Abstract
Pseudomonas aeruginosa and the phytopathogen P. syringae produce the exopolysaccharide alginate, which is a copolymer of d-mannuronic and l-guluronic acids. One of the key regulatory genes controlling alginate biosynthesis in P. aeruginosa is algT, which encodes the alternate sigma factor, ς22. In the present study, the algT gene product from P. syringae pv. syringae showed 90% amino acid identity with its P. aeruginosa counterpart, and sequence analysis of the region flanking algT in P. syringae revealed the presence of nadB, mucA, and mucB in an arrangement virtually identical to that of P. aeruginosa. An algT mutant of P. syringae was defective in alginate production but could be complemented with wild-type algT from P. syringae or P. aeruginosa when expressed in trans. The algT mutant also displayed increased sensitivity to heat, paraquat, and hydrogen peroxide (H2O2); the latter two compounds are known to generate reactive oxygen intermediates. Signals for activation of algT gene expression in P. syringae were investigated with an algT::uidA transcriptional fusion. Like that in P. aeruginosa, algT transcription in P. syringae was activated by heat shock. However, algT expression in P. syringae was also stimulated by osmotic stress and by exposure to paraquat, H2O2, and copper sulfate. The latter two compounds are frequently encountered during colonization of plant tissue and may be unique signals for algT activation in P. syringae.
Many pseudomonads, including the phytopathogen Pseudomonas syringae, produce the exopolysaccharide alginate, a copolymer of O-acetylated β-1,4-linked d-mannuronic acid and its C-5 epimer, l-guluronic acid. P. syringae induces a wide variety of symptoms on plant hosts and can also exist as an epiphyte on plant surfaces without causing disease. Yu et al. (62) used a genetic approach to evaluate the role of alginate in the pathogenicity and epiphytic fitness of P. syringae pv. syringae 3525, which causes bacterial brown spot on beans. Alginate contributed significantly to both virulence and epiphytic survival of P. syringae pv. syringae 3525, perhaps by facilitating colonization and/or dissemination of the bacterium in planta (62).
Alginate has been extensively studied in P. aeruginosa, where it functions as a virulence factor in cystic fibrosis patients (47). An important feature of alginate production by P. aeruginosa is that the alginate biosynthetic genes are normally silent but are activated in the cystic fibrotic lung, which results in a mucoid phenotype. In P. aeruginosa, genes that encode the biosynthesis and regulation of alginate map to four chromosomal locations. With the exception of algC, which is located at 10 min, the structural genes are clustered within an 18-kb region located at 34 min (18, 48). The alginate biosynthetic gene cluster in P. aeruginosa is presumably organized as an operon with transcription initiating at the algD promoter (9).
Genes controlling the regulation of alginate production include algR1 (algR), algR2 (algQ), algR3 (algP), and algB (20, 54). AlgR1 functions as a response regulator and binds to multiple sites upstream of algC and algD (25, 42, 64). The genes which mediate the conversion to constitutive alginate production are located at 68 min on the P. aeruginosa chromosome and include algT (algU), mucA, mucB (algN), mucC (algM), and mucD (algY). The alternative sigma factor encoded by algT, ς22, is required for transcription of algD, algR1, and algT (21, 52, 60). Both the algD and algR1 promoters show a consensus sequence at the −35/10 region, a finding which is consistent with recognition by ς22, suggesting that an RNAP-ς22 complex binds to both promoters and positively regulates transcription (52). MucA is a negative regulator of algT transcription and encodes an anti-ς factor with affinity for ς22 (53, 61). MucB is also a negative regulator and is thought to interact with the periplasmic domain of MucA, thereby altering its conformation so that it binds ς22 and targets it for degradation (39). MucC and MucD also modulate the expression of algT and have been described elsewhere (6, 7).
As in P. aeruginosa, the alginate biosynthetic genes in other pseudomonads are normally silent (19). Interestingly, an indigenous plasmid designated pPSR12 conferred constitutive alginate production to P. syringae pv. syringae FF5 (29). pPSR12 does not contain homologs of the biosynthetic or regulatory genes which control alginate production in P. aeruginosa; instead, this plasmid presumably contains regulatory genes which have not been characterized (29). Mutagenesis of FF5(pPSR12) with Tn5 resulted in the isolation of several alginate defective (Alg−) mutants, including FF5.31 and FF5.32, which contain Tn5 insertions in algL and algR1, respectively (15, 46). The arrangement of the alginate structural gene cluster and the genes flanking algR1 were virtually identical in both P. syringae and P. aeruginosa (15, 46). However, complementation analyses indicated that the structural gene clusters in P. aeruginosa and P. syringae were not functionally interchangeable when expressed from their native promoters (46). Further experiments indicated that P. syringae, unlike P. aeruginosa, does not require a functional copy of algR1 for activation of the algD promoter (15).
In the present study, an Alg− mutant of P. syringae pv. syringae FF5(pPSR12) was shown to contain a Tn5 insertion upstream of the algT-mucABCD gene cluster. This region was cloned from P. syringae, and the role of algT in P. syringae was evaluated. An algT mutant was shown to be defective in alginate production, indicating that algT is essential for alginate biosynthesis in P. syringae. The algT mutant was also more susceptible to killing by heat and superoxide-generating redox cycling compounds, indicating that AlgT (ς22) regulates genes in P. syringae which respond to environmental stress.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Table 1 lists the bacterial strains and plasmids used in the present study. P. syringae was routinely maintained at 28°C on King’s medium B (30), mannitol-glutamate (MG) (26), or MG supplemented with yeast extract at 0.25 g/liter (MGY). Escherichia coli strains were grown on Luria-Bertani medium (41) at 37°C. Antibiotics were added to media at the following concentrations (μg/ml): ampicillin, 100; tetracycline, 12.5; kanamycin, 25; spectinomycin, 25; chloramphenicol, 25; and gentamicin, 2.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | 50 | |
| Pseudomonas syringae pv. syringae | ||
| FF5 | No detectable plasmids, nonmucoid | 29 |
| FF5.36 | Cur Kmr; contains pPSR12, slightly mucoid, nadB::Tn5 | This study |
| FF5.LK1 | Cmr; contains pPSR12, nonmucoid, algT::Cmr | This study |
| Plasmids | ||
| pBluescript SK(+) | Apr; ColEI origin, cloning vehicle | Stratagene |
| pCR2.1 | Apr Kmr; 3.9-kb cloning vector | Invitrogen |
| pRK415 | Tcr; RK2-derived cloning vector | 27 |
| pRK2013 | Kmr; helper plasmid | 17 |
| pRK7813 | Tcr; cosmid vector | 23 |
| pBBR1MCS | Cmr; 4.7-kb broad-host-range cloning vector | 31 |
| pBBR.Gus | Cmr; 6.6-kb promoter probe vector containing uidA in pBBR1MCS | 45 |
| pPSR12 | Cur Smr; 200-kb, confers constitutive alginate production to P. syringae pv. syringae FF5 | 29 |
| pSL1 | Apr Cmr; 650-bp Cmr cassette in pBluescript SK(+) | 33 |
| pMGm | Apr Gmr; 2-kb Gmr cassette | 43 |
| pJG309 | Tcr; contains algT from P. aeruginosa | 19 |
| pFF5.36 | Tcr Kmr; contains Tn5-inactivated alginate genes from FF5.36 in pRK7813 | This study |
| pFF5.36B | Apr Kmr; contains a portion of Tn5, nadB, and algT as a 6-kb BamHI fragment from pFF5.36 in pBluescript SK(+) | This study |
| pLKT5 | Tcr; cosmid clone from FF5(pPSR12) in pRK7813 | This study |
| pBTB6.5 | Apr; contains a 6.5-kb BamHI fragment from pLKT5 in pBluescript SK(+) | This study |
| pBTB.Cm | Apr Cmr; contains algT::Cmr in pBluescript SK(+) | This study |
| pRTB6.5 | Tcr; contains a 6.5-kb BamHI fragment from pLK5T5 in pRK415 | This study |
| pRTB6.5.Cm | Tcr Cmr; contains algT::Cmr in pRK415 | This study |
| pCRalgTA | AprKmr; 1.2-kb PCR fragment in pCR2.1 | This study |
| pCRalgTI | AprKmr; 1.2-kb PCR fragment in pCR2.1 | This study |
| palgTA | Cmr; 1-kb HindIII/PstI fragment containing the algT promoter region in pBBR.Gus; transcriptionally active | This study |
| palgTA.1 | Cmr Gmr; contains Gmr cassette from pMGm in palgTA | This study |
| palgTI | Cmr; 1-kb KpnI/HindIII fragment containing the algT promoter region in pBBR.Gus; transcriptionally inactive | This study |
Molecular genetic techniques.
