sigma E sequence revealed similar sigma factors to be present in phylogenetically diverse bacteria (100). These proteins comprise a distinct subfamily of sigma factors that are shorter than the canonical sigma 70-type sigma factors due to truncations in region 3. These were dubbed the extracytoplasmic function (ECF) sigma factors due to the contribution that many of their founding members made in responding to exogenous stresses. As detailed above, the ECF sigma factors are the largest group of sigma factors in the streptomycetes (12).
The streptomycetes have also been found to have creative transcription factor configurations. BldM and WhiI are developmental regulators belonging to the “orphaned” (no associated kinase) response regulator group of proteins (101, 102). Investigations into the function of these transcription factors in S. venezuelae revealed that BldM governs the expression of one set of genes in its homodimer configuration, and when it heterodimerizes with WhiI, it controls an independent set of target genes (103). Beyond their flexible association with protein partners, work in S. venezuelae has also revealed unusual associations with cyclic nucleotide cofactors. One of the most striking examples of this was seen for BldD, the master regulator of reproductive development in the streptomycetes. From earlier work in S. coelicolor, BldD was known to dimerize (104) and repress the activity of its target genes (105, 106). The nature of the BldD dimer configuration remained mysterious until the crystal structure of BldD was solved in 2014 (107). This revealed that the BldD dimer was not assembled in a conventional way involving protein-protein interactions. Instead, BldD monomers were joined together by a quartet of cyclic-di-GMP molecules arranged in a stacked dimer-of-dimers configuration (107). This study revealed not only a unique mechanism of protein dimerization but also an unexpected conformational flexibility for nucleotide second messengers, as tetramer forms of cyclic di-GMP had not been observed previously.
Despite this unprecedented discovery, protein interactions mediated by cyclic-di-GMP in the streptomycetes now extend beyond BldD. Subsequent investigation into the nature of the interaction between the WhiG sigma factor, a motility-like sigma factor that directs Streptomyces sporulation (108), and its cognate anti-sigma factor RsiG revealed that this is mediated by a dimeric cyclic-di-GMP molecule (109). Individually, the RsiG anti-sigma factor (but not sigma WhiG) binds cyclic-di-GMP. Importantly, this ligand-bound complex is the only form of RsiG that can associate with and sequester sigma WhiG to prevent sporulation. In addition to being an essential cofactor in controlling progression through the Streptomyces life cycle, cyclic-di-GMP was also recently found to bind and stimulate the catalytic activity of a glycogen-degrading enzyme (110). This further expands the role of this second messenger molecule in Streptomyces from modulating the activity of two central transcription factors in the developmental regulatory cascade to mediating energy storage metabolism.
Developmental innovations.
As their regulatory systems suggest, the streptomycetes are complex bacteria, and much of this complexity has been both uncovered and unraveled in the S. coelicolor and S. venezuelae systems. Their fungal-like growth as filamentous hyphae are unusual in the bacterial world, and this is driven by hyphal tip extension. Work in S. coelicolor revealed that this polar growth is mediated by DivIVA (111) and an associated polarisome complex that includes multiple cytoskeletal elements (112–114). It has subsequently been established that polar growth is shared by other actinobacteria as well as members of the Rhizobiales (e.g., Agrobacterium and Sinorhizobium) (115), although the underlying growth mechanism in the latter group has yet to be fully elucidated. Within the context of the Streptomyces life cycle, the DivIVA-polarisome drives the growth of both the branching vegetative hyphae and the nonbranching aerial hyphae (Fig. 1).
The raising of aerial hyphae from the vegetative milieu requires the activity of multiple surfactant proteins, including SapB and the chaplins, which have intriguing structural properties. First identified in S. coelicolor, SapB shares structural similarity with lantipeptide antibiotics (116), but instead of inhibiting the growth of neighboring bacteria, it promotes the raising of aerial hyphae following secretion into the environment during growth under nutrient-replete conditions (19). Like SapB, the chaplin proteins were also identified in S. coelicolor as small proteins with surfactant properties, but they have a very different protein architecture than SapB and are expressed during aerial development under all tested growth conditions (21, 22). After secretion, the chaplins polymerize to form functional amyloid fibrils that coat the surface of the emerging hyphae and promote aerial hyphae formation (21). At the time, this was only the second known example of functional amyloids (after the E. coli curli proteins [117]), and it is now well-established that amyloid proteins play important roles in the community development of many bacteria (117–119).
It has long been postulated that the transition from vegetative to aerial mycelial growth is accompanied by a developmentally regulated cell death process in which part of the vegetative mycelium is dismantled to feed the emerging reproductive aerial hyphae (120–123). However, molecular insights into the basis of this phenomenon have been limited. Recently, two studies reported that in S. coelicolor, hyphal cell death is mediated by a contractile injection system (CIS) (124, 125). CIS are phage-tail-like structures that in other systems predominantly contribute to interspecies interactions (126–129) but seem to function differently in Streptomyces. While the exact molecular mechanism of CIS-mediated cell death remains to be determined, it is important to note that most Streptomyces species encode a cis gene cluster (130, 131), suggesting a conserved role for CIS in intracellular cell death and multicellular development.
Vegetative and aerial hyphae can be distinguished by both their branching status and the nature of their cell surfaces; vegetative hyphae are branched and are hydrophilic, whereas aerial hyphae are unbranched with hydrophobic surfaces. Both cell types are, however, multinucleated. As this would suggest, cell division is not common during vegetative growth and early aerial development, and unlike virtually all other bacteria, cell division is not essential for Streptomyces viability. Deleting the central cell division gene ftsZ in either S. coelicolor or S. venezuelae leads to an inability to form crosswalls and sporulation septa, but the mutation still permits vegetative and aerial growth (132, 133). FtsZ assembly in the reproductive hyphae is a remarkably synchronous process; it assembles into a ladder-like array of rings extending the length of the aerial filaments (Fig. 3B) (71, 134). These FtsZ rings mark the site of future cell division, defining where the aerial hyphae will be subdivided into single-genome prespore compartments. There has been considerable interest in understanding what factors impact FtsZ localization (and the associated chromosome segregation), and work in S. coelicolor and S. venezuelae is revealing this to require the activity of both Streptomyces-specific proteins (73, 135, 136) and proteins found in other bacterial phyla (73–75, 137).
Reproductive growth, encompassing aerial hyphae formation and sporulation, is typically considered to be genetically coordinated with (but spatially segregated from) the specialized metabolite-producing vegetative cells. Recent work in S. coelicolor has established that there is a genetic basis underlying this “division of labor” within a Streptomyces colony (138). It appears that a subset of cells within a colony undergoes significant chromosomal reorganization (amplification or deletion of genomic segments as large as 1 Mb) (138) and a subsequent increase in mutation rate (139). These subpopulations of cells are often highly pigmented, indicating enhanced antibiotic production, and are generally less fit than their sporulating counterparts whose genomic integrity is intact. Notably, however, the presence of these growth-deficient antibiotic-producing cells does not adversely impact the overall fitness of the S. coelicolor colony, at least in the laboratory (140), suggesting that this segregation of cell function into specialized metabolite producer and reproductive cell producer has been evolutionarily advantageous.
