Abstract
To gain insight regarding the mechanisms that extend heteroduplex joints in Escherichia coli recombination, we investigated the effect of recG and ruv genotypes on heteroduplex strand polarity in intramolecular recombination products. We also examined the cumulative effect of mutational inactivation of RecG and single-strand-specific exonucleases on recombination proficiency and the role of Chi sites in RecG-independent recombination. All four strands of the two homologs were incorporated into heteroduplex structures in wild-type cells and in ruv mutants. However, in recG mutants heteroduplexes were generated almost exclusively by pairing the invasive 3′-ending strand with its complementary strand. To explain the dependence of strand exchange reciprocity on RecG activity, we propose that alternative mechanisms may extend the heteroduplex joints after homologous pairing: a reciprocal RecG-mediated mechanism and a nonreciprocal mechanism, mediated by RecA and single-strand-specific exonucleases. The cumulative effect of recG and recJ or xonA mutations on recombination proficiency and the inhibitory effect of recJ and xonA activities on heteroduplex formation by the 5′-ending strands are consistent with this proposal.
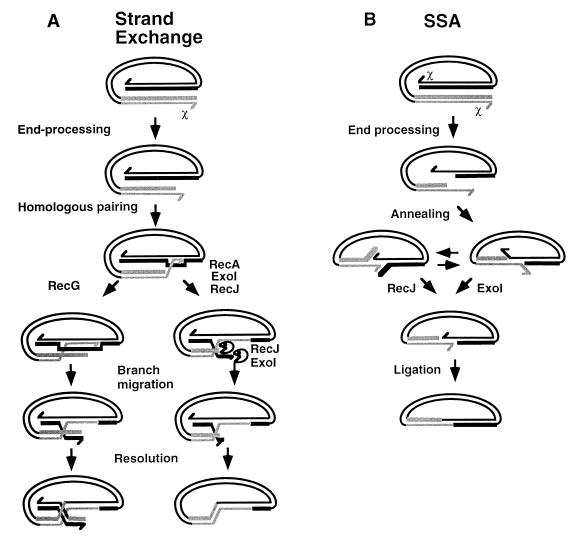
Biochemical and genetic studies indicate that heteroduplex joint formation in Escherichia coli recombination is initiated by pairing of a 3′-ending single-stranded DNA with a double-stranded homolog (5, 9, 29). After pairing, branch migration extends the heteroduplex and, as migration proceeds through a duplex-duplex region, a four-stranded Holliday junction is produced. Resolution of this junction yields products with two complementary heteroduplex structures. One heteroduplex is made by pairing the invasive 3′-ending strand and its complementary strand. The other consists the strands that have been displaced in the primary pairing reaction (Fig. 1A).
FIG. 1.
Hypothetical RecG-dependent and independent branch migration mechanisms. Homologous sequences are designated by black (top) or gray (bottom) parallel lines. 3′-Ending strands of the homologous sequences are narrow and are marked by half arrowheads. 5′-Ending strands are wide. Thus, strands of homoduplex structures have different widths and the same color, and strands of heteroduplex structures have the same width but different colors. (A) After RecA-catalyzed strand invasion, RecG drives the three-stranded junction to a duplex-duplex region, where a Holliday junction is formed. 3′ and 5′ heteroduplexes are then extended by reciprocal strand exchange. In the absence of RecG, polar strand exchange may be catalyzed by RecA, in cooperation with RecJ or exonuclease I (ExoI) that degrades the displaced strands. Both modes of strand exchange depend also on endonucleolytic cleavage of the D-loop. The nonconservative reaction would yield a circular product with a 3′ heteroduplex. (B) A 3′ heteroduplex may be generated by SSA recombination. In this pathway both ends are processed to 3′ overhangs that anneal to a heteroduplex structure.
Branch migration may be driven by RecA-mediated strand exchange or by the junction-specific helicases RuvAB and RecG (reviewed in references 10, 35, 36, and 39). These two helicases bind three- or four-stranded junctions and catalyze branch migration of RecA-generated recombination intermediates (8, 16, 22, 28, 33, 38). However, RecG and RuvAB differentially affect RecA-catalyzed strand exchange. RecG strongly inhibits RecA-mediated heteroduplex formation, whereas RuvAB has little or no effect. (37). Since RecA-catalyzed strand exchange has a 5′→3′ polarity with respect to the invasive strand (24), the inhibitory effect of RecG suggests an opposite polarity for the RecG-catalyzed reaction. This led to the proposition that RecG-mediated migration of three-stranded junctions secures exchanges that have been initiated at 3′-ending single strands (37).
