Abstract
By utilizing reporter transposons, five Providencia stuartii genes that are activated by the accumulation of self-produced extracellular signals have been identified. These genes have been designated cma for conditioned medium activated. The presence of conditioned medium from stationary-phase cultures grown in rich media resulted in the premature activation of each gene in cells at early log phase, with activation values ranging from 6- to 26-fold. Preparation of conditioned medium from an M9 salts medium and fractionation by gel filtration chromatography resulted in fractions within the included volume which activated three of the cma fusions. In addition, depending on the reporter fusion, peak activity was found in different fractions. The partially purified factors activated in a dose-dependent manner. Characterization of the factors activating the cma fusions indicated that they were stable to heat, alkali, and acid. Furthermore, for each cma fusion, factor activity was not reproduced by the addition of homoserine lactone, homocysteine thiolactone, pyruvate, Casamino Acids, or α-ketoglutarate. The identities of three cma genes have been determined and revealed physiological roles in amino acid biosynthesis and nutrient import. To begin to address the pathways for production of or response to the extracellular factors, we have identified a locus, aarA, that is required for the activation of four cma fusions. The AarA product was required for factor activity in extracellular supernatants, indicating a possible role in biosynthesis or export.
The ability of bacteria to sense and monitor population density can be controlled by diffusible chemical signals (20, 26, 28, 45). These self-produced signals mediate changes in gene expression by a process termed quorum sensing (19). Typically, these signals act at intermediate to high cell density due to accumulations above a threshold level. The signals then mediate regulatory changes by a number of mechanisms. They can interact with proteins of the LuxR family (17, 19, 20, 31, 45) or with two-component systems (28), or they can inhibit the activity of phosphatases (32, 39, 47). An additional mechanism involves the ability to stimulate translation (5). Quorum sensing can regulate a variety of cellular events, including antibiotic synthesis, bioluminescence, differentiation, cell division, competence, pigment production, virulence, plasmid transfer, chromosome replication, biofilm maturation, and amino acid metabolism (2–4, 9, 10, 15, 21, 22, 24, 25, 29, 34, 37, 40, 41, 46, 48, 50). These cell-to-cell signals can be N-acyl derivatives of homoserine lactone (7, 16, 38), amino acids (30), peptides (23, 24, 32, 33, 47), proteins (27, 49), or fatty acids (14).
Providencia stuartii is a gram-negative bacterium responsible for urinary tract infections in humans. In P. stuartii, the accumulation of an extracellular factor regulates a chromosomal 2-N-acetyltransferase [AAC(2′)-Ia] involved in both peptidoglycan and aminoglycoside acetylation (44). This factor, designated acetyltransferase-repressing factor (AR-factor), decreases the expression of aac(2′)-Ia in a density-dependent manner (44). In a search for regulatory loci controlling aac(2′)-Ia expression, the aarA gene was identified (40). Mutations in aarA increase aac(2′)-Ia transcription, and this phenotype is more pronounced at high cell densities, indicating a possible role in the response or production of AR-factor (13, 43). Mutations in aarA are pleiotropic and also result in loss of pigment production and abnormal cell division, resulting in a prominent chaining phenotype (43). The deduced AarA protein is very hydrophobic, with several potential membrane-spanning regions, suggesting that it may be an integral membrane protein. AarA does not display significant homology to proteins in the databases.
In order to further understand the role of cell-to-cell signaling in P. stuartii, a search was undertaken to identify additional genes regulated by the accumulation of extracellular signals. By using a strategy previously described for Escherichia coli (2), we identified a total of five lacZ fusions to P. stuartii genes that were activated by the accumulation of extracellular signals. The identification of several genes has revealed potential functions in amino acid biosynthesis and uptake of amino acids and tricarboxylic acid (TCA) cycle intermediates.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains and plasmids used in this study are listed in Table 1. Growth medium was Luria broth (LB) containing 10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter. Minimal M9 medium was modified to contain lower salt concentrations and consisted of 3 g of Na2HPO4, 1.5 g of KH2PO4, 0.25 g of NaCl, and 0.5 g of NH4Cl per liter. After autoclaving, the following were added per liter: 10 ml of 0.01 M CaCl2, 1 ml of 1M MgSO4, and glycerol to a final concentration of 0.4%. Casamino Acids were added to a final concentration of 1.5%. Antibiotics were used at the following concentrations for P. stuartii: kanamycin, 20 μg/ml; tetracycline, 30 μg/ml; and chloramphenicol, 25, 50, and 80 μg/ml. β-Galactosidase assays were done on sodium dodecyl sulfate-chloroform-treated cells by the method of Miller (35).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or relevant markers | Reference or source |
|---|---|---|
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lac-pro) [F′ proAB lacIqlacZΔM15 Tn10] | Stratagene |
| DH5α | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lacZYA-argF)U169 φ80ΔlacZΔM15 | Gibco/BRL |
| SM10 λpir | thi thr leu tonA lacY supE recA RP4-2-Tc::Mu Kmr λpir | 36 |
| P. stuartii strains | ||
| PR50 | Wild type | 44 |
| PR51 | PR50 ΔaarA | 43 |
| XD37 | PR50 cma37::mini-Tn5 lacZ1 | This study |
| XD37.A | XD37 ΔaarA | This study |
| XD67 | PR50 cma67::mini-Tn5 cat/lacZ | This study |
| XD67.A | XD67 ΔaarA | This study |
| XD156 | PR50 cma156::mini-Tn5 cat/lacZ | This study |
| XD156.A | XD156 ΔaarA | This study |
| XD227 | PR50 cma227::mini-Tn5 cat/lacZ | This study |
| XD227.A | XD227 ΔaarA | This study |
| XD230 | PR50 cma230::mini-Tn5 cat/lacZ | This study |
| XD230.A | XD230 ΔaarA | This study |
| Plasmids | ||
| pACYC184Δ1 | pACYC184 containing a ScaI-EcoRI deletion within the chloramphenicol resistance gene | This study |
| pUT::mini-Tn5 lacZ1 | Transposon delivery vector | 11 |
| pUT::mini-Tn5 cat/lacZ | Transposon delivery vector | This study |
Transposon mutagenesis.
