Abstract
The terminal DNA restriction fragments (PstI-D and -B) of Pseudomonas aeruginosa bacteriophage D3 were ligated, cloned, and sequenced. Of the nine open reading frames in this 8.3-kb fragment, four were identified as encoding large-subunit terminase, portal, ClpP protease, and major head proteins. The portal and capsid proteins showed significant homology with proteins of the lambdoid coliphage HK97. Phage D3 was purified by CsCl equilibrium gradient centrifugation (ρ = 1.533 g/ml), and sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed six proteins with molecular masses of 186, 91, 79, 70, 45, and 32 kDa. The pattern was unusual, since a major band corresponding to the expected head protein (43 kDa) was missing and a significant amount of the protein was retained in the stacking gel. The amino terminus of the 186-kDa protein was sequenced, revealing that the D3 head is composed of cross-linked 31-kDa protein subunits, resulting from the proteolysis of the 43-kDa precursor. This is identical to the situation observed with coliphage HK97.
While a significant percentage of all bacteriophages which have been isolated are specific for pseudomonads (1), relatively few have been characterized to the extent of enterobacterial or Bacillus phages or mycobacteriophages. Indeed, the genomes of only two Pseudomonas viruses, cytotoxin-converting phage φCTX (46) and the filamentous bacteriophage Pf3 (41), have been completely sequenced.
Temperate bacteriophage λ is the prototype of a family of phylogenetically related viruses, the lambdoid phages, which show a conserved arrangement of regulatory elements and are able, in certain cases, to form viable recombinants with one another (13). Analysis of available sequence data has indicated that each lambdoid genome is a patchwork of gene segments (cassettes), suggesting extensive recombination (4, 5, 31, 52). Apart from their similarities, lambdoid phages differ in the genetic determinants for repression, integration, packaging, and cell surface recognition, as well as genome size and the nature of phage DNA termini.
We have proposed that Pseudomonas aeruginosa phage D3 is a member of the lambdoid group (25, 26). Electron microscopic studies have shown that D3 is a member of the B1 (isomeric head) morphogroup of the family Siphoviridae. Coliphage λ (head, 60 nm; tail, 150 nm [2]) and Pseudomonas phage D3 (head, 70 nm; tail, 140 nm [38]) are similar in size. The double-stranded DNA (dsDNA) genomes are both linear, with sizes of 48.5 (λ) and 56.4 (D3) kb. Moreover, the DNAs of both phages possess cohesive ends (27), which have salient roles in both the replicative and lysogenic pathways of viral development. With D3 and λ, integration involves genomic circularization and insertion into the chromosome of the host by a Campbell-type model. The recA gene product plays a significant role in the induction of these bacteriophages. Faulty excision generates defective particles capable of transducing markers adjacent to their respective integration (att) sites, gal and bio in the case of λ and met in the case of D3 (13, 15). The organization of the immunity region of P. aeruginosa D3 is highly homologous with that of coliphage λ (25), and with both phages the lysogenic state is maintained by single repressors with similar molecular weights (D3, 24,558; λ, 26,228). Even though these two proteins show poor overall sequence homology, closer examination indicates conservation of functionally important amino acid residues (26). Moreover, like the situation with λ cI transcription, the D3 repressor mRNA originating from PRM lacks the typical prokaryotic ribosome-binding sites (Shine-Dalgarno box) (26). Lastly, analysis of the downstream sequence has revealed the presence of an open reading frame (ORF) encoding a polypeptide of 102 amino acids which has sequence homology to the cII gene product of λ. The cII gene, which is a central effector involved in the lysis-lysogeny decision in λ phage, contains within itself a consensus λ CII-binding site (TTGCN6TTGC) (33, 34, 37a).
Despite their remarkable similarities, D3 and λ phages differ in some notable ways, including their host ranges. The cellular receptor for D3 is the O side chain polysaccharide of lipopolysaccharide, while that of coliphage λ is the LamB protein (39). The genome of coliphage λ exhibits segmented base composition, while that of D3 does not (37b). Furthermore, the ends of the D3 phage genome have 3′-extended termini while lambda possesses 5′-extended termini. In the case of λ, downstream of the cI gene there is a gene (rex) which is transcribed from PRM, whereas sequence analysis by Farinha and colleagues (25) failed to identify any other ORF between D3 cI and the OL-PL complex. It is worth mentioning that Salmonella phage P22, which has a considerable amount of genomic similarity with coliphage λ, also lacks a gene in this region.
So far, studies of P. aeruginosa phage D3 have indicated that D3 is phylogenetically related to the lambdoid phages in its genome organization and gene function. The aim of the research described in this paper has been to add further evidence for this relationship through analysis of the genes involved in DNA packaging and head morphogenesis.
MATERIALS AND METHODS
Bacteria and bacteriophage.
The prototrophic strain of P. aeruginosa, PAO 1, was used for the preparation and titration of the phage lysates and was obtained from B. W. Holloway (Monash University, Melbourne, Australia). Escherichia coli DH5α (Gibco/BRL Life Technologies, Burlington, Canada), genotype F− φ80dlacZ ΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA, was used for the recombinant-DNA experiments.
Bacteriophage D3c (D3c1::IS222), a clear plaque mutant of D3 (28), was obtained from R. E. W. Hancock (University of British Columbia, Vancouver, Canada), and its DNA was isolated by the protocols described for coliphage lambda DNA extraction by Sambrook and colleagues (51).
Media.
Bacteria were grown in Luria broth (LB; Difco Laboratories, Detroit, Mich.) or on LBA plates (LB plus 1.5% [wt/vol] agar) supplemented with 1 mM CaCl2. For phage titrations (3), 3-ml overlays were prepared with LB containing 0.6% (wt/vol) agar and 1 mM CaCl2. For recombinant DNA procedures, LBA was supplemented with ampicillin (100 μg/ml; Sigma Chemical Co., St. Louis, Mo.) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg/ml; VectorBiosystems, Toronto, Canada).