Plasmids were isolated from P. syringae as described by Kado and Liu (24). Restriction enzyme digests, agarose gel electrophoresis, Southern transfers, and isolation of DNA fragments from agarose gels were performed by using standard protocols (50). Genomic DNA was isolated from P. syringae by using established procedures (56), and a genomic library of FF5.36 was constructed in pRK7813 as described previously (2). Clones were mobilized into recipient strains by using a triparental mating procedure and the mobilizer plasmid pRK2013 (4).
DNA fragments were labeled with digoxigenin (Genius Labeling and Detection Kit; Boehringer Mannheim, Indianapolis, Ind.) or with [α-32P]dCTP (Rad Prime DNA Labeling System; Gibco BRL, Gaithersburg, Md.). Hybridizations and posthybridization washes were conducted under high-stringency conditions (57).
Isolation and quantitation of alginate.
Selected strains were inoculated by dilution streaking to MGY agar (three plates per strain) and incubated at 28°C for 96 h. Each plate was handled separately for quantitation of alginate. Cells were washed from each plate and resuspended in 0.9% NaCl. Removal of cellular material from the mucoid growth and estimation of total cellular protein were performed as described previously (40). Alginate production was assessed by the carbazole method, an assay which quantifies the total amount of uronic acid polymers (40). In addition to alginate, other uronic acid polymers are detected by this assay, but we previously demonstrated that these are very minor components of the mucoid material isolated from FF5(pPSR12) (29). Alginic acid from seaweed (Macrocystis pyrifera; Sigma Chemical Co., St. Louis, Mo.) was used as a standard in these experiments. Mean values of three replicates were expressed as micrograms of alginate per milligrams of protein.
DNA sequencing and analysis.
Nucleotide sequencing reactions were performed by the dideoxynucleotide method (50) with AmpliTaq DNA polymerase (Perkin-Elmer, Foster City, Calif.). Automated DNA sequencing was accomplished by using an ABI 373A apparatus and the ABI PRISM Dye Primer Cycle Sequencing Kit (Perkin-Elmer). Automated sequencing was provided by the Oklahoma State University Recombinant DNA-Protein Resource Facility. The Tn5 insertion in FF5.36 was localized by sequencing the DNA flanking the transposon by using the oligonucleotide 5′-GGTTCCGTTCAGGACGCTAC, which is derived from the border region of IS50 (49). Sequence data were aligned and homology searches were executed by using the University of Wisconsin Genetics Computer Group Sequence Analysis Package (version 9.0) or the National Center for Biotechnology Information BLAST network server.
Construction of an algT mutant of P. syringae.
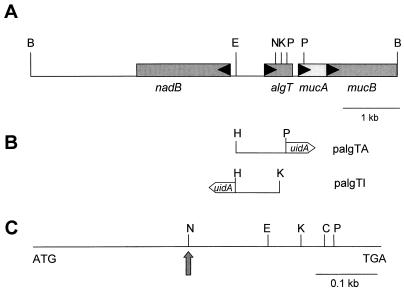
The chloramphenicol resistance (Cmr) gene in pSL1 was used to construct a nonpolar mutation in algT. pBTB6.5, which contains algT in a 6.5-kb BamHI fragment in pBluescript SK(+), was linearized with NruI, which generates a unique site within algT (Fig. 1A). The Cmr cassette in pSL1 was excised as a 0.65-kb SmaI fragment and ligated into linearized pBTB6.5, resulting in pBTB.Cm. The 7.15-kb BamHI fragment in pBTB.Cm was then excised and ligated into BamHI-digested pRK415. pRTB.Cm, the construct containing algT::Cmr in pRK415, was then introduced into P. syringae pv. syringae FF5(pPSR12) by triparental mating, and selection pressure for the vector (Tcr) was removed to facilitate homologous recombination (4).
FIG. 1.
(A) Physical and functional map of the 6.5-kb BamHI fragment in pBTB6.5 and pRTB6.5. The arrows within each open reading frame indicate the direction of translation for each gene. (B) Location and orientation of the algT::uidA transcriptional fusions in palgTA and palgTI. The HindIII site was added during PCR amplification. (C) Expanded view of the 582-bp algT gene from P. syringae FF5(pPSR12). The arrow indicates the location used for insertion of the antibiotic resistance cassette (Cmr). Abbreviations: B, BamHI; C, ClaI; E, EcoRI; K, KpnI; N, NruI; P, PstI.
Heat killing assays.
Bacterial cultures were grown to an A600 of 0.45 at 28°C and then incubated at 43°C for 0, 15, 30, 45, and 60 min; three replicate cultures were sampled at each time point. Cell dilutions were plated onto MGY agar in triplicate, and viable cells were scored as CFU. Survival was expressed as the percentage of input cells which retained viability.
Susceptibility to killing with ROI.
