Abstract
The genetically modified cotton DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 expressing Cry1Ac, Cry1F and Vip3Aa19 from Bacillus thuringiensis Berliner (Bt) has been cultivated in Brazil since the 2020/2021 season. Here, we assessed the performance of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 cotton expressing Cry1Ac, Cry1F and Vip3Aa19 against Helicoverpa armigera (Hübner), Helicoverpa zea (Boddie), and their hybrid progeny. We also carried out evaluations with DAS-21023-5 × DAS-24236-5 cotton containing Cry1Ac and Cry1F. In leaf-disk bioassays, DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 was effective in controlling neonates from laboratory colonies of H. armigera, H. zea and the hybrid progeny (71.9%–100% mortality). On floral bud bioassays using L2 larvae, H. zea presented complete mortality, whereas H. armigera and the hybrid progeny showed <55% mortality. On DAS-21023-5 × DAS-24236-5 cotton, the mortality of H. armigera on leaf-disk and floral buds ranged from 60% to 73%, whereas mortality of hybrids was <46%. This Bt cotton caused complete mortality of H. zea larvae from a laboratory colony in the early growth stages, but mortalities were <55% on advanced growth stages and on floral buds. In field studies conducted from 2014 to 2019, DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 cotton was also effective at protecting plants against H. armigera. In contrast, a population of H. zea collected in western Bahia in 2021/2022 on Bt cotton expressing Cry1 and Vip3Aa proteins, showed 63% mortality after 30 d, with insects developing into fifth and sixth instars, on DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 cotton. We conclude that H. armigera, H. zea, and their hybrid progeny can be managed with DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 cotton; however we found the first evidence in Brazil of a significant reduction in the susceptibility to DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 cotton of a population of H. zea collected from Bt cotton in Bahia in 2021/2022.
Introduction
Helicoverpa spp. is a group of harmful agricultural pests in several regions of the world. In Brazil, the most important Helicoverpa species are the Old World bollworm, Helicoverpa armigera (Hübner)—first recorded in 2013 in soybean in Goiás and Bahia states—, and the corn earworm, Helicoverpa zea (Boddie) (Lepidoptera: Noctuidae) [1–3]. In the current Brazilian crop production systems, outbreaks of Helicoverpa species are favored by the overlapping and simultaneous cultivation of host plants, including soybean, maize, cotton, tomatoes, and dry bean crops [4]. Studies also reported the occurrence of interspecific hybridization between H. armigera and H. zea under laboratory conditions [5–7]. The successful hybridization in a restricted environment suggests that potential for multiple mating events in the field may provide opportunities to select well-adapted hybrid phenotypes [7]. Cases of resistance to Bt proteins in H. zea have been reported in the U.S. and China [8–11] and decreases in susceptibility to Cry proteins by H. armigera have been reported in U.S., India, and China [12, 13].
In Brazil, H. armigera has great importance in soybean and cotton [1, 4], while H. zea was most abundant in maize and cotton [14, 15]. The adoption of genetically-modified (GM) plants expressing insecticidal proteins derived from the soil bacterium Bacillus thuringiensis Berliner (Bt) is the main management tactic used against Helicoverpa species in cotton, soybean and maize fields in Brazil [16, 17]. The successful adoption of Bt crops has enabled more effective management of lepidopteran pests and reduced chemical insecticide applications in these crops worldwide [18–20].
In Brazil, Bt cotton was planted commercially for the first time in 2006. Transgenic cotton technologies are cultivated on approximately 1.4 million hectares (90% of the total area with Bt cotton) in recent cotton seasons [21]. The first generation of commercial Bt plants expressed single Bt proteins, but low compliance with resistance management strategies (e.g. refuge areas) in areas cultivated with Bt maize has favored the cross-crop resistance, affecting the performance of other Bt plants, including Bt cotton varieties [22]. The second and third generations of Bt cotton met the concept of ‘gene pyramiding’ with the expression of two or more Bt proteins with high toxicity and distinct modes of action to the same pest species [23, 24]. For example, the DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 event (commercially named WideStrike® 3) is part of the third generation of Bt cotton cultivated in Brazil and expresses Cry1Ac, Cry1F and Vip3Aa19 insecticidal proteins [25].
Previous studies found that WideStrike® 3 was effective against Spodoptera frugiperda (J. E. Smith, 1797), Spodoptera cosmioides (Walker, 1858), Chloridea virescens (F., 1781), Chrysodeixis includens (Walker, 1858), and Alabama argillacea (Hübner, 1823) (Lepidoptera: Noctuidae) [25]. However, the performance of the WideStrike® 3 was not reported against Helicoverpa species. On the present study, we evaluated the performance of Bt cotton DAS-21023-5 × DAS-24236-5 × SYN-IR102- expressing Cry1Ac, Cry1F and Vip3Aa19 and DAS-21023-5 × DAS-24236-5 that expresses Cry1Ac and Cry1F against H. armigera, H. zea and their hybrid progeny. We performed evaluations in laboratory and field trials across several cotton planting regions in Brazil. We also investigated the susceptibility to Bt cotton expressing Cry and Vip3Aa insecticidal proteins in a population of H. zea collected from Bt cotton expressing the Cry1Ab, Cry2Ae, and Vip3Aa Bt proteins in Bahia, Brazil.
Material and methods
A. Description of pest species and hybridization crosses
The two Helicoverpa species, H. armigera and H. zea, were collected from the field and maintained at Corteva Agriscience insectary (Toledo, Paraná, Brazil). Larvae of H. zea were collected from corn ears and H. armigera from soybean plants in Brasilia, DF during the 2015 crop season. Insects were maintained on an artificial insect diet proposed by Greene et al. [26], being that new field-collected larvae from non-Bt maize (H. zea) and non-Bt cotton or non-Bt soybean (H. armigera) were introduced to the colonies every year (~500 larvae/year). The new field larvae were collected in Paraná, São Paulo or Goiás states from 2016 to 2019. Hereafter, we refer to these populations as Hz-Lab and Ha-Lab. To design hybridization crosses, adults were confined in a PVC cage (30 cm in diameter × 30 cm height). Only the crossing between (500 ♀ Ha-Lab × 500 ♂ Hz-Lab) produced eggs that produced neonates. Hybrid larvae (F1 generation) were tested in confined laboratory conditions, to avoid any escapes to the environment.
In addition, during the 2021/2022 cotton season, a field population of H. zea was collected in Bt cotton expressing Cry1Ab, Cry2Ae and Vip3Aa (variety FM 985, FiberMax® - BASF S.A., São Paulo, SP, Brazil) in Luis Eduardo Magalhães, BA (latitude 12°07’29” S, longitude 46°08’28” W). After the collection, this population was transferred to an artificial diet [26] and maintained in a laboratory at Instituto Mato-Grossense do Algodão (IMAmt), Primavera do Leste, MT, Brazil. We refer to this population as Hz-field.
Before starting bioassays, all species were confirmed by PCR testing according to the methodology described by Perera et al. [27].