The reporter transposon mini-Tn5 cat/lacZ was constructed as follows. First, a promoterless cat gene was excised from pMSG.CAT (Pharmacia) by SalI digestion. This cat cassette was then inserted as a SalI fragment into SalI-digested pQF50 (18), creating pQF50.CAT. A cat/lacZ cassette was then excised from pQF50.CAT as a BamHI/XmnI fragment and treated with the Klenow fragment of DNA polymerase I and deoxynucleoside triphosphates to create blunt ends. Digestion of pUT::mini-Tn5 lacZ1 (11) with SfiI released the lacZ gene, and the remaining vector DNA was treated with T4 DNA polymerase to create blunt ends. The cat/lacZ cassette was inserted in the proper orientation within pUT::mini-Tn5 to allow transcription from exogenous promoters to proceed into the dual reporters. This transposon was designated mini-Tn5 cat/lacZ. A library of mini-Tn5 lacZ (11) or cat/lacZ insertions in P. stuartii PR50 was constructed by a plate mating. Briefly, 100 μl of overnight cultures of PR50 and either S17.1λ pir/pUT::mini-Tn5 lacZ or SM10 λpir/pUT::mini-Tn5 cat/lacZ were mixed together and spotted on an LB agar plate. After growth for 18 h at 37°C, the mating mixture was resuspended in 10 ml of LB and dilutions were plated on LB agar plates containing kanamycin (20 μg/ml), tetracycline (30 μg/ml) to counterselect the donor, and 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) at 100 μg/ml. Plates were then incubated for 48 h at 37°C. To isolate mini-Tn5 cat/lacZ insertions within genes expressed at high density, individual blue colonies were streaked for single colonies by using toothpicks at a density of 30 restreaks per plate on each of the following three plates: LB plus chloramphenicol (25 μg/ml), LB plus chloramphenicol (50 μg/ml), and a master plate of LB plus kanamycin (20 μg/ml), tetracycline (30 μg/ml), and X-Gal. Chloramphenicol sensitivity was scored after 18 h of growth as an obvious defect in single-colony formation.
Preparation of cell-free supernatants.
Conditioned medium was prepared by inoculation of 30 ml of LB with 5 μl of a 10−3 dilution of an overnight culture of P. stuartii PR50 followed by shaking (250 rpm) at 37°C. Cells were typically harvested 3 to 4 h after entering stationary phase, and cells were pelleted at 4,300 × g for 10 min. The resulting supernatants were supplemented with the addition of 20× tryptone-yeast extract to a final concentration of 0.5×, and the pH was brought to 7.5. Conditioned M9 medium was prepared in a similar manner, except for harvesting at an optical density at 600 nm (OD600) of 1.3 unless stated otherwise. Conditioned medium was filter sterilized with a 0.2-μm-pore-size filter and stored at −80°C. Freezing had no apparent effect on activity. Growth phase expression of the cma fusions was done with 30 ml of supplemented conditioned medium or 0.5× LB as a control in 250-ml flasks. Each type of medium was inoculated at a final dilution of 1/1,000 from a low-density culture of the appropriate strain, and flasks were incubated at 37°C with shaking at 300 rpm.
Partial purification of factors from minimal media.
Conditioned medium was prepared from early-stationary-phase cultures of PR50 cells grown in 100 ml of M9 minimal medium. The medium was adjusted to pH 7.5 with NaOH and filter sterilized. The resulting supernatant was concentrated by lyophilization and resuspended at a 20× concentration in 5 ml of sterile water. Five milliliters of material was applied to a Sephadex G-10 column (Pharmacia), and 5-ml fractions were collected by elution with 0.04 M NaCl, pH 7.5. Under these conditions, fraction 12 typically represented the void volume and the M9 salts eluted at fraction 22. Fractions 1 to 11 displayed no activity. Fractions were stored at −80°C. Upon thawing, a white precipitate was occasionally observed and was removed by centrifugation. Fractions were tested for activity by mixing 1 ml of each fraction with 2 ml of 0.5× LB (pH 7.5) and using the appropriate cma fusion as a biosensor. The activating factors were found within the included volume. Purified material was stored at −80°C.
