Abstract
Results from previous studies had suggested that Bacteroides thetaiotaomicron utilizes starch by binding the polysaccharide to the bacterial surface and subsequently degrading the polymer by using cell-associated enzymes. Most of the starch-degrading activity was localized to the periplasm, but a portion appeared to be membrane associated. This raised the possibility that some breakdown might occur in the outer membrane prior to exposure of the polysaccharide to the periplasmic polysaccharide-degrading enzymes. In this study, we show that SusG, an outer membrane protein which has been shown genetically to be essential for starch utilization, has enzymatic activity. Results of protease accessibility experiments support the hypothesis that SusG is exposed on the cell surface. Results of [14C]starch binding assays, however, show that SusG plays a negligible role in binding of starch to the cell surface. Consistent with this, SusG has a relatively high Km for starch and by itself is not sufficient to allow cells to grow on starch or to bind starch. Hence, the main role of SusG is to hydrolyze starch, but the binding of starch to the cell surface is evidently mediated by other proteins presumably interacting with SusG.
Human colonic Bacteroides spp. can utilize a wide range of polysaccharides as sources of carbon and energy. These substrates are broken down by cell-associated enzymes. So far, no extracellular enzymatic activity has been detected, and binding of the polysaccharide to the cell surface appears to be important for utilization. This evidence led us to propose a model in which the polysaccharide is first bound to an outer membrane receptor and then translocated through the outer membrane. Most enzymatic hydrolysis of the polysaccharide probably occurs in the periplasm, but an initial step in breakdown catalyzed by an outer membrane enzyme could not be ruled out. Most studies of outer membrane translocator proteins have focused on porins, which allow small substrates to diffuse through the outer membrane (7). However, Bacteroides spp. can grow on substrates which are much larger than the pore size of outer membrane porins (∼600 Da) and can grow as well on polymeric as on monomeric substrates, suggesting a rapid and efficient uptake process.
We have used the starch utilization system of Bacteroides thetaiotaomicron as a model for defining the various components involved in polysaccharide utilization by Bacteroides spp. B. thetaiotaomicron can utilize all three forms of starch—amylose, amylopectin, and pullulan—as well as component maltooligosaccharides. Amylose consists of α-1,4-linked glucose residues, whereas amylopectin is a branched polymer composed of amylose chains connected by α-1,6 linkages to an amylose backbone. Pullulan is a linear chain of maltotriose residues linked by α-1,6 bonds. We had identified an operon consisting of eight genes (designated sus, for starch utilization system) which encode proteins that catalyze early steps in the breakdown of starch by B. thetaiotaomicron. Two genes, susA and susB, encode a cell-associated starch-degrading enzyme and an α-glucosidase, respectively. A mutant with a disruption in susA grew more slowly than the wild type but was still able to grow on starch. This finding suggested that other starch-degrading enzymes must participate in this pathway. However, only a very low level of starch hydrolysis could be detected in a cell extract from the susA disruption strain (4). Five of the sus genes, susC, -D, -E, -F, and -G, encode outer membrane proteins (OMPs). SusC is essential for starch utilization and is the only OMP necessary for utilization of the smaller oligomers, maltotetraose to maltoheptaose (14). SusC may form an oligomeric channel in the outer membrane of B. thetaiotaomicron. Previous evidence suggested that SusE is not required for starch utilization (15), but whether SusD or SusF is essential has not been established. SusG is essential for starch utilization but not for utilization of maltooligosaccharides.
Because SusG is an OMP and is essential for utilization of full-length starch, we suspected that SusG would have binding or enzymatic activity or both. Sequence analysis had suggested that susG might encode a starch-degrading enzyme because the deduced amino acid sequence of SusG had significant similarity to sequences of a number of α-amylases and had all of the conserved residues and regions shown to be important for catalytic activity (15). Yet, a mutant with a disruption in susG appeared to have the same level of starch-degrading activity as wild type. This could have been due to the fact that the major enzyme activity detectable in cell extracts, SusA (4), overshadowed a much lower starch-degrading activity due to SusG. We report here that SusG has starch-degrading activity and is exposed on the cell surface but plays only a negligible role in starch binding. Thus, the starch utilization system seems to separate binding activity from enzymatic activity.