Recombinant-DNA techniques. (i) Cloning.
The cohesive ends of bacteriophage D3 genomic DNA were ligated with bacteriophage T4 DNA ligase (Gibco/BRL Life Technologies) and subsequently digested with PstI (Gibco/BRL Life Technologies). The digested DNA was electrophoresed through a 0.6% agarose gel with 0.5× TBE buffer (0.045 M Tris-borate, 0.001 M EDTA, pH 8.0) and 0.5 μg of ethidium bromide/ml, and the 8.3-kb PstI-DB fragment was excised and recovered with Prep-A-Gene [Bio-Rad Laboratories (Canada) Ltd., Mississauga, Canada]. The recovered fragment was ligated into PstI-digested pGEM-3Zf(+) (Promega, Madison, Wis.) and electrotransformed into E. coli DH5α cells with a Bio-Rad gene pulser. The cells were recovered in SOC medium (51), and after 1 h of incubation at 37°C, aliquots were plated onto LBA plates containing ampicillin and X-Gal.
(ii) Plasmid isolation.
Recombinant clones were grown overnight in Terrific Broth (Difco) containing ampicillin (100 μg/ml), and plasmid DNA was isolated by a modification of the alkaline lysis technique (51). The plasmid DNA obtained by this procedure was extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol]), precipitated with ethanol, and washed twice with 70% ethanol prior to analysis.
(iii) DNA sequencing, assembly, and analysis.
Fluorescent-dye dideoxy chain-terminating DNA sequencing was carried out at the Guelph Molecular Supercenter (Guelph, Canada) with an Applied Biosystems automated sequencer. The sequence data were assembled with Seqman II software (DNASTAR Inc., Madison, Wis.) and analyzed with DNAMAN (Lynnon BioSoft, Vaudreuil, Canada) and a variety of on-line tools. WebGeneMark.HMM (http://genemark.biology.gatech.edu/GeneMark/whmm.cgigorf/gorf.html) (42) and ORF finder at the National Center for Biotechnology Information (46a) were used to identify potential ORFs. The protein products were checked against the nonredundant GenBank protein database with gapped BLAST version 2.0P (6, 7) for homologs. Protein molecular weights and isoelectric points were determined at ExPASy (http://www.expasy.ch/tools/protparam.html). Protein motifs were examined with PROSITE (9, 10) (http://www.expasy.ch/tools/scnpsit1.html), fingerPRINTScan (http://bioinf.mcc.ac.uk/cgi-bin/dbbrowser/fingerPRINTScan/pval/FPScan.cgi), and the MEME (Multiple Motif Elicitation) and MAST (Motif Alignment and Search Tool) programs (8) (http://www.sdsc.edu/MEME/meme/website/html). Protein alignments were generated with Clustal W at the European Molecular Biology Laboratory-European Bioinformatics Institute (wysiwyg://30/http://www2.ebi.ac.uk/clustalw/) or ALIGN at Genestream-Institut de Génétique Humaine (http://www2.igh.cnrs.fr/bin/align-guess.cgi). Phylogenetic analysis was also carried out with the Windows 95 version of TreeCon (56, 57) obtained from Yves Van de Peer (University of Antwerp, Antwerp, Belgium).
Purification of phage D3.
P. aeruginosa was grown overnight in 25 ml of LB and subcultured in 2 liters of LB at 37°C. While the culture was incubated with shaking, growth was monitored spectrophotometrically. When the optical density of the culture at 650 nm reached 0.25, 5 × 1010 PFU of phage D3 was added. After approximately 6 h, lysis was complete and the phage lysate was treated with 1 μg of DNase I (Boehringer Mannheim Canada, Laval, Canada)/ml for 30 min at 37°C and then centrifuged at 10,000 × g for 15 min. The clear supernatant was retained. Solid NaCl was added to 1 M, and subsequently polyethylene glycol 8000 (BDH Chemicals, Toronto, Canada) was added to a final concentration of 10% (wt/vol) (58). The mixture was cooled on ice and centrifuged at 10,000 × g for 10 min at 4°C. The precipitated phage particles were suspended in 32 ml of buffer (NaCl, 0.58% [wt/vol]; MgSO4 · 7H2O, 0.2% [wt/vol]; 10 mM Tris HCl [pH 7.5]). This suspension was centrifuged at 10,000 × g for 15 min at 4°C, and the pellet was resuspended in fresh buffer containing 5% (vol/vol) Triton X-100 to aid the solubilization of cell debris. The mixture was centrifuged at 10,000 × g for 15 min, and the supernatant was retained. For further purification, the phage was subjected to CsCl step and linear gradient centrifugations (51). For analytical equilibrium gradient centrifugation, the equation of Bruner and Vinograd (12) was used to correlate the refractive index of the CsCl solution and its density.
Electron microscopy.
CsCl-purified phage was dialyzed overnight at 4°C against several changes of 1% (wt/vol) ammonium acetate. A small drop of the dialyzed D3 suspension was placed on a carbon-coated electron microscope grid, and a drop of 1% (wt/vol) phosphotungstic acid (PTA) solution, pH 7.2, was added to it. Excess PTA was removed with the edge of a filter paper, and the remainder was left to air dry. A Hitachi H-7000 electron microscope with an accelerating voltage of 75 kV was used to examine this phage preparation.
Analytical techniques. (i) Protein analysis.
The bicinchoninic protein assay reagent kit (Pierce, Rockford, Ill.) was used to measure the amount of protein in the purified phage preparation.