Identification of the cma fusions.
To clone the cma fusions, the restriction enzyme HindIII, which cuts within mini-Tn5 cat/lacZ immediately after the cat gene, was utilized. Southern blot analysis of cma fusions was performed with a cat-specific probe to determine the sizes of the HindIII fragments containing the cat gene and adjacent upstream chromosomal DNA. Based on the Southern blot analysis, chromosomal DNA was digested with HindIII, and DNA in the appropriate size range was purified by using a Gene-Clean kit (Bio-101) and ligated to HindIII-digested pBluescript SK(−) or to pACYC184Δ1, a pACYC184 (8) derivative created by removing a segment of the chloramphenicol resistance gene. Ligation products were electroporated into XL1 Blue, and transformants were selected on medium containing chloramphenicol (10 μg/ml) to identify plasmids containing the cat gene and upstream P. stuartii DNA. For all plasmids, the sequence of chromosomal DNA upstream from the insertion junction was determined by using the Cy5 labeled primer 5′-GGTTTTCCCAGTCACGACGTTG-3′, which reads outward from the 5′ end of the cat gene. Sequencing reactions were done with a cycle sequencing kit (Amersham), and reactions were run on an ALF sequencer (Pharmacia).
Nucleotide sequence accession numbers.
The GenBank accession numbers of the sequences disrupted by the fusions identified in this work are as follows: cma37, AF148450; cma67, AF148452; cma156, AF148453; and cma227, AF148454.
RESULTS
Identification of P. stuartii genes regulated by cell-to-cell signaling.
A library of random mini-Tn5 lacZ insertions was constructed in P. stuartii PR50 as described in Materials and Methods. Blue colonies were individually screened for β-galactosidase expression in cells at low density grown in LB or in conditioned medium from early-stationary-phase cultures. By using this approach, four fusions, activated at least fourfold by conditioned medium, were identified from screening approximately 300 colonies. One fusion, designated cma37 for conditioned medium activated, was chosen for further study and exhibited a 20- to 30-fold activation by conditioned medium (see below).
The above-described strategy was fairly labor intensive. Therefore, to facilitate the identification of additional fusions activated by extracellular signals, we employed a second strategy similar to that previously described for isolation of quorum-sensing-regulated genes in E. coli (2, 12). The bicistronic reporter transposon mini-Tn5 cat/lacZ was created for this purpose. This transposon contains promoterless chloramphenicol acetyltransferase (cat) and β-galactosidase (lacZ) genes in tandem as reporters of promoter activity. Insertion of the transposon into a transcription unit was identified as a blue colony on X-Gal plates. Blue colonies were then tested for single-colony chloramphenicol resistance. Blue and Cmr colonies indicated an insertion within a constitutively expressed gene. However, a blue Cms colony represented a potential insertion within a gene expressed only at high cell density. Approximately 1,100 blue colonies were screened, and 230 displayed some defect in chloramphenicol resistance. Cells expressing this phenotype were then individually screened at low cell density for β-galactosidase expression in LB and in nutrient-supplemented conditioned medium from stationary-phase cells. A total of five independent fusions in which the presence of conditioned medium prematurely activated the fusions from 6- to 26-fold were identified. Four of these fusions were studied further and designated cma67, cma156, cma227, and cma230. To rule out oxygen depletion as a mechanism for activation, 20×-concentrated conditioned medium was added to fresh LB at a 1/20 dilution. Each cma fusion exhibited activation under these conditions (data not shown). In addition, nutrients were added back to conditioned medium and the pH was corrected to 7.5, suggesting that these variables do not contribute to activation.
Activation of the cma fusions by extracellular signals.
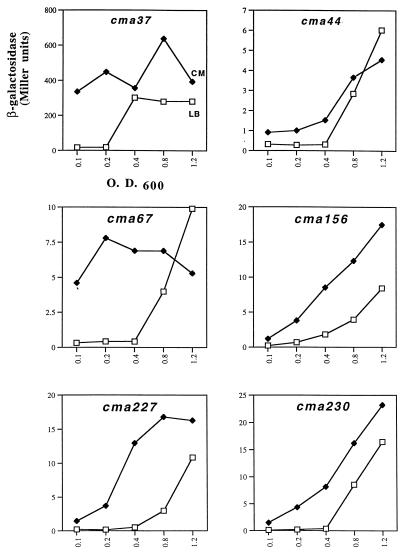
The expression of each cma fusion was examined at different phases of growth in cells grown in LB or in supplemented conditioned medium. In LB alone, the expression of each cma fusion was strongly activated in a growth phase-dependent manner (Fig. 1). The activation values, determined by the ratio of β-galactosidase in cells at stationary phase to that in cells at early log phase, were 17-fold (cma37), 31-fold (cma67), 40-fold (cma156), 56-fold (cma227), and 183-fold (cma230). For each fusion, the presence of conditioned medium resulted in premature activation of each fusion in cells at early to mid-log phase (Fig. 1). The values for activation of each cma fusion by conditioned medium were 26-fold (cma37), 19-fold (cma67), 6-fold (cma156), 25-fold (cma227), and 24-fold (cma230). The addition of conditioned medium did not significantly alter the growth rate of cells until beyond mid-log phase (OD600 = 0.6).