B. Leaf-disk and square bioassays in laboratory trials
Leaf-disk and square bioassays were performed at Corteva Agriscience site located in Mogi-Mirim, SP, Brazil. The cotton technologies evaluated were: 1) A Bt cotton variety (TMG 91WS3 –full Maturity, Tropical Melhoramento & Genética S.A.–Cambé, Paraná, Brazil) containing events DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 (WideStrike® 3 Insect Protection, Corteva Agriscience, Indianapolis, IN) expressing Cry1Ac, Cry1F and Vip3Aa19 Bt proteins; 2) A Bt cotton variety (TMG 81WS–full Maturity) containing events DAS-21023-5 × DAS-24236-5 (WideStrike® Insect Protection, Corteva Agriscience, Indianapolis, IN) expressing Cry1Ac and Cry1F Bt proteins, and 3) A non-Bt cotton variety (FMT 701, Fundação Mato Grosso, Rondonópolis, MT, Brazil). Weekly plantings were done under field conditions for two months to have supply of fresh leaves and squares at the same time. Before the bioassays, Bt and non-Bt plants were checked to confirm Bt protein expression using detection kits for Cry1Ac, Cry1F and Vip3A proteins (Envirologix, São Paulo, SP, Brazil).
Bioassays were performed with leaf disks of Bt and non-Bt cotton varieties previously described from leaves excised of plants in vegetative (growth stage 15 – 5th true leaf unfolded) and reproductive (growth stage 55 –squares distinctly enlarged) stages according to the phenological scale of Munger et al. [28]. Squares (bracts + buds) from a reproductive stage (growth stage 55) were also tested. Leaves and squares were removed from the upper third of the plants when they reached the respective phenological stage. Leaf disks 1.2 cm in diameter were cut using a metallic cutter. Leaf disks and squares were placed over a gelled mixture of water-agar 2.5% (1 ml/well) in 128-well bioassay trays (BIO-BA-128; CD International Inc., Pitman, NJ). The vegetative and reproductive structures tested were separated from the water-agar layer by a filter paper disk. Then, a single neonate (Ha-Lab, Hz-Lab and their F1 hybrid progeny) was placed on each leaf disk or a single L2 larvae was placed on each square. The trays were sealed with self-adhesive plastic sheets (BIO-CV-16; CD International Inc.) that allowed for gas exchange and then placed in a climatic chamber (temperature 25 ± 2°C, 60 ± 5% RH, and 14:10 h photoperiod). The experimental design was completely randomized with 8 replicates/growth stage/cotton technology (each replicate was represented by 16 larvae). Mortality, damage on leaf disks and squares, and larval weight were recorded at 5 d after infestation. The damage on each leaf disk was recorded using two methods. We measured the area (cm2) that was consumed by larva using a transparent grid ruler, and we also estimated the damage as percentage of the disk that was consumed. The damage on squares was classified as a surface feeding on the external side of the square, and as bored squares, when the larvae could feed inside the square. All live larvae in each replicate were pooled to obtain the weight of the replicate and recorded, since some larvae were too small to accurately weigh individually.
C. Survivorship of Helicoverpa species on cotton throughout the larval cycle
Leaves of Bt and non-Bt cotton varieties previously described were excised from greenhouse-grown plants at the growth stage 15, according to the scale of Munger et al. [28]. In the laboratory, leaves were cut into pieces (6–8 cm2) and placed on a gelled mixture of agar-water at 2.5% in 50-mL plastic cups. Then, a single neonate from the Ha-Lab or Hz-Lab populations was placed in each cup, with leaves replaced every 24 h. Cups were placed in a climatic chamber at 25 ± 2°C, 60 ± 5% RH, and 14:10 h photoperiod. The bioassays with Ha-Lab and Hz-Lab populations were performed at Federal University of Santa Maria, RS, Brazil.
Similar bioassays were performed with the Hz-field population at Instituto Mato-Grossense do Algodão, Primavera do Leste, MT, Brazil. In bioassays, leaves were removed from cotton plants at the growth stage 15, cut into pieces and placed in 24-well plastic plates. Then, a single neonate (F1 generation) was placed in each well. When larvae reach 3rd instar, they were individualized in Petri dishes (10 cm diameter × 1.5 cm height), with leaves replaced daily, being maintained in the same environmental conditions previously described. The cotton varieties tested in bioassays using the Hz-field population were: 1) TMG 50WS3 (early Maturity–Tropical Melhoramento & Genética S.A.–Cambé, Paraná, Brazil) containing events DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 expressing Cry1Ac, Cry1F and Vip3Aa19 transgenic proteins; 2) TMG 81WS containing events DAS-21023-5 × DAS-24236-5 expressing Cry1Ac and Cry1F proteins; and 3) non-Bt cotton (IMA 2106 GL–mid Maturity, Instituto Mato-Grossense do Algodão, Cuiabá, MT, Brazil).
Before bioassays, plants were checked for the presence of the expected Bt proteins using detection kits (Envirologix, QuickStix, São Paulo, SP, Brazil) for Cry1Ac, Cry1F and Vip3A proteins. The experimental design was completely randomized with 42 and 26 replicates of 10 neonates for Bt cotton treatments, respectively, and 13 replicates of 10 neonates for the non-Bt cotton in bioassays with Ha-Lab and Hz-Lab, whereas 5 replicates of 20 neonates/cotton variety was used in bioassays with Hz-field. For both species, larval survival was evaluated every 5 d.
D. Field efficacy of Bt cotton against artificial and natural infestations of H. armigera
Ten field experiments were conducted from 2014 to 2019 growing seasons across four different states in Brazil (Table 1). All trials were conducted followed strict adherence to Brazilian regulatory requirements in accredited and certified field research stations operated by Corteva Agriscience or SGS Company. Field trials were performed under regulated permits approved by the Comissão Técnica Nacional de Biossegurança (CTNBio). Treatments included were: 1) A Bt cotton variety containing events DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 (WideStrike® 3 Insect Protection) expressing Cry1Ac, Cry1F and Vip3Aa19 Bt proteins; 2) A Bt cotton variety containing events DAS-21023-5 × DAS-24236-5 (WideStrike® Insect Protection) expressing Cry1Ac and Cry1F Bt proteins, and 3) A non-Bt isoline cotton variety with a similar genotypic background and belonging to the same maturity group as the Bt cotton varieties. The Bt cotton variety used was PHY 440 (Mid-full Maturity, PhytoGen® Cotton Seeds) in all treatments. Each field trial consisted of four replications for each treatment arranged in a randomized complete block design. Plot size varied among locations from 5 to 8 meters in length and 5 or 7 rows wide. Row spacing in all locations varied from 50 to 76 cm.
