Abstract
Background
Serratia marcescens is an opportunistic nosocomial pathogen, and recent reports have highlighted the rapid increase in multidrug resistance in this organism. There is a paucity in genomic data for carbapenem-resistant S. marcescens (CRSM).
Methods
A retrospective cohort study describing laboratory-confirmed CRSM from a tertiary academic hospital in Cape Town, South Africa, for the period 2015–20, was performed. Stored CRSM and contemporary isolates were submitted for WGS using Illumina MiSeq, with the Nextera DNA Flex Library Preparation Kit. Sequence data were analysed in-house using srst2 and Tychus, and CRSM and contemporary isolates were compared.
Results
Twenty-one CRSM and four contemporary isolates were sequenced and analysed. Twenty-four different resistance genes were identified, with all isolates having at least two resistance genes, and seventeen isolates harbouring three or more genes. This correlated well with phenotypic results. The blaOXA-48-like carbapenemase was the most common carbapenemase identified, in 86% (18/21) of CRSM. A core SNP difference tree indicated that the CRSM could be grouped into three clusters. Eleven isolates had shared plasmids. Several genes and SNPs were identified in the CRSM, which may putatively augment virulence, but this requires further functional characterization.
Conclusions
A diverse resistome was observed in CRSM, which was also reflected phenotypically, with blaOXA-48-like the most commonly carbapenemase. Though distinct clusters were observed, no clonality was noted, and a limited number of isolates shared plasmids. This study provides genomic data for emerging CRSM and highlights the importance of ongoing genomic surveillance to inform infection prevention control and antimicrobial stewardship initiatives.
Introduction
Serratia marcescens is a non-spore-forming, Gram-negative bacillus. The genus Serratia consists of more than 20 species, of which S. marcescens is the primary human pathogen. It is a ubiquitous saprophyte that is present in water, soil, plants, animals and insects.1 However, it was not until 1951, when Wheat published a report on cases of nosocomial S. marcescens infection, that it was considered pathogenic.2 It has subsequently been recognized as an important cause of opportunistic infections, especially in hospital settings.
The success of S. marcescens as a pathogen is in part due to its genetic diversity and genome plasticity. Several intrinsic virulence factors have been encountered, contributing to biofilm formation, haemolysins, proteases, siderophores and LPS, which form part of the Gram-negative cell wall.3
Antibiotic resistance is mediated by both intrinsic and acquired mechanisms. The most common mechanism of resistance is β-lactamase production. S. marcescens may harbour chromosomal AmpC β-lactamase genes,4 conferring resistance to penicillins, first-generation cephalosporins, cephamycins and monobactams. This β-lactamase may be hyper-expressed due to induction following exposure to β-lactam antibiotics, or through the selection of derepressed mutants. This results in resistance to all β-lactam antibiotics except fourth-generation cephalosporins and carbapenems.5
Acquired resistance to extended-spectrum β-lactam antibiotics in S. marcescens is plasmid encoded, and confers resistance to third-generation cephalosporins,6 for example blaCTX-M. Other antibiotics affected by acquired resistance determinants include aminoglycosides, fluoroquinolones, macrolides and co-trimoxazole. Other resistance determinants include chromosomally encoded porins, and efflux pumps. β-Lactamases, in combination with decreased cell membrane permeability, can lead to carbapenem resistance.7 Notably, S. marcescens is intrinsically resistant to colistin by modification of LPS, but the exact mechanism is yet to be elucidated.8
The first carbapenem-resistant S. marcescens (CRSM) were described in 1982 and were designated S6, S7 and S8.9 In 1994, Naas et al.10 sequenced the first carbapenemase gene from S. marcescens (blaSME-1). Subsequently, many other carbapenemases have been identified in this organism.
Carbapenem-resistant Enterobacterales (CREs) have been reported from every major centre in South Africa.11,12 The most common organisms causing infections are Klebsiella pneumoniae, Enterobacter spp., S. marcescens, Escherichia coli, Citrobacter spp., Providencia spp. and Morganella spp. The most common carbapenemases identified to date are blaNDM and/or blaOXA-48, with blaVIM, blaGES and blaKPC in the minority of cases.11,12
While molecular studies have focused primarily on K. pneumoniae, genomic data for CRSM are limited, but are important to enable a better understanding of the transmission patterns and prevailing resistance determinants, to inform data-driven local patient management, antimicrobial stewardship (AMS) and infection prevention control (IPC) strategies.
Methods
Ethics
Ethics approval was obtained from the University of Cape Town Human Research Ethics Committee (Ref: 468/2020) and hospital approval obtained from Groote Schuur Hospital for access to patient information and stored samples.