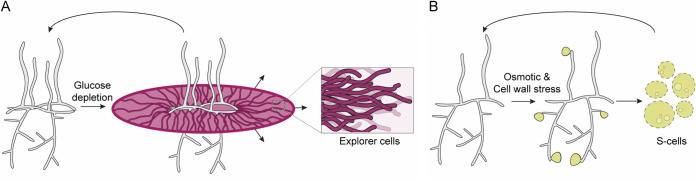
While many of these innovative discoveries have come from investigations using S. coelicolor as a model system, more recent studies in S. venezuelae are revealing that the developmental and growth repertoire of these bacteria is far broader and more flexible than had been previously appreciated. Indeed, changing the conventional growth medium of S. venezuelae or growing S. venezuelae in association with yeast enables an apparent motility-type response in which the colony spreads rapidly over solid surfaces (Fig. 4A). To date, this so-called “exploratory growth” mode appears to be a broadly, but not universally, conserved trait within the streptomycetes (140). Beyond an apparently enhanced growth rate, exploring S. venezuelae is also notable for the release of volatile organic compounds, which serve dual functions in communication and competition; they promote exploration in nearby streptomycetes and at the same time inhibit the growth of other microbes by making iron less bioavailable (141). In addition to this rapid colonial expansion, S. venezuelae has also proven adept at shedding its wall and growing as more individual “S-cells” in response to hyperosmotic conditions (Fig. 4B) (142). Importantly, both S-cells and exploring cells are not terminally differentiated growth states and are capable of resuming their classical developmental growth cycle when conditions permit.
FIG 4.
Alternative Streptomyces growth modes. (A) Exploratory growth is triggered in response to glucose depletion (e.g., when cells are grown beside yeast). Explorer cells grow as nonbranching vegetative hyphae and expand rapidly across solid surfaces. (B) S-cells are extruded from hyphal tips following exposure to hyperosmotic stress or cell wall-targeting antibiotics. S-cells are vesicle-like structures that lack a cell wall and contain all cellular components to replicate. Both explorer and S-cells are transient cell types that can revert to the classic developmental program (Fig. 1).
Interaction innovations.
Cell wall shedding, as observed for S-cells, is now being recognized as a mechanism of phage defense through the removal of many phage receptors (143, 144). As predominantly environmental bacteria, Streptomyces spp. are under constant threat from phage attack, and multiple phage defense systems have been identified through work using S. coelicolor and S. venezuelae. An excellent example of an enzymatic phage defense system is the Pgl system (for phage growth limitation), which was first identified in S. coelicolor in the early 1990s (145). Pgl is a multigene locus whose products include a kinase and methyltransferase (146), with DNA methylation being key to the restriction of phage DNA replication. Notably, Pgl-like systems have been found throughout bacteria where they have been dubbed the “BREX” (for bacteriophage exclusion) systems (147). More recently, the Streptomyces phage defense repertoire has been expanded to include a role for specialized metabolites. Using S. coelicolor as a test system, Kronheim et al. revealed that DNA intercalating compounds have potent antiphage effects (148), whereas Kever et al. observed similar effects for aminoglycoside antibiotics using S. venezuelae (149).
Specialized metabolites have long been known to have key roles in promoting the competitive fitness of their producers given the growth (and now phage) inhibition capabilities of many of these natural products. These compounds are, however, increasingly being recognized for having alternative roles in modulating community dynamics, with the volatile metabolite geosmin being an outstanding example (described above).
FUTURE DIRECTIONS
The decades of work conducted using S. coelicolor, and more recently S. venezuelae, are providing an outstanding platform from which to launch future investigations into this group of fascinating microbes. Given the complexity of the classical Streptomyces sporulating life cycle and newly described growth strategies, there remains much to be understood about the cellular mechanisms and signals underpinning these different developmental stages and the transitions between them.
The first bacterial signaling molecule discovered in Streptomyces was the hormone-like γ-butyrolactone molecule A-factor (150, 151), which induces antibiotic production and morphological development in Streptomyces griseus (reviewed by Horinouchi [152]). While γ-butyrolactones are produced by many Streptomyces species, their biological roles appear to vary (153–155). Given the enormous diversity of specialized metabolites produced by these organisms, it is conceivable that many of these have signaling functions. Determining the nature and activity of these small, diffusible molecules in regulating antibiotic production and development in Streptomyces and facilitating intra- and interspecies communication and competition, including interactions with other microbes, plants, and insects/arthropods, will be a rich area for future investigation.
Innovations in imaging technologies are further revolutionizing our ability to probe cellular dynamics and cell organization. These innovations in turn are providing unprecedented opportunities to address fundamental questions relating to, for example, bacterial growth, cell wall biosynthesis and remodeling, genome dynamics, and the cytoplasmic organization of protein complexes. Thanks to advances in genomic technologies, it is becoming possible to define the structure of the Streptomyces chromosome at different life cycle stages (70). Identifying the proteins responsible for chromosome organization (as well as biosynthetic cluster control) and how their binding and activities change under different conditions will be a major goal moving forward. It is also worth noting that the majority of genome studies to date have been conducted on plasmid-free strains of S. coelicolor. Understanding how plasmid dynamics are coordinated with those of the chromosome, what factors are needed for plasmid organization, segregation, and transmission, and whether these differ for circular versus linear plasmids will all be questions of interest.
Despite a recent flurry of reports on prokaryotic antiphage systems, relatively little is known about how Streptomyces species defend themselves against phages and other selfish mobile genetic elements. Given their unique multicellular lifestyle, genome architecture, and the fact that the products of the majority of the encoded BGCs are unknown, it would not be surprising if Streptomyces genomes harbor an as-of-yet unexplored repertoire of novel defense strategies against parasitic nucleic acid vectors.
In future years, investigations into Streptomyces biology will likely extend well beyond these traditional model systems. The relative ease of generating draft genome sequences, coupled with the fact that many of the genetic tools developed and honed in S. coelicolor and S. venezuelae can be applied to uncharacterized strains, is opening the door to exploring the biology of diverse Streptomyces species. It will be interesting to see how our understanding of Streptomyces biology in the laboratory translates into its native environment and how interactions with other organisms changes their growth, metabolism, and behavior and vice versa.
ACKNOWLEDGMENTS
We thank Phil Robinson for the photograph of the S. coelicolor colony. Research in the Elliot lab is supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant and by a Canadian Institutes of Health Research Project Grant (PJT-162340). Work in the Schlimpert lab is funded by a Royal Society University Research Fellowship (URF\R1\180075) to S.S. and by the BBSRC Institute Strategic Program grant BB/X01097X/1 to the John Innes Centre.