FIG. 1.
Effect of conditioned medium and cell density on expression of the cma fusions. The expression of β-galactosidase from each cma fusion was determined at various stages of growth in LB (□) or conditioned medium (CM) (⧫). β-Galactosidase values are the averages of Miller units from duplicate samples, and standard deviations were less than 10%. Repeat experiments gave values similar to those reported here.
The overall levels of expression from the cma67, cma156, cma227, and cma230 fusions were low. Therefore, as a second method to verify the above results, we examined the expression of each cma fusion by direct measurements of mRNA specific to the cat reporter gene by Northern blot analysis. In each case, the presence of conditioned medium resulted in a large increase in cat-specific message relative to that for cells grown only in LB (data not shown).
The activity of the extracellular factor(s) for each cma fusion was stable to 100°C for 10 min, acid (pH 2.0 for 20 min), and base (pH 12 for 20 min). The ability of certain metabolites to activate each cma fusion was also tested. The addition of pyruvate (2 mM), α-ketoglutarate (2 mM), homoserine lactone (5 mM), homocysteine thiolactone (5 mM), Casamino Acids (1.5%), or acetate (2 mM) had no effect on expression of the cma fusions in cells at low density (data not shown).
Factor production in M9 salts media.
Production of the signals activating the cma fusions was also examined in a defined minimal medium of M9 salts containing either 0.4% glycerol, 1.5% Casamino Acids, or a combination of glycerol and Casamino Acids. For each of these growth conditions, conditioned medium was prepared at the same OD600 of 1.3. Conditioned medium prepared from M9 plus glycerol did not support activation of the fusions, with the exception of cma37, which was weakly activated under these conditions (Table 2). Conditioned medium prepared from M9 plus Casamino Acids or glycerol plus Casamino Acids supported strong activation of the cma37, cma67, and cma230 fusions. However, the cma156 and cma227 fusions were not activated under any of the conditions with M9 medium (Table 2). Control experiments indicated that Casamino Acids supplementation by itself did not activate any of the cma fusions (data not shown).
TABLE 2.
Medium-specific production of the activating factors
| Fusion | Relative activation by the following conditioned mediuma:
|
||
|---|---|---|---|
| M9 + glycerol | M9 + CAAb | M9 + glycerol-CAA | |
| cma37 | 1.9 | 10.6 | 5.5 |
| cma67 | 1.1 | 3.8 | 4.1 |
| cma156 | 0.7 | 1.9 | 0.9 |
| cma227 | 0.8 | 1.4 | 1.2 |
| cma230 | 1.2 | 9.1 | 4.3 |
Ratio of β-galactosidase in the presence of conditioned medium to that in control medium.
CAA, Casamino Acids.
Partial purification of the activating factors.
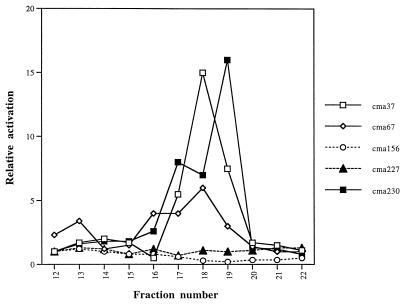
To partially purify the factors activating the cma fusions, conditioned medium was prepared from an M9 salts medium containing 1.5% Casamino Acids as a carbon source. Concentrated conditioned medium was then fractionated by gel filtration with a Sephadex G-10 resin (see Materials and Methods). Fractions were tested at a concentration of 0.33× for activity with each cma fusion as a biosensor. For the cma37, cma67, and cma230 fusions, distinct activation was observed with fractions within the included volume of fractions 12 to 22 (Fig. 2). Peak activation values were 15-fold for cma37 (fraction 18), 6-fold for cma67 (fraction 18), and 16-fold for cma230 (fraction 19). However, the cma156 and cma227 fusions were not activated by any fractions tested (Fig. 2), including fractions 1 to 11, comprising the void volume (data not shown).
FIG. 2.
Fractionation of the activating factors. Concentrated conditioned medium prepared from cells grown in M9 salts plus 1.5% Casamino Acids was fractionated by gel filtration on a Sephadex G-10 column as described in Materials and Methods. Fractions between the void volume and the elution of salts (fractions 12 to 22) were tested for activation of each cma fusion. Fractions 0 to 11 displayed no activity. The same column fraction was used at a concentration of 0.33× in LB to assay each cma fusion. The relative activation values reflect the β-galactosidase activity in each fraction versus the activity in control medium without factor.