Isolate selection
All CRSM isolates cultured from inpatients at Groote Schuur Hospital, Cape Town, and stored at the National Health Laboratory Service (NHLS) laboratory for the study period September 2015 to June 2020, were considered for the study, irrespective of clinical source. Only the first isolate per patient was included and where the first isolate was unavailable for testing, a different isolate from the same patient was used. Isolates were excluded if they were obtained from rectal swabs for CRE screening. Contemporary carbapenem-susceptible strains were selected from samples collected during the study period from the same hospital to serve as a comparative outgroup.
All samples were cultured onto standard microbiological media, organisms identified, and antimicrobial susceptibility testing performed using the VITEK 2 automated system (bioMérieux, France) using standard laboratory protocols. The laboratory information system (Trakcare) was accessed to identify intermediate susceptibility or resistance to carbapenems. Presumptive CRSM were confirmed using the modified Hodge’s test,13 modified carbapenemase inactivation method (mCIM),14 or carbapenemase lateral flow assay (LFA) (Coris BioConcept, Belgium). MICs were determined using ETEST (bioMérieux, France), according to the CLSI guidelines.15
Clinical data were extracted from the laboratory information system and included the date of birth, sex, sample type, ward and associated microbiological test results. The raw data were refined using Stata (StataCorp, USA).
DNA extraction and WGS
Total genomic DNA was extracted using the Zymogen Bacterial/Fungal DNA extraction miniprep kit (Zymo Research, USA) and the resulting quality and yield confirmed using the Nanodrop (Thermo Fisher, USA), as per manufacturer’s instructions.
Libraries were prepared using the Nextera DNA Flex Library Preparation Kit (Illumina, USA), according to the manufacturer’s instructions. WGS was performed on the MiSeq Platform (Illumina) using the v2 300-cycle reagent kit and flow cell, generating 2 × 150 bp, paired-end reads. The raw reads were inspected and reads of poor quality removed using FastQC16 and Trimmomatic.17 Reads were subsequently mapped to a reference genome (S. marcescens ASM351616v1) to produce draft genomes.
There are currently no curated S. marcescens-specific virulence databases for in silico analysis. To identify genetic elements that may have potentially contributed to increased pathogenesis or virulence of the CRSM isolates, a genomic presence/absence comparison was conducted, using Fisher’s exact test with multiple testing correction, comparing the CRSM to the contemporary strains, but excluding the ESBL isolate, and genes with an adjusted P value of ≤0.05 were investigated. The srst218 tool was used to identify resistance elements with ≥80% homology (ARG-ANNOT)19 and plasmid types with 50% coverage and 90% homology (PlasmidFinder)20 based on the short reads.18 A core-genome SNP dendrogram indicating genetic relatedness was constructed using Tychus.21
Results
CRSM
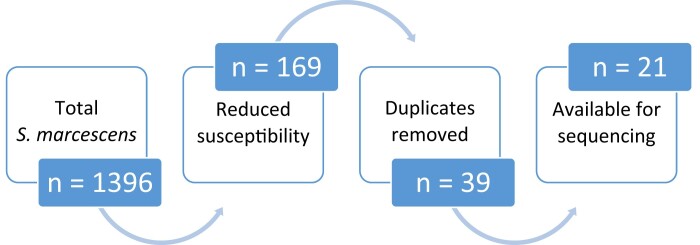
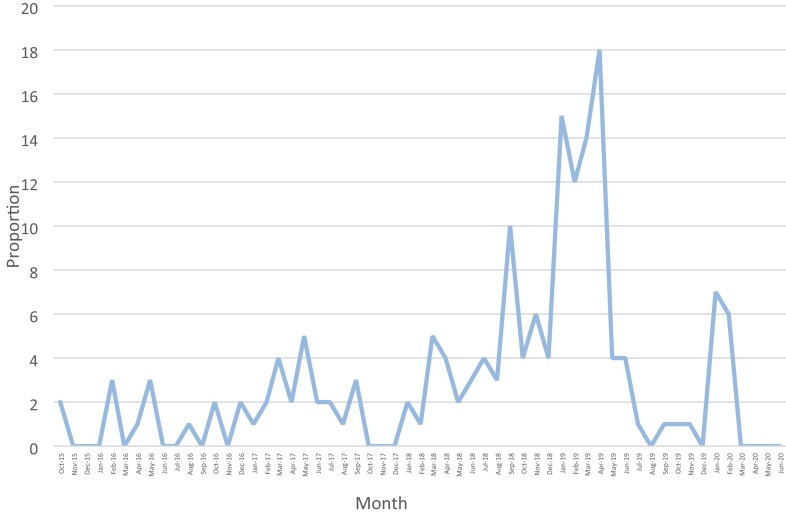
During the study period (September 2015 to June 2020), a total of 128 246 bacterial isolates were collected and analysed by the NHLS laboratory using the VITEK 2 system. Of those, 1396 (1.09%) were identified as S. marcescens. Phenotypic reduced susceptibility to carbapenems was noted in 169/1396 (12.11%) of the isolated S. marcescens, but only 39/169 (23.1%) were non-repeat specimens, with 21/39 (53.85%) available for sequencing and inclusion in the study (Figure 1). The number of CRSM isolates per month remained relatively stable over the study period, except for a noticeable increase in July 2018, peaking during January–April 2019, and again in January–March 2020 (Figure 2).
