ABSTRACT
Streptococcus pneumoniae is an agent of otitis media, septicemia, and meningitis and remains the leading cause of community-acquired pneumonia regardless of vaccine use. Of the various strategies that S. pneumoniae takes to enhance its potential to colonize the human host, quorum sensing (QS) is an intercellular communication process that provides coordination of gene expression at a community level. Numerous putative QS systems are identifiable in the S. pneumoniae genome, but their gene-regulatory activities and contributions to fitness have yet to be fully evaluated. To contribute to assessing regulatory activities of rgg paralogs present in the D39 genome, we conducted transcriptomic analysis of mutants of six QS regulators. Our results find evidence that at least four QS regulators impact the expression of a polycistronic operon (encompassing genes spd_1517 to spd_1513) that is directly controlled by the Rgg/SHP1518 QS system. As an approach to unravel the convergent regulation placed on the spd_1513-1517 operon, we deployed transposon mutagenesis screening in search of upstream regulators of the Rgg/SHP1518 QS system. The screen identified two types of insertion mutants that result in increased activity of Rgg1518-dependent transcription, one type being where the transposon inserted into pepO, an annotated endopeptidase, and the other type being insertions in spxB, a pyruvate oxidase. We demonstrate that pneumococcal PepO degrades SHP1518 to prevent activation of Rgg/SHP1518 QS. Moreover, the glutamic acid residue in the conserved “HExxH” domain is indispensable for the catalytic function of PepO. Finally, we confirmed the metalloendopeptidase property of PepO, which requires zinc ions, but not other ions, to facilitate peptidyl hydrolysis.
IMPORTANCE Streptococcus pneumoniae uses quorum sensing to communicate and regulate virulence. In our study, we focused on one Rgg quorum sensing system (Rgg/SHP1518) and found that multiple other Rgg regulators also control it. We further identified two enzymes that inhibit Rgg/SHP1518 signaling and revealed and validated one enzyme’s mechanisms for breaking down quorum sensing signaling molecules. Our findings shed light on the complex regulatory network of quorum sensing in Streptococcus pneumoniae.
KEYWORDS: Rgg1518, quorum sensing, pepO, spxB, Streptococcus pneumoniae
INTRODUCTION
Streptococcus pneumoniae, a prominent human pathogen, asymptomatically colonizes the mucosal surfaces of the human nasopharynx and upper airway (1). Asymptomatic carriage precedes symptomatic progression, which can lead to serious infections such as bronchitis, otitis media, meningitis, and septicemia (2–4). Implementation of multivalent vaccines directed at pneumococcal polysaccharide capsules and protection from as many as 23 serotypes have decreased disease burdens globally; however, protection is limited, as more than 100 serotypes of S. pneumoniae exist (5). Furthermore, the ability of S. pneumoniae to acquire foreign DNA has facilitated the spread of multidrug-resistant strains and increased antigenic variation. The increased rates of antibiotic resistance and decreased effectiveness of vaccines make it imperative to develop a better understanding of S. pneumoniae and how it regulates virulence.
S. pneumoniae uses chemical communication, known as quorum sensing (QS), to coordinate behaviors such as biofilm formation, competence, and production of antimicrobial peptides (6–9). QS is accomplished through signaling peptides, commonly referred to as pheromones. During QS, pheromones are secreted through membrane transporters to the extracellular space and transmit information to surrounding bacteria (9–12). Secreted pheromones can either be detected at the cell surface through two-component regulatory systems or imported into the cell through generalized oligopeptide transporters, such as Ami. Internalized pheromones interact with cytoplasmic signaling sensors belonging to the family of RRNPP proteins (Rap, Rgg, NprR, PlcR, and PrgX) (13) that are found widespread among Firmicutes (14–23). Prior studies in the regulation of RRNPP QS systems have described the impact of environmental factors and protein regulators. Environmental factors, such as nutrient sources (21, 24–26), metal concentrations (27), and adhesion to epithelial cells (28), can alter the expression of these QS systems. More directly, protein regulators can target Rggs (15, 19, 23, 29), pheromone transporters (30, 31), or QS pheromones (32).
Numerous RRNPP family members are present in the pneumococcal genome, yet only some have been characterized to a limited extent. For example, following Rgg/SHP939 QS activation, a 12-gene cluster regulating capsule synthesis is upregulated, and the rgg939-overexpressing strain displays a thicker capsule and increased biofilm formation (25). Rgg/SHP144 is a CodY-regulated QS system that controls the expression of a virulence peptide (VP1) conserved in pneumococcal strains D39 and TIGR4 (25, 26, 33, 34). Rgg/SHP112 (RtgR/S) regulates peptidase-containing ABC transporters (PCATs) in the Sp9-BS68 strain (20). TprA/PhrA regulates a pneumococcal lantibiotic gene cluster in a carbon-dependent QS activation manner controlled by CcpA and GlnR, two master regulators of carbonate metabolism (21, 35). Several additional RRNPP paralogs exist, and evidence suggests their importance in pathogenesis. For example, in S. pneumoniae serotype 4 strain TIGR4, transposon sequencing (Tn-seq) and signature-tagged mutagenesis (STM) conducted by the Camilli laboratory demonstrated an in vivo role of four Rgg regulators (sp_0141, sp_1115, sp_1946, and sp_2090) (34, 36). Transposon mutants of these Rggs resulted in a significant decrease in fitness in mouse models of pneumonia and nasopharyngeal carriage (34, 36). In S. pneumoniae strain D39, Rgg1952 protects against paraquat-induced oxidative stress (37). Rgg1518 regulates sugar metabolism and capsule expression, and its deletion mutant attenuates colonization and virulence in a mouse model (38). Among these Rgg QS systems, one study proposed an internal QS interaction network. Specifically, Rgg939 upregulates the Rgg144-regulated VP1 locus (spd_145 to spd_147), and Rgg144 negatively regulates the genes spd_1514 to spd_1516 that are directly governed by Rgg1518 (25).
Despite the emerging knowledge of these pneumococcal RRNPP QS systems, they and others remain poorly characterized. We have limited knowledge regarding whether these communication networks are interconnected or regulated. Here, we report transcriptomic analysis of six rgg-mutant strains in S. pneumoniae D39 and find that the recently described Rgg/SHP1518 system (25, 38, 39) is collectively affected by four other Rgg transcriptional regulators. We describe a genetic screen that identified two enzymes, SpxB and PepO, that negatively regulate Rgg1518 signaling. We demonstrate that PepO degrades the pheromone corresponding to Rgg1518 and limits the activity of the signaling system.
RESULTS
The spd_1513-1517 operon is influenced by multiple Rgg transcriptional regulators.
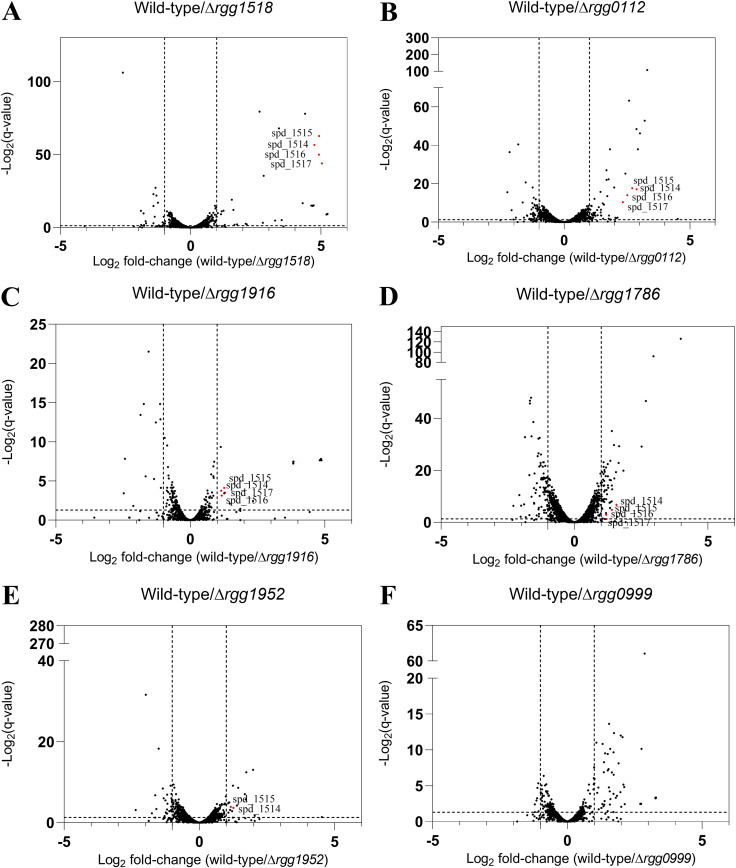
S. pneumoniae D39 encodes nine putative RRNPP-like proteins (Table S3 in the supplemental material), of which four have been reported to be involved in colonization, biofilm synthesis, virulence, and polysaccharide capsule production (Table S3) (21, 25, 38, 40, 41). In an attempt to illuminate the regulatory roles of Rgg proteins that have not been characterized, we constructed individual in-frame deletion mutants of five uncharacterized rgg genes and one gene that was recently studied (rgg1518) (38). The steady-state transcriptional profile of each exponentially growing strain was determined by RNA sequencing. Wild-type D39 and the six rgg mutants were cultured in a chemically defined medium (CDM) supplemented with glucose to mid-log phase (optical density at 600 nm [OD600] of 0.4) for RNA extraction. RNA sequencing and differential expression analysis that compared wild-type to the isogenic deletion of rgg1518 showed that the operon spd_1513 -1517, a putative ABC transporter complex, was downregulated 32-fold in the mutant (Fig. 1A; Table S4). This observation corroborates microarray data from an independent study indicating that spd_1514-1517 is directly regulated by Rgg/SHP1518 (20, 25). Unexpectedly, our transcriptomic analysis also revealed that the genes spd_1514 to spd_1517 were downregulated more than 2-fold in four additional deletion mutants, rgg112, rgg1786, rgg1916, and rgg1952. In contrast to these 5 mutants, Δrgg999 did not elicit changes in the expression of genes controlled by any of those known Rgg QS systems (Fig. 1). Furthermore, a prior microarray study showed that the spd_1514 to spd_1517 genes are also negatively regulated by Rgg/SHP939 and Rgg/SHP144, respectively (25). Together, these results suggest an interplay among the different QS network regulators that manifests in differential regulation of the genes adjacent to rgg1518, possibly working through a common regulatory mechanism.