Table 1. Field trial locations and geographic coordinates for each study performed from 2014 to 2019 in Brazil.
| Location (city, state) | Geographic coordinates | Year | Infestation |
|---|---|---|---|
| Indianópolis, MG | 18°57′29.70′′ S; 47°51′21.10′′ W | 2014 | Helicoverpa armigera artificial infestations |
| 18°57′24.69′′ S; 47°51′12.09′′ W | 2016 | ||
| 18°57′08.60′′ S; 47°51′11.80′′ W | 2016 | ||
| 24°47′14.20′′ S; 49°51′02.00′′ W | 2018 | ||
| Montividiu, GO | 17°22′40.4′′ S; 51°23′39.36′′ W | 2014 | |
| Palotina, PR | 24°21’43.00" S; 53°45’23.70" W | 2016 | |
| Rio Verde, GO | 17°45′02.20″ S; 51°02′18.30″ W | 2016 | |
| 17°45′27.40″ S; 51°02′03.70″ W | 2017 | ||
| 17°45′18.20″ S; 51°02′08.10″ W | 2018 | ||
| Mogi Mirim, SP | 22°26’49.20" S; 47°04’09.90" W | 2019 | |
| Rio Verde, GO | 17°45′02.20″ S; 51°02′18.30″ W | 2016 | Helicoverpa spp. natural infestation |
Field efficacy of Bt cotton against artificial infestations of H. armigera
For this study, only H. armigera was infested in the field trials because previous laboratory trials on leaf disks with H. zea showed complete mortality. Larvae of H. armigera were obtained from laboratory-reared colony maintained by Corteva Agriscience (Mogi Mirim Research Center, Mogi Mirim, São Paulo State, Brazil) or SGS Company (Piracicaba, SP, Brazil). Artificial infestations were conducted at two different cotton reproductive growth stages at all locations to ensure uniform pest pressure across experimental plots (Table 1). The first artificial infestation was conducted at GS6: 65, at the beginning of flowering (“mid bloom”), and the second infestation at GS6: 65+, 10–12 d later [28]. For each plot, ten plants were randomly selected and each one was infested with 10 L1 larvae. Larvae were placed on the growing points of the selected plants, and then covered immediately with mesh cages (150 cm long × 50 cm wide × 150 cm high) to limit larval escape and to avoid mortality caused by natural enemies. Field evaluations consisted of recording the total number of cotton squares on 10 infested plants, the percentage of damaged squares, and the number of live larvae still present. These evaluations were performed 10 d after each infestation.
Field efficacy of Bt cotton against natural infestation of Helicoverpa spp
A natural infestation of Helicoverpa spp. occurred only in one trial at Rio Verde (GO) (Table 1). At this location, plot inspections and evaluations were performed weekly. The data presented in this paper represent the sampling date when the peak number of damaged squares and the number of live larvae were recorded for the non-Bt treatment.
E. Data analysis
The number of insects tested and the number dead in leaf-disk and square bioassays, and survivorship of Ha-Lab, Hz-Lab and Hz-field throughout the larval cycle on Bt and non-Bt cotton leaves were used to estimate 95% confidence intervals (CIs) for the probability of mortality, according to a binomial distribution [29]. To perform these analyses, the function binom.probit from the package binom in R 3.1.0 [30] was used. Percent mortality rate on Bt and non-Bt cotton were declared significantly distinct if 95% CIs did not overlap [31, 32].
The variables of larvae developing on Bt and non-Bt cotton technologies (consumed leaf area, percent of leaf consumption, bud damage, and larval weight) and data from field trials (artificial infestation) on the efficacy of Bt cotton technologies against H. armigera were analyzed using a linear mixed model (PROC MIXED procedure) and statistical significance were obtained by using Tukey’s Honestly Significant Difference test with α = 0.05 in SAS [33]. Prior to analysis, data from each trial were evaluated to ascertain its validity by assessing the injury level in the non-Bt control. Trials selected for analysis only included those in which the non-Bt control averaged ≥10% square injury. Furthermore, prior to the combined analysis, each trial was analyzed individually and the mean square error (MSE) of the residual was used to evaluate the homogeneity of the variance error. Only trials that showed a ratio between the largest and smallest MSE ≤7 were included in the combined cross-trial analyses [34]. This procedure ensured that trials were homogeneous to avoid bias caused by differences among trials (sites and years). The data from the single, naturally-infested location of Helicoverpa spp. was subjected to t-test (PROC TTEST procedure) paired with α = 0.05 in SAS [33]. To improve the normal distribution, data of squares injured were log (x + 1) transformed, while data on number of larvae were transformed using . Non-transformed data are presented in figures.
Results
A. Leaf-disk and square bioassays
Mortality on leaf-disk and square
No significant differences were observed on mortality at 5 d for H. armigera neonates (Ha-Lab) on leaf-disks (67.2%–85.9%) and L2 larvae on squares (54.7%–60.2%) between DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 and DAS-21023-5 × DAS-24236-5 (Table 2). Both GM cotton lines provided significantly greater mortality to H. armigera compared to the non-Bt cotton on all tissue types (Table 2). Mortality observed for the non-Bt cotton treatments averaged 2.3% or lower across tissues, indicating the bioassay test system was robust (Table 2).
Table 2. Percent mortality of neonates (on leaf disk) and L2 larvae (on square) of H. armigera, H. zea and the F1 progeny from ♀H. armigera × ♂H. zea after 5 d on leaves of Bt and non-Bt cotton technologies in laboratory trials.
| Pest species and cotton technologies | n a | Leaf disk (growth stage 15)b | Leaf disk (growth stage 55)b | Square (growth stage 55)b | |||
|---|---|---|---|---|---|---|---|
| Dead | % mortality (95% CI) | Dead | % mortality (95% CI) | Dead | % mortality (95% CI) | ||
| Helicoverpa armigera (Ha-Lab) | |||||||
| DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 | 128 | 110 | 85.9 (79.0–91.1) a | 92 | 71.9 (63.6–79.1) a | 70 | 54.7 (46.0–63.1) a |
| DAS-21023-5 × DAS-24236-5 | 128 | 93 | 72.7 (64.4–79.8) a | 86 | 67.2 (58.7–74.9) a | 77 | 60.2 (51.5–68.3) a |
| Non-Bt cotton | 128 | 3 | 2.3 (0.7–6.5) b | 3 | 2.3 (0.7–6.5) b | 1 | 0.8 (0.1–4.4) b |
| Helicoverpa zea (Hz-Lab) | |||||||
| DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 | 128 | 128 | 100.0 (97.2–100.0) a | 128 | 100.0 (97.2–100.0) a | 128 | 100.0 (97.2–100.0) a |
| DAS-21023-5 × DAS-24236-5 | 128 | 128 | 100.0 (97.2–100.0) a | 53 | 41.4 (33.1–50.0) b | 70 | 54.7 (46.0–63.1) b |
| Non-Bt cotton | 128 | 16 | 12.5 (7.6–19.2) b | 10 | 7.8 (4.1–13.6) c | 14 | 10.9 (6.4–17.3) c |
| F1 hybryd (♀ Ha-Lab × ♂ Hz-Lab)c | |||||||
| DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 | 128 | 128 | 100.0 (97.2–100.0) a | 97 | 75.8 (67.8–82.3) a | 48 | 37.5 (29.5–46.1) a |
| DAS-21023-5 × DAS-24236-5 | 128 | 59 | 46.1 (37.6–54.8) b | 51 | 39.8 (31.7–48.5) b | 15 | 11.7 (7.0–18.2) b |
| Non-Bt cotton | 128 | 14 | 10.9 (6.4–17.3) c | 16 | 12.5 (7.6–19.2) c | 3 | 2.3 (0.7–6.5) c |
aNumber of larvae tested per growth stage.
bGrowth stage number according to Munger et al. [28]. Means (95% CI) followed by the same letter in each column and species are not significantly different due to nonoverlap of 95% CIs.
cOnly the crossing of ♀H. armigera × ♂H. zea produced fertile eggs allowing bioassays to be carried out.