The subtle effect of recG mutations on recombination proficiency suggested the occurrence of a RecG-independent branch migration mechanism. This mechanism may depend on RuvAB, since combinations of ruv and recG mutations have a synergistic effect (13). However, RecA may also catalyze branch migration in the 3′→5′ polarity, provided that the displaced strands are degraded by single-stranded DNA (ssDNA)-specific exonucleases such as RecJ or exonuclease I (1, 4). This polar strand-exchange mechanism is distinguished from the RecG-catalyzed reaction by the structure of the heteroduplex products. The RecG-catalyzed reaction is conservative, and in a duplex-duplex region it is reciprocal. Conversely, the reaction catalyzed by the coupled activities of RecA and ssDNA-specific exonucleases is nonconservative and nonreciprocal. Thus, the RecG-catalyzed reaction would yield products with two complementary heteroduplex structures, whereas the reaction mediated by RecA and ssDNA-specific exonucleases would yield only one. This heteroduplex would consist of the invasive 3′-ending strand and its homolog (Fig. 1A). Another nonconservative mechanism that may produce only one heteroduplex type is single-strand annealing (SSA) (Fig. 1B). SSA recombination is initiated by resectioning of two double-stranded ends and proceeds by annealing of the complementary strands (11, 12).
If the reciprocity of strand exchange depends on RecG activity, recG mutations should inhibit the incorporation of the displaced strands into heteroduplex structures. Here we examine the effect of recG genotype on heteroduplex strand polarity in intramolecular recombination products. We also attempt to discriminate between two hypothetical RecG-independent nonconservative mechanisms: SSA and nonreciprocal strand exchange.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains used in this study are presented in Table 1. Infected cultures harbored pMB4 (2) and were grown in medium supplemented with ampicillin (100 μg/ml). Infection protocols were as described previously (30). The multiplicity of infection (MOI) is indicated in the figure legends. To inhibit replication of phage DNA that escaped restriction, all strains used in this study were lysogenic to λ(ind−).
TABLE 1.
E. coli K-12 strains
| Strain | Relevant genotypea | Reference or source |
|---|---|---|
| N2973 | recG162::Tn10 | 14 |
| N3793 | ΔrecG263::kan | 18 |
| AM561 | ΔruvAC65 eda-51::Tn10 | 19 |
| HRS1200 | ΔruvC200::kan | 25 |
| AC227 | λ (ind−) | 6 |
| AC258 | recD1009 [λ (ind−)] | 6 |
| AC259 | recJ284::Tn10 [λ (ind−)] | 6 |
| AC261 | recD1011 recJ284::Tn10 [λ (ind−)] | 5 |
| AC263 | recD1011 xonA2 [λ (ind−)] | 5 |
| AC267 | xonA2 [λ (ind−)] | 6 |
| AC280 | ΔrecG263::kan [λ (ind−)] | P1 N3793 × AC227 to Kmr |
| AC284 | recD1009 ΔrecG263::kan [λ (ind−)] | P1 N3793 × AC258 to Kmr |
| AC287 | recD1009 recG162::Tn10[λ (ind−)] | P1 N2973 × AC258 to Tcr |
| AC297 | recG162::Tn10 [λ (ind−)] | P1 N2973 × AC227 to Tcr |
| AC301 | ΔruvC200::kan [λ (ind−)] | P1 HRS1200 × AC227 to Kmr |
| AC303 | ΔruvAC65 eda-51::Tn10[λ (ind−)] | P1 AM561 × AC227 to Tcr |
| AC309 | recD1011 recJ284::Tn10 ΔrecG263::kan [λ (ind−)] | P1 N3793 × AC261 to Kmr |
| AC312 | recD1009 ΔruvAC65 eda-51::Tn10[λ (ind−)] | P1 AM561 × AC258 to Tcr |
| AC314 | recD1011 [λ (ind−)] | 6 |
| AC318 | xonA2 ΔrecG263::kan [λ (ind−)] | P1 N3793 × AC267 to Kmr |
| AC319 | recJ284::Tn10 ΔrecG263::kan [λ (ind−)] | P1 N3793 × AC259 to Kmr |
| AC320 | recD1011 recJ284::Tn10 ΔruvC200::kan [λ (ind−)] | P1 HRS1200 × AC261 to Kmr |
| AC321 | recD1011 xonA2 ΔrecG263::kan [λ (ind−)] | P1 N3793 × AC263 to Kmr |
| AC322 | recJ284::Tn10 ΔruvC200::kan [λ (ind−)] | P1 HRS1200 × AC259 to Kmr |
| AC323 | recD1011 ΔruvC:200:kan[λ (ind−)] | P1 HRS1200 × AC258 to Kmr |
| AC324 | recD1011 ΔrecG263::kan [λ (ind−)] | P1 N3793 × AC258 to Kmr |
| AC325 | recD1011 xonA2 ΔruvC200::kan [λ (ind−)] | P1 HRS1200 × AC263 to Kmr |
| AC326 | xonA2 ΔruvC200::kan[λ (ind−)] | P1 HRS1200 × AC267 to Kmr |
All strains are isogenic derivatives of AC227. Other markers are thi-1 his-4 Δ(gpt-proA)62 argE3 thr-1 leuB6 kdgK51 ara14 lacY1 galK2 xyl5 mtl-1 tsx-33 supE44 rpsL31.