FIG 1.
(A to F) Differential gene expression between wild-type (WT) D39 and rgg mutants growing in CDM (glucose). Volcano plots of wild-type D39 versus Δrgg1518 (A), Δrgg0112 (B), Δrgg1916 (C), Δrgg1786 (D), Δrgg1952 (E), and Δrgg0999 (F). Dashed guidelines indicate minimal q value significance (<0.05) and differential expression (>2-fold). Genes spd_1514 to spd_1517 that are differentially expressed are colored red.
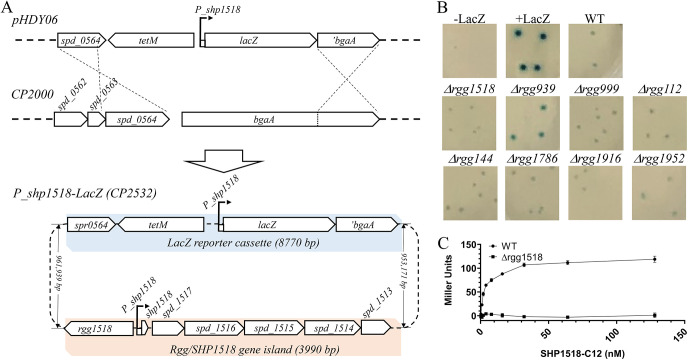
To facilitate directly targeted monitoring of the expression status of the spd_1513-1517 operon, a transcriptional reporter was generated by fusing DNA sequences located between rgg1518 and the small open reading frame (sORF) encoding the pheromone Rgg1518, designated shp1518 (38, 39). This DNA span contains the promoter expressing the operon consisting of shp1518 and spd_1513 to spd_1517. The promoter region was fused to lacZ and incorporated at the bga locus, which is considered an inactive region of the S. pneumoniae genome during laboratory culture (Fig. 2A). β-Galactosidase activity was evaluated in wild-type and rgg-mutant backgrounds in CDM agar (Fig. 2B). The wild-type strain yielded white colonies after 16 h of growth (Fig. S1); however, extending growth to 24 h led to an accumulation of weakly expressed β-galactosidase and a faint blue colony phenotype (Fig. 2B). As Rgg1518 is thought to serve as the primary regulator of the operon (20, 25, 38), the rgg1518 deletion mutant failed, as expected, to express lacZ and yielded white colonies at 24 h (Fig. 2B). The wild-type reporter, cultured in CDM broth and induced with the 12 C-terminal amino acids of SHP1518 (C12), stimulated lacZ expression (Fig. 2C). As little as 2 nM C12 was enough to achieve 50% of the maximum induction of lacZ that was obtained by C12 treatment. Even at the highest concentrations of C12 tested (128 nM), the Δrgg1518 strain was unresponsive and did not produce β-galactosidase activity above unstimulated cultures.
FIG 2.
(A) Genetic map of the Pshp1518-lacZ reporter and β-galactosidase expression in the wild-type strain and Δrgg mutants. The reporter was constructed by homologous recombination of pHDY06 plasmid (tetM: tetracycline resistant) into the bgaA locus of the parental strain CP2000 (−LacZ). (B) Colony color phenotype of wild-type (Pshp1518-lacZ) and Δrgg reporter strains after 40 h of growth in CDM (glucose) sandwich agar supplemented with 400 μg/mL X-Gal; −LacZ: the parental strain without the lacZ gene; +LacZ, a LacZ reporter strain where lacZ expression is driven by a constitutive kanamycin promoter; Δrgg, knockouts of a corresponding rgg regulator. (C) Synthetic C12 activates Rgg/SHP1518 quorum sensing in a dose-dependent manner in CDM. Gradient amounts of C12 peptides were incubated with wild-type and Δrgg1518 strains for 30 min. A Miller assay was performed to monitor LacZ expression of the three strains (recorded as UV420 absorbance). Data represent the average of three technical replicates.
Wild-type expression of lacZ was also evaluated in other types of media absent supplemental C12 to reveal the environmental factors required for QS activation (Fig. S1). Without C12 stimulation, lacZ expression was absent in Todd-Hewitt broth supplemented with yeast extract (THY) and CDM broth culture (data not shown). However, when grown in THY agar, lacZ was strongly expressed (Fig. S1). Among other media types tested (casein hydrolysate yeast extract [CAT], CDM, and tryptic soy broth-sheep blood [TSB-SB] agar), lacZ expression was only observed on CAT agar (Fig. S1). Additional rgg mutants were tested in CDM agar, including those previously found to influence spd_1513-1517 expression (25), and only deletion of rgg939 produced colonies bluer than the wild-type (Fig. 2B), consistent with the prior report (25). Notably, we did not observe visible blue intensity changes in the colonies of rgg mutants that downregulate the expression of spd_1513-1517 in CDM (glucose) broth culture. Because the growth conditions and Rgg QS systems that could affect spd_1513-1517 expression remain unclear, we aimed to identify any Rgg QS system upstream regulators that control spd_1513-1517 expression.
Transposon mutagenesis screenings identify two QS regulators, SpxB and PepO.
Given the confluence of regulation directed at Rgg1518, we used transposon mutagenesis to identify factors that influenced the expression of spd_1513-1517. We reasoned that transposon insertions within genes that negatively impact the expression of the spd_1513-1517 operon would appear blue on CDM plates. Likewise, a transposable element that enhances the transcription of regions adjacent to the transposon insertion site (theoretically possible if transcription extends across the insertion junction) could result in gain-of-function phenotypes. If insertion enhanced transcription of a positive regulator of spd_1513-1517, we would anticipate a blue colony phenotype.
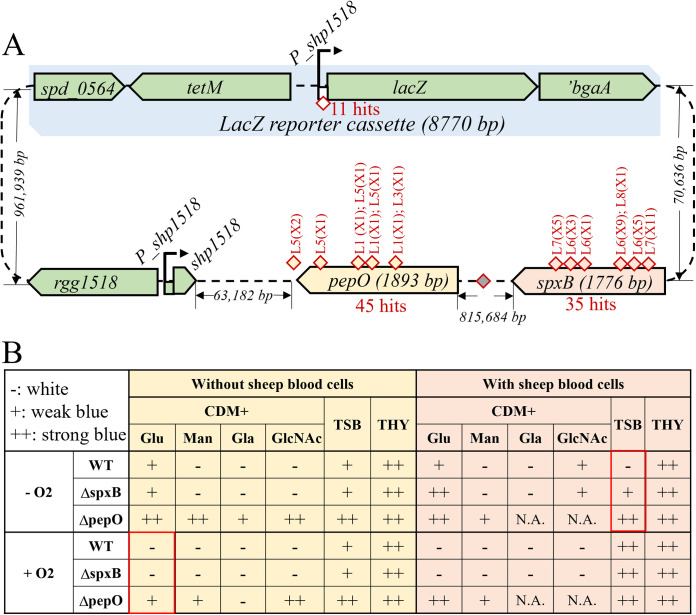
In vitro transposition using a hyperactive variant of Himar1, MarC9 (42), was used to generate insertion mutant libraries in CP2000 genomic DNA (gDNA). The transposon contains a kanamycin resistance marker (KANr) that lacks a transcription terminator; therefore, insertions of the transposon upstream of a gene could result in its expression from the constitutive KANr promoter, while intragenic insertions would inactivate target genes. Transposon-containing gDNA was used to transform the Pshp1518-lacZ reporter strain, and kanamycin-resistant colonies were selected on CDM agar plates containing X-Gal. Then, 30,540 transposon insertion mutants from five independently generated libraries were screened for blue color development, leading to 13 mutants that withstood backcrossing to ensure linkage to the transposon (Fig. 3A; Table S2; Fig. S3). Another 75,000 KANr colonies from 5 additional libraries were screened on TSB-SB X-Gal plates, leading to 151 blue colonies. Forty-four mutants mapped to pepO (8 from CDM and 37 from TSB-SB), 35 insertions mapped to spxB, and 11 were insertions upstream of lacZ (Fig. 3A). Again, backcrossing transformation experiments were performed to ensure transposon insertion linkage to the phenotype (Table S2; Fig. S3). Given the scale of our transposon screening and the frequency of hits mapped to the two genes, we conclude that PepO and SpxB are two novel regulators of Rgg/SHP1518 QS.