No surviving H. zea neonates (Hz-Lab) were also detected at 5 d on leaf-disks harvested at growth stage 15 of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 and DAS-21023-5 × DAS-24236-5 cotton technologies (Table 3). In contrast, on leaf-disk of DAS-21023-5 × DAS-24236-5 from growth stage 55 (neonates) and on squares (L2 larvae) of this same growth stage, mortality (≤54.7%) was significantly lower than on DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 (Table 2). On non-Bt cotton, the mortalities of neonates (leaf-disk) and L2 larvae (squares) of H. zea was ≤12.5%.
Table 3. Damage caused by neonates (on leaf disk) and L2 larvae (on square) of H. armigera, H. zea and the F1 progeny from ♀H. armigera × ♂H. zea after 5 d on leaves of Bt and non-Bt cotton technologies in laboratory trials.
| Pest species and cotton technologies | Damage on leaf disks (growth stage 15)a | Damage on leaf disks (growth stage 55)a | Damage on square (growth stage 55)a | ||||
|---|---|---|---|---|---|---|---|
| Consumed leaf area (cm2) | % leaf consumption | Consumed leaf area (cm2) | % leaf consumption | % squares with surface feeding only | % larvae bored into square | % squares with any damage | |
| Helicoverpa armigera (Ha-Lab) | |||||||
| DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 | 0.03 ± 0.01 b | 1.9 ± 0.5 b | 0.09 ± 0.01 b | 4.6 ± 0.5 b | 39.1 ± 6.4 ab | 28.9 ± 3.5 b | 67.9 ± 6.1 b |
| DAS-21023-5 × DAS-24236-5 | 0.07 ± 0.01 b | 5.0 ± 0.5 b | 0.09 ± 0.02 b | 3.4 ± 0.5 b | 11.7 ± 3.8 c | 57.8 ± 7.1 a | 70.3 ± 4.2 b |
| Non-Bt cotton | 1.13 ± 0.01 a | 60.3 ± 3.0 a | 1.0 ± 0.03 a | 52.2 ± 1.7 a | 30.1 ± 2.2 b | 68.8 ± 2.4 a | 99.2 ± 0.8 a |
| Helicoverpa zea (Hz-Lab) | |||||||
| DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 | 0.0 ± 0.0 b | 0.0 ± 0.0 b | 0.0 ± 0.0 c | 0.0 ± 0.0 c | 0.0 ± 0.0 c | 0.0 ± 0.0 c | 0.0 ± 0.0 c |
| DAS-21023-5 × DAS-24236-5 | 0.0 ± 0.0 b | 0.0 ± 0.0 b | 0.1 ± 0.01 b | 6.4 ± 0.5 b | 48.4 ± 6.1 a | 26.9 ± 6.8 b | 78.1 ± 3.7 b |
| Non-Bt cotton | 0.4 ± 0.1 a | 17.4 ± 1.0 a | 0.3 ± 0.01 a | 16.6 ± 0.7 a | 28.9 ± 5.4 b | 66.4 ± 5.9 a | 94.5 ± 2.2 a |
| F1 hybryd (♀ Ha-Lab × ♂ Hz-Lab) | |||||||
| DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 | 0.0 ± 0.0 b | 0.0 ± 0.0 c | 0.4 ± 0.01 b | 2.4 ± 0.8 c | 49.2 ± 5.2 a | 46.1 ± 4.7 c | 94.5 ± 2.5 a |
| DAS-21023-5 × DAS-24236-5 | 0.12 ± 0.03 b | 9.6 ± 1.6 b | 0.11 ± 0.01 b | 6.3 ± 0.7 b | 32.8 ± 3.1 b | 60.2 ± 4.2 b | 93.0 ± 2.2 a |
| Non-Bt cotton | 0.43 ± 0.03 a | 23.2 ± 1.5 a | 0.41 ± 0.04 a | 21.8 ± 2.0 a | 16.4 ± 3.7 c | 82.8 ± 4.0 a | 99.2 ± 0.8 a |
aGrowth stage number according to Munger et al. [28]. Means ± SE followed by the same letter in each column and species are not significantly different as determined by the Tukey’s test at α = 0.05.
For F1 hybrids from ♀ Ha-Lab × ♂ Hz-Lab, a significantly higher mortality was observed for neonates at 5 d post-exposure on leaf-disks (75.8%–100%) and L2 larvae on floral buds (37.5%) of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 compared to DAS-21023-5 × DAS-24236-5 (39.8%–46.1% on leaf-disks and 11.7% on squares) and non-Bt cotton (≤12.5%) (Table 2).
Damage on leaf-disk and squares
Surviving neonates of H. armigera (Ha-Lab) at 5 d on leaf-disks of both cotton growth stages evaluated of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 and DAS-21023-5 × DAS-24236-5 caused significantly lower damage (≤5% of leaf consumption) than on non-Bt cotton (>52% of leaf consumption) (growth stage 15: F = 359.59; df (Num/Den) = 2/21; P < 0.0001 and growth stage 55: F = 666.26; df (Num/Den) = 2/21; P < 0.0001) (Table 3). At the same time, on DAS-21023-5 × DAS-24236-5 × SYN-IR102-7, a lower number of L2 larvae of H. armigera bored into squares (28.9%) and squares were less damaged (67.9%) than on non-Bt cotton (68.8% of larvae bored into squares with 99.2% of squares presenting any damage) (for both growth stages and each variable df (Num/Den) = 2/21; P < 0.0001) (Table 3). Still, the number of squares with surface feeding was significantly higher for DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 than DAS-21023-5 × DAS-24236-5, indicating that a low number of surviving larvae bored into squares of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 cotton, and remained feeding on the surface (F = 16.44; df (Num/Den) = 2/21; P = 0.0011) (Table 3).
The surviving H. zea neonates (Hz-Lab) on leaf-disks at growth stages 15 and 55 of DAS-21023-5 × DAS-24236-5 (only on this Bt cotton had survivors) had less than 7% of leaf consumption differing significantly from damage caused by larvae on non-Bt cotton (16.6% and 17.4% of leaf consumption) (growth stage 15: F = 357.29; df (Num/Den) = 2/21; P < 0.0001 and growth stage 55: F = 250.06; df (Num/Den) = 2/21; P < 0.0001) (Table 3). Similarly, on DAS-21023-5 × DAS-24236-5 cotton had a lower number of L2 larvae of H. zea bored into squares (26.9%) and less squares presented damage (78%) than on non-Bt cotton (66.4% of larvae bored into squares and 94.5% of squares presented damage) (for each variable df (Num/Den) = 2/21; P < 0.0001) (Table 3). In addition, squares with only surface feeding were higher on DAS-21023-5 × DAS-24236-5 cotton (48.4%) than on non-Bt cotton (28.9%), indicating that surviving L2 H. zea larvae had pronounced growth inhibition on this Bt cotton technology, affecting its capacity to bore into squares (F = 408.31; df (Num/Den) = 2/21; P < 0.0001) (Table 3).