Chimeric phages.
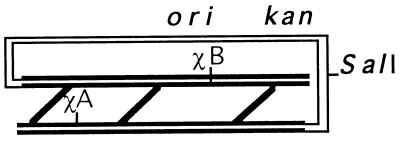
The chimeric phages used in this study are presented in Table 2. All phages harbored intramolecular recombination substrates with a direct terminal repeat, cloned between EcoRI sites. Cloned recombination substrates in all phages, except λZS820 (30), had Chi sites at the indicated loci (5). To facilitate separation of the two heteroduplex structures from each other and from the corresponding homoduplexes, an eight-nucleotide BglII linker was inserted as a heteroallelic marker at the XmnI site on a luxA gene (30).
TABLE 2.
Phages harboring recombination substratesa
| Phage | oripACYC184 | Chi at A | Chi at B |
|---|---|---|---|
| λZS820 | 0 | 0 | 0 |
| λRF949 | + | 0 | + |
| λRF950 | + | + | 0 |
| λRF951 | + | + | + |
| λRF953 | 0 | + | + |
Each Chi site in the schematic representation of the cloned intramolecular recombination substrate consists of three Chi octamers (6). The approximate locations of oripACYC184, the Kanr gene, and Chi insertion sites (A and B) are indicated.
Determination of heteroduplex strand polarity.
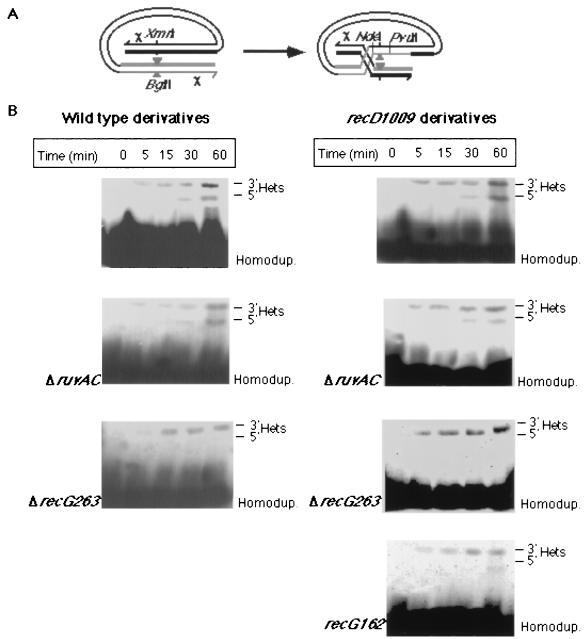
Heteroduplex strand polarity was determined by polyacrylamide gel electrophoresis and Southern hybridization analysis of total cellular DNA preparations digested by PvuII and NdeI (Fig. 2A). The location of heteroduplex fragments, consisting of the strands ending 3′ or 5′ at the break, and of the homoduplex fragments are indicated. These locations were determined by using synthetic heteroduplexes as described previously (30).
FIG. 2.
The effect of recG and ruvAC mutations on heteroduplex strand polarity. (A) A schematic representation of the phage-delivered intramolecular recombination substrate. The location of the Chi octamers (χ), relevant restriction sites, and the eight-nucleotide BglII linker (triangles) are indicated. (B) Total cellular DNA preparations (15 μg/lane) of samples taken at the indicated times after infection (MOI = 3) were subjected to Southern hybridization analysis as described in Materials and Methods. Genotypes of the infected cells are indicated. All wild-type derivatives were infected with λRF953, and all recD1009 derivatives were infected with λZS820. The locations of the electrophoretic bands of the homoduplexes (Homodup.) and the 3′ (3′ Het.) or 5′ (5′ Het.) heteroduplexes are indicated.
Physical monitoring of recombination.
Total cellular DNA preparations of samples taken at the indicated times after infection were digested by SalI and subjected to Southern blot hybridization as described (30). The kinetics of product formation was determined by phosphorimaging analysis. Radioactivity is presented in arbitrary units.
Determination of recombinant frequency and analysis of plasmid recombination products.
To select for Kanr recombinants, cells were infected by the appropriate chimera phage, and samples taken 60 min after infection were plated on kanamycin-supplemented medium as described earlier (21). To minimize the occurrence of intermolecular recombination events or multiple recombination events within the same infected cell, the MOI was 0.2. The recombinant frequency was defined as the ratio of Kanr recombinants to infected cells. To determine the percentage of Chi+ products, isolated colonies were inoculated into kanamycin-supplemented liquid medium. Plasmid DNA preparations of overnight cultures were made by the rapid boiling method (7) and were subjected to restriction endonuclease analysis with NotI and SalI endonucleases. All substrates had a unique SalI site, and all Chi sites were associated with NotI sites (6).
RESULTS
Heteroduplex strand polarity in recG mutants.