Sensitivity to paraquat or hydrogen peroxide (H2O2) was determined by measuring the diameter of the inhibition zone surrounding filters impregnated with reactive oxygen intermediates (ROI)-generating agents. Filter disks (6 mm) were soaked with 5 μl of 1.9% paraquat or 3% H2O2 and placed on a layer of soft agar (2 ml of 0.6% agar) containing 100 μl of an overnight culture of P. syringae; this was allowed to gel on 25 ml of MGY containing 1.5% agar. Cells were incubated at 28°C, and inhibition zones were measured 12 to 16 h after inoculation.
Construction of transcriptional fusions.
pBBR.Gus, which contains a promoterless glucuronidase gene (uidA) downstream of the polylinker in pBBR1MCS, was used to create algT::uidA transcriptional fusions. To obtain the algT promoter region in transcriptionally active and inactive orientations, a 1-kb PCR product was cloned into the HindIII-PstI or KpnI-HindIII sites of pBBR.Gus, respectively. The promoter region was amplified from pBTB6.5 by using the forward primer 5′-CTGAAGCTTCTGCCCTTGGCGACCAC (the HindIII site is underscored and is followed by nucleotides corresponding to −488 to −472 in Fig. 2) and the reverse primer 5′-CTCTTGGGCTATCGCCGCTGTCTC (the complement of nucleotides 580 to 604 in Fig. 2). After amplification of the 1-kb PCR product, ligation in pCR2.1, and transformation into E. coli DH5α, plasmids pCRalgTA and pCRalgT1 were recovered. These were digested with HindIII and PstI (pCRalgTA) or HindIII and KpnI (pCRalgTI) and ligated into pBBR.Gus, resulting in palgTA and palgTI, respectively (Fig. 1B).
FIG. 2.
Nucleotide sequence of algT from P. syringae pv. syringae FF5(pPSR12) containing the 5′ end of nadB and the nadB-algT intergenic region. The vertical arrow shows the Tn5 insertion site in mutant FF5.36. Potential recognition sequences for ς22 are shaded and underlined, and the putative ribosome binding site for algT is underlined. Nucleotides are numbered with respect to the algT translational start site and are indicated on the left; amino acid residues for AlgT are indicated on the right. Translational start sequences are shown in bold italics, and the translational stop codon for algT is indicated by a bold asterisk.
GUS assays.
Transcriptional activity was initially screened by spotting bacterial suspensions (A600 = 0.1) onto MG agar medium amended with chloramphenicol and 20 μg of X-Gluc (5-bromo-4-chloro-3-indolyl glucuronide) per ml; plates were then incubated at 28°C for 48 h. Prior to quantitative glucuronidase (GUS) assays, all strains were grown overnight in MGY broth containing chloramphenicol. Bacterial concentrations were then adjusted to an A600 of 0.1 in MGY broth and incubated at 28°C at 250 rpm. For temporal studies, 1-ml aliquots (three replicates per time point) were removed at 0, 1, 2, 5, 8, 12, 24, and 30 h and analyzed for GUS activity as described previously (44). GUS activity was expressed in units per milligram of protein with 1 U equivalent to 1 nmol of methylumbelliferone formed per min. The effect of salt, sorbitol, and copper sulfate on algT expression was evaluated by adjusting the bacterial concentration to an A600 of 0.1 and incubating the cells for 10 h in MGY broth amended with NaCl (0.15, 0.3, or 0.4 M), sorbitol (0.3, 0.6, or 0.8 M), or CuSO4 (50, 100, or 200 μM). algT transcription was also investigated by preparing bacterial suspensions as described above, growing them to an A600 of 0.5, and incubating them at elevated temperature (50°C) or in media amended with H2O2 or paraquat (0.001 or 0.01%). Bacterial cells (1 ml) were removed at 0, 15, 30, 60, and 120 min for temperature studies and at 0, 15, 30, and 60 min for assays with H2O2 and paraquat.
Nucleotide sequence accession number.
The nucleotide sequence for algT in P. syringae pv. syringae has been deposited in the GenBank database under accession no. AF190580.
RESULTS
Location of Tn5 insertion in FF5.36.
The Tn5 mutant FF5.36 exhibited a leaky phenotype for alginate and produced low amounts of the exopolysaccharide in vitro; this mutant was previously isolated by mutagenesis of FF5(pPSR12), which produces alginate constitutively at high levels (29). To locate the Tn5 insertion in FF5.36, a genomic library of this mutant was constructed in pRK7813, and a cosmid clone containing the Tn5 insertion was recovered and designated pFF5.36. The internal BamHI site in Tn5 and 3 kb of flanking DNA from FF5.36 were cloned from pFF5.36 into pBluescript SK(+), resulting in a clone named pFF5.36B. A primer specific for the border region of IS50 indicated that the Tn5 insertion was located within nadB at nucleotide 61 of the corresponding P. aeruginosa sequence (12). The nadB gene encodes l-aspartate oxidase and is located approximately 400 bp upstream of algT in P. aeruginosa (12). A 600-bp region downstream of the Tn5 insertion was sequenced in pFF5.36B; this region showed 73% nucleotide sequence identity to the first 100 bp of nadB and 65% nucleotide sequence identity to the nadB-algT intergenic region and the 5′ end of algT from P. aeruginosa. These results indicated that the location of nadB and algT was conserved in P. syringae and P. aeruginosa.
Cloning of algT from P. syringae.
A genomic library of P. syringae FF5(pPSR12) was previously constructed in pRK7813 (46). In the current study, the 6-kb BamHI fragment from pFF5.36B, which contains a portion of algT, was used to screen the library for clones containing the complete algT coding region. One clone designated pLKT5 was chosen for further study and contained a 6.5-kb BamHI fragment which hybridized with the probe. This fragment was subcloned in pBluescript SK(+), resulting in pBTB6.5, and partially sequenced by using the T7 and T3 primers. Sequence analysis indicated that the right border of this fragment contained DNA homologous to mucB (Fig. 1A). Since algT is generally associated with the mucABCD gene cluster (20, 38), we suspected that pBTB6.5 contained a functional copy of algT.
Sequence analysis of algT.