We apologize to all colleagues whose important work could not be mentioned due to limitations in space.
We declare no conflict of interest.
We dedicate this review to Sir David Hopwood and Mark Buttner, who through their vision and generosity in sharing material, data, and advice have been instrumental in establishing S. coelicolor and S. venezuelae as model species.
Contributor Information
Susan Schlimpert, Email: susan.schlimpert@jic.ac.uk.
Marie A. Elliot, Email: melliot@mcmaster.ca.
George O’Toole, Geisel School of Medicine at Dartmouth.
REFERENCES
- 1.McDonald BR, Currie CR. 2017. Lateral gene transfer dynamics in the ancient bacterial genus Streptomyces. mBio 8:e00644-17. doi: 10.1128/mBio.00644-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bisset KA, Moore FW. 1949. The relationship of certain branched bacterial genera. J Gen Microbiol 3:387–391. doi: 10.1099/00221287-3-3-387. [DOI] [PubMed] [Google Scholar]
- 3.Jensen PR, Dwight R, Fenical W. 1991. Distribution of actinomycetes in near-shore tropical marine sediments. Appl Environ Microbiol 57:1102–1108. doi: 10.1128/aem.57.4.1102-1108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klausen C, Nicolaisen MH, Strobel BW, Warnecke F, Nielsen JL, Jørgensen NOG. 2005. Abundance of actinobacteria and production of geosmin and 2-methylisoborneol in Danish streams and fish ponds. FEMS Microbiol Ecol 52:265–278. doi: 10.1016/j.femsec.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Kers JA, Cameron KD, Joshi MV, Bukhalid RA, Morello JE, Wach MJ, Gibson DM, Loria R. 2005. A large, mobile pathogenicity island confers plant pathogenicity on Streptomyces species. Mol Microbiol 55:1025–1033. doi: 10.1111/j.1365-2958.2004.04461.x. [DOI] [PubMed] [Google Scholar]
- 6.Quintana ET, Wierzbicka K, Mackiewicz P, Osman A, Fahal AH, Hamid ME, Zakrzewska-Czerwinska J, Maldonado LA, Goodfellow M. 2008. Streptomyces sudanensis sp. nov., a new pathogen isolated from patients with actinomycetoma. Antonie Van Leeuwenhoek 93:305–313. doi: 10.1007/s10482-007-9205-z. [DOI] [PubMed] [Google Scholar]
- 7.Xu M-J, Wang J-H, Bu X-L, Yu H-L, Li P, Ou H-Y, He Y, Xu F-D, Hu X-Y, Zhu X-M, Ao P, Xu J. 2016. Deciphering the streamlined genome of Streptomyces xiamenensis 318 as the producer of the anti-fibrotic drug candidate xiamenmycin. Sci Rep 6:18977. doi: 10.1038/srep18977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baranasic D, Gacesa R, Starcevic A, Zucko J, Blažič M, Horvat M, Gjuračić K, Fujs Š, Hranueli D, Kosec G, Cullum J, Petković H. 2013. Draft genome sequence of Streptomyces rapamycinicus strain NRRL 5491, the producer of the immunosuppressant rapamycin. Genome Announc 1:e00581-13. doi: 10.1128/genomeA.00581-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin YS, Kieser HM, Hopwood DA, Chen CW. 1993. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol Microbiol 10:923–933. doi: 10.1111/j.1365-2958.1993.tb00964.x. [DOI] [PubMed] [Google Scholar]
- 10.Bao K, Cohen SN. 2003. Recruitment of terminal protein to the ends of Streptomyces linear plasmids and chromosomes by a novel telomere-binding protein essential for linear DNA replication. Genes Dev 17:774–785. doi: 10.1101/gad.1060303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang C-C, Huang C-H, Li C-Y, Tsay Y-G, Lee S-C, Chen CW. 2002. The terminal proteins of linear Streptomyces chromosomes and plasmids: a novel class of replication priming proteins. Mol Microbiol 43:297–305. doi: 10.1046/j.1365-2958.2002.02760.x. [DOI] [PubMed] [Google Scholar]
- 12.Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang C-H, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream M-A, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 13.Jeong Y, Kim J-N, Kim MW, Bucca G, Cho S, Yoon YJ, Kim B-G, Roe J-H, Kim SC, Smith CP, Cho B-K. 2016. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2). Nat Commun 7:11605. doi: 10.1038/ncomms11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLean TC, Lo R, Tschowri N, Hoskisson PA, Al Bassam MM, Hutchings MI, Som NFY. 2019. Sensing and responding to diverse extracellular signals: an updated analysis of the sensor kinases and response regulators of Streptomyces species. Microbiology (Reading) 165:929–952. doi: 10.1099/mic.0.000817. [DOI] [PubMed] [Google Scholar]
- 15.Petříčková K, Petříček M. 2003. Eukaryotic-type protein kinases in Streptomyces coelicolor: variations on a common theme. Microbiology (Reading) 149:1609–1621. doi: 10.1099/mic.0.26275-0. [DOI] [PubMed] [Google Scholar]
- 16.Cuthbertson L, Nodwell JR. 2013. The TetR family of regulators. Microbiol Mol Biol Rev 77:440–475. doi: 10.1128/MMBR.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talbot NJ. 2003. Aerial morphogenesis: enter the chaplins. Curr Biol 13:R696–R698. doi: 10.1016/j.cub.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 18.Hempel AM, Cantlay S, Molle V, Wang S-B, Naldrett MJ, Parker JL, Richards DM, Jung Y-G, Buttner MJ, Flärdh K. 2012. The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc Natl Acad Sci USA 109:E2371–E2379. doi: 10.1073/pnas.1207409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willey J, Santamaria R, Guijarro J, Geistlich M, Losick R. 1991. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell 65:641–650. doi: 10.1016/0092-8674(91)90096-h. [DOI] [PubMed] [Google Scholar]
- 20.Claessen D, Wösten HAB, van Keulen G, Faber OG, Alves AMCR, Meijer WG, Dijkhuizen L. 2002. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol Microbiol 44:1483–1492. doi: 10.1046/j.1365-2958.2002.02980.x. [DOI] [PubMed] [Google Scholar]
- 21.Claessen D, Rink R, de Jong W, Siebring J, de Vreugd P, Boersma FGH, Dijkhuizen L, Wosten HAB. 2003. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev 17:1714–1726. doi: 10.1101/gad.264303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliot MA, Karoonuthaisiri N, Huang J, Bibb MJ, Cohen SN, Kao CM, Buttner MJ. 2003. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev 17:1727–1740. doi: 10.1101/gad.264403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becher PG, Verschut V, Bibb MJ, Bush MJ, Molnár BP, Barane E, Al-Bassam MM, Chandra G, Song L, Challis GL, Buttner MJ, Flärdh K. 2020. Developmentally regulated volatiles geosmin and 2-methylisoborneol attract a soil arthropod to Streptomyces bacteria promoting spore dispersal. Nat Microbiol 5:821–829. doi: 10.1038/s41564-020-0697-x. [DOI] [PubMed] [Google Scholar]