Figure 1.
Sample screening for study selection.
Figure 2.
Proportion of CRSM isolated over the study period.
Of the 21 CRSM isolates selected, 20 were confirmed to be carbapenemase positive using either the mCIM (17/21) or the carbapenemase LFA (Coris BioConcept, Belgium) (3/21).
Of the 21 CRSM isolates sequenced, 14/21 (66.7%) were from pure cultures and 7/21 (33.3%) were part of mixed cultures, which included other organisms (data not shown). Based on specimen type, the majority [13/21 (61.9%)] were cultured from blood, 3/21 (14.29%) from urine, 2/21 (9.52%) from intra-abdominal fluids, 2/21 (9.52%) from central venous pressure (CVP) line tips, and 1/21 (4.76%) from a pus aspirate. Most of the patients were male (17/21; 80.95%), with a median age of 42 years (range 14–64 years). The median duration of admission before isolation of a CRSM was 18 days with a range of 1–110 days. Most isolates were obtained from samples submitted while the patient was in ICU (17/21; 80.95%). Of the 21 CRSM-infected patients included, 7/21 (33.3%) had another CRE cultured. These included four K. pneumoniae, two Enterobacter cloacae and one Citrobacter freundii. Other MDR organisms were previously cultured in 15/21 (71.43%) of patients (Table S1, available as Supplementary data at JAC-AMR Online).
All 21 CRSM isolates demonstrated phenotypic resistance to penicillins, second-, third- and fourth-generation cephalosporins, and the β-lactam/β-lactamase inhibitor combinations amoxicillin/clavulanate and piperacillin/tazobactam, with variable resistance to carbapenems (Table S2). CRSM isolates exhibited varying degrees of resistance to the non-β-lactam antibiotics, but no single agent displayed less than 50% resistance, where resistance to ciprofloxacin, gentamicin, tigecycline, amikacin and co-trimoxazole were 90.5%, 85.7%, 66.7%, 61.9% and 57.1%, respectively (data not shown).
Antibiotic consumption data were available for 19/21 (85.71%) of the CRSM-infected patients. All 19 patients were on antibiotics prior to the isolation of CRSM. The most common antibiotics prescribed were β-lactams, specifically piperacillin/tazobactam. The other most frequently prescribed antibiotics were aminoglycosides. Carbapenems were prescribed in 9/19 (47%) of cases. Colistin was administered in four cases prior to isolation of CRSM. Most patients received a median of 2 (range 1–15) antibiotics prior to culturing a CRSM.
WGS
There were approximately 0.8–2.2 million reads per sample, and the mean quality score was Phread >28. Assemblies produced a median value of 75 contigs (range 25–209) and a median N50 of 204 936 bp (range 60 753–530 212), while coverage ranged from 0× to 1242×.
Genetic relatedness
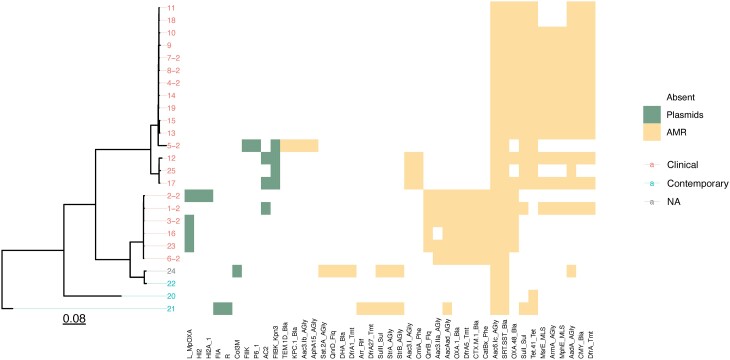
A core SNP difference dendrogram indicated that the isolates could be grouped into three clusters (Figure 3). Cluster 1 (isolates 4, 5, 7–11, 13–15, 18, 19) was the largest, and comprised 12/21 (57%) of CRSM isolates. Cluster 2 (isolates 12, 17, 25) consisted of 3/21 (14%) of CRSM isolates and was closely related to Cluster 1. The third cluster (isolates 1–3, 6, 16, 23) comprised 6 isolates (29%), which harboured the blaOXA-1 gene, the OXA-48 plasmid, and a unique set of resistance genes.
Figure 3.
Dendrogram and heatmap of the sequenced isolates, indicating genetic relatedness, and the presence or absence of plasmids and resistance elements. AMR, antimicrobial resistance. The branch lengths are expressed in terms of changes per number of SNPs, represented by the scale bar.