(ii) Denaturing PAGE (SDS-PAGE).
To prepare the proteins for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), a quantity of phage was mixed with sample buffer (0.0625 M Tris [pH 6.8], 1% [wt/vol] SDS, 10% [vol/vol] glycerol, 2% [vol/vol] 2-mercaptoethanol, 0.001% [wt/vol] bromophenol blue) and heated for 5 min in a boiling-water bath. Bio-Rad high- and low-molecular-weight marker proteins were similarly prepared. To solve the viscosity problem resulting from the release of phage DNA, 0.5 to 1 μl of DNase I (1 mg/ml in 10 mM Tris–10 mM MgCl2–20% [vol/vol] glycerol) was added to 50 μl of phage lysate and incubated at room temperature for 10 min. For most of the analyses, a Bio-Rad Mini-PROTEAN II apparatus was employed with 10 or 12.5% resolving gels and a 4.5% stacking gel and the buffer systems described by Laemmli (40). The gels were stained with Coomassie brilliant blue G250 in perchloric acid (24) or the Bio-Rad Silver Stain or Silver Stain Plus kit.
(iii) Protein sequencing.
A 0.75-mm-thick polyacrylamide gel (10%) was poured, and to ensure complete polymerization, it was stored at 4°C for 24 h. Laemmli running buffer (40) was supplemented with 1 mM thioglycolic acid (Sigma Chemical Co.). The gels were run at 50 to 70 V until the bromophenol blue ran off the bottom of the gel to achieve better separation of bands. The gels were stored in transfer buffer (25 mM Tris [pH 8.3], 192 mM glycine) overnight at 4°C. The proteins were transferred to polyvinylidene difluoride (PVDF) membrane (Boehringer Mannheim) by the protocols provided with the Bio-Rad mini Trans-blot electrophoretic transfer cell. The PVDF membrane was subsequently washed with water and then briefly stained with Coomassie brilliant blue R250 (0.1% Coomassie brilliant blue R250, 40% [vol/vol] methanol, 1% [vol/vol] acetic acid). The stained membrane was then destained with 40% (vol/vol) methanol–5% (vol/vol) acetic acid until bands were visible and then extensively rinsed with distilled water. The excised pieces were submitted for automated sequential Edman degradation sequencing on an Applied Biosystems Procise sequencer at the National Research Council (Ottawa, Canada).
Nucleotide sequence accession number.
The nucleotide sequence described in this paper was deposited in the GenBank database under accession no. AF147978.
RESULTS
Cloning and sequencing.
The cloning strategy for the head morphogenesis genes of phage D3 was based upon the hypothesis that these genes would be in the same relative positions as they are in coliphage lambda. Since no useful restriction sites were available (27), an alternative approach was taken. The genome of D3 contains cohesive ends with a strong inclination to self-ligation (54). The genomic DNA was first incubated with T4 ligase, and the resulting covalently closed circular DNA was digested with PstI. Due to the presence of three PstI sites on the linear genome, digestion results in four fragments, but with the circularized DNA three fragments were found upon gel electrophoresis: A, C, and DB fragments. The last was cloned and sequenced.
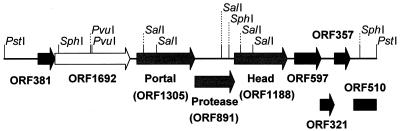
The base composition of this 8,383-bp fragment is 59.4 mol% G+C, which is similar to the mol% G+C content of the whole genome (58%) determined by melting-temperature analysis (37b). The latter value is in conflict with the data of Miller and colleagues (45), who claimed that the overall G+C content of D3 was only 50.4%. A restriction map of the PstI-DB fragment is presented in Fig. 1. No sites were identified for the following common restriction nucleases: EcoRI, BamHI, SmaI, and HindIII. The absence of a HindIII site in this fragment suggests that the published restriction map (27) is imprecise.
FIG. 1.
Physical map of the 8.3-kb D3 PstI-DB fragment showing restriction sites and the locations of the ORFs. In the case of ORF510, only the 5′ end of the gene was identified.
ORFs.
With ORF finder and GeneMark.HMM, nine potential ORFs were identified, and these are also diagrammed in Fig. 1. In all but one case (ORF1305) the initiation codon was AUG, and in each case the ORF was preceded, at an appropriate distance, by a sequence related to a subset of the Shine-Dalgarno motif (TAAGGAGGT [50]). BLAST 2.0 P analysis revealed that four of the putative proteins showed significant homology with proteins in the GenBank database, including enzymes and structural proteins involved in DNA packaging and morphogenesis. The latter are discussed in greater depth below:
(i) ORF1692.
ORF1692 would encode a 63.3-kDa protein with a calculated isoelectric point of 5.2. A BLAST P search showed the greatest homology with a 50.9-kDa hypothetical protein, the product of the E. coli ymfN gene (GenBank accession no. P75978; probability with BLAST, 3 × 10−77). The results, a portion of which are reproduced in Fig. 2, show strong sequence similarity only between D3 residues 200 and 387. Using ALIGN, we found that the overall sequence identity is 31.9%. Other proteins which scored high in the BLAST search included a hypothetical protein from Rhodobacter capsulatus (GenBank accession no. 128374; 2 × 10−69), orf5 of Lactobacillus casei phage A2 (GenBank accession no. X97563; 3 × 10−35); orf2 of the temperate Staphylococcus aureus phage φPVL (GenBank accession no. AB009866; 1 × 10−28), and gp33 of the Streptomyces phage φC31 (GenBank accession no. AJ006589; 5 × 10−16). In the last cases, homology extended for the full length of the D3 protein sequence. Where homology exists with a phage protein, the latter has been demonstrated or proposed to be the terminase large subunit.
FIG. 2.