The intramolecular recombination substrate used in this study is a linear DNA fragment with a direct terminal repeat (Fig. 1). The homologous sequences on this substrate are distinguished from each other by an eight-nucleotide insertion in one of them (see Materials and Methods). Hence, their complementary strands can form two chemically distinct heteroduplex structures: one by pairing the strands ending 3′ at the breaks (Fig. 1, narrow lines) and the other by pairing the strands ending 5′ at the breaks (Fig. 1, wide lines). These heteroduplexes are separable by polyacrylamide gel electrophoresis (30) and will be referred to here as the 3′ heteroduplex and 5′ heteroduplex, respectively. Both heteroduplexes are generated by intramolecular recombination, but only the 3′ heteroduplex is incorporated into circular products (5).
The hypothesis depicted in Fig. 1A postulates that RecG is required for reciprocal strand exchange. Consequently, recG mutations should lower the ratio of 5′ to 3′ heteroduplexes in recombination products. To examine the effect of recG genotype on heteroduplex strand polarity, phage DNA harboring the recombination substrate was delivered into wild-type cells or recG mutants by phage infection, and the substrate was released from its carrier by in vivo EcoRI restriction (30). The accumulation of 3′ and 5′ heteroduplexes in the infected cells was monitored by Southern hybridization analysis of total cellular DNA preparations from samples taken at various time points after infection (Fig. 2B). As observed previously (5), the 3′ heteroduplex was detected earlier, and its initial rate of accumulation was higher than that of the 5′ heteroduplex. Consistent with the hypothesis in Fig. 1A, a recG deletion (ΔrecG263) markedly lowered the ratio of 5′ to 3′ heteroduplexes. In four of six independent experiments, the 5′ heteroduplex was not detected at any time after infection, while in two it was barely detectable in samples taken 60 min after infection (Fig. 3).
FIG. 3.
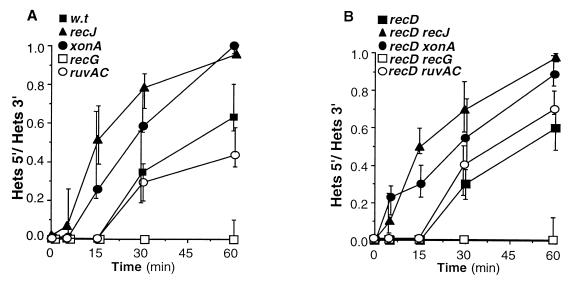
Effect of xonA, recJ, ΔruvAC, and ΔrecG mutations on the ratio of 5′ to 3′ heteroduplexes in intramolecular recombination products. The ratios of 5′ to 3′ heteroduplexes in DNA preparations of samples taken at the indicated times after infection are presented. Radioactivity was determined by phosphorimaging analysis of Southern hybridization patterns like the ones presented in Fig. 2. Relevant genotypes are indicated. Each value is a median of at least three independent experiments. The error bars represent the range.
RecG and RuvAB helicases have similar substrate specificities (39), and genetic evidence suggest that the two enzymes act in overlapping pathways that resolve recombination intermediates (13). It was therefore of interest to determine whether mutational inactivation of the RuvABC-mediated strand exchange and resolution pathway would also affect heteroduplex strand polarity. Unlike the ΔrecG mutation, a ΔruvAC mutation had no detectable effect on heteroduplex strand-polarity (Fig. 2B). The ratio of 5′ to 3′ heteroduplex structures in the ΔruvAC mutant was similar to that in wild-type cells at all time points after infection (Fig. 3A).
Intramolecular recombination occurs in recD mutants at a higher rate than in wild-type cells and is independent of cis-acting Chi sites (6). To determine the effect of recG and ruvAC mutations on heteroduplex strand polarity in recD mutants, we infected the appropriate recD derivatives by use of λZS820 and monitored the accumulation of 3′ and 5′ heteroduplexes (Fig. 2B). As in recD+ cells, a ΔrecG mutation inhibited the accumulation of recombination products with 5′ heteroduplex structures, whereas the ΔruvAC mutation had no detectable effect (Fig. 3B).
The recG162 mutation impairs RecG helicase activity but not its ability to bind DNA and hydrolyze ATP. The mutant enzyme cannot catalyze branch migration (27). To examine whether formation of the 5′ heteroduplex depends on RecG helicase activity, we infected recD1009 recG162 cells with λZS820 and determined heteroduplex strand polarity in recombination products. Like the ΔrecG mutation, the recG162 mutation lowered the ratio of 5′ to 3′ heteroduplexes in recombination products. The 5′ heteroduplex was not detectable at 30 min after infection, and at 60 min it represented ca. 10% of the total heteroduplex DNA. This suggests that RecG helicase activity is required for reciprocal strand exchange.
The effect of xonA, recJ, recG, and ruvAC mutations on the ratio of 5′ to 3′ heteroduplexes in recombination products is summarized in Fig. 3. Mutational inactivation of RecJ or exonuclease I caused an earlier appearance of the 5′ heteroduplex and an increase in the relative rate of its accumulation (5). Conversely, the 5′ heteroduplex was absent or barely detectable after recombination in recG mutants. These data indicate that the invasive 3′-ending strand can be incorporated into heteroduplex products independently of RecG activity but reciprocal strand exchange is RecG dependent. It also suggests that the formation or maintenance of the 5′ heteroduplex is inhibited by RecJ and exonuclease I.