Antibiotic resistance genes
A total of 24 different resistance genes were present in the CRSM isolates and only 9 in the contemporary isolates (Figure 3). These included genes conferring resistance to β-lactams, aminoglycosides, co-trimoxazole, chloramphenicol, fluoroquinolones, macrolides and streptogramins (Table 1). More details are available as Table S3. All isolates had at least two resistance genes, with 17 isolates harbouring at least three or more.
Table 1.
Antibiotic resistance genes detected in CRSM
| Antibiotic enzyme class | Number of isolates, n/N (%) |
Number of different genes detected |
|---|---|---|
| β-Lactamases | 21/21 (100) | 8 |
| Aminoglycosides | 21/21 (100) | 8 |
| Fluoroquinolones | 5/21 (24) | 1 |
| Co-trimoxazole | 21/21 (100) | 3 |
| Macrolides/lincosamines/streptogramins | 12/21 (57) | 2 |
| Tetracyclines | 15/21 (71) | 1 |
Eight different β-lactamase genes were identified. OXA-48-like carbapenemases were detected in 18/21 (85.71%) of the CRSM, of which 14 were blaOXA-181 (78%) and 4 were blaOXA-48 (22%). KPC (blaKPC-2) was found in a single isolate (5), which also harboured blaTEM-206. No known carbapenemases were detected in two of the CRSM isolates. All 21 CRSM isolates harboured one or more genes encoding AmpC β-lactamases (21 blaSRT-1 and 14 blaCMY-4). The rest of the β-lactamases consisted of six (29%) blaCTX-M-15, an ESBL gene, which was present in five of the isolates concurrent to a carbapenemase-encoding gene.
Eight different genes conferring aminoglycoside resistance were identified. All CRSM harboured the aac(6)-Ic gene in combination with one or more of aac(3)-IIa, aac(3)-I, aac(3)-Ib, aacaad, aadA, aphA15 and armA. One or more of dfrA5, dfrA12 and sulI, which confer resistance to co-trimoxazole and its components, were identified in all 21 CRSM isolates. Tet(41), a tetracycline resistance gene was present in 15 isolates, with no genes coding for tigecycline resistance. The macrolide and streptogramin resistance genes mph(E) and msr(E) were present in 12 isolates. The chloramphenicol resistance genes catB4 and cmlA1 were present in nine isolates, and the fluoroquinolone resistance gene qnrB was only present in five isolates. The resistance genes identified correlated with phenotypic resistance profiles, except for aminoglycosides and tigecycline.
In cluster 1, all isolates had tet(41) and the majority had aadA, armA, blaCMY-4, dfrA12, mph(E), blaOXA-181, sulI and blaSRT-1; however, there were no common plasmids found. Cluster 2 isolates all harboured aac(3)-I, aac(6)-Ic, blaSRT-1, sulI, tet(41) and plasmid FIBK_1. Isolates in cluster 3 contained aac(6)-1b, blaCTX-M-15, dfrA14, blaOXA-1 and blaSRT-1. The majority also contained aac(3)-IIa, blaOXA-48 and qnrB. This cluster also included plasmid L-MpOXA identified in four isolates, and is known to carry blaOXA-48 and additional resistance elements22 (Table 1).
Plasmid types
Ten different plasmid replicon types were identified by srst218 using PlasmidFinder. Seven of these were in the CRSM isolates (Table 2): FIBK_1_Kpn3 in 4/21 (19.04%); L_MpOXA in 4/21 (19.04%); AC2 in 3/21 (14.29%); FIIK_1 in 1/21 (4.76%); HI2_1 in 1/21 (4.76%); HI2A_1 in 1/21 (4.76%); and P6_1 in 1/21 (4.76%).
Table 2.
Plasmid replicon types identified
| Plasmid | Number | Percentage of isolates | Cluster |
|---|---|---|---|
| FIBK_1_Kpn3 | 4 | 19 | 1, 3 |
| L__MpOXA-48 | 4 | 19 | 3 |
| A__C2_1 | 3 | 14 | 2, 3 |
| FIIK_1 | 1 | 5 | 1 |
| HI2_1 | 1 | 5 | 3 |
| HI2A_1 | 1 | 5 | 3 |
| P6_1 | 1 | 5 | 1 |
| Col3M | 1 | — | Contemporary |
| FIAHI1_1_HI1 | 1 | — | Contemporary |
| R_1 | 1 | — | Contemporary |
Virulence factors
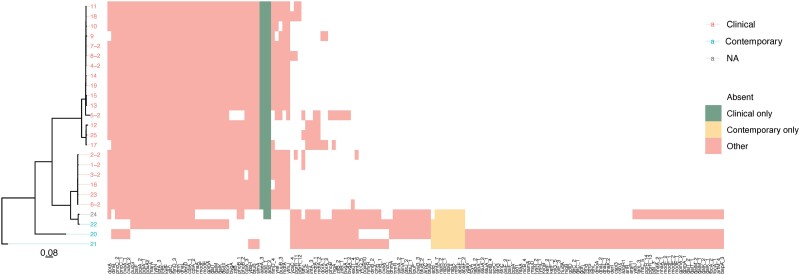
Based on the statistical presence/absence analysis, putative virulence genes, which were only present in CRSM and not in contemporary isolates, included aaeA_3, which encodes an efflux pump and was present in all the CRSM isolates, fimD_4, which encodes fimbriae, puuB3, a gene involved in energy production and conversion, and yfmJ, an NADP-dependent oxidoreductase,23 which were present in the majority of CRSM (Figure 4).