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All Escherichia coli strains used in this study were grown in Luria-Bertani (LB) broth or on LB agar at 37°C. B. thetaiotaomicron 5482, B. thetaiotaomicron 4007, transposon-generated derivatives, and single-disruption mutants used in this study have been described previously (2, 4, 15). When necessary, suicide vectors were used to create additional disruptions in these strains. B. thetaiotaomicron 4007 was used as a control in growth and binding experiments because it is a derivative of 5482 that is tetracycline resistant (tetQ). tetQ is the selectable marker used in the construction of most disruption mutants.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli DH5αMCR | RecA GnS | 6 |
| B. thetaiotaomicron | ||
| 5482 | Wild type, Gnr | Anaerobe Laboratory, Virginia Polytechnic Institute, Blacksburg |
| 4007 | Wild type, Tetr Gnr G7+ Am+ | B. thetaiotaomicron 5482 (Rifr) with CTn DOT |
| Ms-4 | Emr Gnr G7+ Am− | Tn4351-generated mutant of strain 5482 (2) |
| ΩsusA | Emr Gnr G7+ Am+ | Bacteroides suicide vector pGERM containing a PCR-generated 0.65-kbp KpnI-BamHI fragment inserted in B. thetaiotaomicron chromosome in susA (4) |
| ΩsusB | Tetr Gnr G7− Am− | Bacteroides suicide vector pBT-1 containing a PCR-generated 0.61-kbp KpnI-BamHI fragment inserted in the B. thetaiotaomicron chromosome in susB (pBT1-SB) (4) |
| ΩsusC | Tetr Gnr G7− Am− | Bacteroides suicide vector pBT-1 containing a PCR-generated 0.61-kbp KpnI-BamHI fragment inserted in the B. thetaiotaomicron chromosome in susC (pBT1-SC) (15) |
| ΩsusG | Tetr Gnr G7+ Am− | Bacteroides suicide vector pBT-1 containing a PCR-generated 0.61-kbp KpnI-BamHI fragment inserted in the B. thetaiotaomicron chromosome in susG (pBT1-SC) (15) |
| ΩSAB | Emr Tetr Gnr G7− Am− | Double-insertion strain derived from ΩsusA in which pBT1-SB was used to make an insertion in susB (this study) |
| ΩSAC | Emr Tetr Gnr G7− Am− | Double-insertion strain derived from ΩsusA in which pBT1-SC was used to make an insertion in susC (this study) |
| ΩSAG | Emr Tetr Gnr G7+ Am− | Double-insertion strain derived from ΩsusA in which pBT1-SG was used to make an insertion in susG (this study) |
| Plasmids | ||
| pBT-1 | Knr (Tcr) | RSF1010-based suicide vector used to make insertional disruptions (19) |
| pGERM | Apr (Ermr) | pUC19-based suicide vector expressing ermG used to make insertions in the B. thetaiotaomicron chromosome (17; this study) |
| pNLY1::PsusA | Apr Cmr (Cmr) | pACYC-based shuttle vector containing PsusA used to express genes in trans (this study) |
| pSGC23A | Apr Cmr (Cmr) | pNLY1::PsusA containing a PCR-generated susG gene cloned downstream of PsusA (this study) |
Abbreviations: G7, maltoheptaose; Am, amylopectin; Ap, ampicillin; Cm, chloramphenicol; Em, erythromycin; Gn, gentamicin; Tc, tetracycline; CTn, conjugative transposon; CTn DOT, conjugative transposon which contains the tetQ gene. Antibiotic resistances not in parentheses are expressed only in E. coli; antibiotic resistances in parentheses are expressed only in B. thetaiotaomicron.
Cells were grown initially in a prereduced Trypticase-yeast extract-glucose (VPI) medium. For optimal induction of starch utilization genes, cells were transferred to a defined medium containing maltose (0.3%) as the sole carbohydrate source. To test for growth rates on starch, we inoculated cells into a defined medium with amylopectin (0.3%) as the sole carbohydrate source. Antibiotic concentrations were as follows: ampicillin, 200 μg/ml; chloramphenicol, 15 μg/ml (E. coli) or 20 μg/ml (B. thetaiotaomicron); erythromycin, 10 μg/ml; gentamicin, 200 μg/ml; and tetracycline, 1 μg/ml.
DNA methods.
Isolation of plasmids was done by using a Wizard Plus DNA purification system (Promega Corp.). Dephosphorylation reactions and restriction digests were performed as instructed by the manufacturer (Bethesda Research Laboratories, Bethesda, Md., or New England BioLabs, Beverly, Mass.). Transformation of E. coli DH5αMCR was done by the method of Lederberg and Cohen (8). Conjugations, where constructs generated in E. coli were transferred to Bacteroides recipients, were performed as described by Shoemaker et al. (16). Southern blotting was done as described by Maniatis et al. (10) except that a Renaissance detection kit (DuPont-NEN) was used for detection of the bound DNA probe.
Chemicals.
[14C]starch (Nicotiana tobacum L) was purchased from DuPont-NEN. Amylopectin, pullulan, proteinase K, and phenylmethylsulfonyl fluoride (PMSF) were purchased from Sigma Corp. p-Nitrophenyl-α-d-maltoheptaoside was obtained from Boehringer Mannheim Biochemica.
Membrane preparation.