A physical map of pBTB6.5 was constructed to further localize the alginate regulatory genes on this fragment (Fig. 1A). Sequence data for the P. syringae algT gene were initially derived by using a primer based on the nucleotide sequence downstream of the Tn5 insertion located in FF5.36 (see vertical arrow, Fig. 2). Additional sequence data was obtained by primer walking, and both DNA strands were sequenced for verification. The P. syringae algT homologue was 582 bp and was highly related to algT from P. aeruginosa (81 and 90% nucleotide and amino acid identities, respectively) (13). The deduced translational product of algT is a protein consisting of 193 amino acids with a predicted mass of 22.3 kDa. A potential ribosome binding site was identified 7 bp upstream of the predicted translational start site. Two putative AlgT (ς22) recognition sites were located 60 and 248 bp upstream of the algT translational start site (Fig. 2). The location and sequence of the first ς22 recognition site (60 bp upstream of the initiation codon) was conserved in both P. syringae and P. aeruginosa (Fig. 3). The nadB gene in P. syringae was located 516 bp upstream of the algT translational start site and was divergently transcribed with respect to algT (Fig. 1 and 2). Interestingly, nucleotide identity in the 516-bp intergenic region between nadB and algT was only 46% when the P. syringae and P. aeruginosa regions were compared (Fig. 3). Additional sequencing downstream of algT revealed mucA and mucB homologues (Fig. 1) which showed 68 and 70% nucleotide identities, respectively, to the genes previously sequenced in P. aeruginosa (19, 35). In summary, the arrangement of nadB, algT, mucA, and mucB is conserved in P. syringae, P. aeruginosa, and A. vinelandii (12, 35, 38).
FIG. 3.
Alignment of the algT promoter sequences from P. syringae pv. syringae FF5(pPSR12) (Ps algT) and P. aeruginosa (Pa algT). The nucleotides for the P. aeruginosa sequence are shown on the left with +1 (see asterisk) corresponding to the transcriptional start site. Nucleotides for the P. syringae pv. syringae algT promoter are shown on the right with +1 corresponding to the translational start site. Gaps (--) were used to maximize the alignment, and identical bases are shaded. The ς22 recognition sequences in both species are indicated in boldface type and are double-underscored. The nadB and algT translational start sites are in boldface, and the direction of translation is indicated with an arrow.
Construction of an algT mutant.
FF5.36, which contains a Tn5 insertion in nadB, was unstable with respect to alginate production. To avoid potential polar effects on adjacent genes, we constructed an algT mutant with a Cmr cassette which lacks transcriptional terminators. Recombination of the Cmr cassette into algT was verified by PCR and sequence analysis. FF5.LK1, the algT mutant resulting from this experiment, produced 61 μg of uronic acid polymers/mg of protein, a level approximately 43-fold lower and significantly less (P = 0.01) than FF5(pPSR12), which synthesized 2,652 μg of uronic acid polymers/mg of protein. Previous work indicated that most of the uronic acid polymers synthesized by FF5(pPSR12) were alginate (29). Furthermore, alginate-defective strains of FF5(pPSR12) still synthesized low amounts of uronic acid polymers in the carbazole assay (15, 29, 46); therefore, we concluded that the algT mutant, FF5.LK1, was defective in alginate production.
Complementation experiments.
pRTB6.5, which contains algT, mucA, and mucB in pRK415, was evaluated for its ability to complement P. syringae pv. syringae FF5.LK1 for alginate production. pRTB6.5 did not restore alginate production to FF5.LK1, possibly because this plasmid also contains the negative regulators, mucA and mucB, which could suppress the conversion to mucoidy (19, 36). Consequently, we examined whether palgTA.1, which contains algT but lacks extraneous flanking DNA, could restore alginate production to FF5.LK1. Transconjugants of FF5.LK1 containing palgTA.1 were visibly mucoid and produced 1,086 μg of alginate/mg of protein; this amount was significantly higher (P = 0.01) than the level synthesized by FF5.LK1, indicating that palgTA.1 could partially complement the algT mutant. We also investigated whether pJG309, which contains algT from P. aeruginosa, could complement FF5.LK1 for alginate production. FF5.LK1(pJG309) transconjugants produced 1,081 μg of alginate per mg of protein, a level equivalent to that obtained with FF5.LK1(palgTA.1), which suggests that the two genes may be functionally interchangeable.
Effects of algT on susceptibility to heat and ROI.
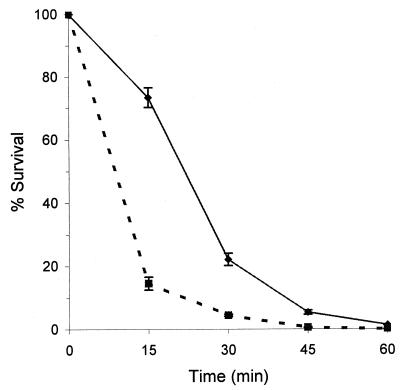
Previous reports indicate that algT functions as an alternative sigma factor in P. aeruginosa and is involved in the transcriptional activation of heat shock genes (37, 52). Therefore, we evaluated whether the algT mutation in FF5.LK1 resulted in an increased sensitivity to heat killing when compared to the wild-type FF5(pPSR12). Survival after exposure to 43°C was significantly reduced in the algT mutant compared to the wild-type strain (Fig. 4). Within 15 min there was an 85% reduction in the viability of the mutant compared with only 27% in the wild-type (Fig. 4). No difference in viability between the wild-type and mutant strains was apparent after a 60-min incubation at 43°C (Fig. 4).
FIG. 4.
Heat-killing curves for P. syringae pv. syringae FF5(pPSR12) (wild-type [⧫]) and FF5.LK1 (algT mutant [■]). The strains were incubated at 43°C for 15, 30, 45, and 60 min, and surviving cells were counted as CFU. Bars indicate standard errors of the means, and survival is expressed as the percentage of input CFU at time zero. The experiment was repeated with similar results.
To determine whether algT is involved in tolerance to compounds that generate ROIs, the wild-type FF5(pPSR12) and algT mutant FF5.LK1 were exposed to H2O2 and paraquat, a superoxide-generating redox cycling compound (16). FF5.LK1 was significantly more sensitive to paraquat and H2O2 than FF5(pPSR12) (Table 2), indicating that algT has a role in mediating resistance to ROIs in P. syringae. Both FF5(pPSR12) and FF5.LK1 grew at identical rates in vitro (data not shown), indicating that the algT mutation did not significantly affect growth.
TABLE 2.
Sensitivity to killing by paraquat and H2O2 in the wild-type and algT mutant of P. syringae pv. syringae
| Strain | Characteristics | Growth inhibition zone (mean diam [mm] ± SE)a
|
|
|---|---|---|---|
| Paraquat (1.9%) | H2O2 (3%) | ||
| FF5(pPSR12) | Mucoid, wild-type | 24.4 ± 0.2 | 13.3 ± 0.1 |
| FF5.LK1 | Nonmucoid, algT::Cmr | 32.2 ± 0.2 | 16.5 ± 0.1 |
Sensitivities to paraquat and H2O2 are expressed as diameters of growth inhibition zones surrounding filter disks impregnated with 5 μl of the indicated solutions. The experiment was repeated with similar results.