Processing of only one end is sufficient for RecG-independent recombination.
To account for the results seen in Fig. 3, we considered two RecG-independent pathways that would not incorporate 5′-ending strands into heteroduplex structures. One pathway involves invasion of double-stranded DNA by a 3′-ending single strand and nonreciprocal strand exchange, catalyzed by RecA and ssDNA exonucleases (Fig. 1A). The other pathway involves pairing by the SSA mechanism (Fig. 1B). Processing of only one end is required for strand invasion, but both ends must be processed to single-stranded overhangs in SSA recombination (11, 12). Chi participates in RecBCD-mediated end processing (10, 32) and is essential for intramolecular recombination by the substrates used in this study (6). Thus, recombination dependence on Chi sites in both homologs would suggest an SSA mechanism. Furthermore, since active Chi sites are invariably lost in end processing, Chi sites on both homologs would be lost in SSA recombination, but only one Chi would be lost in pairing by a strand invasion mechanism. To test these predictions, cells were infected by chimeric phages harboring a linear fragment with a direct terminal repeat and Chi sites on either one or on both homologs. To facilitate isolation of cells that harbor circular recombination products and determination of recombinant frequencies, a pACYC184 replication origin and a kanr gene were located between the repeated sequences (31). Consistent with earlier results (6), the recG mutation lowered recombinant frequency, and recombination proficiencies of substrates with a single Chi site were lower than that of the substrate with two Chi sites. However, the recG mutation did not abolish recombination of substrates with a single Chi site. The ratio of recombinant frequency of substrates with a single Chi site to that of substrates with two Chi sites was similar in wild-type cells and ruvAC and recG mutants (Table 3). We analyzed plasmid recombination products of substrates with two Chi sites for the maintenance of Chi in intramolecular recombination. This was accomplished by restriction analysis of isolated plasmid recombination products with SalI and NotI endonucleases. In all substrates employed for this study, SalI had a unique site and NotI sites were associated with all Chi sequences (6). The percentage of products that maintained a single Chi site in recombination was not affected by the recG or ruvAC genotypes (Table 4). These results suggest a recG-independent recombination mechanism that involves processing of only one end and are therefore inconsistent with SSA recombination.
TABLE 3.
A Chi site on one homolog is sufficient for intramolecular recombination in recG and ruvAC mutantsa
| Strain | Genotype | % Frequency (range) of Kanr recombinants
|
||
|---|---|---|---|---|
| Chi at A and B | Chi at A | Chi at B | ||
| AC227 | Wild type | 10.3 (10–12.1) [1.0] | 3.0 (2.4–3.7) [0.3] | 1.7 (1.2–2.2) [0.2] |
| AC297 | recG162 | 3.5 (2.5–3.7) [1.0] | 0.8 (0.6–0.9) [0.2] | 0.4 (0.3–0.4) [0.1] |
| AC303 | ΔruvAC | 4.1 (3.7–6.7) [1.0] | 1.4 (1.1–1.6) [0.3] | 0.6 (0.5–0.9) [0.1] |
The approximate locations of Chi sites, oripACYC184 (ori), and the Kanr gene are indicated. Kanr recombinants were scored as described in Materials and Methods (MOI = 0.2). The median and range (in parentheses) of at least three independent experiments are presented. For each genotype, values in brackets represent the ratios of recombinant frequencies for the indicated substrate to that for the substrate with two Chi sites.
TABLE 4.
A Chi site is incorporated into intramolecular recombination products in recG and ruvAC mutantsa
| Strain | Genotype | % Chi+ products (range) |
|---|---|---|
| AC227 | Wild type | 36 (35–36) |
| AC297 | recG162 | 44 (42–45) |
| AC303 | ΔruvAC | 39 (35–45) |
λRF953 (see Table 2) was used in all experiments. The percentage of Chi+ recombination products was determined as described in Materials and Methods and is presented as the median and range of at least three independent experiments.
A cumulative effect of recG and xonA or recJ mutations on recombination kinetics.