Comparison of the sequences of D3 ORF1692 and hypothetical E. coli protein YFMN_ECOLI (GenBank accession no. P75978). The homology between these proteins is particularly evident in the region shown, and the results suggest that the latter protein is truncated at its 5′ end. Identical residues are indicated by colons, while conservative changes are indicated by periods.
(ii) ORF1305 (portal protein).
ORF1305, which begins with a GUG start codon, lies downstream of ORF1692 and has a molecular mass of 48.0 kDa and a calculated isoelectric point of 9.0. A BLAST P search showed significant sequence homology with the portal protein of coliphage HK97, which is a lambdoid phage (GenBank accession no. P49859; probability, 2 × 10−56). ALIGN indicated an overall 31% sequence identity. Two other proteins showed significant homology, gp34 from φC31 (GenBank accession no. AJ006589; 5 × 10−35) and the portal protein of φPVL (GenBank accession no. AB009866; 10−15). Portal proteins are minor structural proteins with molecular masses of 42 to 60 kDa and variable isoelectric points (λ, 5.7; HK97, 7.7; φPVL, 5.9), which appears to be universal among dsDNA phages. Each portal protein makes a 12- to 13-subunit annular structure (11, 19, 29, 35, 49) at one corner of the icosahedral shell, where it serves as the entrance and exit port for the DNA, the site for head assembly, and the attachment site for the tail.
(iii) ORF891 (protease).
The protein predicted from the DNA sequence of ORF891 has a molecular weight of 31,900 and an isoelectric point of 4.7. Following a comparison between the predicted protein and all sequences in the GenBank sequence database, a low-level sequence similarity to bacterial proteases, including Streptomyces coelicolor ClpP protease (GenBank accession no. 383445; 2 × 10−10) was noted. While PROSITE failed to reveal conserved motifs within this protein, FingerPRINTScan did. The latter program identified two conserved motifs, which strongly suggested that this ORF encodes an ATP-dependent Clp protease. The ClpP family of proteinases is usually composed of two subunits: the ClpP subunit, which contains both serine and histidine residues as active sites, is responsible for proteolytic activity, while the regulatory subunit, ClpA, has ATPase activity (16, 43). Given that the portal (ORF1305) and the major capsid (ORF1187) proteins show considerable homology with analogous proteins in coliphage HK97, it is surprising that the intervening genes, encoding proteases, show no homology.
(iv) ORF1187 (major head protein).
ORF1187 would encode a predicted protein with a molecular mass of 42.9 kDa and an isoelectric point of 4.8. A BLAST P search indicated sequence homology with only the major head protein (gp5) of coliphage HK97 (GenBank accession no. P49861; 10−69). An alignment between the predicted D3 major head protein and that of HK97 (Fig. 3) shows 43% overall identity, which is particularly evident in the latter two-thirds of the sequence.
FIG. 3.
A comparison (ALIGN) of the sequences of the major head proteins of D3 and HK97 showing the protease cleavage points (▴▾) and the residues involved in cross-linking (✖). Identical residues are indicated by colons, while conservative changes are designated by periods.
Analysis of phage and its proteins.
In an effort to correlate phage structural proteins with proteins identified on the basis of DNA sequence analysis, phage D3 was purified by polyethylene glycol precipitation and two CsCl gradient centrifugations. The purified D3 particles exhibited a density of 1.5244 g/ml in CsCl, a value significantly greater than that (1.504 g/ml) reported for coliphage λ (48).
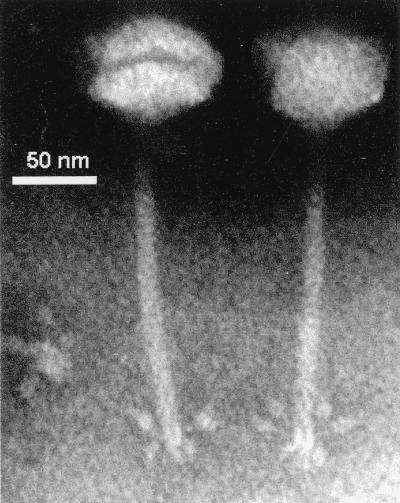
Electron microscopy.
Electron-microscopic studies showed that bacteriophage D3 has an icosahedral head, a long flexible striated tail, and six visible tail fibers with terminal knobs (Fig. 4). The head diameter was 55 ± 3 nm, and the tail was 113 ± 21.5 nm long and 7.0 ± 0.80 nm wide. While the overall morphology is identical to that in the electron micrographs published by Miller and coworkers (45), they noted a head diameter of 70 nm and a tail length of 150 nm. The reason for this discrepancy is not known.
FIG. 4.
Electron micrograph of D3 negatively stained with PTA. The tail fibers with terminal knobs are clearly seen.
Protein analysis.
D3 virions were analyzed by denaturing PAGE, and the protein was revealed by Coomassie blue and silver staining (Fig. 5). Five clearly visible bands with molecular weights of 186,000, 91,000, 79,000, 70,000, and 45,000 were observed. In addition, a large amount of material did not enter the gel matrix and remained trapped in the wells. At the bottom of the gel, a 32-kDa smear was noted. Because the DNase I ran at approximately this Mr, no inference could be made concerning phage proteins which might also be present. Based on DNA sequence analysis, a major band corresponding to the 43-kDa capsid protein was expected on the gel, but it was missing. Instead, there was a diffuse high-molecular-weight band with an Mr of 186,000. In order to identify the molecular nature of this material, the protein was resolved by SDS-PAGE and electroblotted onto a PVDF membrane and was submitted for automated sequential Edman degradation. The sequence of this protein was Ala-Ile-Thr-Ser-Ile-Glu-Gly-Ser-Gly-Gly. This sequence matches the predicted sequence of the major head protein starting with A-112, suggesting that the actual head protein is cleaved from a precursor protein and has lost 111 amino acids from its amino terminus (Fig. 3). Based upon sequence analysis, the portal protein with a calculated mass of 48 kDa may be represented by the 45-kDa band in the protein gel.