Kinetics of algT transcription.
palgTA, palgTI, and pBBR.Gus were mobilized into P. syringae pv. syringae FF5 and assayed for GUS activity. Colonies of FF5(palgTA) turned blue on media containing X-Gluc, indicating that algT was transcribed at physiological temperatures (28°C). FF5 transconjugants containing palgTI (algT in the transcriptionally inactive orientation) or pBBR.Gus (vector control) remained colorless on X-Gluc media. When FF5 transconjugants containing palgTA, palgTI, or pBBR.Gus were grown in MGY broth at 28°C, growth curves were similar, indicating that the transcriptional fusions had no significant effect on growth (data not shown). A time course experiment at 28°C indicated that algT transcriptional activity increased steadily over time with 960 U of GUS/mg of protein at 30 h; this gradual increase in expression is similar to observations made for algT in P. aeruginosa (13). GUS activity in FF5(palgTI) and FF5(pBBR.Gus) remained low (1 to 14 U) throughout the sampling period and was not significantly different between the two transconjugants; consequently, FF5(pBBR.Gus) was used as a negative control in all subsequent experiments.
algT expression in response to selected factors.
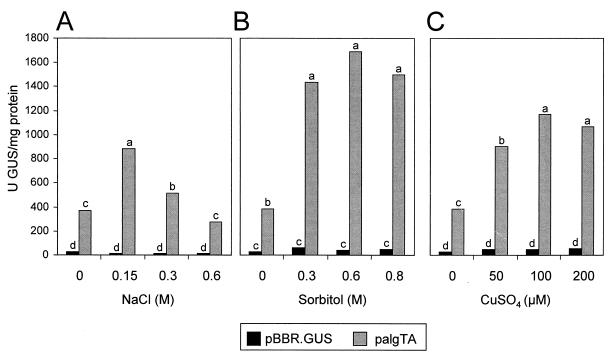
GUS activity in FF5(palgTA) was significantly higher (P = 0.01) when the growth medium was amended with 0.15 or 0.3 M NaCl; in contrast, FF5(pBBR.Gus) showed no response to the addition of NaCl (Fig. 5A). To determine whether the effect of NaCl was ionic or osmotic, sorbitol (a nonionic, nonmetabolizable solute) was examined for its effect on algT expression. Sorbitol was added to MGY broth at 0.3, 0.6, and 0.8 M, concentrations which are osmotically equivalent to 0.15, 0.3, and 0.4 M NaCl, respectively. The transcriptional activity of algT was significantly higher (P = 0.01) than the nonsupplemented control when sorbitol was added at all concentrations tested (Fig. 5B). Therefore, the stimulation of algT gene expression by NaCl is due to increased osmolarity rather than an ionic effect.
FIG. 5.
GUS activity in P. syringae pv. syringae FF5 derivatives grown in MGY broth containing sodium chloride (A), sorbitol (B), or copper sulfate (C). Prior to the GUS assays, all strains were grown for 20 h in MGY broth containing chloramphenicol. The bacterial concentration was adjusted to an A600 of 0.1, and the cells were incubated at 250 rpm for 10 h at 28°C in MGY broth amended with NaCl, sorbitol, or copper sulfate. palgTA contains the algT promoter in the transcriptionally active orientation, and pBBR.Gus contains a promoterless glucuronidase gene. Values are the mean from one experiment containing three replicates, and the experiment was repeated with similar results. Treatments accompanied by the same lowercase letter were not significantly different at a P of 0.01 as shown by Duncan’s multiple-range test.
We previously demonstrated that the addition of copper sulfate to the growth medium increased both alginate production and algD transcriptional activity in P. syringae pv. syringae FF5 (29, 46). In the present study, we found that algT gene expression was significantly higher (P = 0.01) than the nonamended control when copper sulfate was added at all concentrations tested (Fig. 5C). Since the algT gene product, ς22, binds to the algD promoter and activates transcription, these results suggest that copper sulfate may stimulate alginate production via the algT signal transduction pathway.
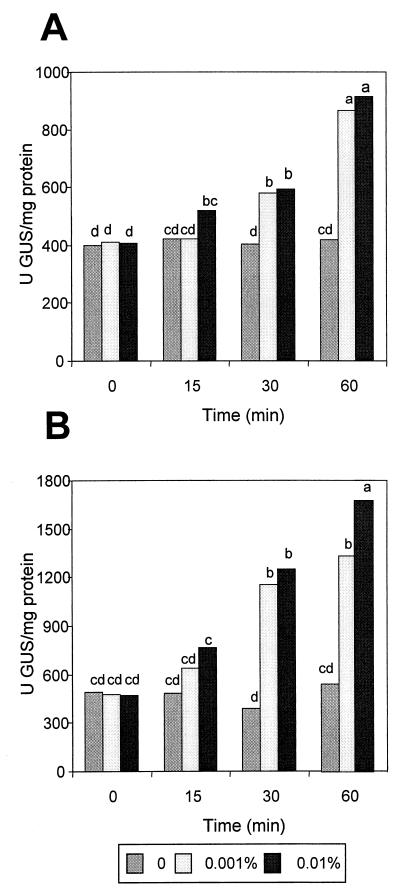
The addition of H2O2 or paraquat to actively growing cultures of FF5(palgTA) at 0.001 and 0.01% stimulated algT expression 30 min after each compound was added (Fig. 6). When FF5(palgTA) was incubated for 60 min in H2O2 or paraquat, a 2.3- to 3.7-fold increase in algT expression was observed, respectively (Fig. 6). Longer incubation periods (4 h) did not result in further stimulation of algT gene expression (data not shown). Furthermore, a basal level of algT transcriptional activity (ca. 400 U of GUS/mg of protein) was necessary to see further induction of the algT promoter when ROI-generating compounds were added; otherwise both H2O2 and paraquat were toxic (data not shown).
FIG. 6.
GUS activity in P. syringae pv. syringae FF5 derivatives grown in MGY broth containing H2O2 (A) or paraquat (B). Prior to the GUS assays, all strains were initially grown as described in Fig. 5; bacterial concentrations were then adjusted to an A600 of 0.5 and incubated in MGY broth amended with H2O2 or paraquat. palgTA is described in Fig. 5. Values represent the mean from one experiment containing three replicates; the experiment was repeated with similar results. Treatments accompanied by the same lowercase letter were not significantly different at a P of 0.01 as determined by Duncan’s multiple-range test.
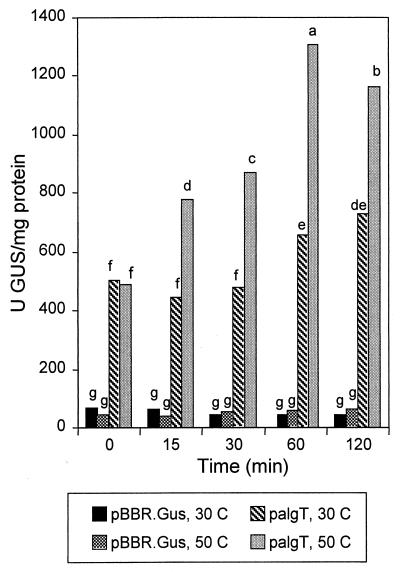
When actively growing FF5(palgTA) cells were subjected to a temperature upshift (30 to 50°C), a significant increase in algT transcriptional activity was apparent within 15 min and was twofold higher than in control cells (which were not heat shocked) at 60 min (Fig. 7). GUS activity in FF5(pBBR.Gus) remained low, regardless of temperature (Fig. 7).