The partially purified fractions were also tested in dose-response assays for activation of the cma37, cma67, and cma230 fusions. When used at concentrations of 0.33×, 0.16×, and 0.08× the original concentration, the fraction exhibiting peak activity on the cma37 fusion was capable of activation values of 13-, 14-, and 2-fold, relative to that with LB (data not shown). For the cma67 fusion, activation values of five-, four-, and onefold were observed at concentrations of 0.33×, 0.16×, and 0.08× that of the active fraction. Activation values for cma230 were 8-, 4-, and 0.5-fold at 0.33×, 0.16×, and 0.08× the concentration of the active fraction.
Identification of the cma fusions.
Each of the cma::mini-Tn5 lacZ or mini-Tn5 cat/lacZ fusions was cloned out along with flanking upstream DNA. The sequence of DNA upstream of the insertion site was determined, and open reading frames which were positioned to drive expression into the reporter genes were subjected to BLAST searches. To date, we have been unable to clone the cma230 fusion.
(i) cma37.
The cma37::mini-Tn5 lacZ insertion disrupted an open reading frame encoding a protein with 50% identity and 64% similarity over 122 amino acids to the YaeE protein of E. coli (accession no. P31547). YaeE is similar to the permease component of ABC transporter complexes involved in the uptake of amino acids, including E. coli ProW (P14176) and GltK (P41075).
(ii) cma67.
The cma67 fusion disrupts the beginning of a sequence encoding putative CysK homolog. The region upstream of this insertion encoded a putative 46-amino-acid protein with 89% identity and 91% similarity to the first 46 amino acids of the CysK protein of E. coli (6). This protein encodes O-acetylserine lyase A, which is involved in cysteine biosynthesis.
(iii) cma156.
The cma156 fusion is at a position corresponding to 34 amino acids downstream from the beginning of the product of an open reading frame which encodes a putative protein with 64% amino acid identity to a hypothetical protein (U32743) in Haemophilus influenzae. The function of this protein is unknown. However, the entire H. influenzae protein exhibited 30 to 40% amino acid identity to putative sodium-dependent dicarboxylate transporters from several different bacteria and from eukaryotic cells. These proteins function in the uptake of TCA cycle intermediates such as succinate and citrate.
(iv) cma227.
The cma227 insertion is within an open reading frame encoding 108 amino acids upstream of the insertion site. The product of this open reading frame did not exhibit significant homology to previously identified proteins.
AarA is required for production of an extracellular factor at mid-log phase.
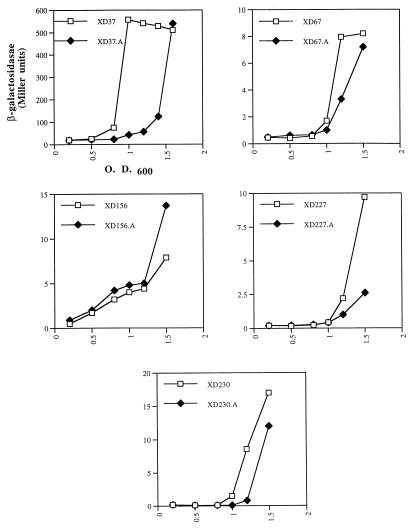
To identify P. stuartii genes involved in the cell-to-cell signaling, the strain XD37 cma37::mini-Tn5 lacZ was mutagenized with mini-Tn5Cm (11). Mutants that were defective in activation of the cma37 fusion were selected with a pale blue or white phenotype. Analysis of several mutants indicated that mini-Tn5Cm had inserted into the previously identified aarA gene, encoding a protein involved in negative regulation of the chromosomal 2′-N-acetyltransferase gene [aac(2′)-Ia] (43). To further investigate the requirement for AarA in expression of the cma37 fusion, allelic replacement was used to create an in-frame aarA deletion in the cma37::mini-Tn5 lacZ background by previously described procedures (43). The ΔaarA allele was verified by Southern blot analysis. The expression of cma37::lacZ was then examined at various stages of growth in both wild-type (XD37) and ΔaarA (XD37.A) backgrounds. In a wild-type background, the cma37 fusion was activated in cells at an OD600 of between 0.5 and 0.8, with a peak induction occurring at an OD600 of 1.0 (Fig. 3). In contrast, expression of cma37 was significantly delayed in the ΔaarA background, and induction did not occur until an OD600 of 1.2 to 1.5 (Fig. 3). Based on this result, the ΔaarA allele was introduced into P. stuartii strains containing the cma67, cma156, cma227, and cma230 fusions. The density-dependent activation of the cma67, cma227, and cma230 fusions was also delayed in the aarA mutant (Fig. 3). In contrast, the expression of cma156 was not delayed in the aarA mutant.
FIG. 3.
Role of AarA in activation of the cma::lacZ fusions. The expression of β-galactosidase from each of the cma::lacZ fusion is shown at various cell densities in the wild-type (□) or ΔaarA (⧫) background is shown.