- 24.Hopwood DA. 1988. The Leeuwenhoek lecture, 1987. Towards an understanding of gene switching in Streptomyces, the basis of sporulation and antibiotic production. Proc R Soc Lond B Biol Sci 235:121–138. doi: 10.1098/rspb.1988.0067. [DOI] [PubMed] [Google Scholar]
- 25.Bentley SD, Brown S, Murphy LD, Harris DE, Quail MA, Parkhill J, Barrell BG, McCormick JR, Santamaria RI, Losick R, Yamasaki M, Kinashi H, Chen CW, Chandra G, Jakimowicz D, Kieser HM, Kieser T, Chater KF. 2004. SCP1, a 356,023 bp linear plasmid adapted to the ecology and developmental biology of its host, Streptomyces coelicolor A3(2). Mol Microbiol 51:1615–1628. doi: 10.1111/j.1365-2958.2003.03949.x. [DOI] [PubMed] [Google Scholar]
- 26.Haug I, Weissenborn A, Brolle D, Bentley S, Kieser T, Altenbuchner J. 2003. Streptomyces coelicolor A3(2) plasmid SCP2*: deductions from the complete sequence. Microbiology (Reading) 149:505–513. doi: 10.1099/mic.0.25751-0. [DOI] [PubMed] [Google Scholar]
- 27.Chakraburtty R, Bibb M. 1997. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J Bacteriol 179:5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopwood DA, Bibb MJ, Chater KF, Bruton CJ, Lydiate DJ, Smith CP, Ward JM, Schrempf H. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, UK. [Google Scholar]
- 29.Ochi K, Hosoya Y. 1998. Genetic mapping and characterization of novel mutations which suppress the effect of a relC mutation on antibiotic production in Streptomyces coelicolor A3(2). J Antibiot (Tokyo) 51:592–595. doi: 10.7164/antibiotics.51.592. [DOI] [PubMed] [Google Scholar]
- 30.Weaver D, Karoonuthaisiri N, Tsai H-H, Huang C-H, Ho M-L, Gai S, Patel KG, Huang J, Cohen SN, Hopwood DA, Chen CW, Kao CM. 2004. Genome plasticity in Streptomyces: identification of 1 Mb TIRs in the S. coelicolor A3(2) chromosome. Mol Microbiol 51:1535–1550. doi: 10.1111/j.1365-2958.2003.03920.x. [DOI] [PubMed] [Google Scholar]
- 31.Sermonti G, Spada-Sermonti I. 1955. Genetic recombination in Streptomyces. Nature 176:121. doi: 10.1038/176121a0. [DOI] [PubMed] [Google Scholar]
- 32.Braendle DH, Szybalski W. 1957. Genetic interaction among streptomycetes: heterokaryosis and synkaryosis. Proc Natl Acad Sci USA 43:947–955. doi: 10.1073/pnas.43.11.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alikhanian SI, Mindlin SZ. 1957. Recombinations in Streptomyces rimosus. Nature 180:1208–1209. doi: 10.1038/1801208a0. [DOI] [PubMed] [Google Scholar]
- 34.Saito H. 1957. Genetic recombination in Streptomyces griseoflavus. Microb Gen Bull 15:25–26. [DOI] [PubMed] [Google Scholar]
- 35.Hopwood DA. 1965. A circular linkage map in the actinomycete Streptomyces coelicolor. J Mol Biol 12:514–516. doi: 10.1016/s0022-2836(65)80274-1. [DOI] [PubMed] [Google Scholar]
- 36.Bibb MJ, Ward JM, Kieser T, Cohen SN, Hopwood DA. 1981. Excision of chromosomal DNA sequences from Streptomyces coelicolor forms a novel family of plasmids detectable in Streptomyces lividans. Mol Gen Genet 184:230–240. doi: 10.1007/BF00272910. [DOI] [PubMed] [Google Scholar]
- 37.Gregory MA, Till R, Smith MCM. 2003. Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J Bacteriol 185:5320–5323. doi: 10.1128/JB.185.17.5320-5323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lomovskaya ND, Mkrtumian NM, Gostimskaya NL, Danilenko VN. 1972. Characterization of temperate actinophage phi C31 isolated from Streptomyces coelicolor A3(2). J Virol 9:258–262. doi: 10.1128/JVI.9.2.258-262.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flett F, Mersinias V, Smith CP. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett 155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 40.Redenbach M, Kieser HM, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood DA. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol 21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 41.Bibb M, Schottel JL, Cohen SN. 1980. A DNA cloning system for interspecies gene transfer in antibiotic-producing Streptomyces. Nature 284:526–531. doi: 10.1038/284526a0. [DOI] [PubMed] [Google Scholar]
- 42.Thompson CJ, Ward JM, Hopwood DA. 1980. DNA cloning in Streptomyces: resistance genes from antibiotic-producing species. Nature 286:525–527. doi: 10.1038/286525a0. [DOI] [PubMed] [Google Scholar]
- 43.Bystrykh LV, Fernández-Moreno MA, Herrema JK, Malpartida F, Hopwood DA, Dijkhuizen L. 1996. Production of actinorhodin-related “blue pigments” by Streptomyces coelicolor A3(2). J Bacteriol 178:2238–2244. doi: 10.1128/jb.178.8.2238-2244.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsao SW, Rudd BA, He XG, Chang CJ, Floss HG. 1985. Identification of a red pigment from Streptomyces coelicolor A3(2) as a mixture of prodigiosin derivatives. J Antibiot (Tokyo) 38:128–131. doi: 10.7164/antibiotics.38.128. [DOI] [PubMed] [Google Scholar]
- 45.Davis NK, Chater KF. 1990. Spore colour in Streptomyces coelicolor A3(2) involves the developmentally regulated synthesis of a compound biosynthetically related to polyketide antibiotics. Mol Microbiol 4:1679–1691. doi: 10.1111/j.1365-2958.1990.tb00545.x. [DOI] [PubMed] [Google Scholar]
- 46.Pawlik K, Kotowska M, Kolesiński P. 2010. Streptomyces coelicolor A3(2) produces a new yellow pigment associated with the polyketide synthase Cpk. J Mol Microbiol Biotechnol 19:147–151. doi: 10.1159/000321501. [DOI] [PubMed] [Google Scholar]
- 47.Mak S, Nodwell JR. 2017. Actinorhodin is a redox-active antibiotic with a complex mode of action against Gram-positive cells. Mol Microbiol 106:597–613. doi: 10.1111/mmi.13837. [DOI] [PubMed] [Google Scholar]
- 48.Rudd BA, Hopwood DA. 1979. Genetics of actinorhodin biosynthesis by Streptomyces coelicolor A3 (2). J Gen Microbiol 114:35–43. doi: 10.1099/00221287-114-1-35. [DOI] [PubMed] [Google Scholar]
- 49.Malpartida F, Hopwood DA. 1984. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. Nature 309:462–464. doi: 10.1038/309462a0. [DOI] [PubMed] [Google Scholar]
- 50.Das A, Khosla C. 2009. Biosynthesis of aromatic polyketides in bacteria. Acc Chem Res 42:631–639. doi: 10.1021/ar8002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hopwood DA, Malpartida F, Kieser HM, Ikeda H, Duncan J, Fujii I, Rudd BA, Floss HG, Omura S. 1985. Production of “hybrid” antibiotics by genetic engineering. Nature 314:642–644. doi: 10.1038/314642a0. [DOI] [PubMed] [Google Scholar]