FIG. 7.
GUS activity in P. syringae pv. syringae FF5 subjected to heat shock (temperature shift from 30 to 50°C). Prior to GUS assays, all strains were grown at 30°C as described in Fig. 5. The bacteria were incubated at 30°C until the concentration had an A600 of 0.5 and were then exposed to heat shock by rapid transfer to 50°C. palgTA and pBBR.Gus are described in Fig. 5. Values represent the mean from one experiment containing three replicates, and the experiment was repeated with similar results. Treatments accompanied by the same lowercase letter were not significantly different at a P of 0.01 as determined by Duncan’s multiple-range test.
DISCUSSION
In the present study, the Alg− mutant FF5.36 contained a Tn5 insertion in nadB, which encodes l-aspartate oxidase, a flavoprotein in the pathway for NAD biosynthesis (12). The nadB gene in both P. syringae and P. aeruginosa (12) is encoded upstream of algT and divergently transcribed with respect to algT. In P. aeruginosa, nadB was not essential for NAD production, and a Tn501 insertion in nadB did not affect alginate biosynthesis (12). However, in the present study, the nadB::Tn5 insertion in FF5.36 was unstable with respect to alginate production. Although we could not identify additional Tn5 insertions in FF5.36, it remains possible that additional point mutations may have occurred, leading to a nonmucoid phenotype. Therefore, the algT mutant FF5.LK1 was constructed in the present study, and all subsequent experiments were conducted with this mutant.
The algT genes in P. syringae and P. aeruginosa are highly homologous (81% nucleotide identity) and closely related to rpoE, which encodes ςE, an alternate sigma factor involved in high-temperature gene expression in E. coli (11, 13, 14). Our results show that the algT promoter region in P. syringae contained two motifs conserved in promoters transcribed by the RNAP-ςE complex (11) (Fig. 3). In P. aeruginosa, these two promoters were AlgT-dependent and designated P1 and P3 (52). The conservation of these promoters in P. syringae and P. aeruginosa and the complementation of FF5.LK1 with algT from both species suggest that the algT homologs in these two pseudomonads may be functionally interchangeable.
In P. aeruginosa, the negative regulatory genes mucA and mucB suppress alginate production, and mutagenesis of these genes results in a mucoid phenotype (19, 36). In the current study, pRTB6.5, which contains algT, mucA, and mucB, did not restore alginate production to the algT mutant FF5.LK1. However, palgTA.1, which contains algT without extraneous flanking DNA, partially restored alginate production to FF5.LK1. Previous studies have shown that MucA physically binds AlgT (ς22) and functions as an anti-ς factor (53, 61), whereas MucB is presumed to alter the conformation of MucA in such a way that it targets ς22 for degradation (39). Therefore, a stoichiometric relationship exists between these three proteins and may explain why alginate production was not fully restored to wild-type levels in FF5.LK1(palgTA.1).
There is growing evidence that the algT-mucABCD gene cluster forms a signal transduction system that modulates algT activity in response to environmental stress (13, 37, 51, 52, 63). The algT gene fusion from P. syringae was transcriptionally activated in response to both NaCl and sorbitol (Fig. 5), indicating that osmotic stress is a stimulus for algT activation in both P. syringae and P. aeruginosa (52). Phytopathogenic bacteria are exposed to high osmolarities on the leaf surface (3), and the increased synthesis of alginate is critical to survival during epiphytic colonization (62); therefore, transcriptional activation of algT may enhance epiphytic fitness.
The algT mutant of P. syringae was more sensitive to H2O2 and paraquat, and algT expression was activated in response to both compounds. Although an algT mutant of P. aeruginosa showed increased susceptibility to paraquat, no difference in sensitivity to H2O2 was detected between the mutant and wild-type strains (37). These results suggest that P. syringae and P. aeruginosa differ in their response to ROIs. Although aspects of the oxidative burst are similar in animal and plant hosts (32), plant cells produce ROIs (mainly H2O2) constitutively throughout the defense response (5), and H2O2 has an important role in plant disease resistance (1, 8, 59). In animals, alginate production by P. aeruginosa may suppress the oxidative burst in neutrophils and scavenge the ROIs produced by phagocytic cells (22, 55). Therefore, the activation of algT by ROIs and the subsequent production of alginate may help P. syringae evade the plant defense response.
In previous studies, copper sulfate stimulated algD transcriptional activity and alginate production in P. syringae (29, 46). However, copper sulfate was not a signal for algD gene expression or alginate production in clinical strains of P. aeruginosa, possibly because these strains are not repeatedly exposed to toxic levels of copper sulfate (29, 46). In the current study, the algT promoter in P. syringae pv. syringae FF5 was stimulated by exposure to copper sulfate (Fig. 5C), which is consistent with earlier studies showing algD activation in response to copper sulfate (46). In agriculture, bactericidal sprays containing copper sulfate are frequently used for the control of P. syringae and other phytopathogenic bacteria, and copper-mediated stress is high. Because copper is known to generate free radicals (58), the increased production of alginate in response to copper sulfate may be caused by oxidative stress. Alternatively, the sequence divergence in the nadB-algT intergenic regions of P. syringae and P. aeruginosa may reflect the unique activation of the algT promoter in P. syringae by copper sulfate.
The algT mutant of P. syringae was more sensitive to elevated temperature (Fig. 4), and algT expression was activated in response to heat shock (Fig. 7). In contrast to human and animal pathogens, little is known about how phytopathogenic bacteria respond to temperature stress. We recently demonstrated that P. syringae responds to heat shock by producing DnaK (28), a molecular chaperone that facilitates the disassembly of proteins that have been damaged by heat stress (34). The present study expands our knowledge of the temperature stress response in P. syringae and clearly shows that algT increases the heat tolerance of this bacterium. The increased production of alginate in response to elevated temperatures could be advantageous since the alginate capsule could provide some protection from the dehydration and desiccation which develop during heat stress.
In P. syringae, algT is required for alginate production and increases the survival of the bacterium during environmental stress. Copper and H2O2 are toxic compounds that P. syringae encounters during colonization of host plant tissues, and these substances may be unique signals for algT activation in this bacterium. However, heat shock is a conserved signal for activation of algT expression in both P. aeruginosa (52) and P. syringae. In P. aeruginosa, AlgT (ς22) is required for transcription of algD, which encodes GDP-mannose dehydrogenase, the first committed step in the alginate biosynthetic pathway (10). In P. syringae, the algD promoter region contains a putative recognition site for ς22 (15), but the requirement of ς22 for algD transcription has not yet been demonstrated. However, the transcriptional activation of the algT and algD (46) promoters in response to heat, osmotic stress, and copper sulfate supports the hypothesis that algT may control activation of algD transcription in P. syringae. Studies are currently under way to examine this hypothesis and other possible roles for algT in the pathogenicity and fitness of P. syringae.