FIG. 5.
SDS-PAGE analysis (lanes A and C) of the D3 structural proteins showing the 186-kDa material (∗∗) as well as higher-molecular-weight material trapped in the wells (∗). Lane B contains DNase I, while lanes D and E contain the Bio-Rad low- and high-molecular-weight markers.
DISCUSSION
We have successfully cloned the region of the P. aeruginosa D3 genome responsible for packaging and head morphogenesis, and in the following paragraphs we will discuss the relationship between these genes and those of other phages, particularly coliphage HK97.
Major head protein.
Capsid assembly has been investigated for many dsDNA viruses, and the common theme includes progressive transitions in the capsid subunits as assembly proceeds and the use of accessory proteins to aid assembly (2, 37). Bacteriophages such as phages P22, T7, ΨM2, c2, AR1, CP1, φCTX, P2, and 186 follow a very common mechanism of head assembly by employing an accessory factor known as scaffolding protein. The latter interacts with the coat subunits to cause correct assembly into the shell of the prohead. In some cases, such as that of P22, the scaffolding protein leaves the head during DNA packaging and may be reused in subsequent rounds of capsid assembly (23, 47), while in others, including that of T4, it is proteolytically removed from the prohead (44). Despite similarities among the phages in the involvement of scaffolding proteins in capsid assembly, they show considerable evolutionary distance (23).
The lambdoid coliphage HK97 has been shown to possess a unique mechanism of head assembly, since its capsids are assembled from head protein pentamers and hexamers without a scaffold (22). Phage HK97 head assembly involves an extended series of transitions (17, 21, 32). In head assembly, prohead I is the first shell structure and is composed of 420 copies of the 42-kDa major head protein and 12 copies of the portal protein. In the first transition, a 25-kDa protease removes 102 amino acids from the amino terminus of the capsid proteins, giving rise to prohead II. Prohead II undergoes a conformational change that results in shell expansion, leading to an angular structure, head I (22). The last transition involves the formation of the mature virus head (head II) and the autocatalytic formation of covalent bonds between subunits. Duda and colleagues have demonstrated that the head protein subunits become cross-linked through the side chains of Lys-169 and Asn-356. They have recently demonstrated that the bridged capsid hexamers and pentamers are associated in a complex catenated structure that has been termed protein chain mail (20). Collectively, these changes allow for the formation of a very thin but extremely stable capsid structure. The head of mycobacteriophage L5, which is also a member of the family Siphoviridae, also undergoes conformational changes leading to cross-linking of the capsid subunits (30). Streptomyces phage φC31 also lacks a scaffold homolog and has a protease gene preceding its capsid gene and thus may also follow a similar pathway of capsid morphogenesis. Removal of 116 amino acids from the head gene product of staphylococcal bacteriophage φPVL has been reported, suggesting that proteolytic cleavage of structural proteins during capsid assembly is not uncommon (36), and indeed, it was originally discovered in T4 capsid morphogenesis (11a).
The data presented in this paper strongly suggest that P. aeruginosa phage D3 head assembly mimics that of HK97. This is supported by the following observations. First, the layout of D3 genes is identical to that of HK97 genes, with portal, protease, and capsid genes arranged without a scaffold. The proteins of D3 virions display an unusual electrophoretic pattern which is reminiscent of that of HK97 in the lack of a major 43-kDa head band and the appearance of high-molecular-weight material, including protein, which failed to enter the gel matrix. The amino acid sequence of the 186-kDa protein revealed that it was a derivative of the major head subunit protein starting at Ala-112. This suggests that the actual D3 head protein is cleaved from a precursor protein and has lost 111 amino acids from its amino terminus, a value remarkably similar to the situation that occurs during HK97 head morphogenesis. In both cases, the cleaved major head proteins have a molecular weight of 31,000, and both proteins show high sequence identity in this part of the protein sequence. While the cleavage sites in the HK97 and D3 capsid proteins are poorly conserved in sequence, they are strongly conserved in position. Since the molecular mass of the processed capsid protein is 31 kDa and that of the aggregate is 186 kDa, we can calculate that the latter probably represents cross-linked hexamers. In HK97, cross-links are arranged in such a way that they join subunits that surround fivefold or sixfold symmetry axes of the icosahedron. Each subunit joins its Lys-169 to the Asn-356 of its neighbor on one side and joins its Asn-356 to the Lys-169 of its neighbor on the other side. We hypothesize that in D3 head assembly the conserved residues Lys-178 and Asn-363 are also involved in cross-linking.
The fraction of material which failed to enter the resolving gel can also be explained by reference to the work done on HK97 capsid assembly. In the chain mail model, all of the subunits within each hexamer and pentamer are joined into closed protein rings and adjacent rings are interlocked topologically (20, 32).
ORF1692.
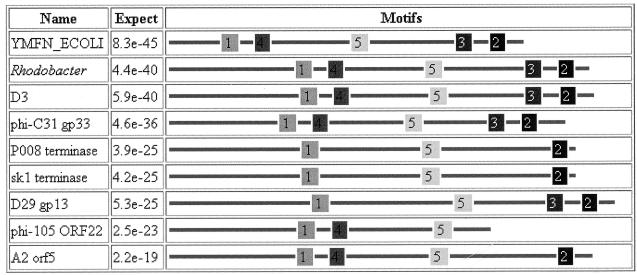
The DNA replication cycle for many viruses, including lambdoid phages, involves the synthesis of concatemers, which are the substrate for the packaging enzymes. This involves a protein complex called terminase, which in the case of coliphage λ, binds at cosB sites and introduces staggered nicks at an adjacent site (cosN), generating the cohesive ends of the encapsidated DNA (18). Terminase also forms the ternary DNA-terminase-prohead complex that leads to the packaging of DNA. Phylogenetically, phage terminases, like integrases, are a very diverse group of proteins (Fig. 6).