Figure 4.
Dendrogram and heatmap of the CRSM and contemporary isolates, indicating the presence or absence of potential virulence and pathogenicity elements. The branch lengths are expressed in terms of changes per number of SNPs, represented by the scale bar.
Genes detected in the majority of CRSM, and in a minority of contemporary strains, included: fimD_3, which encodes fimbriae; dsdX, a d-serine transporter protein; dntK, which in conjunction with pdxA2 is involved in carbohydrate metabolism; and IcfB, which is part of long chain fatty acid metabolism. MarA is a transcriptional activator of antibiotic resistance genes and ybbH is another transcription regulator23 (Figure 4).
There were variations detected in the following genes, which were present in both CRSM and contemporary isolates, namely: add, which is involved in nucleotide metabolism; bvgA, which activates the transcription of virulence genes; cdiA, a component of a cellular contact-dependent growth inhibitor, which inhibits growth of closely related bacteria; adtA, which encodes an efflux pump; phoB, which is involved in swarming motility; and zitB, which is a zinc transporter23 (Figure 4).
Several genes of interest in the contemporary isolates also encoded putative virulence factors. These included genes related to efflux pumps (abaF, arpC, oqxB, kefF), fimbria (yfcQ) and biofilm formation (tabA), as well as arlR, which regulates adhesion, autolysis, multidrug resistance and virulence factors, and hudA, a proven virulence attenuator.23 The roles of these genes in infection and immune evasion should be further investigated.
Discussion
Carbapenem-resistant Enterobacterales are an increasing global problem, including in South Africa. S. marcescens was previously shown to be the third most common CRE isolated in this country by two studies undertaken by the National Institute of Communicable diseases (NICD).11,12 Despite this, limited genomic data for CRSM are available for South Africa. WGS is an invaluable tool, especially in outbreak and epidemiological investigations, but is underutilized in our setting due to resource constraints, despite a progressive drop in price and availability. This study is one of the few to use WGS to examine CRSM and the first to examine a cohort of CRSM in South Africa.
Carbapenem resistance is usually attributed to a combination of factors including defined antibiotic resistance genes, efflux pumps and altered cell membrane permeability.7 This study investigated the genetic occurrence of resistance and potential virulence elements in a collection of CRSM. Most of the CRSM isolates in this study harboured blaOXA-48-like enzymes, with one carrying blaKPC-2. In contrast, Xu et al.24 reported CRSM from a tertiary hospital in China, and identified blaKPC-2 as the only carbapenemase. KPC was also the most common carbapenemase identified in two CRSM studies from Brazil25 and the USA.26 Notably, in Madrid, blaVIM-1 and blaOXA-48 predominated,27 and blaNDM caused an CRSM outbreak in Romania.28
There were two isolates where no known carbapenemase genes were identified. One of these isolates harboured blaSRT-1, and was blaOXA-48 positive using the carbapenemase LFA. This isolate likely harboured a novel carbapenemase gene or genotype. Since the blaOXA-48 family is large and diverse, new genotypes are frequently described. Since no blaOXA-48-like gene was identified, the presence of a novel carbapenemase gene cannot be excluded, and should be investigated in future work.
The second isolate harboured blaSRT-1, blaOXA-1 and blaCTX-M, and was mCIM negative, yet phenotypically resistant to ertapenem. This isolate harboured an ESBL gene (blaOXA-1), which has been shown to confer varying resistance to the carbapenems with phenotypic resistance to ertapenem more common than to meropenem, albeit in K. pneumoniae.29 Since the mCIM is performed using meropenem, this isolate was likely mCIM negative but phenotypically ertapenem resistant based on the specific activity of this blaOXA-1 gene.
Multiple virulence genes were identified in this study, including those promoting resistance such as efflux pumps, which export antimicrobials out of the bacterium, and transcriptional activators of resistance genes. Other virulence genes include those for fimbriae, which facilitate adherence and subsequent colonization, and those involved in metabolism, either through nutrient transport or energy production. Variations in common genes were found and these genes may provide a colonization advantage through inhibiting growth of closely related bacteria, or by increased motility, although their significance is yet to be confirmed.