Membranes were prepared by the method of Valentine and Salyers (20). Cells were grown in a defined medium with maltose (0.3%) as the sole carbohydrate source to late log phase (optical density at 650 nm of 0.6 to 0.8). The cells were washed once with 20 mM potassium phosphate buffer (pH 7.2) and resuspended in 5 ml of the same buffer. These cells were disrupted by sonication. After separation of the cell extract from insoluble material by centrifugation, the whole membranes (both inner and outer membranes) were pelleted from the cell extract by ultracentrifugation (200,000 × g for 2.5 h at 4°C). The soluble fraction was collected, and the membrane pellet was washed once with 20 mM potassium phosphate buffer and pelleted again by ultracentrifugation under the same conditions. The membrane pellet was resuspended in 20 mM potassium phosphate buffer, and the membranes were dispersed by sonication. For analysis of enzymatic activity, the membrane preparations were resuspended in a 50 mM potassium phosphate buffer–20% glycerol solution instead of 20 mM potassium phosphate buffer. Glycerol was added to allow storage of the enzymes at −80°C and did not affect specific activity.
[14C]starch binding experiments.
[14C]starch binding assays on wild-type and mutant B. thetaiotaomicron were performed by a modification of the procedure of Anderson and Salyers (1). Cells were grown in maltose-minimal medium to an optical density at 650 nm of 0.5 to 0.6. The cells were washed twice with 0.1 M phosphate-buffered saline (PBS; pH 7.4) to dissociate any loose capsular material. Cells were resuspended in PBS to an optical density at 650 nm of 0.4. The cell suspension was incubated with either radiolabeled starch or a mixture of labeled and unlabeled starch for 5 min at room temperature under aerobic conditions. Under aerobic conditions, no starch uptake occurs. That is, after the first binding step occurs, no further accumulation of starch is seen even after an hour (1). After centrifugation of the cells, the supernatant fluid was discarded and the cells were washed twice with 0.5 ml of PBS. All of the cell pellets were then resuspended in 100 μl of PBS. This solution was transferred to 2.0 ml of scintillation fluid, and the amount of bound label was determined with a Beckman model 5000TD scintillation counter.
In binding studies, higher concentrations of starch contained a mixture of [14C]starch and unlabeled amylopectin in PBS to reach the desired concentration. As a control for nonspecific binding of starch, we tested binding to mutant ΩsusC, which expresses none of the starch-associated OMPs and has been shown to have only a very low residual binding activity (15). Values were reported in micrograms of starch bound per milligram of cell protein. These values were obtained by multiplying the total counts per minute by a dilution factor, which was the ratio of labeled starch to total starch in each assay. That number was converted by an empirical constant (based on observed counts per minute per given amount of starch) to disintegrations per minute, which allowed the total micrograms of starch bound to be calculated by using the reported values of 2.2 × 106 dpm per μg of starch. Experimental values were standardized by assaying whole-cell protein concentration, using a modification of the method of Lowry et al. (9) with bovine serum albumin as a standard.
Assay for enzyme activity in membrane fractions of wild-type and various susA and susG disruption strains.
In the original studies of Smith and Salyers (18), the enzyme assay conditions were optimized for the major activity detectable in cell extracts, which we now know is due to SusA. These conditions might not be optimal for the residual membrane-associated activity found in the susA disruption mutant (4). To determine the optimum conditions for the other enzyme(s), we used p-nitrophenyl-α-d-maltoheptaoside as the substrate (final concentration of 0.5 mM). We found that adding detergent (e.g., n-octyl-β-d-glucoside) to solubilize the protein from the membrane or adding cofactors such as Ca2+ did not enhance the observed enzyme activity. Consequently, we used the original conditions of Smith and Salyers (18), 50 mM potassium phosphate buffer with no additional detergents or cofactors, to measure enzyme activity. This mixture was used in all subsequent assays of enzyme activity.
We used the chromogenic substrate p-nitrophenyl-α-d-maltoheptaoside to assay α-1,4-amylase activity of whole-membrane protein extracts. Amylase activity was calculated as instructed by the manufacturer (Boehringer Mannheim Biochemica). We determined the Km for both SusA and SusG by using resuspended membranes from mutants ΩsusB and ΩSAB(pSGC23A), respectively. These strains expressed only one of the two starch-degrading enzymes, SusA and SusG, respectively. Resuspended membranes were used to measure SusA activity since much of the activity appears to be membrane associated (1), and this would give us a more valid comparison of activity with that of membrane-associated SusG. In these strains, SusB, an α-glucosidase which might interfere with the measurement of each enzyme by hydrolyzing its products, was not expressed. Specific activity was assayed at various concentrations, and Km was determined from Lineweaver-Burke plots. Bacteroides membranes were obtained by ultracentrifugation as described by Valentine and Salyers (see above and reference 20).
Protease treatment of cells.