FIG 3.
Transposon mutants that elevate expression from Pshp1518-lacZ. (A) Summary of the two rounds of transposon mutagenesis. The lacZ reporter cassette (light blue), the Rgg/SHP1518 genes, pepO (yellow), and spxB (pink) are presented with labeled distances. Yellow and pink diamonds symbolize transposon insertions in pepO and spxB, respectively. The white diamond indicates transposon insertions in Pshp1518 that drive lacZ expression. (B) Evaluation of ΔspxB and ΔpepO colony color phenotypes under various growth conditions. Red boxes indicate the two conditions used in the transposon screenings. Wild-type and mutant cells were grown in different media under the following conditions: 37°C incubator with 5% CO2, with/without O2 (in a candle jar or not), and with/without sheep blood cells; −, white; +, weak blue; ++ strong blue; N.A., not available.
SpxB-regulated repression of Pshp1518 is growth medium dependent.
As spxB mutants turned blue only on TSB-SB agar and not on CDM agar, it is suggested that SpxB repressed Pshp1518 via a condition-dependent mechanism. We evaluated additional environmental factors to reveal conditions in which spxB repressed pheromone and spd_1513-1517 gene expression, including three media types (CDM, TSB-SB, and THY), four carbon sources (glucose, mannose, galactose, and N-acetylglucosamine), different oxygen levels, and with or without sheep blood supplement (Fig. 3B). Results indicated that only TSB-SB medium grown in a 5% CO2-supplemented incubator led to ΔspxB-specific activation of Pshp1518 (Fig. 3B). The mechanism by which SpxB represses Pshp1518 remains unknown, but several possibilities are considered in the Discussion.
PepO degrades SHP1518.
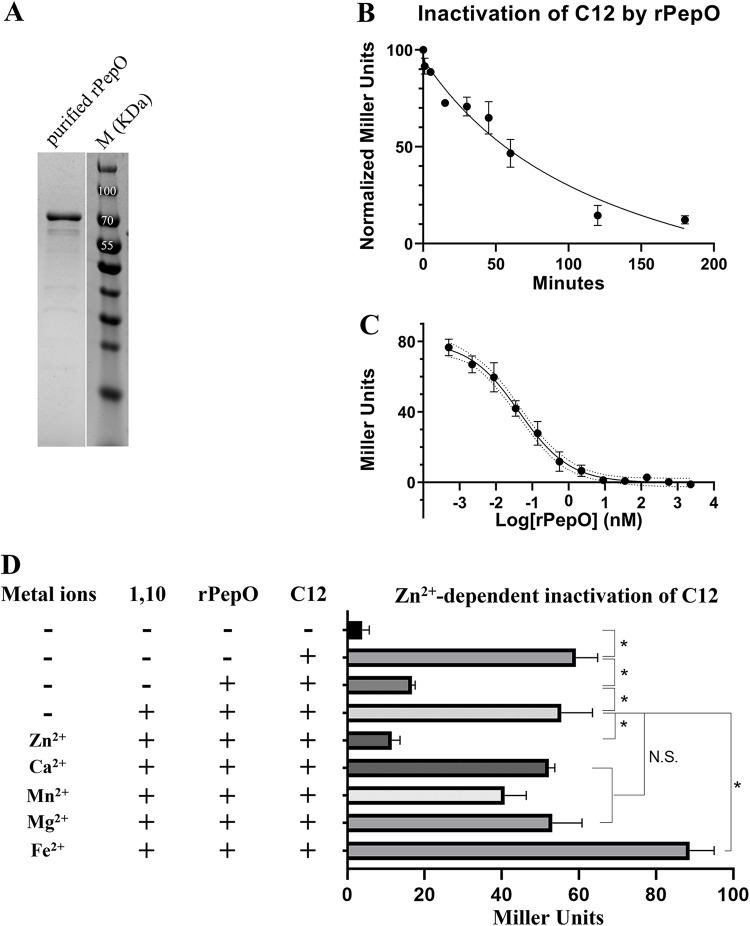
In S. pneumoniae, PepO is annotated as an endopeptidase (28, 43), as homology and structure prediction analyses suggest that PepO belongs to the M13 family of metallopeptidases present in all life forms except fungi, including bacteria, archaea, protozoa, plants, and animals and (https://www.ebi.ac.uk/merops/cgi-bin/famsum?family=M13). However, no study has tested pneumococcal PepO’s metallopeptidase properties. The amino acid sequence of S. pneumoniae PepO is 65% identical to that of PepO in Streptococcus pyogenes. Notably, the PepO of Streptococcus pyogenes degrades the SHP2 and SHP3 pheromones of the Rgg2/3 quorum sensing system (32). We, therefore, hypothesized that PepO might degrade SHP1518, thereby interfering with the ability of Rgg1518 to activate transcription of the spd_1513-1517 operon. To explore this possibility, we expressed and purified pneumococcal PepO in Escherichia coli (Fig. 4A) and evaluated its enzymatic potential to degrade SHP1518. Purified recombinant PepO (rPepO) was incubated with C12 for different amounts of time, and reaction products were evaluated for induction of the Pshp1518-lacZ reporter strain. As shown in Fig. 4B, in reactions with 50 nM rPepO and 5 μM C12, a 50% loss of pheromone activity occurred after 45 min, and a 90% reduction occurred after 2 h. We also titrated the concentration of rPepO and evaluated C12 activity after incubation with the enzyme for a fixed time of 180 min (Fig. 4C). Reaction products were diluted 100-fold and were applied to the Pshp1518-lacZ strain. A 50% reduction of 5 μM C12 activity required 50 pM rPepO (Fig. 4C), comparable to the SHP-degrading activity of S. pyogenes rPepO (50% activity loss of 0.5 μM SHP3 when it was incubated with 10 pM rPepO) (32). Our results suggest that PepO directly degrades SHP1518 and that it may act to abrogate QS activation.
FIG 4.
rPepO degrades C12 in a Zn2+-dependent manner. (A) SDS-PAGE image of purified rPepO (75.74 kDa). (B and C) C12 degradation by rPepO is dependent on reaction time (B) and concentration (C). (D) Comparison of rPepO activity in native, metal-chelated, and metal-supplemented conditions. Data in B to D represent the average of three biological repeats; *, P < 0.05. CDM (glucose) medium was used in the assays.
As PepO is predicted to belong to the M13 family of metallopeptidases whose members are mostly zinc dependent (44, 45), we tested whether metal ions were required for SHP turnover. The metal chelator 1,10-phenanthroline was applied to rPepO before and during incubation with C12 (46, 47). The addition led to the protection of SHP activity (Fig. 4D). By contrast, when amounts of individual metals were added to the reactions in 10-fold excess over the chelation potential of 1,10-phenanthroline, only Zn2+ supplementation restored proteolytic activity to rPepO. (Fig. 4D).
The enzymatic activity of PepO depends on the glutamic acid residue of the HExxH domain.
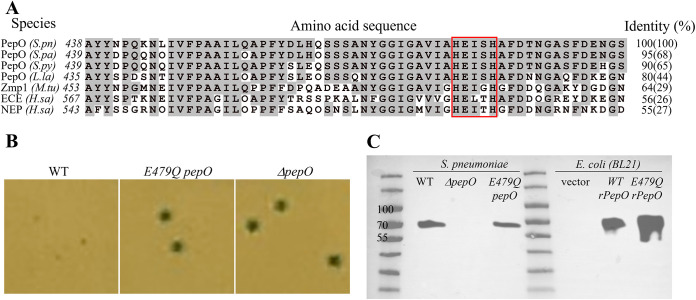
The canonical C-terminal motif of M13 metallopeptidases, HExxH, is a critical motif for zinc binding (Fig. 5A) (48, 49). However, the importance of this motif in streptococcal PepO has not been tested. Alignment of pneumococcal PepO with closest homologs from other streptococci finds the highest sequence identities to Streptococcus parasanguinis PepO (68%) and Streptococcus pyogenes PepO (66%), and these proteins even share substantial levels of homology with mammalian neutral endopeptidase (NEP) (50–54) and mammalian endothelin-converting enzyme (ECE) (55–57). As illustrated in Fig. 5A, all aligned PepO homologs contain the HExxH motif, and flanking residues are highly conserved across prokaryotic and eukaryotic examples (58, 59).