Core SNP analysis indicated that all CRSM isolates grouped into one of three clusters, with significant homology within the clusters although no clonality was observed, as was originally suspected. Since these isolates were spread over a 4 year period, there may have been an environmental reservoir rather than a point source contributing to the dissemination of these isolates, or the resistance and putative virulence factors identified. This is even more likely when considering that some CRSM were part of a mixed culture including other CRE, which may have acted as reservoirs. The patients were in different ICUs and/or wards, which may imply a common ancestor that evolved in the specific environments or be due to other genetic elements that could not be identified. Alternatively, patient referral from other hospitals may have introduced different strains.
Interestingly, the third cluster on the core SNP difference tree comprised six isolates, which all harboured the blaOXA-1 gene. These isolates also carried several resistance genes against several classes of antibiotics, which were not detected in the other strains in the study. Previous studies have indicated that blaOXA-1 and aac(6)-Ib are often co-carried.30 This is further compounded by the fact that four of these strains carried the MpOXA-48 plasmid, known to carry blaOXA-48 and ESBLs, such as blaCTX-M-15 identified in our study. These findings likely indicate a shared resistance plasmid between the strains, which confers an MDR phenotype. Other plasmid replicons were also detected, but their relevance is unclear.
S. marcescens accounted for approximately 1% of organisms isolated in our laboratory during the study period, but exhibited a high rate of carbapenem resistance compared with contemporary surveillance studies elsewhere.31 Of the CRSM isolates collected, the majority were from patients in ICU, particularly the surgical ICU, which is in accordance with previous reports.24,27,32 Our CRSM patients had several MDR organisms, including other CRE, ESBL-producing organisms, carbapenem-resistant Acinetobacter baumannii (CRAB), carbapenem-resistant Pseudomonas aeruginosa (CRPA) and MRSA, isolated previously. This may have represented an opportunity for genetic exchange between the organisms, possibly plasmid mediated, or may reflect the high environmental burden of difficult-to-treat resistance in Gram-negative pathogens in our setting.33,34
Antibiotic utilization data showed extensive use of broad-spectrum antibiotics prior to isolating a CRSM. This was not surprising, since prior antibiotic exposure is a major risk factor for colonization and infection with CRE.35 Considering the range of resistance genes identified in our study, any of these antibiotics may have contributed to selection for MDR strains. The administering of colistin, to which S. marcescens is intrinsically resistant, may have played a role in selecting for CRSM, although it was only prescribed in the minority of our patients, prior to isolating a CRSM. Notably, overall colistin consumption data over the study period did show an increase, which coincided with the CRSM isolation peaks noted.
Based on antimicrobial susceptibility results, extremely limited treatment options remain for CRSM in our institution. For non-β-lactam antibiotics, tigecycline resistance was phenotypically detected in isolates that contained the tet(41) gene, which encodes a tetracycline efflux pump; however, it does not confer resistance, but may be associated with other resistance determinants that lead to tigecycline resistance. With regard to the aminoglycosides, phenotypic non-susceptibility did not correlate with genotypic aminoglycoside resistance determinants. Therefore, due to the presence of multiple resistance genes and a highly resistant phenotypic profile, treatment of CRSM, particularly in ICU patients in our setting, remains challenging. In this regard, since blaOXA-48-like carbapenemases predominated, ceftazidime/avibactam may represent a therapeutic option. However, it is currently expensive and difficult to access in our public sector setting.36
Limitations
This was a small descriptive retrospective study and not all isolates identified were available for sequencing. By its nature it is underpowered to draw any statistically significant conclusions. Notably, there are no genomic databases for S. marcescens, which hindered the search for virulence factors and other in silico analyses. Future WGS studies should include more non-CRSM isolates, to expand the comparison for virulence factors. We also did not include rectal screening isolates, which may have had IPC implications. Furthermore, environmental sampling to exclude institutional contamination was not performed.
Due to the nature of high-throughput short-read sequencing, plasmid reconstruction could not be conducted. However, of the 10 different plasmid types identified, all isolates with the L_MpOXA plasmid also carried the blaOXA-48 gene, indicating that the carbapenemase genes were likely plasmid encoded. Although this plasmid was found in a minority of isolates, this mobile genetic element does have the potential to spread to other organisms, including other strains of S. marcescens, and may therefore prove a transmission risk.
Conclusions
This study is the first in South Africa, to our knowledge, and one of only a handful internationally to examine the genomic profile of CRSM using WGS. The isolates clustered into three groups and since there may have been limited shared plasmids between them, this could suggest a common lineage with differentiation within specific environments over time. Extensive prior use of broad-spectrum antibiotics, as well as a complex resistome in our isolates, highlights both AMS and therapeutic challenges in our setting. Resistance elements in combination with putative virulence factors may have provided a survival advantage. This study highlights the importance of ongoing WGS studies on S. marcescens, which could potentially inform data-driven local patient management principles, AMS and IPC strategies.
Supplementary Material
Acknowledgements
We would like to acknowledge the staff of Groote Schuur Hospital pharmacy for providing the electronic antibiotic prescription data.