To determine if SusG was exposed on the cell surface, we tested whether it was accessible to proteolytic activity. We used a relatively nonspecific protease, proteinase K, to increase the likelihood that a surface-exposed protein would be cleaved. For this experiment, cells were transferred from an overnight culture of cells grown in VPI to 100 ml of defined medium containing 0.3% maltose. These cells were grown to late exponential phase (optical density at 650 nm of 0.8) and harvested by centrifugation at room temperature. The cell pellet was washed twice with 100 mM potassium phosphate buffer (pH 7.2) to dissociate any loose capsular material from the cells. Subsequently, the cells were resuspended in 9 ml of 100 mM potassium phosphate buffer and fresh proteinase K (20 mg/ml) was added to a final concentration of 2 mg/ml. The cells were incubated at 37°C with occasional mixing. Samples of 2 ml were removed at 0, 0.5, 1, 2, and 4 h. To each sample, PMSF (10 mg/ml) was added to a final concentration of 10 mM to stop proteinase K activity. The cells were harvested by centrifugation and washed once with 2 ml of a 100 mM potassium phosphate buffer-PMSF solution. The final cell pellet was resuspended in 100 mM potassium phosphate buffer containing 1 mM PMSF. After disruption of the cells by sonication, protein concentration of the cell extract was determined by a modification of the method of Lowry et al. (9). Approximately 100 μg of protein from each sample was solubilized in Laemmli buffer and electrophoresed on a sodium dodecyl sulfate-polyacrylamide gel. The gel was transferred to a Bio-Rad Trans-Blot nitrocellulose membrane. SusG protein was detected by using a Bio-Rad goat anti-mouse-horseradish peroxidase Opti-4CN kit, using antisera directed against the protein. As a control, we repeated the above procedure except that cells were incubated in buffer with no added proteinase K. As a control to ensure that the proteinase K treatment had not disrupted the outer membrane, we used Western blotting to confirm that SusA was not degraded during the proteinase K treatment period. SusA antibody was detected by using a Bio-Rad goat anti-rabbit-horseradish peroxidase Opti-4CN substrate kit, using antisera directed against the protein.
susG expression in trans.
susG was amplified by PCR using primers GAATGGCCGTCGCGGATCCAATGAAATC, which is 30 bp upstream of the putative start codon, and AAGGTTTCTTGGAGCTCGATAGAAAC, which lies 30 bp downstream of the putative transcriptional stop site. This product was cloned into the multiple cloning site of the expression vector pNLY1::PsusA (17) (Fig. 1). In this construct, the gene of interest is cloned downstream of a 392-bp PCR fragment containing the susA promoter (PsusA). This plasmid was called pSGC23A. pSGC23A was mated to ΩsusG, ΩSAB, ΩSAC, ΩSAG, and ΩsusC. These transconjugants were tested by immunoblotting to confirm SusG expression as well as expression of other starch proteins. In addition, these strains were tested for growth on starch.
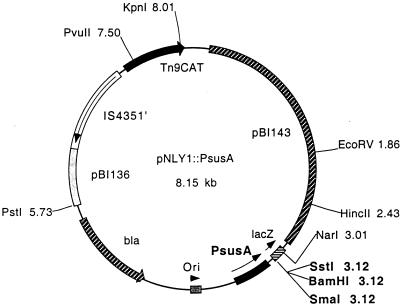
FIG. 1.
Partial restriction map of pNLY1::PsusA, the expression vector used to express susG in trans in B. thetaiotaomicron. The selectable marker in E. coli is ampicillin resistance (bla) plus chloramphenicol resistance (CAT). The selectable marker in B. thetaiotaomicron is chloramphenicol resistance. The E. coli replication region (pBI136) and B. thetaiotaomicron replication and mobilization region (pBI143) are shown. The CAT gene is cloned downstream of the IS4351 promoter. The promoter to control expression of cloned genes was obtained by PCR of a 392-bp fragment from the B. thetaiotaomicron chromosome containing PsusA.
For all relevant strains, we used the assay described above to test for polysaccharidase activity of membrane extracts. We also compared starch binding ability by a strain expressing susG in trans but not the other starch-associated OMPs [ΩsusC(pSGC23A)] with a strain which expresses none of the starch-associated OMPs (ΩsusC). Furthermore, we tested whether SusG was still exposed on the cell surface when expressed independently of the other starch-associated OMPs.
RESULTS
susG expression in trans.
From previous studies in which susG was inactivated by an insertional disruption or a transposon insertion, we knew that SusG was essential for growth on starch (15). To determine the role of SusG itself, we needed to express this gene in trans to determine the properties of a strain producing SusG independently of the other OMPs. In this case, it was important to be particularly careful about the promoter used to drive susG expression and the effect of that promoter on expression of chromosomal genes, because we had found previously that PsusA and susB PsusB could titrate the regulatory protein, SusR, thus reducing transcription of the entire susB-susG operon when either promoter region was provided in trans on a multicopy plasmid (5). We used PsusA because it is 30-fold weaker and had much less effect in trans than PsusB. The original shuttle vector used to clone susG for in trans complementation, pNLY1, is present at 8 to 10 copies/cell (17). Consequently, taking into account the lower expression level of PsusA than of PsusB and the copy number of the plasmid, expression of susG from this plasmid should be close to wild-type levels. The susG expression plasmid, designated pSGC23A, was transferred into an ΩsusG background to test for complementation.