FIG. 6.
Phylogenetic analysis of bacteriophage putative and actual large-subunit terminases. The sequences were aligned with ClustalW and then analyzed with TreeCon, with the putative terminase from Methanobacterium phage psiM2 (GenBank accession no. AF65411) used to root the tree. The symbol at the upper left indicates 10% change in sequence. Shown are phi-105 orf22 (AB016282), phi-C31 gp33 (AJ006589), A2 orf5 (X97563), hypothetical Rhodobacter (AF010496) and E. coli (YMFN_ECOLI; P75978) proteins, D29 gp13 (AF022214), sk1 (AF011378), SPP1 (AB023396), P21 (P36693), lambda (P03708), 933W (AAD25456.1), DT1 (AAD21880.1), P2 (AAD03269.1), P008 (AAC63033.1), 186 (AAC34148.1), bIL170 (AAC27181.1), N15 gp2 (AAC19038.1), HP1 (P51718), phi41 (AAB41467.1), TM4 gp4 (AAD17572.1), phiCTX orf2 (BAA36228.1), and PBSX (P39786).
A comparison between the D3 late operon and that of other members of the family Siphoviridae indicates that ORF1692 is located at the same relative position as the large-subunit terminase gene in phages such as λ. The BLAST P search indicated that the product of this ORF showed the greatest sequence similarity to a hypothetical protein encoded by a gene of unknown function, ymfN, in E. coli K-12. This gene lies centrally in the intE (integrase)-to-pin (invertase) gene cluster. Both of these genes are remnants of the cryptic lamboid prophage e14 (14). In addition, the homology search revealed that the D3 gene product had sequence similarity to Bacillus subtilis phi-105 ORF22 (30% identity by ALIGN), L. casei bacteriophage A2 (27%), and Streptomyces phage φC31 gp33 (23%) and to the putative terminases of Lactobacillus lactis phages P008 and SK1 (20%). In each case where homology with a phage protein was found, it was to a putative or actual terminase from a member of the family Siphoviridae infectious for a gram-positive bacterium. By using MEME and MAST (8) to identify conserved protein motifs, five motifs were identified as being shared by this group of proteins (Fig. 7).
FIG. 7.
MEME analysis of a selection of putative and actual terminases showing the arrangement of five conserved motifs: 1, DGYNPHCAIVDEYH; 2, KIDPAVALIMA; 3, HDGNPVMTWHIGN; 4, QPLMWIITTAG; and 5, LGFDLSWKGDLTAIV.
Based upon the sequence homology between gpORF1692 and YMFN_ECOLI, it could be theorized that the progenitor of D3 was originally a coliphage related to e14 or that recombination occurred between the D3 progenitor and cryptic e14 on the E. coli chromosome. While this theory is interesting, one has to explain the homology with the terminases from phages of gram-positive bacteria and the presence of 3′-extended termini in the D3 genome (54). These termini are characteristic of phages infecting gram-positive cells. Indeed, it would make intuitive sense that a change in the nature of the termini might also be associated with a fundamental change in the terminase protein.
It is possible that ORF381, which lies upstream of ORF1692, represents the small subunit of terminase. If this is true, it may partially explain why our cosmid packaging strain, P. aeruginosa(D3Δcos), which lacked a portion of this gene, was inefficient in packaging (53, 55).
Hendrix and colleagues have recently proposed that not merely the lambdoid phages but possibly all members of the order Caudovirales have evolved through extensive exchange of genetic information from a pool of informational cassettes. This has occurred not only within phages active on members of a given bacterial family but also between families. This is best exemplified by the observation that coliphage HK97 and Streptomyces phage φC31 terminase, portal, protease, and capsid proteins have 28, 29, 28, and 20% sequence identity, respectively (31). The results reported here for D3 extend and support the ideas developed by Hendrix and colleagues. As noted, P. aeruginosa phage D3 shares many remarkable features with lambdoid phages, including the immunity (25, 26), replicative, and int-att regions (37b), while also having other genes (Fig. 1) which have no homology to existing phage genes in the databases.
ACKNOWLEDGMENTS
This research was supported by a grant from the Natural Sciences and Engineering Research Council of Canada.
We particularly acknowledge the contributions of Bob Tomkin (Electron Microscope Facility at Queen’s University) for the electron micrographs, Brad Cooney (Guelph Molecular Supercenter) for the DNA sequencing, and David Watson (National Research Council) for the protein sequencing.