When F1 hybrids from ♀ Ha-Lab × ♂ Hz-Lab were evaluated, significantly lower damage from surviving larvae on leaf-disks of both growth stages of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 and DAS-21023-5 × DAS-24236-5 cotton events (≤9.6% leaf consumption) than on non-Bt cotton (21.8%–23.3% of leaf consumption) were detected (growth stage 15: F = 87.25; df (Num/Den) = 2/21; P < 0.0001 and growth stage 55: F = 59.48; df (Num/Den) = 2/21; P < 0.0001) (Table 3). Similarly, DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 had significantly higher squares with surface feeding (49.2%) and a lower number of larvae bored into squares (46.1%) compared to DAS-21023-5 × DAS-24236-5 (32.8% and 60.2%, respectively) and non-Bt cotton (16.4% and 82.8%, respectively), demonstrating that in three-gene Bt cotton, fewer larvae were able to bore into squares (for each variable df (Num/Den) = 2/21; P < 0.0001) (Table 3). Regardless, no significant differences in the percentage of squares with any damage among Bt and non-Bt cotton treatments evaluated (93%–99.2%) were detected (F = 2.74; df (Num/Den) = 2/21; P = 0.0879) (Table 3).
Surviving neonates (on leaf-disk) and L2 larvae (on squares) of H. armigera (Ha-Lab) and F1 hybrids on DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 and DAS-21023-5 × DAS-24236-5 cotton technologies had significant lower larval weights (<0.90 mg/larva on leaf-disk and <3.7mg/larva on floral buds) than on non-Bt cotton (>4.6 mg/larva on leaf-disk and >12.6 mg/larva on floral buds) (for each pest, growth stage and plant structure df (Num/Den) = 2/21; P < 0.0001). These results indicate that surviving larvae on Bt cotton technologies had more than 80% (on leaf-disk) and 70% (on squares) growth inhibition relative to those fed on non-Bt cotton. For H. zea (Hz-Lab), the larval weights were not computed due to the limited number of insects surviving on both Bt cotton technologies.
B. Survivorship of Helicoverpa species on Bt cotton throughout the larval cycle
The survivorship of H. armigera (Ha-Lab) decreased with increased feeding time on leaves of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 and DAS-21023-5 × DAS-24236-5 cotton technologies (Table 4). For H. armigera, significant lower survivorship from 5 d onwards on DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 and DAS-21023-5 × DAS-24236-5 DAS-21023-5 × DAS-24236-5 than on non-Bt cotton, with <1% of larvae surviving until 30 d on leaves of both Bt cotton technologies. In the same period, on non-Bt cotton 52.2% of larvae developed into L5 and L6 stages (Table 4).
Table 4. Survivorship of H. armigera (Ha-Lab) and H. zea (Hz-Lab) on leaves of Bt and non-Bt cotton technologies throughout the larval cycle in laboratory trials.
| Feeding time (d) | DAS-21023-5 × DAS-24236-5 × SYN-IR102-7a | DAS-21023-5 × DAS-24236-5a | Non-Bt cottona | |||
|---|---|---|---|---|---|---|
| n | % survival (95% CI) | n | % survival (95% CI) | n | % survival (95% CI) | |
| Helicoverpa armigera (Ha-Lab) | ||||||
| 0 | 420 | 100.0 (99.2–100.0) a | 260 | 100.0 (98.6–100.0) a | 130 | 100.0 (97.3–100.0) a |
| 5 | 104 | 25.0 (21.0–29.3) b | 112 | 42.4 (36.6–48.4) b | 121 | 89.0 (82.8–93.4) b |
| 10 | 46 | 11.1 (8.3–14.4) c | 76 | 28.8 (23.6–34.5) c | 112 | 82.3 (75.2–88.0) b |
| 15 | 23 | 5.5 (3.7–8.1) d | 25 | 9.5 (6.4–13.5) d | 91 | 67.0 (58.7–74.4) c |
| 20 | 13 | 3.1 (1.8–5.2) de | 10 | 3.8 (2.0–6.7) de | 84 | 61.8 (53.4–69.6) c |
| 25 | 8 | 2.0 (0.9–3.7) de | 3 | 1.1 (0.3–3.2) ef | 75 | 55.1 (46.7–63.3) c |
| 30 | 3 | 0.7 (0.2–2.1) e | 0 | 0.0 (0.0–1.4) f | 71 | 52.2 (43.8–60.5) c |
| Helicoverpa zea (Hz-Lab) b | ||||||
| 0 | 420 | 100.0 (99.2–100.0) a | 260 | 100.0 (98.6–100.0) a | 130 | 100.0 (97.3–100.0) a |
| 5 | 0 | 0.0 (0.0–0.01) b | 151 | 58.1 (52.0–63.9) b | 111 | 85.4 (78.5–90.6) b |
aMeans (95% CI) followed by the same letter in each column for each species and cotton variety are not significantly different due to nonoverlap of 95% CIs.
bThe evaluations were performed only up to 5 d, as there was complete mortality of the Hz-Lab population on leaves of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 cotton.
For larvae of H. zea (Hz-Lab), no survivors after 5 d on leaves of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 were observed (Table 4). Nonetheless, surviving larvae on DAS-21023-5 × DAS-24236-5 presented 81% stunting (larvae that did not reach L2 stage) when compared to those developed on non-Bt cotton. At the same time point, 58.1 and 85.4% survival were verified on DAS-21023-5 × DAS-24236-5 and non-Bt cotton, respectively.
In contrast, the population of H. zea (Hz-field) collected during 2021/22 season surviving on Bt cotton expressing Cry1Ab, Cry2Ae and Vip3Aa proteins, at F1 generation, showed 37% survival on leaves of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 and 68% on DAS-21023-5 × DAS-24236-5 at 30 d, developing into L5 and L6 stages (Table 5). On non-Bt cotton, the survival rate was 84% at the same period.
Table 5. Survivorship of H. zea from Bahia (Hz-field) on leaves of Bt and non-Bt cotton technologies throughout the larval cycle in laboratory trials.
| Feeding time (d) | DAS-21023-5 × DAS-24236-5 × SYN-IR102-7a | DAS-21023-5 × DAS-24236-5a | Non-Bt cottona | |||
|---|---|---|---|---|---|---|
| n | % survival (95% CI) | n | % survival (95% CI) | n | % survival (95% CI) | |
| Helicoverpa zea (Hz-field from Bahia) | ||||||
| 0 | 100 | 100.0 (96.4–100.0) a | 100 | 100.0 (96.4–100.0) a | 100 | 100.0 (96.4–100.0) a |
| 5 | 100 | 100.0 (96.4–100.0) a | 100 | 100.0 (96.4–100.0) a | 100 | 100.0 (96.4–100.0) a |
| 10 | 89 | 89.0 (81.6–94.0) a | 100 | 100.0 (96.4–100.0) a | 100 | 100.0 (96.4–100.0) a |
| 15 | 79 | 79.0 (70.2–86.1) a | 100 | 100.0 (96.4–100.0) a | 100 | 100.0 (96.4–100.0) a |
| 20 | 73 | 73.0 (63.7–81.0) b | 98 | 98.0 (93.1–99.6) a | 100 | 100.0 (96.4–100.0) a |
| 25 | 62 | 62.0 (52.2–71.1) b | 84 | 84.0 (75.8–90.1) b | 96 | 96.0 (90.4–98.6) ab |
| 30 | 37 | 37.0 (28.0–46.8) c | 68 | 68.0 (58.4–76.5) b | 84 | 84.0 (75.8–90.1) b |
aMeans (95% CI) followed by the same letter in each column for each species and cotton variety are not significantly different due to nonoverlap of 95% CIs.