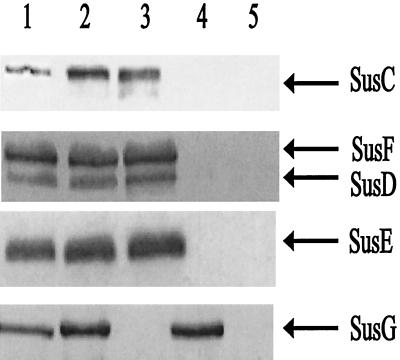
Strain ΩsusG(pSGC23A) had a generation time of 1.7 h on amylopectin, whereas a strain that had wild-type susG, B. thetaiotaomicron 4007(pNLY1::PsusA), had a generation time of 1.4 h. Thus, the cloned gene complemented the mutation, allowing it to grow on starch at about the same rate as the wild type. In addition, it appeared that the cloned susG was fully functional and that PsusA did not reduce significantly expression of the chromosomal sus genes. In Western blots of membranes from cells grown on minimal medium plus maltose, we noted a slight overexpression of SusG in the membrane fraction compared to the wild type, but the amount appeared to be at most twofold higher than the wild-type level. We also confirmed by Western blotting that the other OMPs (SusC to SusF) were being expressed at wild-type levels (Fig. 2). The fact that a slight overexpression of SusG was not deleterious to the cell suggested that SusG may not need to achieve a tight stoichiometric balance with the other OMPs for effective starch degradation. We used this susG expression plasmid in subsequent experiments to test SusG’s role in starch utilization.
FIG. 2.
Immunoblot showing SusG expression from a multicopy plasmid. Approximately 50 μg of protein was loaded onto each lane. All membrane fractions were obtained from cells grown on minimal medium with maltose as the sole carbohydrate source. Lanes: 1, membrane fraction from B. thetaiotaomicron 5482; 2, membrane fraction from B. thetaiotaomicron ΩsusG(pSGC23A); 3, membrane fraction from B. thetaiotaomicron ΩsusG; 4, membrane fraction from B. thetaiotaomicron ΩsusC(pSGC23A); 5, membrane fraction from B. thetaiotaomicron ΩsusC. The arrows mark the known starch OMPs.
susG encodes a starch-hydrolyzing enzyme.
Previous experiments had shown that cell extracts from B. thetaiotaomicron mutant Ms-4, which had a transposon inserted in susG, appeared to have wild-type levels of starch-hydrolyzing enzyme activity (1), but the high activity of SusA in these extracts could have been masking another enzyme activity. To eliminate the major starch-degrading activity detected in our assay system (SusA), we constructed a mutant with a disruption in susA (ΩsusA) and compared its enzymatic activity with that of a double mutant that had disruptions in both susA and susG (ΩSAG). We used Western blotting to confirm that both mutants lacked SusA but still expressed SusB to SusF normally and that ΩsusA produced SusG whereas ΩSAG lacked SusG.
To determine if SusG had enzymatic activity, we used membranes as the enzyme source, both because we knew that SusG was an OMP and because using membrane fractions would concentrate the activity being assayed to give us a higher sensitivity of detection. SusA, the major neopullulanase in our system, partitions mostly (∼65%) with the membrane (2). Disruption of susA in mutant ΩsusA decreased the membrane-associated amylase activity sixfold (Table 2), yet there was still detectable membrane-associated activity in this strain. When susG was disrupted to create the double mutant ΩSAG, this activity decreased over 13-fold to below detectable levels (Table 2). Thus, SusG accounts for the residual activity in the ΩsusA mutant. To support this claim, we determined if SusG provided in trans would restore the enzyme activity. We transferred the multicopy plasmid pSGC23A into the ΩSAG strain and tested for starch-degrading activity. In the resulting strain, starch-degrading activity increased from below detectable levels to over two times that of the ΩsusA strain (Table 2). This increase in activity over ΩsusA was probably due to the slight overexpression of SusG from the multicopy plasmid that we had observed on Western blots (Fig. 2).
TABLE 2.
Amylase activities of whole-membrane extracts of various strainsa
| B. thetaiotaomicron strain | Phenotypeb | Amylase activity (U/mg of cell protein [10−3]c) |
|---|---|---|
| 5482 (wild type) | A+ B+ C+ D+ E+ F+ G+ | 4,204 |
| 5482 grown on glucose | A+ B+ C+ D+ E+ F+ G+ | <50 |
| ΩsusA | A− B+ C+ D+ E+ F+ G+ | 690 |
| ΩSAG | A− B+ C+ D+ E+ F+ G− | <50 |
| ΩSAG(pSGC23A) | A− B+ C+ D+ E+ F+ G++ | 1,665 |
| ΩSAC | A− B+ C− D− E− F− G− | |
| ΩSAC(pSGC23A) | A− B+ C− D− E− F− G++ | 1,113 |
For this assay, all strains were grown on minimal medium plus maltose except as indicated. Variation between replicate samples was less than 10%.