ACKNOWLEDGMENTS
C.B. acknowledges support from the Oklahoma Agricultural Experiment Station and Public Health Service grant AI 43311-01 from the National Institutes of Health.
We thank A. M. Chakrabarty and A. Peñaloza-Vázquez for reviewing this manuscript prior to publication.
REFERENCES
- 1.Alvarez M E, Pennel R I, Meijer P J, Ishikawa A, Dixon R A, Lamb C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell. 1998;92:773–784. doi: 10.1016/s0092-8674(00)81405-1. [DOI] [PubMed] [Google Scholar]
- 2.Barta T M, Kinscherf T G, Willis D K. Regulation of tabtoxin production by the lemA gene in Pseudomonas syringae. J Bacteriol. 1992;174:3021–3029. doi: 10.1128/jb.174.9.3021-3029.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beattie G A, Lindow S E. The secret life of foliar bacterial pathogens on leaves. Annu Rev Phytopathol. 1995;33:145–172. doi: 10.1146/annurev.py.33.090195.001045. [DOI] [PubMed] [Google Scholar]
- 4.Bender C L, Young S A, Mitchell R E. Conservation of plasmid DNA sequences in coronatine-producing pathovars of Pseudomonas syringae. Appl Environ Microbiol. 1991;57:993–999. doi: 10.1128/aem.57.4.993-999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolwell G P, Wojtaszek P. Mechanisms for the generation of reactive oxygen species in plant defence—a broad perspective. Physiol Mol Plant Pathol. 1997;51:347–366. [Google Scholar]
- 6.Boucher J C, Martinez-Salazar J, Schurr M J, Mudd M H, Yu H, Deretic V. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boucher J C, Schurr M J, Yu H, Rowen D W, Deretic V. Pseudomonas aeruginosa in cystic fibrosis: role of mucC in the regulation of alginate production and stress sensitivity. Microbiology. 1997;143:3473–3480. doi: 10.1099/00221287-143-11-3473. [DOI] [PubMed] [Google Scholar]
- 8.Chamnongpol S, Willekens H, Moeder W, Langebartels C, Sanderman H, Van Montagu M, Inzé D, Van Camp W. Defense activation and enhanced pathogen tolerance induced by H2O2 in transgenic tobacco. Proc Natl Acad Sci USA. 1998;95:5818–5823. doi: 10.1073/pnas.95.10.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chitnis C E, Ohman D E. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol Microbiol. 1993;8:583–590. doi: 10.1111/j.1365-2958.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 10.Deretic V, Gill J F, Chakrabarty A M. Pseudomonas aeruginosa infection in cystic fibrosis: nucleotide sequence and transcriptional regulation of the algD gene. Nucleic Acids Res. 1987;15:4567–4581. doi: 10.1093/nar/15.11.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deretic V, Schurr M J, Boucher J C, Martin D W. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J Bacteriol. 1994;176:2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeVries C A, Hassett D J, Flynn J L, Ohman D E. Genetic linkage in Pseudomonas aeruginosa of algT and nadB: mutation in nadB does not affect NAD biosynthesis or alginate production. Gene. 1995;156:63–67. doi: 10.1016/0378-1119(95)00028-5. [DOI] [PubMed] [Google Scholar]
- 13.DeVries C A, Ohman D E. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternative sigma factor, and shows evidence of autoregulation. J Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson J W, Gross C A. Identification of the ςE subunit of Escherichia coli RNA polymerase: a second alternate ς factor involved in high-temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 15.Fakhr M K, Peñaloza-Vázquez A, Chakrabarty A M, Bender C L. Regulation of alginate biosynthesis in Pseudomonas syringae pv. syringae. J Bacteriol. 1999;181:3478–3485. doi: 10.1128/jb.181.11.3478-3485.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farr S, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figurski D, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gacesa P. Bacterial alginate biosynthesis—recent progress and future prospects. Microbiology. 1998;144:1133–1143. doi: 10.1099/00221287-144-5-1133. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg J B, Gorman W L, Flynn J L, Ohman D E. A mutation in algN permits trans activation of alginate production by algT in Pseudomonas species. J Bacteriol. 1993;175:1303–1308. doi: 10.1128/jb.175.5.1303-1308.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hershberger C D, Ye R W, Parsek M R, Xie Z-D, Chakrabarty A M. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative ς factor (ςE) Proc Natl Acad Sci USA. 1995;92:7941–7945. doi: 10.1073/pnas.92.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen E T, Kharazmi A, Lam K, Costerton J W, Høiby N. Human polymorphonuclear leucocyte response to Pseudomonas aeruginosa grown in biofilms. Infect Immun. 1990;58:2382–2385. doi: 10.1128/iai.58.7.2383-2385.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones J D G, Gutterson N. An efficient mobilizable cosmid vector, pRK7813, and its use in a rapid method for marker exchange in Pseudomonas fluorescens strain HV37a. Gene. 1987;61:299–306. doi: 10.1016/0378-1119(87)90193-4. [DOI] [PubMed] [Google Scholar]
- 24.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato J, Chakrabarty A M. Purification of the regulatory protein AlgR1 and its binding in the far upstream region of the algD promoter in P. aeruginosa. Proc Natl Acad Sci USA. 1991;88:1760–1764. doi: 10.1073/pnas.88.5.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keane P J, Kerr A, New P B. Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Aust J Biol Sci. 1970;23:585–595. [Google Scholar]
- 27.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 28.Keith L M W, Partridge J E, Bender C L. dnaK and the heat stress response of Pseudomonas syringae pv. glycinea. Mol Plant-Microbe Interact. 1999;12:563–574. doi: 10.1094/MPMI.1999.12.7.563. [DOI] [PubMed] [Google Scholar]
- 29.Kidambi S P, Sundin G W, Palmer D A, Chakrabarty A M, Bender C L. Copper as a signal for alginate synthesis in Pseudomonas syringae pv. syringae. Appl Environ Microbiol. 1995;61:2172–2179. doi: 10.1128/aem.61.6.2172-2179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:2172–2179. [PubMed] [Google Scholar]
- 31.Kovach M E, Phillips R W, Elzer P H, Roop III R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 32.Lamb C, Dixon R A. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 33.Lukomski S, Hull R A, Hull S I. Identification of the O antigen polymerase (rfc) gene in Escherichia coli O4 by insertional mutagenesis using a nonpolar chloramphenicol resistance cassette. J Bacteriol. 1996;178:240–247. doi: 10.1128/jb.178.1.240-247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mager W H, Kruijff A J J. Stress-induced transcriptional activation. Microbiol Rev. 1995;59:506–531. doi: 10.1128/mr.59.3.506-531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin D W, Schurr M J, Mudd M H, Deretic V. Differentiation of Pseudomonas aeruginosa into the alginate-producing form: inactivation of mucB causes conversion to mucoidy. Mol Microbiol. 1993;9:497–506. doi: 10.1111/j.1365-2958.1993.tb01711.x. [DOI] [PubMed] [Google Scholar]