C. Field trials
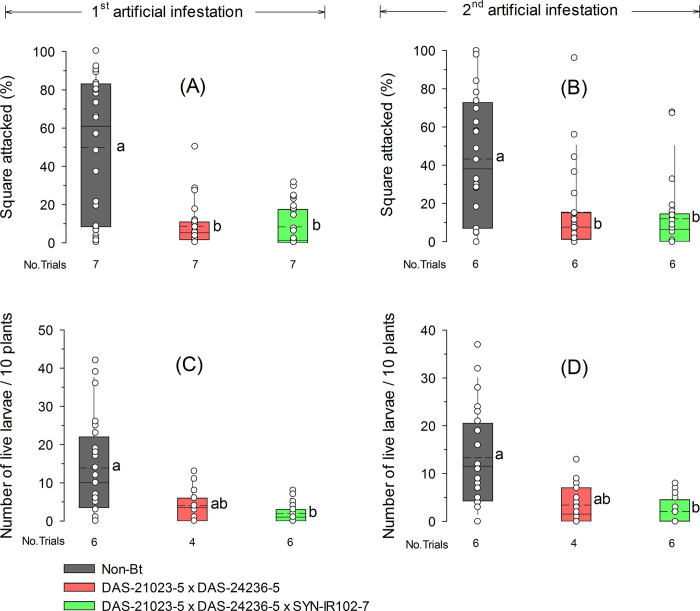
The Bt cotton technologies with the events DAS-21023-5 × DAS-24236-5 ×SYN-IR102-7 and DAS-21023-5 × DAS-24236-5 provided similar efficacy against H. armigera after artificial infestations under field conditions (Fig 1). Both Bt technologies significantly reduced the damaged squares compared to non-Bt cotton (1st infestation: F = 13.3; df (Num/Den) = 2/12; P < 0.001; 2nd infestation: F = 10.5; df (Num/Den) = 2/10; P = 0.004). In non-Bt cotton, the mean percentage of squares attacked by H. armigera was 49.9% after first infestation and 43.2% after second infestation, while in Bt cotton, squares attacked reached 8.6% and 15.5% for DAS-21023-5 × DAS-24236-5 and 8.4% and 11.9% for DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 in the first and second infestation (Fig 1A and 1B). The mean number of H. armigera live larvae found on DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 after the first (1.9 larvae/10 plants) and second infestation (2.0 larvae/10 plants) were significantly lower than on non-Bt cotton (13.9 and 13.3 larvae/10 plants, respectively) (1st infestation: F = 7.5; df (Num/Den) = 2/14.5; P = 0.005; 2nd infestation: F = 8.4; df (Num/Den) = 2/9.6; P = 0.007). However, both treatments did not differ from DAS-21023-5 × DAS-24236-5 (4.1 and 3.4 larvae/10 plants) in the two infestations (Fig 1C and 1D).
Fig 1.
Squares attacked (A—B) and live larvae (C—D) of Helicoverpa armigera in Bt and non-Bt cotton technologies after two field artificial infestations (L1 stage) during the cotton reproductive growth stage. The dashed and solid lines in boxplots show the mean and median across trials, respectively. Dot markers indicate values from individual field trial plots. Boxplots with different letters were significant different by a Tukey’s test (α = 0.05).
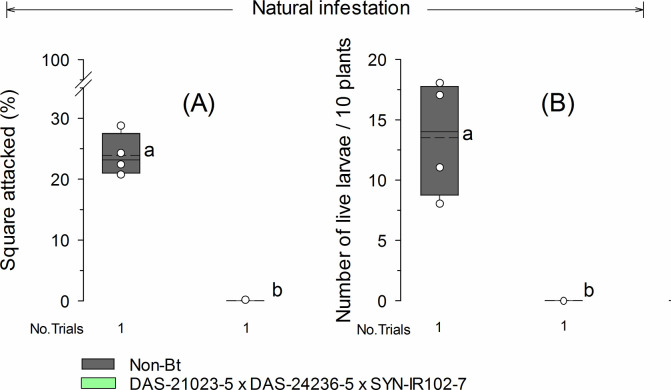
Under a natural infestation of Helicoverpa spp., Bt cotton DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 had significantly lower damage to squares (t = 47.2; df = 3; P < 0.001) and a lower number of live larvae (t = 8.5; df = 3; P = 0.003) compared to the non-Bt cotton (Fig 2). No square damage and live larvae of Helicoverpa spp. were observed on DAS-21023-5 × DAS-24236-5 × SYN-IR102-7, while in non-Bt cotton the mean percentage of square attack by natural infestation of Helicoverpa spp. was 23.8%, with 13.5 live larvae/10 plants (Fig 2A and 2B).
Fig 2.
Squares attacked (A) and live larvae (B) of Helicoverpa spp. in Bt cotton technology DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 and non-Bt cotton under field natural infestation during the cotton reproductive growth stage. The dashed and solid lines in boxplots show the mean and median, respectively. Dot markers indicate values from individual field trial plots. Boxplots with different letters were significant different by a paired t-test (α = 0.05).
Discussion
The susceptibility of Helicoverpa species to Cry1F and Vip3Aa19 in Brazil have not been fully characterized; susceptibility to another Vip protein (e.g. Vip3Aa20) was reported for H. armigera and H. zea and to Cry1Ac for H. armigera, indicating that H. armigera was more tolerant to Vip3Aa20 than H. zea, but had high susceptibility to Cy1Ac [35, 36]. To assess the performance of Cry1Ac, Cry1F and Vip3Aa19 Bt proteins expressed in DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 cotton technology against the Heliothine species complex in Brazil, we performed laboratory and field studies with H. armigera and H. zea and the hybrid progeny of the species. In most experiments presented here, insects from laboratory colonies that were annually augmented with field-collected insects were used.
The relative high mortality levels observed for Helicoverpa species from laboratory-reared colonies and hybrids on leaves of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 cotton indicate that Cry1Ac, Cry1F and Vip3Aa19 are effective in protecting cotton plants against damage from these target pests. This 3-gene Bt cotton technology also improved protection against square damage caused by Helicoverpa species and hybrids compared to 2-gene Bt cotton and should improve yield protection, since larvae may prefer to attack reproductive structures of cotton [37, 38]. According to Rios et al. [7], there is chance of interspecific crosses in Helicoverpa species, with interspecific crossing probably occurring when pest populations are at high densities, suggesting that populations of these pests should be maintained low levels in the field. Therefore, the effectiveness of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 cotton in controlling Helicoverpa species may reduce population levels across the landscape, favoring that the ‘species identities’ will be maintained under field conditions.
When comparing survival of H. zea colonies throughout the larval life cycle in two different studies (Tables 4 and 5), the field population of H. zea collected in Bt cotton expressing Cry1Ab, Cry2Ae and Vip3A in western Bahia, Brazil during the 2021/2022 season had lower mortality on DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 cotton technology than was observed for the laboratory H. zea population. During this season, it was verified a high occurrence of larvae of the genus Helicoverpa spp. in the western Bahia, Brazil causing considerable damage in reproductive structures of Bt cotton expressing Cry1Ab, Cry2Ae and Vip3Aa [39]. These larvae were later identified by PCR tests as H. zea and was the source of Hz-field population here tested. Our findings suggest that the Hz-field population collected from a Bt cotton field expressing Cry1, Cry2 and Vip3 in Bahia has lower susceptibility to Cry1 and Vip3Aa in comparison to the Hz-Lab. While not a definitive test for resistance to the Bt proteins (a known laboratory-susceptible H. zea population was not tested in the same study as the Bahia-collected Hz-field population), the current evidence (collection of survivors in a Bt field and the laboratory bioassay findings relative to other findings with a H. zea laboratory population) implies that reduced control is possible in the field. Diminished susceptibility to Bt proteins in H. zea and H. armigera has been previously reported in several states of U.S., India, Pakistan, and China [8–13, 40–42]. These observations reinforce the need for compliance with non-Bt refuge guidelines when planting Bt cotton technologies to retard the evolution of resistance in Helicoverpa species in Brazil.