The delay in expression of the cma37, cma67, cma227, and cma230 fusions could be due to either decreased production of the extracellular signal activating these fusions or a defect in response to the signal. To examine the first possibility, conditioned medium was prepared from PR50 (wild type) and PR51 (ΔaarA) at an OD600 of either 0.8 or 1.5. The ability of each conditioned medium to prematurely activate the cma fusions was examined in cells at early log phase (Table 3). The cma37, cma67, and cma230 fusions were activated by conditioned medium prepared from wild-type PR50 cells at an OD of 0.8 but not by conditioned medium prepared from PR51 cells at the same OD. The cma156 and cma227 fusions were not activated by conditioned medium prepared at an OD of 0.8 from either strain (Table 3). When conditioned medium was prepared at an OD of 1.5, the cma37, cma67, and cma230 fusions were again activated by conditioned medium prepared from wild-type PR50 (Table 3). In addition, the conditioned medium prepared from PR51 ΔaarA at an OD of 1.5 now supported significant activation of these fusions (Table 3). The cma156 and cma227 fusions displayed slightly higher levels of activation with conditioned medium prepared from wild-type PR50 relative to PR51. The low level of activation of the cma156 and cma227 fusions by conditioned medium prepared at an OD of 1.5 suggested a requirement for conditioned medium prepared from higher-density cultures (Fig. 1). This was tested by using conditioned medium prepared from cells at an OD of 1.8. Under these conditions, both the cma156 and cma227 fusions exhibited activation with conditioned medium prepared from wild-type PR50 but not from PR51 ΔaarA (Table 3).
TABLE 3.
AarA is required for factor production
| Conditioned medium used for growth
|
Relative activation by conditioned mediuma
|
|||||
|---|---|---|---|---|---|---|
| Strain | OD at harvest | cma37 | cma67 | cma156 | cma227 | cma230 |
| PR50 | 0.8 | 12.0 | 4.7 | 1.7 | 1.4 | 4.9 |
| PR51 | 0.8 | 0.6 | 1.2 | 1.5 | 1.7 | 1.7 |
| PR50 | 1.5 | 15.6 | 12.4 | 2.3 | 2.0 | 65.8 |
| PR51 | 1.5 | 16.6 | 8.4 | 1.7 | 1.8 | 21.5 |
| PR50 | 1.8 | NDb | ND | 5.2 | 3.6 | ND |
| PR51 | 1.8 | ND | ND | 1.7 | 1.6 | ND |
Ratio of Miller units in cells harvested at early log phase (OD = 0.25) in conditioned medium to that with LB.
ND, not determined.
The requirement for aarA in the response to the extracellular signal was also examined with the cma37 fusion. In a wild-type background, the cma37 fusion was activated 26-, 36-, and 12-fold in response to undiluted, 0.5×, and 0.25× conditioned medium, respectively. The cma37 fusion in the ΔaarA background was activated 40-fold in undiluted conditioned medium and 39-fold in 0.5× conditioned medium (data not shown). However, at a conditioned medium concentration of 0.25×, the cma37 fusion was not activated in the ΔaarA background (data not shown). In these experiments, cells were inoculated into media at a very low density (1/1,000 of the culture) and were allowed to grow to an OD600 of 0.3 before testing. Since the observed defect in factor response was seen only at the lowest dilution of conditioned medium (0.25×), the observed defect in factor response may have been due to a lack of factor production in ΔaarA cells. For example, during growth, cells will secrete factor and contribute to the amount of factor already present in conditioned medium. To minimize this interference, cells were assayed in 0.25× conditioned medium at an OD600 of 0.1. At this OD, there was no activation of the cma37 fusion in wild-type or ΔaarA mutant cells (not shown). Thus, the apparent defect in factor response by the aarA mutant at an OD600 of 0.3 is likely to have resulted from a lack of production to supplement the low factor amounts in 0.25× conditioned medium.
DISCUSSION
In order to further understand the role of cell-to-cell signaling in the stationary-phase physiology of bacteria, quorum-sensing-activated genes in P. stuartii have been identified. The predicted products of some of these genes are involved in diverse cellular processes. The cma37 fusion is within a gene whose product is highly similar to YaeE of E. coli, a protein homologous to the permease component of ABC transporter complexes involved in amino acid and peptide uptake. The cma156 fusion is within a putative transporter for dicarboxylic acids such as malate, succinate, and citrate. The activation of both of these genes by the accumulation of extracellular signals may indicate a requirement for uptake of potential nutrients such as amino acids or TCA cycle intermediates. The cysK gene, encoding O-acetylserine lyase A, was also identified. Interestingly, cysK was previously identified in a search for E. coli genes regulated by quorum sensing (2). The activation of cysK may reflect a requirement for increased cysteine in stationary-phase cells. At present, we are investigating whether additional genes required for cysteine biosynthesis are also regulated by quorum sensing or if this regulation is unique to cysK.