FIG 5.
The glutamic acid residue in the conserved “HExxH” catalytic motif is critical for PepO’s quorum quenching activity. (A) Sequence and alignment of S. pneumoniae PepO and its homologs surrounding the HExxH motif (red box). Gray-highlighted residues are identical to those of the S. pneumoniae PepO. Sequences and NCBI accession numbers are as follows: S.pn, Streptococcus pneumoniae PepO, WP_054394657.1; S.pa, Streptococcus parasanguinis PepO, MBF1717056.1; S.py, Streptococcus pyogenes PepO, WP_136303158.1; L.la, Lactococcus lactis PepO, WP_143457511.1; M.tu, Mycobacterium tuberculosis Zmp1, KBK61972.1; H.sa, Homo sapiens ECE isoform 1, BAG59124.1; H.sa, Homo sapiens NEP, AAI43466.1. Sequence identities are compared for the 58-amino-acid stretch surrounding the HExxH motif (and full-length protein). (B) Blue/white colony phenotypes of pneumococcal strains (Pshp1518-lacZ) expressing wild-type PepO, E479Q PepO, or ΔpepO in CDM (glucose) agar. (C) Western blotting of wild-type and mutant PepO expressed in S. pneumoniae or E. coli BL21. Samples are prepared from whole-cell lysates of Pshp1518-lacZ S. pneumoniae strains containing the indicated genotype or E. coli BL21 strains. For blotting, the membrane was incubated overnight at 4°C with a rabbit anti-PepO antibody (1:1,000) raised against pneumococcal PepO (43) followed by a 30-min incubation with a goat anti-rabbit horseradish peroxidase (HRP) secondary antibody (1:10,000). The sizes of native PepO and rPepO are 71.9 kDa and 74.5 kDa, respectively.
The structural data of PepO are limited, as only one bacterial PepO crystal structure is available in Protein Data Bank (PDB) from Mycobacterium tuberculosis, annotated as Zmp1 (49); it indicates that the zinc ion is coordinated by the nitrogen of the two histidines and the oxygen of glutamate, and the glutamate residue polarizes the water molecule that facilitates the nucleophilic attack on the substrate peptide bond (32, 49). Given the similar activities of rPepO between S. pneumoniae and S. pyogenes and the high sequence identity around the HExxH catalytic domain of S. pneumoniae, S. pyogenes, and M. tuberculosis, we aimed to understand if the HExxH catalytic domain is crucial for the endopeptidase activity of PepO in quenching Rgg/SHP1518 QS (49). The substitution of glutamate for glutamine was designed to retain the enzyme’s shape but disrupt catalytic activity. Expressing PepO-E479Q in S. pneumoniae did not alter protein amounts detected in cell lysates (Fig. 5C). However, the E479Q mutant produced blue colonies, resembling the phenotype of the ΔpepO mutant (Fig. 5B), indicating that a single amino acid change in the HExxH domain was sufficient to abolish PepO’s activity toward SHP1518.
Here, we confirmed the indispensable role of HExxH in PepO’s catalytic function. Based on this, we reason that any PepO inhibitor targeting the HExxH domain could disrupt pneumococcal PepO’s regulation of Rgg/SHP1518 QS.
DISCUSSION
By studying S. pneumoniae transcriptomes under various conditions, researchers have obtained invaluable data regarding pneumococcal regulatory networks, virulence factors, and immune evasion strategies (28, 60–65). The transcriptomic analysis in this study focused on regulatory system mutants, which helped map QS regulons and their cross-talk (Fig. 6). Our initial objective was to inform on regulatory networks governed by Rgg/pheromone QS systems. Our finding echoed a previous report indicating that more than one Rgg QS system controls the spd_1513-1517 system (25). Furthermore, our result extends the number of Rgg regulators of the cross-talk network regulating spd_1513-1517 from three to seven. However, this approach was limited by a lack of knowledge pertaining to the optimization of growth conditions that allow for Rgg1518 QS activation to occur, since most differences of spd_1513-1517 expression were minor and only Δrgg939 is validated by the Pshp1518-lacZ reporter.
FIG 6.
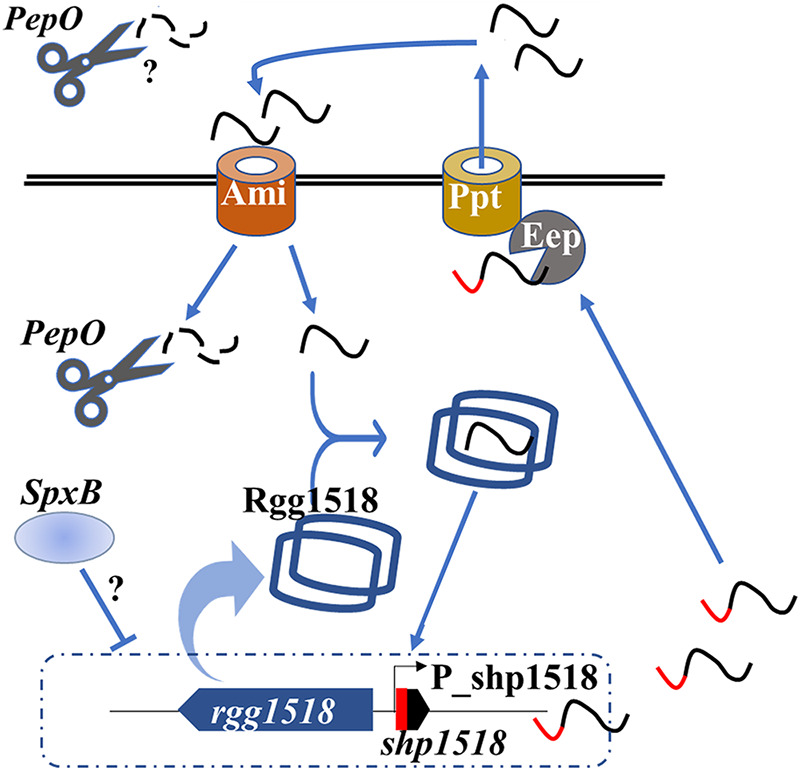
A hypothetical model of PepO repressing Rgg/SHP1518 quorum sensing. The precursor SHP expressed from shp1518 is exported via PptAB (15, 20, 30) and imported via the AmiABC transporter (20). Mature SHP binds to and activates Rgg/SHP1518 (blue dimer shaped) to regulate the expression of shp1518 and the spd_1513-1517 operon. Precursor (red and black) or mature (black) SHP1518 peptide is degraded during the process by PepO (black scissors) intracellularly or extracellularly. SpxB represses (indicated by a T-bar) Rgg/SHP1518 in an unknown acetyl phosphate pathway. Arrows indicate transportation or binding to an indicated target.
The observed cross-talk among Rgg systems inspired us to use transposon mutagenesis to identify factors that impact the expression of the Rgg1518 target operon. We established a Tn mutagenesis platform and screened a total of approximately 105,000 independent Tn insertion mutants to search for upstream regulators of spd_1513-1517. We identified two classes of loss-of-function mutants in distant genes, pepO::Tn and spxB::Tn, that resulted in Rgg/SHP1518 QS activation. In the screening, Rgg939 was not identified as an Rgg/SHP1518 negative regulator, as suggested by Fig. 2B. We believe the reason is the differences between the length of growth of the two experiments. The incubation time of the transposon screening that identified PepO was 16 h, whereas the cells from Fig. 2B were incubated under the same conditions for 40 h because we did not observe a distinct difference in the colony phenotypes after 16 h of growth. We believe the phenotype from Δrgg939 reveals its role in the negative regulation of Rgg1518 QS, but it was not strong enough to be identified during our screening.
The spxB gene (spd_0636) has a high degree of conservation among many streptococci (66, 67). The expression of spxB is regulated by catabolite control protein A (CcpA) in many streptococcal species, as the CcpA binding motif is common upstream of spxB (68, 69). In S. pneumoniae, a consensus CcpA binding motif is found and located at position −63 to −47 upstream of the spxB translational start site, suggesting that pneumococcal spxB is also regulated by CcpA. This interpretation is strengthened by our observations of Rgg/SHP1518 QS activation in certain growth media with different carbon sources (Fig. 3B). SpxB is a pyruvate oxidase that converts pyruvate to acetyl phosphate (AcP), carbon dioxide, and hydrogen peroxide. This reaction requires oxygen, water, and vitamin B1 as a cofactor. AcP is one of the major sources of phosphate in the cell. Evidence has shown that inactivating the genes of SpxB could result in deficient AcP levels and strongly reduced gene expression mediated by a two-component system (70). S. pneumoniae has 13 two-component regulatory systems that differentially regulate gene expression as a function of phosphorylation of response regulator proteins by the sensor kinase (71–73). Therefore, an SpxB-dependent two-component system might explain the phenotypes observed in the ΔspxB mutant.