- 52.McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. 1993. Engineered biosynthesis of novel polyketides. Science 262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- 53.Craney A, Ahmed S, Nodwell J. 2013. Towards a new science of secondary metabolism. J Antibiot (Tokyo) 66:387–400. doi: 10.1038/ja.2013.25. [DOI] [PubMed] [Google Scholar]
- 54.Martín J-F, Liras P. 2010. Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr Opin Microbiol 13:263–273. doi: 10.1016/j.mib.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 55.Olano C, Lombó F, Méndez C, Salas JA. 2008. Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng 10:281–292. doi: 10.1016/j.ymben.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 56.Sola-Landa A, Moura RS, Martín JF. 2003. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc Natl Acad Sci USA 100:6133–6138. doi: 10.1073/pnas.0931429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allenby NEE, Laing E, Bucca G, Kierzek AM, Smith CP. 2012. Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: genome-wide identification of in vivo targets. Nucleic Acids Res 40:9543–9556. doi: 10.1093/nar/gks766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang R, Mast Y, Wang J, Zhang W, Zhao G, Wohlleben W, Lu Y, Jiang W. 2013. Identification of two-component system AfsQ1/Q2 regulon and its cross-regulation with GlnR in Streptomyces coelicolor. Mol Microbiol 87:30–48. doi: 10.1111/mmi.12080. [DOI] [PubMed] [Google Scholar]
- 59.Hopwood DA, Wildermuth H, Palmer HM. 1970. Mutants of Streptomyces coelicolor defective in sporulation. J Gen Microbiol 61:397–408. doi: 10.1099/00221287-61-3-397. [DOI] [PubMed] [Google Scholar]
- 60.Merrick MJ. 1976. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J Gen Microbiol 96:299–315. doi: 10.1099/00221287-96-2-299. [DOI] [PubMed] [Google Scholar]
- 61.Ehrlich J, Gottlieb D, Burkholder PR, Anderson LE, Pridham TG. 1948. Streptomyces venezuelae, n. sp., the source of chloromycetin. J Bacteriol 56:467–477. doi: 10.1128/jb.56.4.467-477.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ehrlich J, Bartz QR, Smith RM, Joslyn DA, Burkholder PR. 1947. Chloromycetin, a new antibiotic from a soil actinomycete. Science 106:417. doi: 10.1126/science.106.2757.417. [DOI] [PubMed] [Google Scholar]
- 63.Gomez-Escribano JP, Holmes NA, Schlimpert S, Bibb MJ, Chandra G, Wilkinson B, Buttner MJ, Bibb MJ. 2021. Streptomyces venezuelae NRRL B-65442: genome sequence of a model strain used to study morphological differentiation in filamentous actinobacteria. J Ind Microbiol Biotechnol 48:kuab035. doi: 10.1093/jimb/kuab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glazebrook MA, Doull JL, Stuttard C, Vining LC. 1990. Sporulation of Streptomyces venezuelae in submerged cultures. J Gen Microbiol 136:581–588. doi: 10.1099/00221287-136-3-581. [DOI] [PubMed] [Google Scholar]
- 65.Stuttard C. 1979. Transduction of auxotrophic markers in a chloramphenicol-producing strain of Streptomyces. J Gen Microbiol 110:479–482. doi: 10.1099/00221287-110-2-479. [DOI] [PubMed] [Google Scholar]
- 66.Bush MJ, Tschowri N, Schlimpert S, Flärdh K, Buttner MJ. 2015. c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat Rev Microbiol 13:749–760. doi: 10.1038/nrmicro3546. [DOI] [PubMed] [Google Scholar]
- 67.Zambri MP, Williams MA, Elliot MA. 2022. How Streptomyces thrive: advancing our understanding of classical development and uncovering new behaviors. Adv Microb Physiol 80:203–236. doi: 10.1016/bs.ampbs.2022.01.004. [DOI] [PubMed] [Google Scholar]
- 68.Bibb MJ, Domonkos Á, Chandra G, Buttner MJ. 2012. Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by σBldN and a cognate anti-sigma factor, RsbN. Mol Microbiol 84:1033–1049. doi: 10.1111/j.1365-2958.2012.08070.x. [DOI] [PubMed] [Google Scholar]
- 69.Bush MJ, Bibb MJ, Chandra G, Findlay KC, Buttner MJ. 2013. Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae. mBio 4:e00684-13. doi: 10.1128/mBio.00684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bush MJ, Chandra G, Bibb MJ, Findlay KC, Buttner MJ. 2016. Genome-wide chromatin immunoprecipitation sequencing analysis shows that WhiB is a transcription factor that cocontrols its regulon with WhiA to initiate developmental cell division in Streptomyces. mBio 7:e00523-16. doi: 10.1128/mBio.00523-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schlimpert S, Flärdh K, Buttner MJ. 2016. Fluorescence time-lapse imaging of the complete S. venezuelae life cycle using a microfluidic device. J Vis Exp. doi: 10.3791/53863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fröjd MJ, Flärdh K. 2019. Apical assemblies of intermediate filament-like protein FilP are highly dynamic and affect polar growth determinant DivIVA in Streptomyces venezuelae. Mol Microbiol 112:47–61. doi: 10.1111/mmi.14253. [DOI] [PubMed] [Google Scholar]
- 73.Schlimpert S, Wasserstrom S, Chandra G, Bibb MJ, Findlay KC, Flärdh K, Buttner MJ. 2017. Two dynamin-like proteins stabilize FtsZ rings during Streptomyces sporulation. Proc Natl Acad Sci USA 114:E6176–E6183. doi: 10.1073/pnas.1704612114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bush MJ, Gallagher KA, Chandra G, Findlay KC, Schlimpert S. 2022. Hyphal compartmentalization and sporulation in Streptomyces require the conserved cell division protein SepX. Nat Commun 13:71. doi: 10.1038/s41467-021-27638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramos-León F, Bush MJ, Sallmen JW, Chandra G, Richardson J, Findlay KC, McCormick JR, Schlimpert S. 2021. A conserved cell division protein directly regulates FtsZ dynamics in filamentous and unicellular actinobacteria. eLife 10:e63387. doi: 10.7554/eLife.63387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Szafran MJ, Małecki T, Strzałka A, Pawlikiewicz K, Duława J, Zarek A, Kois-Ostrowska A, Findlay KC, Le TBK, Jakimowicz D. 2021. Spatial rearrangement of the Streptomyces venezuelae linear chromosome during sporogenic development. Nat Commun 12:5222. doi: 10.1038/s41467-021-25461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Falguera JVT, Stratton KJ, Bush MJ, Jani C, Findlay KC, Schlimpert S, Nodwell JRY. 2022. DNA damage-induced block of sporulation in Streptomyces venezuelae involves downregulation of ssgB. Microbiology 168:e001198. doi: 10.1099/mic.0.001222. [DOI] [PubMed] [Google Scholar]