- 36.Martin D W, Schurr M J, Mudd M H, Govan J R, Holloway B W, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin D W, Schurr M J, Yu H, Deretic V. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to stress response. J Bacteriol. 1994;176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martínez-Salazar J M, Moreno S, Nájera R, Boucher J C, Espin G, Soberon-Chávez G, Deretic V. Characterization of the genes coding for the putative sigma factor AlgU and its regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their roles in alginate biosynthesis. J Bacteriol. 1996;178:1800–1808. doi: 10.1128/jb.178.7.1800-1808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathee K, McPherson C J, Ohman D E. Posttranslational control of the algT (algU)-encoded ς22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN) J Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.May T B, Chakrabarty A M. Isolation and assay of Pseudomonas aeruginosa alginate. Methods Enzymol. 1994;235:295–304. doi: 10.1016/0076-6879(94)35148-1. [DOI] [PubMed] [Google Scholar]
- 41.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 42.Mohr C D, Leveau J H J, Krieg D P, Hibler N S, Deretic V. AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J Bacteriol. 1992;174:6624–6633. doi: 10.1128/jb.174.20.6624-6633.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murillo J, Shen H, Gerold D, Sharma A K, Cooksey D A, Keen N T. Characterization of pPT23B, the plasmid involved in syringolide production by Pseudomonas syringae pv. tomato. Plasmid. 1994;31:275–287. doi: 10.1006/plas.1994.1029. [DOI] [PubMed] [Google Scholar]
- 44.Palmer D A, Bender C L, Sharma S B. Use of Tn5-gusA5 to investigate environmental and nutritional effects on gene expression in the coronatine biosynthetic gene cluster of Pseudomonas syringae pv. glycinea. Can J Microbiol. 1997;43:517–525. doi: 10.1139/m97-074. [DOI] [PubMed] [Google Scholar]
- 45.Peñaloza-Vázquez A, Bender C L. Characterization of CorR, a transcriptional activator which is required for biosynthesis of the phytotoxin coronatine. J Bacteriol. 1998;180:6252–6259. doi: 10.1128/jb.180.23.6252-6259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peñaloza-Vázquez A, Kidambi S P, Chakrabarty A M, Bender C L. Characterization of the alginate biosynthetic gene cluster in Pseudomonas syringae pv. syringae. J Bacteriol. 1997;179:4464–4472. doi: 10.1128/jb.179.14.4464-4472.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pier G B. Pseudomonas aeruginosa: a key problem in cystic fibrosis. ASM News. 1998;64:339–347. [Google Scholar]
- 48.Rehm B H A, Valla S. Bacterial alginates: biosynthesis and applications. Appl Microbiol Biotechnol. 1997;48:281–288. doi: 10.1007/s002530051051. [DOI] [PubMed] [Google Scholar]
- 49.Rich J J, Willis D K. A single oligonucleotide can be used to rapidly isolate DNA sequences flanking a transposon Tn5 insertion by the polymerase chain reaction. Nucleic Acids Res. 1990;18:6673–6676. doi: 10.1093/nar/18.22.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 51.Schurr M J, Deretic V. Microbial pathogenesis in cystic fibrosis: co-ordinate regulation of heat-shock response and conversion to mucoidy in Pseudomonas aeruginosa. Mol Microbiol. 1997;24:411–420. doi: 10.1046/j.1365-2958.1997.3411711.x. [DOI] [PubMed] [Google Scholar]
- 52.Schurr M J, Yu H, Boucher J C, Hibler N S, Deretic V. Multiple promoters and induction by heat shock of the gene encoding the alternative sigma factor AlgU (ςE) which controls mucoidy in cystic fibrosis isolates of Pseudomonas aeruginosa. J Bacteriol. 1995;177:5670–5679. doi: 10.1128/jb.177.19.5670-5679.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schurr M J, Yu H, Martinez-Salazar J M, Boucher J C, Deretic V. Control of AlgU, a member of the ςE-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shankar S, Ye R, Schlictman D, Chakrabarty A M. Exopolysaccharide alginate synthesis in Pseudomonas aeruginosa: enzymology and regulation of gene expression. Adv Enzymol. 1995;70:221–255. doi: 10.1002/9780470123164.ch4. [DOI] [PubMed] [Google Scholar]
- 55.Simpson J A, Smith S E, Dean R T. Scavenging by alginate of free radicals released by macrophages. Free Radical Biol Med. 1989;6:347–353. doi: 10.1016/0891-5849(89)90078-6. [DOI] [PubMed] [Google Scholar]
- 56.Staskawicz B J, Dahlbeck D, Keen N T. Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race-specific incompatibility on Glycine max (L.) Merr. Proc Natl Acad Sci USA. 1984;81:6024–6028. doi: 10.1073/pnas.81.19.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sundin G W, Bender C L. Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae pv. syringae. Appl Environ Microbiol. 1993;59:1018–1024. doi: 10.1128/aem.59.4.1018-1024.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thurman R B, Gerba C P. The molecular mechanisms of copper and silver ion disinfection of bacteria and viruses. Crit Rev Environ Control. 1989;18:295–315. [Google Scholar]
- 59.Van Camp W, Van Montagu M, Inzé D. H2O2 and NO: redox signals in disease resistance. Trends Plant Sci. 1998;3:330–334. [Google Scholar]
- 60.Wozniak D J, Ohman D E. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie Z, Hershberger C D, Shankar S, Ye R W, Chakrabarty A M. Sigma factor-anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol. 1996;178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu J, Peñaloza-Vázquez A, Chakrabarty A M, Bender C L. Involvement of the exopolysaccharide alginate in the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Mol Microbiol. 1999;33:712–720. doi: 10.1046/j.1365-2958.1999.01516.x. [DOI] [PubMed] [Google Scholar]
- 63.Yu H, Schurr M J, Deretic V. Functional equivalence of Escherichia coli ςE and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J Bacteriol. 1995;177:3259–3268. doi: 10.1128/jb.177.11.3259-3268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zielinski N A, Maharaj R, Roychoudhury S, Danganan C E, Hendrickson W, Chakrabarty A M. Alginate synthesis in Pseudomonas aeruginosa: environmental regulation of the algC promoter. J Bacteriol. 1992;174:7680–7688. doi: 10.1128/jb.174.23.7680-7688.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]