PepO (spd_1460) is annotated as an endopeptidase that belongs to the M13 family proteins (74). In the MEROPS database, the M13 family comprises metalloendopeptidases that are confined to acting on peptides with no more than 40 residues (75). Functions of the M13 family proteins include posttranslational modification, protein turnover, and chaperones (52, 53, 59, 76). Unlike ΔspxB, the phenotype observed for ΔpepO is not growth media dependent. We tested and compared the blue colony phenotype of ΔpepO with that of wild-type in CDM, CAT, and THY agar, and ΔpepO showed a much stronger blue intensity than the wild-type in all three media (data not shown). However, the CAT and THY media lead to some level of autoinduction of Pshp1518, which results in a weak (CAT) and strong (THY) blue phenotype of wild-type cells (Fig. S1 in the supplemental material). We reason that PepO is capable of degrading SHP1518 and, therefore, may reduce the concentration of SHP1518 in S. pneumoniae cultures. Still, large amounts of peptides in rich media like CAT or THY may partially affect its function. If levels of SHP1518 remain below the concentration needed to bind and stimulate Rgg1518, transcription of the Rgg/SHP1518 target genes could be reduced. Considering that multiple Rgg QS systems were seen to regulate the spd_1513-1517 operon and the fact that all QS systems require a threshold concentration of their cognate pheromones to be activated, we suspect that QS cross-talk may be impacted by the PepO endopeptidase. This interpretation could be tested by in vitro assays mixing pneumococcal rPepO with different synthetic SHPs intraspecifically and interspecifically. We infer that PepO can affect QS, but whether it interferes with pheromone production, pheromone sensing, or both remains an open question.
The capacity of PepO to degrade intercellular signals is complicated by contradictory findings pertaining to the cellular location of the enzyme. S. pyogenes PepO resides in the cytoplasm (32). Similarly, PepO is also found in the cytoplasm of S. parasanguinis (54). However, Agarwal et al. claim that S. pneumoniae PepO is a moonlighting protein that functions as a cytosolic and membrane-bound protease that works on extracellular polypeptides (43, 77). This study does not explore the cellular localization of PepO when it interacts with the peptides, so we are unclear if PepO degrades the mature form of SHP1518 intra- or extracellularly. Also, we did not investigate the ability of PepO to digest the prepeptide form of SHP1518. Yet, we understand that knowing this could further facilitate interpreting its role in regulating QS.
In S. pneumoniae, the endopeptidase activity of PepO was evaluated in two in vitro studies assessing the degradation of competence-stimulating peptide CSP, which is the essential peptide regulating pneumococcal competence QS (78, 79). Their results indicated that PepO degrades CSP in vitro; however, the ΔpepO mutant showed no change in transformation efficiency (79). Therefore, these findings do not support a hypothesis that PepO participates in competence QS regulation. In this study, we revealed, for the first time, the role of pneumococcal PepO in regulating QS and demonstrated that both the enzymatic profile of PepO and its in vitro activity are consistent with its QS-regulatory properties.
Besides its endopeptidase nature, PepO is also regarded as a virulence factor. For example, purified pneumococcal rPepO elicited a robust innate immune response in mice via Toll-like receptor 2 (TLR2)/TLR4 signaling pathways (80). These responses include particle uptake by macrophages (81), promoting host anti-infection responses by autophagy (82), and eliciting interleukin-8 (IL-8) and interferon-γ-inducible protein 10 (IP-10) expression from human bronchial epithelial cells (83). In an in vivo pneumococcal-host infection transcriptomic study, pepO is highly expressed under infection conditions, and deletion of pepO resulted in a significant decline of bacterial burden at its corresponding anatomical site (65). While none of these virulence studies considered the role of PepO in regulating S. pneumoniae QS, results from our study suggest a possible mechanism of action of PepO in regulating the RRNPP QS systems by targeting their pheromones to govern virulence functions.
MATERIALS AND METHODS
Bacterial strains and culture media.
S. pneumoniae strains used in this study are descendants of the laboratory strain Rx (84), an avirulent, unencapsulated derivative of strain D39 (85). CP2000 (86) is an Rx-derived strain that contains a complete knockout of the capsule synthesis locus (Δcps), a mutation in the β-galactosidase gene, and other mutations listed in Table S1 in the supplemental material. Other pneumococcal strains listed in Table S1 are derivatives of CP2000 constructed by homologous recombination of either integrative plasmids or PCR donors containing homologous regions flanking the insert or deletion (Table S1; Appendix SA and SB). E. coli strains used in this study are derivatives of DH5α and BL21 purchased from Thermo Scientific. S. pneumoniae strain construction details are listed in Appendix SB. Four types of media were used for S. pneumoniae growth: casein hydrolysate yeast extract medium (CAT) (87), Todd-Hewitt yeast extract medium (THY) (88), tryptic soy broth-sheep blood medium (TSB-SB) (89), and a chemically defined medium (CDM) (90). TSB-SB agar plates contained 1.5% agar. CDM, CAT, and THY sandwich agar plates were prepared by adding 3 mL of each of the following fractions from the bottom to the top: 1.5% agar as a base (55°C), 0.75% agar-cell mixture (37°), 1.5% agar as a buffer layer (55°C), and 1.5% agar supplemented with/without selective drugs with 4× concentration (55°C). Except for the results related to SpxB, glucose is the only carbon source for CDM.
RNA isolation and sequencing.
Three independent cultures of wild-type S. pneumoniae D39 and rgg-mutant strains were cultured in 10 mL of CDM supplemented with 0.1% choline and 0.5% Oxyrase and grown to an OD600 of 0.4 at 37°C and 5% CO2. Cultures were chilled on ice and harvested by centrifugation. Cell pellets were suspended in 1 mL of RNALater (Ambion), incubated at room temperature for 10 min, repelleted, and stored at −80°C. Total RNA was extracted using an Ambion RiboPure RNA purification bacteria kit following the manufacturer’s instructions. RNA was assessed using Tapestation 2200 (Agilent) and was used to prepare cDNA libraries sequenced on an Illumina HiSeq 4000 with 100-bp single reads. mRNA sequencing and mapping were conducted by the University of Illinois at Chicago Research Informatics Core.
Transposase expression and purification.
100 mL mid-log-phase cultures of E. coli BL21 carrying the plasmid pMarC9 (a gift from the van Opijnen lab) were used for transposase purification.. The plasmid encodes a maltose-binding protein (MBP) fused to Himar1 Mariner MarC9 transposase (91). The fusion protein was induced with 0.3 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG; Goldbio) and purified by affinity chromatography using amylose resin (Biolabs). The enzyme was eluted with 10 mM maltose in 20 mM Tris (pH 7.4), 200 mM NaCl, 1 mM EDTA, cOmplete EDTA-free protease inhibitor cocktail (Roche), 2 mM dithiothreitol (DTT), and 10% (vol/vol) glycerol. Ten micrograms of purified MBP-MarC9 transposase was obtained and stored at −80°C as 20 ng/μL aliquots.
In vitro transposon mutagenesis and blue/white screening.
The protocol for in vitro transposon mutagenesis was adapted from van Opijnen (91) (Fig. S2). To mutate pneumococcal genomic DNA (gDNA), 15 μg of S. pneumoniae gDNA, 15 μg of pMagellan6 transposon plasmid (a gift from the van Opijnen lab [91]), and 15 μL of 20 ng/μL purified MarC9 transposase were mixed in 0.75 mL of buffer A (91) and incubated for 2 h at 30°C. The reaction mixture was heat inactivated for 10 min at 75°C before being placed on ice for 5 min. Transposon-mutagenized gDNA contains single-strand nicks, which were repaired as described previously (91). The repaired DNA was used directly to transform S. pneumoniae to generate mutant libraries. Two transposon libraries were generated by slightly different parameters. For the first setup, 400 mL of wild-type (CP2532, Pshp1518-lacZ reporter; Table S1) cells at an OD550 of 0.05 were transformed with 17.5 μg of Tn gDNA in THY. After transformation and incubation, KANr CFU (transposon insertion transformants) and total cell counts were recorded to evaluate the overall transformation efficiency. A total of 30,540 KANr CFU were obtained. For the second library, 400 mL of CP2532 cells at an OD550 of 0.05 were transformed with 22.5 μg of Tn gDNA in THY, and 75,000 transformants were obtained. Mutant cells from each round were divided into five groups (libraries) to screen for blue colonies. Colonies from each library (approximately 4,000 colonies/plate, 5 plates/library) were collected by scraping KANr cells from the TSB-SB agar surface. Cells were resuspended, diluted, regrown to an OD550 of 0.1 in THY broth, and stored at −80°C. Two rounds of screening were performed: in CDM sandwich X-Gal agar, screening was performed aerobically at 37°C for 16 h, and in TSB-SB X-Gal agar, screening was performed at 37°C with 5% CO2 for 16 h. Blue colonies from each library were isolated, regrown to an OD550 of 0.1, and stored in 12% glycerol at −80°C.