Contributor Information
Amanda Julia Overmeyer, Division of Medical Microbiology, Department of Pathology, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa; Microbiology Laboratory, National Health Laboratory Service, Groote Schuur Hospital, Cape Town, South Africa.
Elizabeth Prentice, Division of Medical Microbiology, Department of Pathology, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa; Microbiology Laboratory, National Health Laboratory Service, Groote Schuur Hospital, Cape Town, South Africa.
Adrian Brink, Division of Medical Microbiology, Department of Pathology, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa; Microbiology Laboratory, National Health Laboratory Service, Groote Schuur Hospital, Cape Town, South Africa; Institute of Infectious Disease and Molecular Medicine, University of Cape Town, Cape Town, South Africa.
Katie Lennard, Institute of Infectious Disease and Molecular Medicine, University of Cape Town, Cape Town, South Africa.
Clinton Moodley, Division of Medical Microbiology, Department of Pathology, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa; Microbiology Laboratory, National Health Laboratory Service, Groote Schuur Hospital, Cape Town, South Africa.
Funding
This work was supported by a development grant from the National Health Laboratory Service Research Trust (PR20990).
Transparency declarations
No conflicts to declare.
Author contributions
C.M., E.P. and A.B. conceptualized the study. C.M. and A.O. conducted the laboratory testing, data analysis and interpretation. K.L. performed all bioinformatic analyses. C.M., E.P., K.L., A.B. and A.O. contributed to the manuscript preparation.
Supplementary data
Tables S1 to S3 are available as Supplementary data at JAC-AMR Online.
References
- 1. Grimont F, Grimont PA. The genus Serratia. In: Dworkin M, Falkow S, Rosenberg Eet al. eds. The Prokaryotes. Springer, 2006, 219–44. [Google Scholar]
- 2. Wheat RP, Zuckerman A, Rantz LA. Infection due to chromobacteria: report of eleven cases. AMA Arch Intern Med 1951; 88: 461–6. 10.1001/archinte.1951.03810100045004 [DOI] [PubMed] [Google Scholar]
- 3. Kurz CL, Chauvet S, Andrès Eet al. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J 2003; 22: 1451–60. 10.1093/emboj/cdg159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mahlen SD, Morrow SS, Abdalhamid Bet al. Analyses of ampC gene expression in Serratia marcescens reveal new regulatory properties. J Antimicrob Chemother 2003; 51: 791–802. 10.1093/jac/dkg133 [DOI] [PubMed] [Google Scholar]
- 5. Harris P. Clinical management of infections caused by Enterobacteriaceae that express extended-spectrum β-lactamase and AmpC enzymes. Semin Respir Crit Care Med 2015; 36: 56–73. 10.1055/s-0034-1398387 [DOI] [PubMed] [Google Scholar]
- 6. Thomson KS. Extended-spectrum-β-lactamase, AmpC, and carbapenemase issues. J Clin Microbiol 2010; 48: 1019–25. 10.1128/JCM.00219-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Suh B, Bae IK, Kim Jet al. Outbreak of meropenem-resistant Serratia marcescens comediated by chromosomal AmpC β-lactamase overproduction and outer membrane protein loss. Antimicrob Agents Chemother 2010; 54: 5057–61. 10.1128/AAC.00768-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin QY, Tsai Y-L, Liu M-Cet al. Serratia marcescens arn, a PhoP-regulated locus necessary for polymyxin B resistance. Antimicrob Agents Chemother 2014; 58: 5181–90. 10.1128/AAC.00013-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang Y, Wu P, Livermore D. Biochemical characterization of a beta-lactamase that hydrolyzes penems and carbapenems from two Serratia marcescens isolates. Antimicrob Agents Chemother 1990; 34: 755–8. 10.1128/AAC.34.5.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Naas T, Vandel L, Sougakoff Wet al. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A beta-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob Agents Chemother 1994; 38: 1262–70. 10.1128/AAC.38.6.1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perovic O, Britz E, Chetty Vet al. Molecular detection of carbapenemase-producing genes in referral Enterobacteriaceae in South Africa: a short report: clinical update. S Afr Med J 2016; 106: 975–7. 10.7196/SAMJ.2016.v106i10.11300 [DOI] [PubMed] [Google Scholar]
- 12. Perovic O, Ismail H, Quan Vet al. Carbapenem-resistant Enterobacteriaceae in patients with bacteraemia at tertiary hospitals in South Africa, 2015 to 2018. E J Clin Microbiol Infect Dis 2020; 39: 1287–94. 10.1007/s10096-020-03845-4 [DOI] [PubMed] [Google Scholar]
- 13. Girlich D, Poirel L, Nordmann P. Value of the modified hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol 2012; 50: 477–9. 10.1128/JCM.05247-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pierce VM, Simner PJ, Lonsway DRet al. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J Clin Microbiol 2017; 55: 2321–33. 10.1128/JCM.00193-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. CLSI . Performance Standards for Antimicrobial Susceptibility Testing —Thirty-First Edition:M100. 2021. [Google Scholar]