Further investigation of Helicoverpa populations from the Bahia cotton-producing region are needed to fully understand the long-term implications for the durability of both Bt cotton and Bt maize against these species. For example, it will be important to confirm whether the Hz-field population is resistant to Cry1, Cry2 and/or Vip3Aa Bt proteins and to understand the performance of Hz-field on Bt maize hybrids. The ability of H. zea and H. armigera to produce hybrid offspring may also have implications for the development of resistance to Bt proteins in cotton-infesting populations of H. armigera that are not currently understood.
Currently, relevant exposure to Cry and Vip3Aa proteins by Helicoverpa species, which is part of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7, is ongoing in Brazil, because of the widespread planting of Bt soybean, Bt cotton and Bt maize expressing a single-mode-of-action Bt proteins. Although H. armigera was first identified in soybean in Brazil [1], this species prefers cotton as a host plant [4]. Despite the relative high level of adoption of Bt cotton technologies expressing Cry proteins (~90% of the total cotton area), these technologies continue to provide reasonable levels of control against Helicoverpa species. The abundance of Helicoverpa species has not been quantified in cotton in Brazil, but studies stated a low abundance of H. armigera in soybean fields [20]. The low abundance of this pest in soybean is a positive aspect for the IPM, since same Bt proteins are expressed in cotton and soybean against H. armigera, indicating that broad adoption of Bt plants reduced population levels of this species.
From pest management perspective, the cotton technology DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 beyond reasonable levels of mortality against H. armigera, also reduced the damage to squares. These results expand those previously reported by Marques et al. [25], showing this same Bt cotton technology is highly effective against other lepidopteran pests of cotton. It is noteworthy that, the non-Vip3Aa containing products usually did not provide full control of S. frugiperda, and growers implemented multiple insecticide sprays that probably contributed to controlling Helicoverpa spp. as well. As growers are moving to Vip3Aa-containing varieties, less insecticide sprays are needed for lepidopteran control, reducing the monitoring of lepidopterans and increasing potential for escapes of Helicoverpa spp. in the field [39]. Therefore, to maintain the benefits of DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 cotton for pest control in Brazil, where Helicoverpa species have numerous generations per year and are exposed to same mode-of-action Bt proteins in other crops, this GM cotton technology should be integrated with other control tactics, including the adoption of at least 20% of structured refuge (non-Bt cotton) for resistance management.
Acknowledgments
We gratefully acknowledge the efforts and assistance of our many colleagues at Corteva Agriscience.
Data Availability
All relevant data are within the paper.
Funding Statement
Oderlei Bernardi received funding from the National Council for Technological and Scientific Development (CNPq) (Process # 305464/2020-5) to perform the study. Corteva Agriscience provided salary to the authors who are employees of Corteva Agriscience. Corteva Agriscience authors participated in the study design which was submitted and approved by the Brazilian government authority. Also, authors participated in the decision to publish, and the preparation of the manuscript. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
References
- 1.Czepak C, Albernaz KC, Vivan LM, Guimarães HO, Carvalhais T. First reported occurrence of Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae) in Brazil. Pesqui Agropecu Trop. 2013; 43: 110–113. [Google Scholar]
- 2.Pinto FA, Mattos MVV, Silva FWS, Rocha SL, Elliot SL. The spread of Helicoverpa armigera (Lepidoptera: Noctuidae) and coexistence with Helicoverpa zea in Southeastern Brazil. Insects. 2017; 8: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonçalves RM, Mastrangelo TA, Rodrigues JCV, Paulo DF, Omoto C, Corrêa AS, et al. Invasion origin, rapid population expansion, and the lack of genetic structure of cotton bollworm (Helicoverpa armigera) in the Americas. Ecol Evol. 2019; 9: 7378–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dourado PM, Pantoja-Gomez LM, Horikoshi RJ, Carvalho RA, Omoto C, Corrêa AS, et al. Host plant use of Helicoverpa spp. (Lepidoptera: Noctuidae) in the Brazilian agricultural landscape. Pest Manag Sci. 2021; 77: 780–794. [DOI] [PubMed] [Google Scholar]
- 5.Anderson CJ, Oakeshott JG, Tay WT, Gordon KHJ, Zwick A, Walsh TK. Hybridization and gene flow in the mega-pest lineage of moth, Helicoverpa. Proc Natl Acad Sci USA. 2018; 115: 5034–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cordeiro EMG, Pantoja-Gomez LM, Paiva JB, Nascimento ARB, Omoto C, Michel AP, et al. Hybridization and introgression between Helicoverpa armigera and H. zea: an adaptational bridge. BMC Evol Biol. 2020; 20: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rios DAM, Specht A, Roque-Specht VF, Sosa-Gómez DR, Fochezato J, Malaquias JV, et al. Helicoverpa armigera and Helicoverpa zea hybridization: constraints, heterosis, and implications for pest management. Pest Manag Sci. 2021; 78: 955–964. [DOI] [PubMed] [Google Scholar]
- 8.Tabashnik BE. ABCs of insect resistance to Bt. PLoS Genet. 2015; 11: e1005646. doi: 10.1371/journal.pgen.1005646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dively GP, Venugopal PD, Finkenbinder C. Field-evolved resistance in corn earworm to Cry proteins expressed by transgenic sweet corn. PLoS One. 2016; 11: e0169115. doi: 10.1371/journal.pone.0169115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reisig DD, Huseth AS, Bacheler JS, Aghaee MA, Braswell L, Burrack HJ, et al. Long-term empirical and observational evidence of practical Helicoverpa zea resistance to cotton with pyramided Bt toxins. J Econ Entomol. 2018; 111: 1824–1833. [DOI] [PubMed] [Google Scholar]
- 11.Yang F, Kerns DL, Little N, Brown SA, Stewart SD, Catchotd AL, et al. Practical resistance to Cry toxins and efficacy of Vip3Aa in Bt cotton against Helicoverpa zea. Pest Manag Sci. 2022; 78: 5234–5242. [DOI] [PubMed] [Google Scholar]
- 12.Dandan Z, Yutao X, Wenbo C, Yanhui L, Kongming W. Field monitoring of Helicoverpa armigera (Lepidoptera: Noctuidae) Cry1Ac insecticidal protein resistance in China (2005–2017). Pest Manag Sci. 2019; 75: 753–759. [DOI] [PubMed] [Google Scholar]