- 78.Donczew M, Mackiewicz P, Wróbel A, Flärdh K, Zakrzewska-Czerwińska J, Jakimowicz D. 2016. ParA and ParB coordinate chromosome segregation with cell elongation and division during Streptomyces sporulation. Open Biol 6:150263. doi: 10.1098/rsob.150263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces Genetics. John Innes Foundation, Norwich, UK. [Google Scholar]
- 80.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA 100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gust B, Chandra G, Jakimowicz D, Yuqing T, Bruton CJ, Chater KF. 2004. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv Appl Microbiol 54:107–128. doi: 10.1016/s0065-2164(04)54004-2. [DOI] [PubMed] [Google Scholar]
- 82.Tong Y, Charusanti P, Zhang L, Weber T, Lee SY. 2015. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth Biol 4:1020–1029. doi: 10.1021/acssynbio.5b00038. [DOI] [PubMed] [Google Scholar]
- 83.Tong Y, Whitford CM, Blin K, Jørgensen TS, Weber T, Lee SY. 2020. CRISPR–Cas9, CRISPRi and CRISPR-BEST-mediated genetic manipulation in streptomycetes. Nat Protoc 15:2470–2502. doi: 10.1038/s41596-020-0339-z. [DOI] [PubMed] [Google Scholar]
- 84.Blin K, Shaw S, Tong Y, Weber T. 2020. Designing sgRNAs for CRISPR-BEST base editing applications with CRISPy-web 2.0. Synth Syst Biotechnol 5:99–102. doi: 10.1016/j.synbio.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mitousis L, Thoma Y, Musiol-Kroll EM. 2020. An update on molecular tools for genetic engineering of actinomycetes—the source of important antibiotics and other valuable compounds. Antibiotics (Basel) 9:494. doi: 10.3390/antibiotics9080494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rudolph MM, Vockenhuber M-P, Suess B. 2013. Synthetic riboswitches for the conditional control of gene expression in Streptomyces coelicolor. Microbiology (Reading) 159:1416–1422. doi: 10.1099/mic.0.067322-0. [DOI] [PubMed] [Google Scholar]
- 87.Rodríguez-García A, Combes P, Pérez-Redondo R, Smith MCA, Smith MCM. 2005. Natural and synthetic tetracycline-inducible promoters for use in the antibiotic-producing bacteria Streptomyces. Nucleic Acids Res 33:e87. doi: 10.1093/nar/gni086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ji C-H, Kim J-P, Kang H-S. 2018. Library of synthetic Streptomyces regulatory sequences for use in promoter engineering of natural product biosynthetic gene clusters. ACS Synth Biol 7:1946–1955. doi: 10.1021/acssynbio.8b00175. [DOI] [PubMed] [Google Scholar]
- 89.Siegl T, Tokovenko B, Myronovskyi M, Luzhetskyy A. 2013. Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab Eng 19:98–106. doi: 10.1016/j.ymben.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 90.Phelan RM, Sachs D, Petkiewicz SJ, Barajas JF, Blake-Hedges JM, Thompson MG, Reider Apel A, Rasor BJ, Katz L, Keasling JD. 2017. Development of next generation synthetic biology tools for use in Streptomyces venezuelae. ACS Synth Biol 6:159–166. doi: 10.1021/acssynbio.6b00202. [DOI] [PubMed] [Google Scholar]
- 91.Blin K, Shaw S, Augustijn HE, Reitz ZL, Biermann F, Alanjary M, Fetter A, Terlouw BR, Metcalf WW, Helfrich EJN, van Wezel GP, Medema MH, Weber T. 2023. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. doi: 10.1093/nar/gkad344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gomez-Escribano JP, Bibb MJ. 2011. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol 4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Feeney MA, Newitt JT, Addington E, Algora-Gallardo L, Allan C, Balis L, Birke AS, Castaño-Espriu L, Charkoudian LK, Devine R, Gayrard D, Hamilton J, Hennrich O, Hoskisson PA, Keith-Baker M, Klein JG, Kruasuwan W, Mark DR, Mast Y, McHugh RE, McLean TC, Mohit E, Munnoch JT, Murray J, Noble K, Otani H, Parra J, Pereira CF, Perry L, Pintor-Escobar L, Pritchard L, Prudence SMM, Russell AH, Schniete JK, Seipke RF, Sélem-Mojica N, Undabarrena A, Vind K, van Wezel GP, Wilkinson B, Worsley SF, Duncan KR, Fernández-Martínez LT, Hutchings MI. 2022. ActinoBase: tools and protocols for researchers working on Streptomyces and other filamentous actinobacteria. Microb Genom 8:mgen000824. doi: 10.1099/mgen.0.000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hopwood DA. 2019. Highlights of Streptomyces genetics. Heredity 123:23–32. doi: 10.1038/s41437-019-0196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Piret JM, Chater KF. 1985. Phage-mediated cloning of bldA, a region involved in Streptomyces coelicolor morphological development, and its analysis by genetic complementation. J Bacteriol 163:965–972. doi: 10.1128/jb.163.3.965-972.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lawlor EJ, Baylis HA, Chater KF. 1987. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev 1:1305–1310. doi: 10.1101/gad.1.10.1305. [DOI] [PubMed] [Google Scholar]
- 97.Nguyen KT, Tenor J, Stettler H, Nguyen LT, Nguyen LD, Thompson CJ. 2003. Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J Bacteriol 185:7291–7296. doi: 10.1128/JB.185.24.7291-7296.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Takano E, Tao M, Long F, Bibb MJ, Wang L, Li W, Buttner MJ, Bibb MJ, Deng ZX, Chater KF. 2003. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol Microbiol 50:475–486. doi: 10.1046/j.1365-2958.2003.03728.x. [DOI] [PubMed] [Google Scholar]
- 99.Kalan L, Gessner A, Thaker MN, Waglechner N, Zhu X, Szawiola A, Bechthold A, Wright GD, Zechel DL. 2013. A cryptic polyene biosynthetic gene cluster in Streptomyces calvus is expressed upon complementation with a functional bldA gene. Chem Biol 20:1214–1224. doi: 10.1016/j.chembiol.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 100.Lonetto MA, Brown KL, Rudd KE, Buttner MJ. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA 91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aínsa JA, Parry HD, Chater KF. 1999. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2). Mol Microbiol 34:607–619. doi: 10.1046/j.1365-2958.1999.01630.x. [DOI] [PubMed] [Google Scholar]
- 102.Molle V, Buttner MJ. 2000. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol Microbiol 36:1265–1278. doi: 10.1046/j.1365-2958.2000.01977.x. [DOI] [PubMed] [Google Scholar]