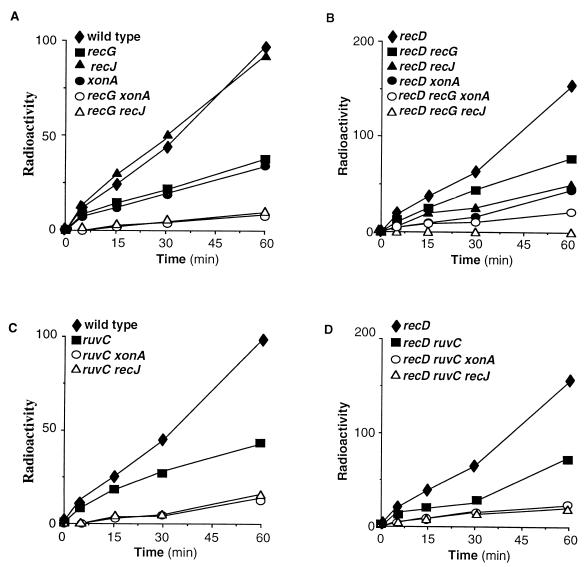
If RecJ and exonuclease I participate in a mechanism that functionally overlaps RecG activity, xonA or recJ mutations should lower recombination proficiency of recG mutants. To test this prediction, we compared recombination proficiencies in xonA recG or recJ recG double mutants to those in recG, recJ, and xonA mutants. EcoRI-expressing cells of the appropriate genotypes were infected by λRF953, and the accumulation of recombination products was monitored by Southern hybridization as described before (30). Consistent with earlier results (6), a recJ mutation did not affect recombination kinetics, while xonA or recG mutations had only a partial effect. However, rates of accumulation of recombination products in recJ recG or xonA recG double mutants were markedly lower than those in any of the single mutants (Fig. 4A). We also examined the combined effect of a recG with xonA or recJ mutations in recD mutants (Fig. 4B). RecJ plays a role in RecD-independent recombination, presumably by generating the invasive 3′-ending strand at the presynaptic stage (5, 15, 17). recG, xonA, and recJ mutations lowered the recombination proficiency in recD mutants, and a combination of recG and xonA mutations had a cumulative effect. Significantly, recombination products were not detectable in recD recG recJ triple mutants at any time after infection (three independent experiments). We also examined the combined effect of ruvC with recJ or xonA mutations on recombination kinetics. Inactivation of either one of the two exonucleases lowered the recombination proficiency in ruvC mutants (Fig. 4C and D).
FIG. 4.
The effect of xonA, recJ, recG, and ruvC mutations, and the indicated double mutations, on intramolecular recombination proficiency. Substrates were released from infecting Chimera phage by in vivo restriction and samples were taken at the indicated times after infection. All wild-type derivatives were infected with λRF953, and all recD1011 derivatives were infected with λZS820 (MOI = 2). SalI-digested total cellular DNA preparations were subjected to Southern hybridization as described in Material and Methods. The kinetics of product formation were determined by phosphorimaging analysis of Southern hybridization patterns. The radioactivity is presented in arbitrary units.
DISCUSSION
We tested predictions of a hypothesis that postulates alternative branch migration mechanisms in E. coli recombination: a conservative mechanism catalyzed by RecG helicase and a nonconservative mechanism catalyzed by RecA and ssDNA-specific exonucleases (Fig. 1A). Consistent with this hypothesis, the recG genotype affected heteroduplex strand polarity in intramolecular recombination products. All four strands of the recombining homologs were incorporated into heteroduplex products in recG+ cells, whereas in recG mutants the 5′-ending strands were absent or barely detectable from heteroduplex structures. Unlike the recG mutations, a ΔruvAC mutation did not affect heteroduplex strand polarity. This phenotypic difference between recG and ruv mutants supports the proposition that RuvAB and RecG are not redundant activities fulfilling the same function in recombination (39). It also suggests that, in vivo, the reciprocity of strand exchange depends on RecG but not on RuvAB. One intriguing possibility is that RecG acts on three-stranded intermediates, generated by RecA-mediated strand invasion, and promotes branch migration into a duplex-duplex region. Subsequently, RuvAB drives the four-stranded Holliday junction to a preferred RuvC cleavage site (37).
What mechanism drives the primary three-stranded junction in the absence of RecG activity? Indirect evidence suggested that the polar strand exchange reactions catalyzed by the coupled activities of RecA and ssDNA-specific exonucleases (1, 4) have a biological relevance. A role for RecJ and exonuclease I in recombination was suggested by the inhibitory effect of recJ and xonA mutations on “short homology” transduction (20), phage DNA recombination (23), conjugational recombination (34), and intramolecular recombination of linear substrates (6). These exonucleases may act at the presynaptic stage of the recombination pathway by processing the double-stranded DNA ends to single-stranded overhangs or at the postsynaptic stage by conferring a 3′→5′ polarity on RecA strand exchange activity (20, 23). Since synapsis involves pairing of a 3′-ending strand, it is unlikely that exonuclease I acts at the presynaptic stage. Furthermore, a role for RecJ and exonuclease I in a nonconservative branch migration mechanism is suggested by the observation that mutational inactivation of these enzymes increases the ratio of 5′ to 3′ heteroduplexes in intramolecular recombination products (reference 5 and Fig. 3). These findings are consistent with a mechanism that involves cleavage of the anticomplementary strand in the D-loop intermediate by a junction-specific endonuclease (3) and migration of the three-stranded junction by the combined activity of RecA and ssDNA-specific exonucleases. The effect of recJ and xonA mutations on heteroduplex strand polarity (5) suggests that this mechanism may play a role in recG+ cells.