REFERENCES
- 1.Ackermann H-W. Frequency of morphological phage descriptions in 1995. Arch Virol. 1996;141:209–218. doi: 10.1007/BF01718394. [DOI] [PubMed] [Google Scholar]
- 2.Ackermann H-W. Tailed bacteriophages: the order caudovirales. Adv Virus Res. 1999;51:135–201. doi: 10.1016/S0065-3527(08)60785-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams M D. Bacteriophages. New York, N.Y: Interscience Publishers, Inc.; 1959. [Google Scholar]
- 4.Akhverdian V Z, Khrenova E A, Lobanov A O, Krylov V N. Role of divergence in evolution of group B3 Pseudomonas aeruginosa transposable phage evolution. Genetika. 1998;34:846–849. [PubMed] [Google Scholar]
- 5.Akhverdian V Z, Lobanov A O, Khrenova E A, Krylov V N. Recombinational origin of natural transposable phages of related species belonging to group B3, active in Pseudomonas aeruginosa species. Genetika. 1998;34:697–700. [PubMed] [Google Scholar]
- 6.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 7.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–4022. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey T L, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 9.Bairoch A. The PROSITE dictionary of sites and patterns in proteins, its current status. Nucleic Acids Res. 1993;21:3097–3103. doi: 10.1093/nar/21.13.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bazinet C, Villafane R, King J. Novel second-site suppression of a cold-sensitive defect in phage P22 procapsid assembly. J Mol Biol. 1990;216:701–716. doi: 10.1016/0022-2836(90)90393-Z. [DOI] [PubMed] [Google Scholar]
- 11a.Black L W, Shumankov I L. Morphogenesis of the T4 head. In: Mathews C K, Kutter E M, Mosig G, Berget P B, editors. Bacteriophage T4. Washington, D.C.: American Society for Microbiology; 1983. pp. 219–245. [Google Scholar]
- 12.Bruner R, Vinograd J. The evaluation of standard sedimentation coefficients of sodium RNA and sodium DNA from sedimentation velocity data in concentrated NaCl and CsCl solutions. Biochim Biophys Acta. 1965;108:18–29. doi: 10.1016/0005-2787(65)90104-8. [DOI] [PubMed] [Google Scholar]
- 13.Campbell A. Comparative molecular biology of lambdoid phages. Annu Rev Microbiol. 1994;48:193–222. doi: 10.1146/annurev.mi.48.100194.001205. [DOI] [PubMed] [Google Scholar]
- 14.Campbell A M. Cryptic prophages. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 2041–2046. [Google Scholar]
- 15.Cavenagh M M, Miller R V. Specialized transduction of Pseudomonas aeruginosa PAO by bacteriophage D3. J Bacteriol. 1986;165:448–452. doi: 10.1128/jb.165.2.448-452.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung C H, Seol J H, Kang M S. Protease Ti (Clp), a multicomponent ATP-dependent protease in Escherichia coli. Biol Chem. 1996;377:549–554. [PubMed] [Google Scholar]
- 17.Conway J F, Duda R L, Cheng N, Hendrix R W, Steven A C. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J Mol Biol. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- 18.Cue D, Feiss M. Genetic evidence that recognition of cosQ, the signal for termination of phage lambda DNA packaging, depends on the extent of head filling. Genetics. 1997;147:7–17. doi: 10.1093/genetics/147.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dube P, Tavares P, Lurz R, van Heel M. The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J. 1993;12:1303–1309. doi: 10.1002/j.1460-2075.1993.tb05775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duda R L. Protein chainmail: catenated protein in viral capsids. Cell. 1998;94:55–60. doi: 10.1016/s0092-8674(00)81221-0. [DOI] [PubMed] [Google Scholar]
- 21.Duda R L, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix R W. Structural transitions during bacteriophage HK97 head assembly. J Mol Biol. 1995;247:618–635. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]
- 22.Duda R L, Martincic K, Hendrix R W. Genetic basis of bacteriophage HK97 prohead assembly. J Mol Biol. 1995;247:636–647. doi: 10.1006/jmbi.1994.0169. [DOI] [PubMed] [Google Scholar]
- 23.Eppler K, Wyckoff E, Goates J, Parr R, Casjens S. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology. 1991;183:519–538. doi: 10.1016/0042-6822(91)90981-g. [DOI] [PubMed] [Google Scholar]
- 24.Fagay D M, Bayley D P, Kostyukova A S, Thomas N, Jarrell K. Isolation and characterization of flagella and flagellin proteins from the theromoacidophilic archaea Thermoplasma volcanium and Sulfolobus shibatae. J Bacteriol. 1996;178:902–905. doi: 10.1128/jb.178.3.902-905.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farinha M A, Allan B J, Gertman E M, Ronald S L, Kropinski A M. Cloning of the early promoters of Pseudomonas aeruginosa bacteriophage D3: sequence of the immunity region of D3. J Bacteriol. 1994;176:4809–4815. doi: 10.1128/jb.176.16.4809-4815.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farinha M A, Kropinski A M. Overexpression, purification, and analysis of the c1 repressor protein of Pseudomonas aeruginosa bacteriophage D3. Can J Microbiol. 1997;43:220–226. doi: 10.1139/m97-030. [DOI] [PubMed] [Google Scholar]
- 27.Gertman E, Berry D, Kropinski A M. Serotype-converting bacteriophage D3 of Pseudomonas aeruginosa: vegetative and prophage restriction maps. Gene. 1987;52:51–57. doi: 10.1016/0378-1119(87)90394-5. [DOI] [PubMed] [Google Scholar]
- 28.Gertman E, White B N, Berry D, Kropinski A M. IS222, a new insertion element associated with the genome of Pseudomonas aeruginosa. J Bacteriol. 1986;166:1134–1136. doi: 10.1128/jb.166.3.1134-1136.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guasch A, Pous J, Parraga A, Valpuesta J M, Carrascosa J L, Coll M. Crystallographic analysis reveals the 12-fold symmetry of the bacteriophage phi29 connector particle. J Mol Biol. 1998;281:219–225. doi: 10.1006/jmbi.1998.1928. [DOI] [PubMed] [Google Scholar]
- 30.Hatfull G F, Sarkis G J. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol Microbiol. 1993;7:395–405. doi: 10.1111/j.1365-2958.1993.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 31.Hendrix R, Smith M C, Burns R N, Ford M E, Hatfull G F. Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendrix R W, Duda R L. Bacteriophage HK97 head assembly: a protein ballet. Adv Virus Res. 1998;50:235–288. doi: 10.1016/s0065-3527(08)60810-6. [DOI] [PubMed] [Google Scholar]
- 33.Ho Y S, Mahoney M E, Wulff D L, Rosenberg M. Identification of the DNA binding domain of the phage lambda cII transcriptional activator and the direct correlation of cII protein stability with its oligomeric forms. Genes Dev. 1988;2:184–195. doi: 10.1101/gad.2.2.184. [DOI] [PubMed] [Google Scholar]
- 34.Ho Y S, Wulff D L, Rosenberg M. Bacteriophage lambda protein cII binds promoters on the opposite face of the DNA helix from RNA polymerase. Nature. 1983;304:703–708. doi: 10.1038/304703a0. [DOI] [PubMed] [Google Scholar]
- 35.Jardine P J, Coombs D H. Capsid expansion follows the initiation of DNA packaging in bacteriophage T4. J Mol Biol. 1998;284:661–672. doi: 10.1006/jmbi.1998.2179. [DOI] [PubMed] [Google Scholar]
- 36.Kaneko J, Kimura T, Narita S, Tomita T, Kamio Y. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage φPVL carrying Panton-Valentine leukocidin genes. Gene. 1998;215:57–67. doi: 10.1016/s0378-1119(98)00278-9. [DOI] [PubMed] [Google Scholar]
- 37.Kellenberger E. Form determination of the heads of bacteriophages. Eur J Biochem. 1990;190:233–248. doi: 10.1111/j.1432-1033.1990.tb15568.x. [DOI] [PubMed] [Google Scholar]
- 37a.Kropinski, A. M., et al. Unpublished data.