Colony PCR counterscreening.
Colony PCR was used as a method to identify transposon insertions in the promoter of lacZ. Lysates of isolated colonies suspended in 10 μL of Y-PER buffer (Thermo Scientific) for 10 min were diluted 10-fold and used at 1/20 volume of PCRs with primers DH030 and DH026.
Transposon insertion mapping.
To sequence regions flanking the transposon insertions, genomic DNA was digested with MmeI, which cuts 20 bp beyond the magellan6 termini, leaving a 2-base overhang for adapter ligation (DH058 + DH059; Table S1) (91). Sequences between the adapter and KANr gene were amplified using primers DH060 and DH061. The resulting 700-bp PCR products contained 20-bp mutant-specific DNA used to map transposon insertion sites by Sanger sequencing using primer DH061.
Assay of beta-galactosidase.
A previously published protocol was used to determine the β-galactosidase activity in lysates of culture samples (92). Samples were untreated or induced by adding synthetic pheromone SHP1518-C12 (the 12 C-terminal amino acids of SHP1518; Abclonal) for 30 min, lysed with Triton X-100, and monitored in a microplate reader (Biotek Synergy 2) at 420 nm. Reaction kinetics were recorded for 1.5 to 2.5 h unless otherwise specified. Specific β-galactosidase activities were calculated using the following equation: Miller units = 1,000 × (OD420)/(T × V × OD550), where T indicates the reaction time in minutes (duration of the enzymatic reaction), and V indicates the volume of culture used in the assay (in milliliters). One Miller unit represents nanomoles of o-nitrophenol/minute/milliliter of cells/OD550.
Recombinant pneumococcal PepO expression and purification.
pET28a was the backbone of the plasmids expressing wild-type N′-6×His-PepO (pHDY13) or N′-6×His-E479Q PepO (pHDY16) (Appendix SA). E. coli BL21 cells carrying pHDY13 were grown in 1 L of LB supplemented with 100 μg/mL ampicillin. Cells were incubated at 28°C and 225 rpm until the OD600 reached 0.4 to 0.6. IPTG was added to 0.3 mM, and induced cells were grown for 3 to 4 h. After induction, cells were harvested and lysed by two freeze-thaw cycles followed by sonication (Branson Digital Sonifier; 200 W, 50% amplitude, 10/15 s on/off, 12 cycles). Twenty-five milliliters of clarified lysates was loaded onto 3 mL of equilibrated nickel-nitrilotriacetic acid (Ni-NTA) resin (HisPur, Thermo Scientific). The 6×His-tagged rPepO was eluted with 250 mM imidazole and further purified by size-exclusion chromatography (SEC; Superdex 200 increase 10/300 GL; buffer: 50 mM NaH2PO4, 200 mM NaCl, 5 mM 2-mercaptoethanol [pH 7.0]; 0.6 to 0.8 mL/min; below 3 MPa). A 30-kDa-cutoff spin column (Vivaspin) was used to concentrate purified rPepO samples, and protein concentration was determined by measuring UV absorbance at 280 nm (Nanodrop ND-1000). Approximately 3 mg of purified rPepO was yielded and diluted to 4.6 μM aliquots in 50% glycerol stored at −80°C.
In vitro rPepO inactivation of SHP1518-C12 assay.
Two assays were performed to test the ability of rPepO to degrade C12 (the 12 C-terminal amino acids of SHP1518). First, 50 nM rPepO was incubated with 5 μM C12 for different amounts of time, ranging from 0 to 180 min. Alternatively, gradient amounts of rPepO were incubated with 5 μM C12 for 3 h. In both assays, after incubation, the reaction mixtures were diluted 100-fold and added to Pshp1518-lacZ cells to assay the remaining activity of C12 (Assay of beta-galactosidase). To test if the activity of rPepO depends on Zn2+ or other metal ions, 200 nM rPepO was mixed with/without 1 mM 1,10-phenanthroline and with/without 10 mM excess metal ions for 30 min. Then, C12 was added to sample mixtures to 5 μM and incubated for 3 h. Lastly, the reaction mixtures were diluted 100-fold and added to Pshp1518-lacZ cells to assay the remaining activity of C12 (Assay of beta-galactosidase).
ACKNOWLEDGMENTS
This study is funded, in part, by an NIAID grant to M.J.F. (AI091779) and by departmental fundings to D.A.M. and M.J.F. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
D.H. and I.L. conducted the experiments, performed data analysis, and drafted the original manuscript. M.J.F. and D.A.M. conceptualized and supervised this study, and they reviewed and edited the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Donald A. Morrison, Email: damorris@uic.edu.
Tina M. Henkin, Ohio State University
REFERENCES
- 1.Ghaffar F, Friedland IR, McCracken GH, Jr. 1999. Dynamics of nasopharyngeal colonization by Streptococcus pneumoniae. Pediatr Infect Dis J 18:638–646. doi: 10.1097/00006454-199907000-00016. [DOI] [PubMed] [Google Scholar]
- 2.Weiser JN, Ferreira DM, Paton JC. 2018. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol 16:355–367. doi: 10.1038/s41579-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogaert D, De Groot R, Hermans PW. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 4.Regev-Yochay G, Raz M, Dagan R, Porat N, Shainberg B, Pinco E, Keller N, Rubinstein E. 2004. Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin Infect Dis 38:632–639. doi: 10.1086/381547. [DOI] [PubMed] [Google Scholar]
- 5.Ganaie F, Saad JS, McGee L, van Tonder AJ, Bentley SD, Lo SW, Gladstone RA, Turner P, Keenan JD, Breiman RF, Nahm MH. 2020. A new pneumococcal capsule type, 10D, is the 100th serotype and has a large cps fragment from an oral Streptococcus. mBio 11:e00937-20. doi: 10.1128/mBio.00937-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan K, Sibley CD, Davidson CJ, Surette MG. 2009. Chemical interactions between organisms in microbial communities, p 1–17. In Collin M, Schuch R (ed), Bacterial sensing and signaling, vol 16. Karger, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- 7.Harapanahalli AK, Younes JA, Allan E, van der Mei HC, Busscher HJ. 2015. Chemical signals and mechanosensing in bacterial responses to their environment. PLoS Pathogens 11:e1005057. doi: 10.1371/journal.ppat.1005057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozada-Chavez I, Janga SC, Collado-Vides J. 2006. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res 34:3434–3445. doi: 10.1093/nar/gkl423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoch JA. 2000. Two-component and phosphorelay signal transduction. Curr Opin Microbiol 3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 11.Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 12.Stock JB, Stock AM, Mottonen JM. 1990. Signal transduction in bacteria. Nature 344:395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Pascual D, Monnet V, Gardan R. 2016. Bacterial cell-cell communication in the host via RRNPP peptide-binding regulators. Front Microbiol 7:706. doi: 10.3389/fmicb.2016.00706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyon WR, Gibson CM, Caparon MG. 1998. A role for trigger factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J 17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Pascual D, Gaudu P, Fleuchot B, Besset C, Rosinski-Chupin I, Guillot A, Monnet V, Gardan R. 2015. RovS and its associated signaling peptide form a cell-to-cell communication system required for Streptococcus agalactiae pathogenesis. mBio 6:e02306-14. doi: 10.1128/mBio.02306-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samen UM, Eikmanns BJ, Reinscheid DJ. 2006. The transcriptional regulator RovS controls the attachment of Streptococcus agalactiae to human epithelial cells and the expression of virulence genes. Infect Immun 74:5625–5635. doi: 10.1128/IAI.00667-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez A, Borges F, Gintz B, Decaris B, Leblond-Bourget N. 2006. The rggC locus, with a frameshift mutation, is involved in oxidative stress response by Streptococcus thermophilus. Arch Microbiol 186:161–169. doi: 10.1007/s00203-006-0130-8. [DOI] [PubMed] [Google Scholar]