An alternative interpretation of the data presented in Fig. 2 is that, in the absence of RecG, intramolecular recombination is by the SSA mechanism that incorporates only the 3′-ending strands into a heteroduplex structures. This possibility seems unlikely since processing of only one end is sufficient for intramolecular recombination (Tables 3 and 4).
Mutations in ruvC, recG, xonA, or recJ genes have only a partial effect on recombination, whereas mutational inactivation of any pair of these genes causes recombination deficiency (references 5, 13, 20, 23, and 34 and the present study). These data are consistent with the notion of a functional overlap between the activities of the respective gene products in recombination. Thus, RecG or a combination of RecA and ssDNA-specific exonucleases may act on a RecA-generated D-loop to extend the heteroduplex structure. The synergistic effect observed in recG ruv double mutants and the cumulative effect of ruvC with recJ or xonA mutations suggest that RuvABC acts in both pathways. RuvABC may act in both pathways by resolving branched recombination intermediates. It is also possible that, in the absence of RecG, RuvAB acts in concert with ssDNA-specific exonucleases to promote unidirectional strand exchange. The partial effect of ruvABC mutations on recombination proficiency suggests an alternative mechanism that resolves three- or four-stranded recombination intermediates. Such mechanism may involve a functional analog of the λ Rap protein (26).
ACKNOWLEDGMENT
We thank Wilfried Wackernagel, Hideo Shinagawa, and Robert G. Lloyd for bacterial strains and helpful scientific discussions and Robert G. Lloyd for critical review of the manuscript.
This work was supported by grants from the United States-Israel Binational Science Foundation (grant 97-00058) and The Volkswagen Foundation and the Ministry of Science and Art of the State of Niedersachsen, Niedersachsen, Germany.
REFERENCES
- 1.Bedale W A, Inman R B, Cox M M. A reverse DNA strand exchange mediated by recA protein and exonuclease I. J Biol Chem. 1993;268:15004–15016. [PubMed] [Google Scholar]
- 2.Betlach M C, Herschfield V, Chan L, Brown W, Goodman H, Boyer H W. A restriction endonuclease analysis of the bacterial plasmid controling EcoRI restriction and modification of DNA. FASEB J. 1976;35:2037–2043. [PubMed] [Google Scholar]
- 3.Chiu S K, Low K B, Yuan A, Radding C M. Resolution of an early RecA-recombination intermediate by a junction-specific endonuclease. Proc Natl Acad Sci USA. 1997;94:6079–6083. doi: 10.1073/pnas.94.12.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corrette-Bennett S E, Lovett S T. Enhancement of RecA strand-transfer activity by the RecJ exonuclease of Escherichia coli. J Biol Chem. 1995;270:6881–6885. doi: 10.1074/jbc.270.12.6881. [DOI] [PubMed] [Google Scholar]
- 5.Friedman-Ohana R, Cohen A. Heteroduplex joint formation in Escherichia coli recombination is initiated by pairing of a 3′-ending strand. Proc Natl Acad Sci USA. 1998;95:6909–6914. doi: 10.1073/pnas.95.12.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman-Ohana R, Karunker I, Cohen A. Chi-dependent intramolecular recombination in Escherichia coli. Genetics. 1998;148:545–557. doi: 10.1093/genetics/148.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–198. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 8.Iwasaki H, Takahagi M, Nakata A, Shingawa H. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 1992;6:2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- 9.Konforti B B, Davis R W. The preference for a 3′ homologous end is intrinsic to recA-promoted strand exchange. J Biol Chem. 1990;265:6916–6920. [PubMed] [Google Scholar]
- 10.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin F L, Sperle K, Sternberg N. Intramolecular recombination between DNAs introduced into mouse L cells by a nonconservative pathway that leads to crossover products. Mol Cell Biol. 1990;10:103–112. doi: 10.1128/mcb.10.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin F L, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloyd R G. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J Bacteriol. 1991;173:5414–5418. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd R G, Buckman C. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J Bacteriol. 1991;173:1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lloyd R G, Buckman C. Overlapping functions of recD, recJ, and recN provide evidence of three epistatic groups of genes in Escherichia coli recombination and DNA repair. Biochimie. 1991;73:313–320. doi: 10.1016/0300-9084(91)90218-p. [DOI] [PubMed] [Google Scholar]
- 16.Lloyd R G, Sharp G J. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 1993;12:17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lovett S T, Luisi-DeLuca C, Kolodner R D. The genetic dependence of recombination in recD mutants of Escherichia coli. Genetics. 1988;120:37–45. doi: 10.1093/genetics/120.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahdi A A, Sharples G J, Mandel T N, Lloyd R G. Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J Mol Biol. 1996;257:561–573. doi: 10.1006/jmbi.1996.0185. [DOI] [PubMed] [Google Scholar]