- 103.Al-Bassam MM, Bibb MJ, Bush MJ, Chandra G, Buttner MJ. 2014. Response regulator heterodimer formation controls a key stage in Streptomyces development. PLoS Genet 10:e1004554. doi: 10.1371/journal.pgen.1004554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Elliot MA, Locke TR, Galibois CM, Leskiw BK. 2003. BldD from Streptomyces coelicolor is a non-essential global regulator that binds its own promoter as a dimer. FEMS Microbiol Lett 225:35–40. doi: 10.1016/S0378-1097(03)00474-9. [DOI] [PubMed] [Google Scholar]
- 105.Elliot MA, Bibb MJ, Buttner MJ, Leskiw BK. 2001. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol Microbiol 40:257–269. doi: 10.1046/j.1365-2958.2001.02387.x. [DOI] [PubMed] [Google Scholar]
- 106.den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ. 2010. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol Microbiol 78:361–379. doi: 10.1111/j.1365-2958.2010.07338.x. [DOI] [PubMed] [Google Scholar]
- 107.Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, Buttner MJ. 2014. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158:1136–1147. doi: 10.1016/j.cell.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chater KF, Bruton CJ, Plaskitt KA, Buttner MJ, Méndez C, Helmann JD. 1989. The developmental fate of S. coelicolor hyphae depends upon a gene product homologous with the motility sigma factor of B. subtilis. Cell 59:133–143. doi: 10.1016/0092-8674(89)90876-3. [DOI] [PubMed] [Google Scholar]
- 109.Gallagher KA, Schumacher MA, Bush MJ, Bibb MJ, Chandra G, Holmes NA, Zeng W, Henderson M, Zhang H, Findlay KC, Brennan RG, Buttner MJ. 2020. c-di-GMP arms an anti-σ to control progression of multicellular differentiation in Streptomyces. Mol Cell 77:586–599. doi: 10.1016/j.molcel.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schumacher MA, Wörmann ME, Henderson M, Salinas R, Latoscha A, Al-Bassam MM, Singh KS, Barclay E, Gunka K, Tschowri N. 2022. Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP. Nat Commun 13:5834. doi: 10.1038/s41467-022-33537-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Flärdh K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol Microbiol 49:1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- 112.Fuchino K, Bagchi S, Cantlay S, Sandblad L, Wu D, Bergman J, Kamali-Moghaddam M, Flärdh K, Ausmees N. 2013. Dynamic gradients of an intermediate filament-like cytoskeleton are recruited by a polarity landmark during apical growth. Proc Natl Acad Sci USA 110:E1889–1897. doi: 10.1073/pnas.1305358110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ditkowski B, Holmes N, Rydzak J, Donczew M, Bezulska M, Ginda K, Kedzierski P, Zakrzewska-Czerwińska J, Kelemen GH, Jakimowicz D. 2013. Dynamic interplay of ParA with the polarity protein, Scy, coordinates the growth with chromosome segregation in Streptomyces coelicolor. Open Biol 3:130006. doi: 10.1098/rsob.130006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Holmes NA, Walshaw J, Leggett RM, Thibessard A, Dalton KA, Gillespie MD, Hemmings AM, Gust B, Kelemen GH. 2013. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proc Natl Acad Sci USA 110:E397–E406. doi: 10.1073/pnas.1210657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brown PJB, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. 2012. Polar growth in the Alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci USA 109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kodani S, Hudson ME, Durrant MC, Buttner MJ, Nodwell JR, Willey JM. 2004. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc Natl Acad Sci USA 101:11448–11453. doi: 10.1073/pnas.0404220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci USA 107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schwartz K, Syed AK, Stephenson RE, Rickard AH, Boles BR. 2012. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog 8:e1002744. doi: 10.1371/journal.ppat.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wildermuth H. 1970. Development and organization of the aerial mycelium in Streptomyces coelicolor. J Gen Microbiol 60:43–50. doi: 10.1099/00221287-60-1-43. [DOI] [PubMed] [Google Scholar]
- 121.Filippova SN, Vinogradova KA. 2017. Programmed cell death as one of the stages of streptomycete differentiation. Microbiology 86:439–454. doi: 10.1134/S0026261717040075. [DOI] [Google Scholar]
- 122.Manteca Á, Fernández M, Sánchez J. 2005. A death round affecting a young compartmentalized mycelium precedes aerial mycelium dismantling in confluent surface cultures of Streptomyces antibioticus. Microbiology (Reading) 151:3689–3697. doi: 10.1099/mic.0.28045-0. [DOI] [PubMed] [Google Scholar]
- 123.Tenconi E, Traxler M, Tellatin D, van Wezel GP, Rigali S. 2020. Prodiginines postpone the onset of sporulation in Streptomyces coelicolor. Antibiotics (Basel) 9:847. doi: 10.3390/antibiotics9120847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vladimirov M, Zhang RX, Mak S, Nodwell JR, Davidson AR. 2023. A contractile injection system is required for developmentally regulated cell death in Streptomyces coelicolor. Nat Commun 14:1469. doi: 10.1038/s41467-023-37087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Casu B, Sallmen JW, Schlimpert S, Pilhofer M. 2023. Cytoplasmic contractile injection systems mediate cell death in Streptomyces. Nat Microbiol 8:711–726. doi: 10.1038/s41564-023-01341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Leiman PG, Shneider MM. 2012. Contractile tail machines of bacteriophages. Adv Exp Med Biol 726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- 127.Taylor NMI, van Raaij MJ, Leiman PG. 2018. Contractile injection systems of bacteriophages and related systems. Mol Microbiol 108:6–15. doi: 10.1111/mmi.13921. [DOI] [PubMed] [Google Scholar]
- 128.Brackmann M, Nazarov S, Wang J, Basler M. 2017. Using force to punch holes: mechanics of contractile nanomachines. Trends Cell Biol 27:623–632. doi: 10.1016/j.tcb.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 129.Coulthurst S. 2019. The type VI secretion system: a versatile bacterial weapon. Microbiology (Reading) 165:503–515. doi: 10.1099/mic.0.000789. [DOI] [PubMed] [Google Scholar]