Production of Sus proteins in each strain. For example, “A” corresponds to production of SusA. ++ indicates twofold-higher expression than in the wild type.
Units are in micromoles per minute. The substrate used to detect amylase activity was p-nitrophenyl-α-d-maltoheptaoside.
We also wanted to determine if this observed enzyme activity required any of the other starch-associated OMPs. Accordingly, we constructed a double mutant with disruptions in both susA and susC (ΩSAC) and introduced pSGC23A. This strain expressed neither SusA nor any of the starch-associated OMPs except SusG. The original ΩSAC strain without the plasmid had no detectable starch-degrading activity. When the plasmid expressing susG (pSGC23A) was introduced, starch-degrading activity of this strain was comparable to that observed for ΩSAG(pSGC23A) (Table 2), confirming that SusG has enzymatic activity even when the other starch-associated OMPs are not present. It is interesting that this strain, ΩSAC(pSGC23A), which expresses SusG but not SusC to SusF, could not grow at all on starch but could grow on maltose and maltotriose. Thus, if SusG by itself were degrading starch in vivo to maltose and maltotriose, this strain should have been able to grow on starch. Data from our assay (where amylose was the substrate) support the hypothesis that SusG hydrolyzes amylose. The fact that a susG disruption mutant could not grow on pullulan suggests that SusG also degrades pullulan and is thus a neopullulanase.
SusG is exposed on the cell surface.
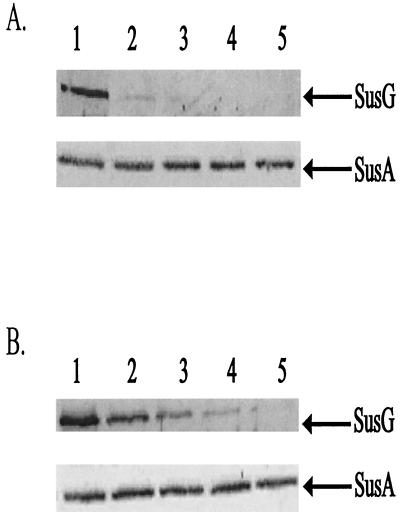
Since SusG had enzymatic activity, we wanted to determine if SusG was exposed on the cell surface. If the protein is surface exposed, enzymatic cleavage might occur on the cell surface. We tested for exposure on the cell surface by determining the protease accessibility of SusG in intact cells. After 2 h of proteinase K treatment, SusG was no longer detectable in these cells (Fig. 3). The complete degradation of SusG shows that proteinase K was able to attack SusG on the cell surface. We performed control experiments to confirm that SusG was stable in the absence of proteinase K and that proteinase K degradation of surface-exposed proteins was not destroying the outer membrane. In a control experiment where proteinase K was not added, SusG was stable throughout the course of the experiment. To confirm that the cells treated with proteinase K remained intact during the digestion period, we determined whether SusA, a periplasmic enzyme, was present at the time points where SusG was not present. According to the results of immunoblotting, SusA was present at a constant level at all time points in this experiment (Fig. 3). This confirms that we were seeing surface degradation of OMPs but not disruption of the outer membrane. In a separate experiment, to rule out the possibility that SusA was unusually resistant to proteases, we disrupted cells to release SusA and then added proteinase K. Under these conditions, SusA disappeared within 30 min after addition of proteinase K, confirming that SusA is readily degraded by proteinase K if it is accessible to the enzyme.
FIG. 3.
Immunoblots showing proteolytic sensitivity of SusG in intact cells. Cells were treated with proteinase K (2 mg/ml), and degradation of SusG was observed over time. SusA, a periplasmic marker, is shown to be stable during the course of each experiment. Approximately 100 μg of protein from whole-cell extracts was loaded onto each lane. Lanes 1 to 5 represent whole-cell extracts of B. thetaiotaomicron wild-type (A) and ΩsusC(pSGC23A) (B) strains at 0, 30, 60, 120, and 240 min, respectively.
Additionally, we tested whether SusG was exposed on the cell surface when expressed independently of the other starch-associated OMPs. Since these OMPs are involved in starch utilization, one or more of these proteins could be necessary for SusG to be properly localized and stable in the outer membrane. For this experiment, we assayed proteinase K accessibility of SusG in strain ΩsusC(pSGC23A), which produced SusG but not SusC to SusF. In intact cells of this strain, SusG was eliminated by proteinase K treatment after 4 h, somewhat more slowly than SusG in a wild-type background (Fig. 3). The apparent slower disappearance is probably due to the fact that there is more SusG in the mutant. Meanwhile, in a control experiment, SusG was present at a constant level when proteinase K was not added. Thus, SusG localizes to and is stable in the outer membrane independent of the other starch-associated OMPs.
SusG exhibits very low affinity for starch and does not play a significant role in starch binding.