Previous work on the regulation of the peptidoglycan/aminoglycoside 2′-N-acetyltransferase [aac(2′)-Ia] gene identified a role for an extracellular factor (AR-factor) in the downregulation of aac(2′)-Ia at high density (44). This decrease in aac(2′)-Ia expression was blocked in aarA mutants (43). In light of the present work demonstrating a requirement for aarA in production of an extracellular factor which activates the cma37, cma67, cma156, cma227, and cma230 fusions, it seems possible that AR-factor may regulate these cma fusions. In support of this, the fractionation of conditioned medium has shown that the peak fraction for activation of cma37 is also the peak fraction for repression of aac(2′)-Ia (13).
At present, the chemical nature of the factor(s) that activates the cma fusions is unknown. The factor which activates the cma37 fusion has polar properties and does not bind to a C18 column (data not shown). Based on fractionation studies using Sephadex G-10, the factor(s) activating each cma fusion partitioned into the included volume, suggesting a molecular mass of below 700 Da. For the cma156 and cma227 fusions, production of the factor was observed only in LB media. There was no activation with conditioned medium prepared from cells grown in M9 minimal media. For the cma37, cma67, and cma230 fusions, factor production was observed both in rich LB media and in M9 minimal media, but only when Casamino Acids were used as a carbon source.
The preferential production of quorum-sensing signals in rich medium has been reported for at least one other bacterial system (42). In P. stuartii, the physiological significance of increased factor production in the presence of rich LB medium or M9 medium supplemented with Casamino Acids is unclear. It may reflect a response directed at utilizing peptides or specific amino acids as energy sources when more preferred nutrients become depleted at high density. If such a system exists, it would be advantageous for cells to activate the quorum-sensing systems only when the amino acids or peptides are present. Activation of the YaeE homolog (cma37), which has a putative function in amino acid transport, provides support for this hypothesis. In addition, a subset of quorum-sensing-activated genes in E. coli appear to have a role in the use of amino acids as energy sources (2).
An important aspect of this work is the identification of a new gene required for production of an extracellular activating factor. An in-frame deletion of the aarA gene resulted in a delay in the density-dependent induction of the cma37, cma67, cma227, and cma230 fusions (Fig. 3). Our data suggests that this occurs due to a lack of factor production at mid-log phase (Table 3). The AarA protein is hydrophobic and is predicted to be an integral membrane protein (43). Although AarA does not exhibit significant homology to previously identified proteins, it is possible that it is involved in a transport process which exports the signal from the cell. However, the possibility of a direct role for AarA in biosynthesis of the factor cannot be ruled out. It is important to note that even in the aarA null background, there is eventually enough factor present at stationary phase to permit activation of the cma fusions. The factor activity at high cell density in the aarA null background could be due to a secondary biosynthetic or export system. Alternatively, if the factor remains internalized, the activity could be due to cell lysis. Although the AarA protein does not display significant homology to other bacterial proteins, there appear to be functional homologs of AarA present in both E. coli and Proteus mirabilis. Representative plasmids from genomic libraries of either bacterium have been found to completely restore factor production when introduced into the ΔaarA mutant strain. The identification of these homologs, particularly in E. coli, may provide additional information on the role of AarA in the production of an extracellular signaling molecule.
ACKNOWLEDGMENT
This work was supported by grant MCB 9405882 from the National Science Foundation.
REFERENCES
- 1.Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 2.Baca-DeLancey R R, South M M T, Ding X, Rather P N. Escherichia coli genes regulated by cell-to-cell signaling. Proc Natl Acad Sci USA. 1999;96:4610–4614. doi: 10.1073/pnas.96.8.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bainton N J, Bycroft B W, Chhabra S R, Stead P, Gledhill L, Hill P J, Rees C E D, Winson M K, Salmond G P C, Stewart G S A B, Williams P. A general role for the lux autoinducer in bacterial cell signalling: control of antibiotic biosynthesis in Erwinia. Gene. 1992;116:87–91. doi: 10.1016/0378-1119(92)90633-z. [DOI] [PubMed] [Google Scholar]
- 4.Bassler B L, Wright M, Showalter R E, Silverman M R. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 5.Bensing B A, Manias D A, Dunny G M. Pheromone cCF10 and plasmid pCF10-encoded regulatory molecules act post-transcriptionally to activate expression of downstream conjugation functions. Mol Microbiol. 1997;24:285–294. doi: 10.1046/j.1365-2958.1997.3301710.x. [DOI] [PubMed] [Google Scholar]
- 6.Byrne C R, Monroe R S, Ward K A, Kredich N M. DNA sequences of the cysK regions of Salmonella typhimurium and Escherichia coli and linkage of the cysK regions to ptsH. J Bacteriol. 1988;170:3150–3157. doi: 10.1128/jb.170.7.3150-3157.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao J G, Meighen E A. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- 8.Chang A C Y, Cohen S N. Construction and characterization of amplifiable DNA cloning vehicles derived from the p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Q, Campbell E A, Naughton A M, Johnson S, Masure H R. The com locus controls genetic transformation in Streptococcus pneumoniae. Mol Microbiol. 1997;23:683–692. doi: 10.1046/j.1365-2958.1997.2481617.x. [DOI] [PubMed] [Google Scholar]
- 10.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 11.deLorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.deLorenzo V, Cases, Herrero I M, Timmis K N. Early and late responses of TOL promoters to pathway inducers: identification of postexponential promoters in Pseudomonas putida with lacZ-tet bicistronic reporters. J Bacteriol. 1993;175:6902–6907. doi: 10.1128/jb.175.21.6902-6907.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding, X., and P. N. Rather. 1999. Unpublished data.