- 13.Singh TVK, Kukanur VS, Supriya GB. Frequency of resistance alleles to Cry1Ac toxin from cotton bollworm, Helicoverpa armigera (Hübner) collected from Bt-cotton growing areas of Telangana state of India. J Invertebr Pathol. 2021; 183: 107559. [DOI] [PubMed] [Google Scholar]
- 14.Leite NA, Alves-Pereira A, Corrêa AS, Zucchi MI, Omoto C. Demographics and genetic variability of the New World bollworm (Helicoverpa zea) and the Old World bollworm (Helicoverpa armigera) in Brazil. PLoS One. 2014; 9: e113286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bentivenha JPF, Paula-Moraes SV, Baldin ELL, Specht A, Silva IF, Hunt TE. Battle in the New World: Helicoverpa armigera versus Helicoverpa zea (Lepidoptera: Noctuidae). PLoS One. 2016; 11: e0167182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanco CA, Chiaravalle C, Dalla-Rizza M, Farias JR, García-Degano MF, Gataminza G, et al. Current situation of pests targeted by Bt crops in Latin America. Curr Opin Insect Sci. 2016; 15: 131–138. doi: 10.1016/j.cois.2016.04.012 [DOI] [PubMed] [Google Scholar]
- 17.Silva FR, Trujillo D, Bernardi O, Rodrigues JCV, Bailey WD, Gilligan TM. et al. Comparative toxicity of Helicoverpa armigera and Helicoverpa zea (Lepidoptera: Noctuidae) to selected insecticides. Insects. 2020; 11: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y, Wu K, Jiang Y, Guo Y, Desneux N. Widespread adoption of Bt cotton and insecticide decrease promotes biocontrol services. Nature. 2012; 487: 362–365. doi: 10.1038/nature11153 [DOI] [PubMed] [Google Scholar]
- 19.Brookes G, Barfoot P. Farm income and production impacts of using GM crop technology 1996–2015. GM Crops & Food. 2018; 8: 156–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horikoshi RJ, Dourado PM, Berger GU, Fernandes DS, Omoto C, Wilse A, et al. Large-scale assessment of lepidopteran soybean pests and efficacy of Cry1Ac soybean in Brazil. Sci Rep. 2021; 11: 15956. doi: 10.1038/s41598-021-95483-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Céleres. 2019. Informativo de Biotecnologia Céleres. IB 19.01. Novembro de 2019. Available from: http://www.celeres.com.br/wp-content/uploads/2019/11/BoletimBiotecnologiaC%C3%A9leres_Novembro2019-2.pdf [Google Scholar]
- 22.Horikoshi RJ, Bernardi D, Bernardi O, Malaquias JB, Okuma DM, Miraldo LL. et al. Effective dominance of resistance of Spodoptera frugiperda to Bt maize and cotton varieties: implications for resistance management. Sci Rep. 2016; 6: 34864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brévault T, Heuberger S, Zhang M, Ellers-Kirk C, Ni X, Masson L, et al. Potential shortfall of pyramided transgenic cotton for insect resistance management. Proc Natl Acad Sci USA. 2013; 110: 5806–5811. doi: 10.1073/pnas.1216719110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrière Y, Crickmore N, Tabashnik BE. Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat Biotechnol. 2015; 33: 161–168. doi: 10.1038/nbt.3099 [DOI] [PubMed] [Google Scholar]
- 25.Marques LH, Lepping M, Castro BA, Santos AC, Rossetto J, Nunes MZ, et al. Field efficacy of Bt cotton containing events DAS-21023-5 × DAS-24236-5 × SYN-IR102-7 against lepidopteran pests and impact on the non-target arthropod community in Brazil. PLoS One. 2021; 16: e0251134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greene GL, Leppla NC, Dickerson WA. Velvetbean caterpillar: a rearing procedure and artificial medium. J Econ Entomol. 1976; 69: 487–488. [Google Scholar]
- 27.Perera OP, Allen KC, Jain D, Purcell M, Little NS, Luttrell RG. Rapid identification of Helicoverpa armigera and Helicoverpa zea (Lepidoptera: Noctuidae) using ribosomal RNA internal transcribed spacer 1. J Insect Sci. 2015; 15: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munger P, Bleiholder H, Hack H, Hess M, Stauß R, Van den Boom T. et al. Phenological growth stages of the cotton plant (Gossypium hirsutum L.) codification and description according to the BBCH scale. J Agron Crop Sci. 2018; 180: 143–149. [Google Scholar]
- 29.Dorai-Raj S. binom: Binomial confidence intervals for several parameterizations. R package version. 2009; 1.0–5. Available from: http://CRAN.R-project.org/package_binom [Google Scholar]
- 30.R Development Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria; 2018. [Google Scholar]
- 31.Savin NE, Robertson JL, Russell RM. A critical evaluation of bioassay in insecticide research: likelihood ratio tests of dose-mortality regression. Bull Entomol Soc Am. 1977; 23: 257–266. [Google Scholar]
- 32.Robertson JL, Russel RM, Preisler HK, Savin NE. Bioassays with arthropods, 2nd ed. CRC Press, Boca Raton, FL; 2007. [Google Scholar]
- 33.SAS Institute. Statistical analysis system: getting started with the SAS learning. SAS Institute, Cary, NC; 2008. [Google Scholar]
- 34.Pimentel-Gomes F. Curso de estatística experimental. FEALQ, Piracicaba; 2009. [Google Scholar]
- 35.Dourado PM, Bacalhau FB, Amado D, Carvalho RA, Martinelli S, Head GP, et al. High susceptibility to Cry1Ac and low resistance allele frequency reduce the risk of resistance of Helicoverpa armigera to Bt soybean in Brazil. PLoS One 2016; 11: e0161388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leite NA, Pereira RM, Durigan MR, Amado D, Fatoretto J, Medeiros FCL, et al. Susceptibility of Brazilian populations of Helicoverpa armigera and Helicoverpa zea (Lepidoptera: Noctuidae) to Vip3Aa20. J Econ Entomol. 2018; 111: 399–404. [DOI] [PubMed] [Google Scholar]
- 37.Kumar S, Saini RK. Feeding preference and damage potential of Helicoverpa armigera (Hübner) on different promising cotton genotypes/hybrid. J Agr Sci Tech. 2008; 10: 411–420. [Google Scholar]
- 38.Sivasupramaniam S, Moar WJ, Ruschke LG, Osborn JA, Jiang C, Sebaugh JL, et al. 2008. Toxicity and characterization of cotton expressing Bacillus thuringiensis Cry1Ac and Cry2Ab2 proteins for control of lepidopteran pests. J Econ Entomol. 2008; 101: 546–554. [DOI] [PubMed] [Google Scholar]
- 39.Instituto Mato-Grossense do Algodão (IMAmt). Helicoverpa spp. alerta de monitoramento em lavouras de algodão Vip. 2022; Circular Técnica n.° 52. Available from: https://imamt.org.br/wp-content/uploads/2022/07/circular_tecnica_edicao52.pdf [Google Scholar]
- 40.Ahmad S, Cheema HMN, Khan AA, Khan RSA, Ahmad JN. Resistance status of Helicoverpa armigera against Bt cotton in Pakistan. Transgenic Res. 2019; 28: 199–212. [DOI] [PubMed] [Google Scholar]
- 41.Yang F, Gonzáles JCS, Sword GA, Kerns DL. Genetic basis of resistance to the Vip3Aa Bt protein in Helicoverpa zea. Pest Manag Sci. 2020; 77: 1530–1535. [DOI] [PubMed] [Google Scholar]
- 42.Santiago-González JC, Kerns DL, Yang F. Resistance allele frequency of Helicoverpa zea to Vip3Aa Bacillus thuringiensis protein in the Southeastern U.S. Insects. 2023; 14: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.