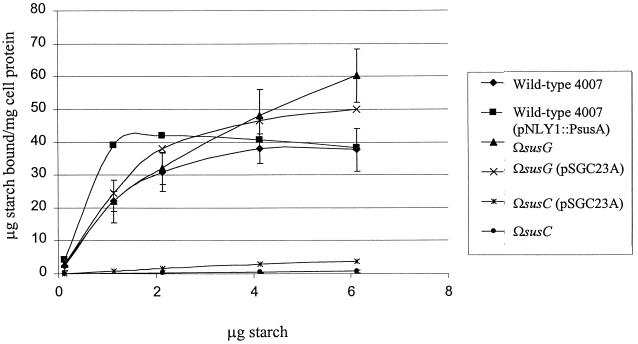
We used membrane fractions from two different mutants to determine the Km of SusG for starch hydrolysis and compare it to that of SusA. In the strain ΩsusB, the only membrane-associated starch-degrading enzyme present is SusA, because the polarity of the insertion prevents the expression of SusG. In strain ΩSAB(pSGC23A), only SusG is produced. The Km of SusG was 3.1 mM. By comparison, the Km of SusA was 0.125 mM, almost 20 times lower than that of SusG. These results suggest that SusG has a low affinity for starch. To test the contribution of SusG to starch binding, we expressed SusG independently of the other OMPs. This was achieved by placing pSGC23A in an ΩsusC background. The ΩsusC(pSGC23A) strain produced SusA, SusB, and SusG but not SusC, SusD, SusE, or SusF. This strain did not exhibit significant binding above the background binding of ΩsusC (Fig. 4). Other proteins seem to be more directly involved in binding than SusG.
FIG. 4.
Starch binding of B. thetaiotaomicron mutants at various starch concentrations. Cells bound increasing amounts of starch ([14C]starch plus unlabeled amylopectin) proportional to starch concentration until the binding sites were saturated, at which point binding was independent of starch concentration. Error bars are shown for B. thetaiotaomicron 4007, ΩsusG, and ΩsusC only.
To degrade starch sufficiently for cell growth, SusG’s activity could be enhanced by starch binding proteins which bring the substrate closer to it. This possibility suggested that SusG may closely interact with starch binding proteins. Whether this is a direct or indirect interaction has not been determined. If there is a direct interaction, the possibility exists that SusG’s absence (e.g., in ΩsusG) may affect binding by these potentially closely associated starch binding proteins. We examined this possibility by using a starch binding assay. Initially, in doing the assay that measures binding of [14C]starch to intact cells, we merely pelleted the cells and did not wash them because we assumed that binding would be reversible. Subsequently, we found that most of the [14C]starch remained associated with the cells even after washing and was thus irreversibly bound. Since the wash step eliminated some of the variation, we used this procedure to compare mutants with the wild type. Under the aerobic assay conditions used, there was no uptake and metabolism of the starch. Thus, our assay should measure only tight binding to the cell surface and not transport.
When a range of starch concentrations was tested for irreversible binding of starch, it became clear that a susG disruption mutant (ΩsusG) bound amounts similar to those bound by the wild type at saturating conditions of starch. Binding was saturated at approximately 40 μg of bound starch/mg of cell protein for both wild-type and ΩsusG strains. When SusG expression was restored by expression in trans [ΩsusG(pSGC23A)], starch binding was again equal to that of the wild type (Fig. 4). It appears that lack of SusG may have increased the KD of binding somewhat, but if so, this difference was barely significant. SusG appears not to play a significant role in starch binding, and its absence has little effect on starch binding.
DISCUSSION
Our results demonstrate that SusG plays an important role in starch breakdown. One function is enzymatic hydrolysis of starch. There are a number of lines of evidence supporting the hypothesis that SusG is a starch-degrading enzyme and not just a protein that activates an enzyme. Smith and Salyers (18) partially purified membrane-bound starch-degrading proteins by subjecting Triton X-100 extracts of B. thetaiotaomicron membranes to isoelectric focusing. SusG is the only one of the Sus OMPs that can be solubilized by Triton X-100 (15). Therefore, we would expect SusG to have been present in these extracts. Smith and Salyers found a fraction of amylase activity with pI of 4.4 to 4.9. SusG has a predicted pI of 4.8 to 4.9, once the putative signal sequence is removed. In this range, the enzymatic activity was low relative to the other band, now known to be SusA, but it was still detectable. SusG has significant sequence similarity to α-amylases in the databases, is responsible for the residual membrane-bound activity in a susA disruption strain, and has enzyme activity when expressed independently of the other starch-associated OMPs. Thus, based on current genetic and previous biochemical evidence, it is reasonable to suggest that SusG has starch-hydrolyzing activity. Another possible role for SusG is binding of starch to the cell surface. Our results suggest, however, that SusG makes little contribution to starch binding. If SusG has little or no role in binding starch, the binding of starch must be mediated by other starch OMPs which cooperate with SusG and even enhance SusG’s enzymatic activity.