- 14.Downard J, Toal D. Branched-chain fatty acids: the case for a novel form of cell-cell signalling during Myxococcus xanthus development. Mol Microbiol. 1995;16:171–175. doi: 10.1111/j.1365-2958.1995.tb02290.x. [DOI] [PubMed] [Google Scholar]
- 15.Dunny G, Brown B L, Clewell D B. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci USA. 1978;75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberhard A, Burglingame A L, Eberhard C, Kenyon G L, Nealson K H, Oppenheimer N J. Structural identification of an autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 17.Engebrecht J, Nealson K, Silverman M. Bacterial luminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 18.Farinha M A, Kropinski A J. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol. 1990;172:3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuqua W C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 21.García-Lara J, Shang L H, Rothfield L I. An extracellular factor regulates expression of sdiA, a transcriptional activator of cell division genes in Escherichia coli. J Bacteriol. 1996;178:2742–2748. doi: 10.1128/jb.178.10.2742-2748.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grossman A D, Losick R. Extracellular control of spore formation in Bacillus subtilis. Proc Natl Acad Sci USA. 1988;85:4369–4373. doi: 10.1073/pnas.85.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Håvarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji G, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones S, Yu B, Bainton N J, Birdsall M, Bycroft B W, Chhabra S R, Cox A J R, Golby P, Reeves P J, Stephens S, Winson M K, Salmond G P C, Stewart G S A B, Williams P. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser D, Losick R. How and why bacteria talk to each other. Cell. 1993;73:873–885. doi: 10.1016/0092-8674(93)90268-u. [DOI] [PubMed] [Google Scholar]
- 27.Kim S K, Kaiser D. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of Myxococcus xanthus. Cell. 1990;61:19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- 28.Kleerebezem M, Quadri L E N, Kuipers O P, deVos W M. Quorum sensing by peptide pheromones and two-component signal transduction systems in gram positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 29.Kuspa A, Kross L, Kaiser D. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev Biol. 1986;117:267–276. doi: 10.1016/0012-1606(86)90369-6. [DOI] [PubMed] [Google Scholar]
- 30.Kuspa A, Plamann L, Kaiser D. Identification of heat stable A-factor from Myxococcus xanthus. J Bacteriol. 1992;174:3319–3326. doi: 10.1128/jb.174.10.3319-3326.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S A B, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 32.Lazazzera B A, Solomon J M, Grossman A D. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell. 1997;89:917–925. doi: 10.1016/s0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 33.Magnuson R, Solomon J, Grossman A D. Biochemical and genetic characterization of a competence pheromone from Bacillus subtilis. Cell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 34.McClean K H, Winson M K, Fish L, Taylor A, Chhabra S R, Camara M, Daykin M, Lamb J H, Swift S, Bycroft B W, Stewart G S, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 35.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 36.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell to cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 38.Pearson J P, Gray K G, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perego M, Hoch J A. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:1549–1553. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piper K R, Beck von Bodman S, Farrand S K. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature (London) 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 41.Pirhonen M, Flego D, Heikinheimo R, Palva E T. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 1993;12:2467–2476. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puskas A, Greenberg E P, Kaplan S, Schaefer A L. A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J Bacteriol. 1997;179:7530–7537. doi: 10.1128/jb.179.23.7530-7537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rather P N, Orosz E. Characterization of aarA, a pleiotrophic negative regulator of the 2′-N-acetyltransferase in Providencia stuartii. J Bacteriol. 1994;176:5140–5144. doi: 10.1128/jb.176.16.5140-5144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rather P N, Parojcic M M, Paradise M R. An extracellular factor regulating expression of the chromosomal aminoglycoside 2′-N-acetyltransferase of Providencia stuartii. Antimicrob Agents Chemother. 1997;41:1749–1754. doi: 10.1128/aac.41.8.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salmond G P C, Bycroft B W, Stewart G S A B, Williams P. The bacterial enigma: cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 46.Sitnikov D M, Schineller J B, Baldwin T O. Control of cell division in Escherichia coli: regulation of transcription of ftsQA involves both rpoS and SdiA mediated autoinduction. Proc Natl Acad Sci USA. 1996;93:336–341. doi: 10.1073/pnas.93.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solomon J M, Lazazzera B A, Grossman A D. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 48.Surette M G, Bassler B L. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiley J, Santamaria R, Guijarro J, Geistlich M, Losick R. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell. 1991;65:641–650. doi: 10.1016/0092-8674(91)90096-h. [DOI] [PubMed] [Google Scholar]
- 50.Zhang L, Murphy P J, Kerr A, Tate M E. Agrobacterium conjugation and gene regulation by N-acyl-homoserine lactones. Nature (London) 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]