- 18.Neely MN, Lyon WR, Runft DL, Caparon M. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J Bacteriol 185:5166–5174. doi: 10.1128/JB.185.17.5166-5174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loughman JA, Caparon MG. 2006. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J 25:5414–5422. doi: 10.1038/sj.emboj.7601393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang CY, Medlin JS, Nguyen DR, Disbennett WM, Dawid S. 2020. Molecular determinants of substrate selectivity of a pneumococcal Rgg-regulated peptidase-containing ABC transporter. mBio 11:e02502-19. doi: 10.1128/mBio.02502-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoover SE, Perez AJ, Tsui HC, Sinha D, Smiley DL, DiMarchi RD, Winkler ME, Lazazzera BA. 2015. A new quorum-sensing system (TprA/PhrA) for Streptococcus pneumoniae D39 that regulates a lantibiotic biosynthesis gene cluster. Mol Microbiol 97:229–243. doi: 10.1111/mmi.13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Declerck N, Bouillaut L, Chaix D, Rugani N, Slamti L, Hoh F, Lereclus D, Arold ST. 2007. Structure of PlcR: insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria. Proc Natl Acad Sci USA 104:18490–18495. doi: 10.1073/pnas.0704501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. 2011. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog 7:e1002190. doi: 10.1371/journal.ppat.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo JKK, McIver KS, Federle MJ. 2021. Carbon catabolite repression on the Rgg2/3 quorum sensing system in Streptococcus pyogenes is mediated by PTSMan and Mga. Mol Microbiol 117:525–538. doi: 10.1111/mmi.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhi X, Abdullah IT, Gazioglu O, Manzoor I, Shafeeq S, Kuipers OP, Hiller NL, Andrew PW, Yesilkaya H. 2018. Rgg-Shp regulators are important for pneumococcal colonization and invasion through their effect on mannose utilization and capsule synthesis. Sci Rep 8:6369. doi: 10.1038/s41598-018-24910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendriksen WT, Bootsma HJ, Estevao S, Hoogenboezem T, de Jong A, de Groot R, Kuipers OP, Hermans PWM. 2008. CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J Bacteriol 190:590–601. doi: 10.1128/JB.00917-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang JC, Jimenez JC, Federle MJ. 2015. Induction of a quorum sensing pathway by environmental signals enhances group A streptococcal resistance to lysozyme. Mol Microbiol 97:1097–1113. doi: 10.1111/mmi.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aprianto R, Slager J, Holsappel S, Veening JW. 2018. High-resolution analysis of the pneumococcal transcriptome under a wide range of infection-relevant conditions. Nucleic Acids Res 46:9990–10006. doi: 10.1093/nar/gky750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aggarwal C, Jimenez JC, Lee H, Chlipala GE, Ratia K, Federle MJ. 2015. Identification of quorum-sensing inhibitors disrupting signaling between Rgg and short hydrophobic peptides in streptococci. mBio 6:e00393-15. doi: 10.1128/mBio.00393-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang JC, Federle MJ. 2016. PptAB exports Rgg quorum-sensing peptides in Streptococcus. PLoS One 11:e0168461. doi: 10.1371/journal.pone.0168461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleuchot B, Gitton C, Guillot A, Vidic J, Nicolas P, Besset C, Fontaine L, Hols P, Leblond-Bourget N, Monnet V, Gardan R. 2011. Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol Microbiol 80:1102–1119. doi: 10.1111/j.1365-2958.2011.07633.x. [DOI] [PubMed] [Google Scholar]
- 32.Wilkening RV, Chang JC, Federle MJ. 2016. PepO, a CovRS-controlled endopeptidase, disrupts Streptococcus pyogenes quorum sensing. Mol Microbiol 99:71–87. doi: 10.1111/mmi.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuevas RA, Eutsey R, Kadam A, West-Roberts JA, Woolford CA, Mitchell AP, Mason KM, Hiller NL. 2017. A novel streptococcal cell-cell communication peptide promotes pneumococcal virulence and biofilm formation. Mol Microbiol 105:554–571. doi: 10.1111/mmi.13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hava D, Camilli A. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 45:1389–1406. doi: 10.1046/j.1365-2958.2002.03106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motib AS, Al-Bayati FAY, Manzoor I, Shafeeq S, Kadam A, Kuipers OP, Hiller NL, Andrew PW, Yesilkaya H. 2019. TprA/PhrA quorum sensing system has a major effect on pneumococcal survival in respiratory tract and blood, and its activity is controlled by CcpA and GInR. Front Cell Infect Microbiol 9:326. doi: 10.3389/fcimb.2019.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Opijnen T, Camilli A. 2012. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res 22:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bortoni ME, Terra VS, Hinds J, Andrew PW, Yesilkaya H. 2009. The pneumococcal response to oxidative stress includes a role for Rgg. Microbiology (Reading) 155:4123–4134. doi: 10.1099/mic.0.028282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shlla B, Gazioglu O, Shafeeq S, Manzoor I, Kuipers OP, Ulijasz A, Hiller NL, Andrew PW, Yesilkaya H. 2021. The Rgg1518 transcriptional regulator is a necessary facet of sugar metabolism and virulence in Streptococcus pneumoniae. Mol Microbiol 116:996–1008. doi: 10.1111/mmi.14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laczkovich I, Mangano K, Shao X, Hockenberry AJ, Gao Y, Mankin A, Vazquez-Laslop N, Federle MJ. 2022. Discovery of unannotated small open reading frames in Streptococcus pneumoniae D39 involved in quorum sensing and virulence using ribosome profiling. mBio 13:e0124722. doi: 10.1128/mbio.01247-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Junges R, Salvadori G, Shekhar S, Amdal HA, Periselneris JN, Chen T, Brown JS, Petersen FC. 2017. A quorum-sensing system that regulates Streptococcus pneumoniae biofilm formation and surface polysaccharide production. mSphere 2:e00324-17. doi: 10.1128/mSphere.00324-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carvalho SM, Kloosterman TG, Kuipers OP, Neves AR. 2011. CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PLoS One 6:e26707. doi: 10.1371/journal.pone.0026707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lampe DJ, Churchill MEA, Robertson HM. 1996. Purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J 15:5470–5479. doi: 10.1002/j.1460-2075.1996.tb00930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agarwal V, Kuchipudi A, Fulde M, Riesbeck K, Bergmann S, Blom AM. 2013. Streptococcus pneumoniae endopeptidase O (PepO) is a multifunctional plasminogen- and fibronectin-binding protein, facilitating evasion of innate immunity and invasion of host cells. J Biol Chem 288:6849–6863. doi: 10.1074/jbc.M112.405530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bland ND, Pinney JW, Thomas JE, Turner AJ, Isaac RE. 2008. Bioinformatic analysis of the neprilysin (M13) family of peptidases reveals complex evolutionary and functional relationships. BMC Evol Biol 8:16. doi: 10.1186/1471-2148-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang JY, Wang P, Li CY, Dong S, Song XY, Zhang XY, Xie BB, Zhou BC, Zhang YZ, Chen XL. 2015. Characterization of a new M13 metallopeptidase from deep-sea Shewanella sp. E525-6 and mechanistic insight into its catalysis. Front Microbiol 6:1498. doi: 10.3389/fmicb.2015.01498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song Y, Li S, Ray A, Das DS, Qi J, Samur MK, Tai YT, Munshi N, Carrasco RD, Chauhan D, Anderson KC. 2017. Blockade of deubiquitylating enzyme Rpn11 triggers apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Oncogene 36:5631–5638. doi: 10.1038/onc.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Felber JP, Coombs TL, Vallee BL. 1962. The mechanism of inhibition of carboxypeptidase A by 1,10-phenanthroline. Biochemistry 1:231–238. doi: 10.1021/bi00908a006. [DOI] [PubMed] [Google Scholar]
- 48.Turner AJ, Isaac RE, Coates D. 2001. The neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. Bioessays 23:261–269. doi:. [DOI] [PubMed] [Google Scholar]
- 49.Ferraris DM, Sbardella D, Petrera A, Marini S, Amstutz B, Coletta M, Sander P, Rizzi M. 2011. Crystal structure of Mycobacterium tuberculosis zinc-dependent metalloprotease-1 (Zmp1), a metalloprotease involved in pathogenicity. J Biol Chem 286:32475–32482. doi: 10.1074/jbc.M111.271809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen CY, Salles G, Seldin MF, Kister AE, Reinherz EL, Shipp MA. 1992. Murine common acute lymphoblastic leukemia antigen (CD10 neutral endopeptidase 24.11). Molecular characterization, chromosomal localization, and modeling of the active site. J Immunol 148:2817–2825. doi: 10.4049/jimmunol.148.9.2817. [DOI] [PubMed] [Google Scholar]
- 51.Devault A, Lazure C, Nault C, Le Moual H, Seidah NG, Chretien M, Kahn P, Powell J, Mallet J, Beaumont A. 1987. Amino acid sequence of rabbit kidney neutral endopeptidase 24.11 (enkephalinase) deduced from a complementary DNA. EMBO J 6:1317–1322. doi: 10.1002/j.1460-2075.1987.tb02370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malfroy B, Kuang WJ, Seeburg PH, Mason AJ, Schofield PR. 1988. Molecular cloning and amino acid sequence of human enkephalinase (neutral endopeptidase). FEBS Lett 229:206–210. doi: 10.1016/0014-5793(88)80828-7. [DOI] [PubMed] [Google Scholar]
- 53.Malfroy B, Schofield PR, Kuang WJ, Seeburg PH, Mason AJ, Henzel WJ. 1987. Molecular cloning and amino acid sequence of rat enkephalinase. Biochem Biophys Res Commun 144:59–66. doi: 10.1016/s0006-291x(87)80475-8. [DOI] [PubMed] [Google Scholar]