- 130.Chen L, Song N, Liu B, Zhang N, Alikhan N-F, Zhou Z, Zhou Y, Zhou S, Zheng D, Chen M, Hapeshi A, Healey J, Waterfield NR, Yang J, Yang G. 2019. Genome-wide identification and characterization of a superfamily of bacterial extracellular contractile injection systems. Cell Rep 29:511–521. doi: 10.1016/j.celrep.2019.08.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Geller AM, Pollin I, Zlotkin D, Danov A, Nachmias N, Andreopoulos WB, Shemesh K, Levy A. 2021. The extracellular contractile injection system is enriched in environmental microbes and associates with numerous toxins. Nat Commun 12:3743. doi: 10.1038/s41467-021-23777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McCormick JR, Su EP, Driks A, Losick R. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol Microbiol 14:243–254. doi: 10.1111/j.1365-2958.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 133.Santos-Beneit F, Roberts DM, Cantlay S, McCormick JR, Errington J. 2017. A mechanism for FtsZ-independent proliferation in Streptomyces. Nat Commun 8:1378. doi: 10.1038/s41467-017-01596-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schwedock J, McCormick JR, Angert ER, Nodwell JR, Losick R. 1997. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol Microbiol 25:847–858. doi: 10.1111/j.1365-2958.1997.mmi507.x. [DOI] [PubMed] [Google Scholar]
- 135.Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. 2011. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev 25:89–99. doi: 10.1101/gad.600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang L, Willemse J, Yagüe P, de Waal E, Claessen D, van Wezel GP. 2023. The SepF-like proteins SflA and SflB prevent ectopic localization of FtsZ and DivIVA during sporulation of Streptomyces coelicolor. Biochem Biophys Res Commun 645:79–87. doi: 10.1016/j.bbrc.2023.01.021. [DOI] [PubMed] [Google Scholar]
- 137.Zhang L, Willemse J, Claessen D, van Wezel GP. 2016. SepG coordinates sporulation-specific cell division and nucleoid organization in Streptomyces coelicolor. Open Biol 6:150164. doi: 10.1098/rsob.150164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang Z, Du C, de Barsy F, Liem M, Liakopoulos A, van Wezel GP, Choi YH, Claessen D, Rozen DE. 2020. Antibiotic production in Streptomyces is organized by a division of labor through terminal genomic differentiation. Sci Adv 6:eaay5781. doi: 10.1126/sciadv.aay5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang Z, Shitut S, Claushuis B, Claessen D, Rozen DE. 2022. Mutational meltdown of putative microbial altruists in Streptomyces coelicolor colonies. Nat Commun 13:2266. doi: 10.1038/s41467-022-29924-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jones SE, Ho L, Rees CA, Hill JE, Nodwell JR, Elliot MA. 2017. Streptomyces exploration is triggered by fungal interactions and volatile signals. eLife 6:e21738. doi: 10.7554/eLife.21738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jones SE, Pham CA, Zambri MP, McKillip J, Carlson EE, Elliot MA. 2019. Streptomyces volatile compounds influence exploration and microbial community dynamics by altering iron availability. mBio 10:e00171-19. doi: 10.1128/mBio.00171-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ramijan K, Ultee E, Willemse J, Zhang Z, Wondergem JAJ, van der Meij A, Heinrich D, Briegel A, van Wezel GP, Claessen D. 2018. Stress-induced formation of cell wall-deficient cells in filamentous actinomycetes. Nat Commun 9:5164. doi: 10.1038/s41467-018-07560-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ongenae V, Mabrouk AS, Crooijmans M, Rozen D, Briegel A, Claessen D. 2022. Reversible bacteriophage resistance by shedding the bacterial cell wall. Open Biol 12:210379. doi: 10.1098/rsob.210379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wohlfarth JC, Feldmüller M, Schneller A, Kilcher S, Burkolter M, Meile S, Pilhofer M, Schuppler M, Loessner MJ. 2023. l-Form conversion in Gram-positive bacteria enables escape from phage infection. Nat Microbiol 8:387–399. doi: 10.1038/s41564-022-01317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Laity C, Chater KF, Lewis CG, Buttner MJ. 1993. Genetic analysis of the phi C31-specific phage growth limitation (Pgl) system of Streptomyces coelicolor A3(2). Mol Microbiol 7:329–336. doi: 10.1111/j.1365-2958.1993.tb01124.x. [DOI] [PubMed] [Google Scholar]
- 146.Hoskisson PA, Sumby P, Smith MCM. 2015. The phage growth limitation system in Streptomyces coelicolor A(3)2 is a toxin/antitoxin system, comprising enzymes with DNA methyltransferase, protein kinase and ATPase activity. Virology 477:100–109. doi: 10.1016/j.virol.2014.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Goldfarb T, Sberro H, Weinstock E, Cohen O, Doron S, Charpak-Amikam Y, Afik S, Ofir G, Sorek R. 2015. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J 34:169–183. doi: 10.15252/embj.201489455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kronheim S, Daniel-Ivad M, Duan Z, Hwang S, Wong AI, Mantel I, Nodwell JR, Maxwell KL. 2018. A chemical defence against phage infection. Nature 564:283–286. doi: 10.1038/s41586-018-0767-x. [DOI] [PubMed] [Google Scholar]
- 149.Kever L, Hardy A, Luthe T, Hünnefeld M, Gätgens C, Milke L, Wiechert J, Wittmann J, Moraru C, Marienhagen J, Frunzke J. 2022. Aminoglycoside antibiotics inhibit phage infection by blocking an early step of the infection cycle. mBio 13:e0078322. doi: 10.1128/mbio.00783-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Khokhlov AS, Tovarova II, Borisova LN, Pliner SA, Shevchenko LN, Kornitskaia EI, Ivkina NS, Rapoport IA. 1967. The A-factor, responsible for streptomycin biosynthesis by mutant strains of Actinomyces streptomycini. Dokl Akad Nauk SSSR 177:232–235. [PubMed] [Google Scholar]
- 151.Ohnishi Y, Kameyama S, Onaka H, Horinouchi S. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol Microbiol 34:102–111. doi: 10.1046/j.1365-2958.1999.01579.x. [DOI] [PubMed] [Google Scholar]
- 152.Horinouchi S. 2007. Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Biosci Biotechnol Biochem 71:283–299. doi: 10.1271/bbb.60627. [DOI] [PubMed] [Google Scholar]
- 153.Nishida H, Ohnishi Y, Beppu T, Horinouchi S. 2007. Evolution of gamma-butyrolactone synthases and receptors in Streptomyces. Environ Microbiol 9:1986–1994. doi: 10.1111/j.1462-2920.2007.01314.x. [DOI] [PubMed] [Google Scholar]
- 154.Takano E. 2006. Gamma-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr Opin Microbiol 9:287–294. doi: 10.1016/j.mib.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 155.Chater KF, Horinouchi S. 2003. Signalling early developmental events in two highly diverged Streptomyces species. Mol Microbiol 48:9–15. doi: 10.1046/j.1365-2958.2003.03476.x. [DOI] [PubMed] [Google Scholar]