- 19.Mandal T N, Mahdi A A, Sharples G J, Lloyd R G. Resolution of Holliday intermediates in recombination and DNA repair: indirect suppression of ruvA, ruvB, and ruvC mutations. J Bacteriol. 1993;175:4325–4334. doi: 10.1128/jb.175.14.4325-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miesel L, Roth J R. Evidence that SbcB and RecF pathway functions contribute to RecBCD-dependent transductional recombination. J Bacteriol. 1996;178:3146–3155. doi: 10.1128/jb.178.11.3146-3155.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nussbaum A, Shalit M, Cohen A. Restriction-stimulated homologous recombination of plasmids by the RecE pathway of Escherichia coli. Genetics. 1992;130:37–49. doi: 10.1093/genetics/130.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsons C A, Tsaneva I, Lloyd R G, West S C. Interaction of Escherichia coli RuvA and RuvB proteins with synthetic Holliday junctions. Proc Natl Acad Sci USA. 1992;89:5452–5456. doi: 10.1073/pnas.89.12.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Razavy H, Szigety S K, Rosenberg S M. Evidence for both 3′ and 5′ single-strand DNA ends in intermediates in Chi-stimulated recombination in vivo. Genetics. 1996;142:333–339. doi: 10.1093/genetics/142.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Register J C I, Griffith J. The directionality of RecA protein assembly onto single-strand DNA is the same as the direction of strand assimilation during strand exchange. J Biol Chem. 1985;260:12308–12312. [PubMed] [Google Scholar]
- 25.Saito A, Iwasaki H, Ariyoshi M, Morikawa K, Shinagawa H. Identification of four acidic amino acids that constitute the catalytic center of RuvC Holliday junction resolvase. Proc Natl Acad Sci USA. 1995;92:7470–7474. doi: 10.1073/pnas.92.16.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharples G J, Corbbett L M, Graham I R. λRap protein is a structure-specific endonuclease involved in phage recombination. Proc Natl Acad Sci USA. 1998;95:13507–13512. doi: 10.1073/pnas.95.23.13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharples G J, Whitby M C, Ryder L, Lloyd R G. A mutation in helicase motif III of E. coli RecG protein abolishes branch migration of Holliday junctions. Nucleic Acids Res. 1994;22:308–313. doi: 10.1093/nar/22.3.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiba T, Iwasak H, Nakata A, Shinagawa H. SOS-inducible DNA repair proteins, RuvA and RuvB, of Escherichia coli: functional interactions between RuvA and RuvB for ATP hydrolysis and renaturation of the cruciform structure in supercoiled DNA. Proc Natl Acad Sci USA. 1991;88:8445–8449. doi: 10.1073/pnas.88.19.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siddiqi I, Stahl M M, Stahl F W. Heteroduplex chain polarity in recombination of phage λ by the Red, RecBCD, RecBC(D−) and RecF pathways. Genetics. 1991;128:7–22. doi: 10.1093/genetics/128.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silberstein Z, Shalit M, Cohen A. Heteroduplex strand-specificity in restriction-stimulated recombination by the RecE pathway of Escherichia coli. Genetics. 1993;133:439–448. doi: 10.1093/genetics/133.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silberstein Z, Tzfati Y, Cohen A. Primary products of break-induced recombination by Escherichia coli RecE pathway. J Bacteriol. 1995;177:1692–1698. doi: 10.1128/jb.177.7.1692-1698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith G R. Hotspots of homologous recombination. Experientia. 1994;50:234–241. doi: 10.1007/BF01924006. [DOI] [PubMed] [Google Scholar]
- 33.Tsaneva I R, Muller B, West S C. ATP-dependent branch migration of Holliday junctions promoted by RuvA and RuvB proteins of E. coli. Cell. 1992;69:1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- 34.Viswanathan M, Lovett S T. Single-strand DNA-specific exonucleases in Escherichia coli: roles in repair and mutation avoidance. Genetics. 1998;149:7–16. doi: 10.1093/genetics/149.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West S C. Enzymes and molecular mechanisms of genetic recombination. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- 36.West S C. The processing of recombination intermediates: mechanistic insights from studies of bacterial proteins. Cell. 1994;76:9–15. doi: 10.1016/0092-8674(94)90168-6. [DOI] [PubMed] [Google Scholar]
- 37.Whitby M C, Lloyd R G. Branch migration of three-strand recombination intermediates by RecG, a possible pathway for securing exchanges initiated by 3′-tailed duplex DNA. EMBO J. 1995;14:3302–3310. doi: 10.1002/j.1460-2075.1995.tb07337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitby M C, Ryder L, Lloyd R G. Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell. 1993;75:341–350. doi: 10.1016/0092-8674(93)80075-p. [DOI] [PubMed] [Google Scholar]
- 39.Whitby M C, Sharples G J, Lloyd R G. The RuvAB and RecG proteins of Escherichia coli. In: Eckstein F, Lilley O M J, editors. Nucleic acids and molecular biology. Vol. 9. Berlin, Germany: Springer-Verlag; 1995. pp. 66–83. [Google Scholar]