- 54.Froeliger EH, Oetjen J, Bond JP, Fives-Taylor P. 1999. Streptococcus parasanguis pepO encodes an endopeptidase with structure and activity similar to those of enzymes that modulate peptide receptor signaling in eukaryotic cells. Infect Immun 67:5206–5214. doi: 10.1128/IAI.67.10.5206-5214.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt M, Kroger B, Jacob E, Seulberger H, Subkowski T, Otter R, Meyer T, Schmalzing G, Hillen H. 1994. Molecular characterization of human and bovine endothelin converting enzyme (ECE-1). FEBS Lett 356:238–243. doi: 10.1016/0014-5793(94)01277-6. [DOI] [PubMed] [Google Scholar]
- 56.Shima H, Yamanouchi M, Omori K, Sugiura M, Kawashima K, Sato T. 1995. Endothelin-1 production and endothelin converting enzyme expression by guinea pig airway epithelial cells. Biochem Mol Biol Int 37:1001–1010. [PubMed] [Google Scholar]
- 57.Shimada K, Takahashi M, Tanzawa K. 1994. Cloning and functional expression of endothelin-converting enzyme from rat endothelial cells. J Biol Chem 269:18275–18278. doi: 10.1016/S0021-9258(17)32298-6. [DOI] [PubMed] [Google Scholar]
- 58.Hase CC, Finkelstein RA. 1993. Bacterial extracellular zinc-containing metalloproteases. Microbiol Rev 57:823–837. doi: 10.1128/mr.57.4.823-837.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rawlings ND, Barrett AJ. 1995. Evolutionary families of metallopeptidases. Methods Enzymol 248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 60.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Härtig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RAT, Nannapaneni P, Noone D, Pohl S, Rinn B, Rügheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, Bessières P, et al. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 61.Mader U, Nicolas P, Depke M, Pane-Farre J, Debarbouille M, van der Kooi-Pol MM, Guerin C, Derozier S, Hiron A, Jarmer H, Leduc A, Michalik S, Reilman E, Schaffer M, Schmidt F, Bessieres P, Noirot P, Hecker M, Msadek T, Volker U, van Dijl JM. 2016. Staphylococcus aureus transcriptome architecture: from laboratory to infection-mimicking conditions. PLoS Genet 12:e1005962. doi: 10.1371/journal.pgen.1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JCD. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 63.Chang X, Li Y, Ping J, Xing X-B, Sun H, Jia P, Wang C, Li Y-Y, Li Y-X. 2011. EcoBrowser: a web-based tool for visualizing transcriptome data of Escherichia coli. BMC Res Notes 4:405. doi: 10.1186/1756-0500-4-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolf T, Kammer P, Brunke S, Linde J. 2018. Two’s company: studying interspecies relationships with dual RNA-seq. Curr Opin Microbiol 42:7–12. doi: 10.1016/j.mib.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 65.D’Mello A, Riegler AN, Martinez E, Beno SM, Ricketts TD, Foxman EF, Orihuela CJ, Tettelin H. 2020. An in vivo atlas of host-pathogen transcriptomes during Streptococcus pneumoniae colonization and disease. Proc Natl Acad Sci USA 117:33507–33518. doi: 10.1073/pnas.2010428117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu L, Kreth J. 2012. The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid Med Cell Longev 2012:717843. doi: 10.1155/2012/717843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu L, Xu Y, Ferretti JJ, Kreth J. 2014. Probing oral microbial functionality—expression of spxB in plaque samples. PLoS One 9:e86685. doi: 10.1371/journal.pone.0086685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Warner JB, Lolkema JS. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol Mol Biol Rev 67:475–490. doi: 10.1128/MMBR.67.4.475-490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iyer R, Baliga NS, Camilli A. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol 187:8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marx P, Meiers M, Bruckner R. 2014. Activity of the response regulator CiaR in mutants of Streptococcus pneumoniae R6 altered in acetyl phosphate production. Front Microbiol 5:772. doi: 10.3389/fmicb.2014.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gamez G, Castro A, Gomez-Mejia A, Gallego M, Bedoya A, Camargo M, Hammerschmidt S. 2018. The variome of pneumococcal virulence factors and regulators. BMC Genomics 19:10. doi: 10.1186/s12864-017-4376-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grebe TW, Stock JB. 1999. The histidine protein kinase superfamily. Adv Microb Physiol 41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- 73.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 74.Slager J, Aprianto R, Veening JW. 2018. Deep genome annotation of the opportunistic human pathogen Streptococcus pneumoniae D39. Nucleic Acids Res 46:9971–9989. doi: 10.1093/nar/gky725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. 2018. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res 46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turner AJ, Brown CD, Carson JA, Barnes K. 2000. The neprilysin family in health and disease. Adv Exp Med Biol 477:229–240. doi: 10.1007/0-306-46826-3_25. [DOI] [PubMed] [Google Scholar]
- 77.Jeffery C. 2018. Intracellular proteins moonlighting as bacterial adhesion factors. AIMS Microbiol 4:362–376. doi: 10.3934/microbiol.2018.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Horne DP, Tomasz A. S.1977. Cell surface components implicated as attachment sites for the pneumococcal competence activator, p 11–34. In Portolès A, López R, Espinosa M (ed), Modern trends in bacterial transformation and transfection. Elsevier/North-Holland Biomedical Press, Amsterdam, the Netherlands. [Google Scholar]
- 79.Berge M, Langen H, Claverys JP, Martin B. 2002. Identification of a protein that inactivates the competence-stimulating peptide of Streptococcus pneumoniae. J Bacteriol 184:610–613. doi: 10.1128/JB.184.2.610-613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang H, Kang L, Yao H, He Y, Wang X, Xu W, Song Z, Yin Y, Zhang X. 2016. Streptococcus pneumoniae endopeptidase O (PepO) elicits a strong innate immune response in mice via TLR2 and TLR4 signaling pathways. Front Cell Infect Microbiol 6:23. doi: 10.3389/fcimb.2016.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yao H, Zhang H, Lan K, Wang H, Su Y, Li D, Song Z, Cui F, Yin Y, Zhang X. 2017. Purified Streptococcus pneumoniae endopeptidase O (PepO) enhances particle uptake by macrophages in a Toll-like receptor 2- and miR-155-dependent manner. Infect Immun 85:e01012-16. doi: 10.1128/IAI.01012-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shu Z, Yuan J, Wang H, Zhang J, Li S, Zhang H, Liu Y, Yin Y, Zhang X. 2020. Streptococcus pneumoniae PepO promotes host anti-infection defense via autophagy in a Toll-like receptor 2/4 dependent manner. Virulence 11:270–282. doi: 10.1080/21505594.2020.1739411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zou J, Zhou L, Hu C, Jing P, Guo X, Liu S, Lei Y, Yang S, Deng J, Zhang H. 2017. IL-8 and IP-10 expression from human bronchial epithelial cells BEAS-2B are promoted by Streptococcus pneumoniae endopeptidase O (PepO). BMC Microbiol 17:187. doi: 10.1186/s12866-017-1081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cuppone AM, Colombini L, Fox V, Pinzauti D, Santoro F, Pozzi G, Iannelli F. 2021. Complete genome sequence of Streptococcus pneumoniae strain Rx1, a Hex mismatch repair-deficient standard transformation recipient. Microbiol Resour Announc 10:e0079921. doi: 10.1128/MRA.00799-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weng L, Piotrowski A, Morrison DA. 2013. Exit from competence for genetic transformation in Streptococcus pneumoniae is regulated at multiple levels. PLoS One 8:e64197. doi: 10.1371/journal.pone.0064197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Porter RD, Guild WR. 1976. Characterization of some pneumococcal bacteriophages. J Virol 19:659–667. doi: 10.1128/JVI.19.2.659-667.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Todd EW, Hewitt LF. 1932. A new culture medium for the production of antigenic streptococcal hæmolysin. J Pathol 35:973–974. doi: 10.1002/path.1700350614. [DOI] [Google Scholar]
- 89.Schmid RE, Washington JA, II, Anhalt JP. 1978. Gentamicin-blood agar for isolation of Streptococcus pneumoniae from respiratory secretions. J Clin Microbiol 7:426–427. doi: 10.1128/jcm.7.5.426-427.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van de Rijn I, Kessler RE. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun 27:444–448. doi: 10.1128/iai.27.2.444-448.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van Opijnen T, Lazinski DW, Camilli A. 2014. Genome-wide fitness and genetic interactions determined by Tn-seq, a high-throughput massively parallel sequencing method for microorganisms. Curr Protoc Mol Biol 106:7.16.1–7.16.24. doi: 10.1002/0471142727.mb0716s106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lafleur L, Miller RG, Phillips RA. 1972. A quantitative assay for the progenitors of bone marrow-associated lymphocytes. J Exp Med 135:1363–1374. doi: 10.1084/jem.135.6.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S4, Fig. S1 to S4, and Appendices SA and SB. Download jb.00087-23-s0001.pdf, PDF file, 0.8 MB (883.6KB, pdf)