The cellulosomes of clostridia and the pullulanase system of Klebsiella pneumoniae have cell-associated enzymes that degrade polysaccharides on the cell surface (3, 13). The difference between the two, aside from the outer membrane, is that the cellulosome also contains cellodextrin binding proteins which are in close proximity to the cellulases (11). As a relevant example, in Clostridium cellulolyticum, the cellulose binding domains of CipC are hypothesized to enhance hydrolysis of cellulose by keeping the substrate in a favorable position for enzymatic attack (12). This hypothesis has been difficult to test rigorously because of the lack of a genetic Clostridium system. In our system, however, we can test this hypothesis in living cells that have been genetically modified to produce only some of the proteins involved. B. thetaiotaomicron appears also to have starch binding proteins separate from the enzymes that work in concert with them to cleave the substrate and sequester the protein. Evidence presented here suggests that SusG has a very low affinity for starch, necessitating starch binding proteins to promote hydrolytic attack, which would be similar to the cellulosome’s proposed mechanism of polysaccharide hydrolysis.
Taken together, results of our previous and present studies show that OMPs, including a putative porin SusC, are involved in the binding of starch to the outer membrane of B. thetaiotaomicron. Starch binding by OMPs has been shown to be an essential step for this process (2). SusG appears to act primarily as an enzyme cleaving the starch bound by those proteins to smaller oligomers which can then traverse the membrane.
ACKNOWLEDGMENTS
This study was supported by grant AI17876 from the National Institutes of Health.
We thank Nadja B. Shoemaker for constructing pNLY1::PsusA and pGERM as well as providing excellent technical advice. We also thank Jorge Frias-Lopez for superb technical advice on the proteinase K accessibility experiments.
REFERENCES
- 1.Anderson K L, Salyers A A. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J Bacteriol. 1989;171:3192–3198. doi: 10.1128/jb.171.6.3192-3198.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson K L, Salyers A A. Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1989;171:3199–3204. doi: 10.1128/jb.171.6.3199-3204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beguin P, Lemaire M. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit Rev Biochem Mol Biol. 1996;31:201–236. doi: 10.3109/10409239609106584. [DOI] [PubMed] [Google Scholar]
- 4.D’elia J N, Salyers A A. Contribution of a neopullulanase, a pullulanase and an α-glucosidase to growth of Bacteroides thetaiotaomicron on starch. J Bacteriol. 1996;178:7173–7179. doi: 10.1128/jb.178.24.7173-7179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’elia J N, Salyers A A. Effect of regulatory levels on utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol. 1996;178:7180–7186. doi: 10.1128/jb.178.24.7180-7186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D. Studies on the transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 7.Kanazawa K, Kobayashi Y, Nakano M, Sakurai M, Gotoh N, Nishino T. Identification of three porins in the outer membrane of Bacteroides fragilis. FEMS Microbiol Lett. 1995;127:181–186. [Google Scholar]
- 8.Lederberg E M, Cohen S M. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974;119:1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 10.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 11.Pages S, Belaich A, Tardif C, Reverbel-Leroy C, Gaudin C, Belaich P. Interaction between the endoglucanase CelA and the scaffolding protein CipC of the Clostridium cellulolyticum cellulosome. J Bacteriol. 1996;178:2279–2286. doi: 10.1128/jb.178.8.2279-2286.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pages S, Gal L, Belaich A, Gaudin C, Tardif C, Belaich J-P. Role of scaffolding protein CipC of Clostridium cellulolyticum in cellulose degradation. Appl Environ Microbiol. 1997;179:2810–2816. doi: 10.1128/jb.179.9.2810-2816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeves A R, D’elia J N, Frias J, Salyers A A. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J Bacteriol. 1996;178:823–830. doi: 10.1128/jb.178.3.823-830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reeves A R, Wang G-R, Salyers A A. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol. 1997;179:643–649. doi: 10.1128/jb.179.3.643-649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoemaker N B, Getty C, Guthrie E P, Salyers A A. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J Bacteriol. 1986;166:959–965. doi: 10.1128/jb.166.3.959-965.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoemaker, N. B., G.-R. Wang, and A. A. Salyers. Multiple gene products and an excision required sequence (XRS) necessary for the excision of the mobilizable Bacteroides element NBU1. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 18.Smith K A, Salyers A A. Characterization of a neopullulanase and an α-glucosidase from Bacteroides thetaiotaomicron 95-1. J Bacteriol. 1991;173:2962–2968. doi: 10.1128/jb.173.9.2962-2968.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tancula E, Feldhaus M J, Bedzyk L A, Salyers A A. Location and characterization of genes involved in binding of starch to the surface of Bacteroides thetaiotaomicron. J Bacteriol. 1992;174:5609–5616. doi: 10.1128/jb.174.17.5609-5616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valentine P, Salyers A A. Analysis of proteins associated with growth of Bacteroides ovatus on the branched galactomannan guar gum. Appl Environ Microbiol. 1992;58:1534–1540. doi: 10.1128/aem.58.5.1534-1540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]