- 16. Andrews S. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 17. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinform 2014; 30: 2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Inouye M, Dashnow H, Raven L-Aet al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 2014; 6: 90. 10.1186/s13073-014-0090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gupta SK, Padmanabhan BR, Diene SMet al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 2014; 58: 212–20. 10.1128/AAC.01310-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carattoli A, Zankari E, García-Fernández Aet al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 2014; 58: 3895–903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dean C, Noyes N, Lakin Set al. Tychus: a whole genome sequencing pipeline for assembly, annotation and phylogenetics of bacterial genomes. bioRxiv 2018: 283101. 10.1101/283101 [DOI] [Google Scholar]
- 22. Poirel L, Bonnin RA, Nordmann P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother 2012; 56: 559–62. 10.1128/AAC.05289-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. The UniProt Consortium . UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res 2023; 51: D523–D31. 10.1093/nar/gkac1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu Q, Fu Y, Zhao Fet al. Molecular characterization of carbapenem-resistant Serratia marcescens clinical isolates in a tertiary hospital in Hangzhou, China. Infect Drug Resist 2020; 13: 999–1008. 10.2147/IDR.S243197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Prado GV, Mendes ET, Martins RCet al. Carbapenem-resistant Serratia marcescens bloodstream infection in hematopoietic stem cell transplantation patients: will it be the next challenge? Transpl Infect Dis 2021; 23: e13630. 10.1111/tid.13630 [DOI] [PubMed] [Google Scholar]
- 26. Jimenez A, Abbo LM, Martinez Oet al. KPC-3-producing Serratia marcescens outbreak between acute and long-term care facilities, Florida, USA. Emerg Infect Dis 2020; 26: 2746–50. 10.3201/eid2611.202203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pérez-Viso B, Hernández-García M, Ponce-Alonso Met al. Characterization of carbapenemase-producing Serratia marcescens and whole-genome sequencing for plasmid typing in a hospital in Madrid, Spain (2016–18). J Antimicrob Chemother 2021; 76: 110–6. 10.1093/jac/dkaa398 [DOI] [PubMed] [Google Scholar]
- 28. Phan H, Stoesser N, Maciuca Iet al. Illumina short-read and MinION long-read WGS to characterize the molecular epidemiology of an NDM-1 Serratia marcescens outbreak in Romania. J Antimicrob Chemother 2018; 73: 672–9. 10.1093/jac/dkx456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sugumar M, Kumar K, Manoharan Aet al. Detection of OXA-1 β-lactamase gene of Klebsiella pneumoniae from blood stream infections (BSI) by conventional PCR and in-silico analysis to understand the mechanism of OXA mediated resistance. PLoS One 2014; 9: e91800. 10.1371/journal.pone.0091800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Livermore DM, Day M, Cleary Pet al. OXA-1 β-lactamase and non-susceptibility to penicillin/β-lactamase inhibitor combinations among ESBL-producing Escherichia coli. J Antimicrob Chemother 2019; 74: 326–33. 10.1093/jac/dky453 [DOI] [PubMed] [Google Scholar]
- 31. González GM, Treviño-Rangel RDL, Campos CLet al. Surveillance of antimicrobial resistance in Serratia marcescens in Mexico. New Microbiol 2020; 43: 34–7. [PubMed] [Google Scholar]
- 32. Saralegui C, Ponce-Alonso M, Pérez-Viso Bet al. Genomics of Serratia marcescens isolates causing outbreaks in the same pediatric unit 47 years apart: position in an updated phylogeny of the species. Front Microbiol 2020; 11: 451. 10.3389/fmicb.2020.00451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jacobson RK, Manesen MR, Moodley Cet al. Molecular characterisation and epidemiological investigation of an outbreak of blaOXA-181 carbapenemase producing isolates of Klebsiella pneumoniae in South Africa. S Afr Med J 2015; 105:1030. 10.7196/SAMJ.2015.v105i12.9926 [DOI] [PubMed] [Google Scholar]
- 34. Mudau M, Jacobson R, Minenza Net al. Outbreak of multi-drug resistant Pseudomonas aeruginosa bloodstream infection in the haematology unit of a South African academic hospital. PLoS One 2013; 8: e55985. 10.1371/journal.pone.0055985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tacconelli E. Antimicrobial use: risk driver of multidrug resistant microorganisms in healthcare settings. Curr Opin Infect Dis 2009; 22: 352–8. 10.1097/QCO.0b013e32832d52e0 [DOI] [PubMed] [Google Scholar]
- 36. Tootla H, Copelyn J, Botha Aet al. Using ceftazidime-avibactam for persistent carbapenem-resistant Serratia marcescens infection highlights antimicrobial stewardship challenges with new beta-lactam-inhibitor combination antibiotics. S Afr Med J 2021; 111: 729–31. 10.7196/SAMJ.2021.v111i8.15762 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.