- 37b.Kropinski, A. M. Unpublished results.
- 38.Krylov V N, Tolmachova T O, Akhverdian V Z. DNA homology in species of bacteriophages active on Pseudomonas aeruginosa. Arch Virol. 1993;131:141–151. doi: 10.1007/BF01379086. [DOI] [PubMed] [Google Scholar]
- 39.Kuzio J, Kropinski A M. O-antigen conversion in Pseudomonas aeruginosa PAO1 by bacteriophage D3. J Bacteriol. 1983;155:203–212. doi: 10.1128/jb.155.1.203-212.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 41.Luiten R G, Putterman D G, Schoenmakers J G, Konings R N, Day L A. Nucleotide sequence of the genome of Pf3, an IncP-1 plasmid-specific filamentous bacteriophage of Pseudomonas aeruginosa. J Virol. 1985;56:268–276. doi: 10.1128/jvi.56.1.268-276.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukashin A, Borodovsky M. GeneMark.hmm: a new solution for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurizi M R, Singh S K, Thompson M W, Kessel M, Ginsburg A. Molecular properties of ClpAP protease of Escherichia coli: ATP-dependent association of ClpA and ClpP. Biochemistry. 1998;37:7778–7786. doi: 10.1021/bi973093e. [DOI] [PubMed] [Google Scholar]
- 44.Mesyanzhinov V V, Sobolev B N, Marusich E I, Prilipov A G, Efimov V P. A proposed structure of bacteriophage T4 gene product 22—a major prohead scaffolding core protein. J Struct Biol. 1990;104:24–31. doi: 10.1016/1047-8477(90)90053-f. [DOI] [PubMed] [Google Scholar]
- 45.Miller R V, Pemberton J M, Richards K E. F116, D3, and G101: temperate bacteriophages of Pseudomonas aeruginosa. Virology. 1974;59:566–569. doi: 10.1016/0042-6822(74)90466-8. [DOI] [PubMed] [Google Scholar]
- 46.Nakayama, K., and T. Hayashi. Unpublished data.
- 46a.National Center for Biotechnology Information. ORF finder. National Center for Biotechnology Information, Bethesda, Md. http://www.ncbi.nlm.nih.gov/. [18 October 1999, last date accessed.]
- 47.Parker M H, Casjens S, Prevelige P E J. Functional domains of bacteriophage P22 scaffolding protein. J Mol Biol. 1998;281:69–79. doi: 10.1006/jmbi.1998.1917. [DOI] [PubMed] [Google Scholar]
- 48.Pica L, Calef E. Polymorphism in sedimentation velocity and density of lambda bacteriophage. J Mol Biol. 1968;32:513–520. doi: 10.1016/0022-2836(68)90340-9. [DOI] [PubMed] [Google Scholar]
- 49.Rishovd S, Marvik O J, Jacobsen E, Lindqvist B H. Bacteriophage P2 and P4 morphogenesis: identification and characterization of the portal protein. Virology. 1994;200:744–751. doi: 10.1006/viro.1994.1238. [DOI] [PubMed] [Google Scholar]
- 50.Rudd K E, Schneider T D. Compilation of E. coli ribosome binding sites. In: Miller J H, editor. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 17.19–17.45. [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Schmieger H. Molecular survey of the Salmonella phage typing system of Anderson. J Bacteriol. 1999;181:1630–1635. doi: 10.1128/jb.181.5.1630-1635.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharp R, Gertman E, Farinha M A, Kropinski A M. Transduction of a plasmid containing the bacteriophage D3 cos site in Pseudomonas aeruginosa. J Bacteriol. 1990;172:3509–3511. doi: 10.1128/jb.172.6.3509-3511.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharp R, Jansons I S, Gertman E, Kropinski A M. Genetic and sequence analysis of the cos region of the temperate Pseudomonas aeruginosa bacteriophage, D3. Gene. 1996;177:47–53. doi: 10.1016/0378-1119(96)00268-5. [DOI] [PubMed] [Google Scholar]
- 55.Sharp R W. Development of a cosmid cloning system for Pseudomonas aeruginosa. Ph.D. thesis. Kingston, Ontario, Canada: Queen’s University; 1991. [Google Scholar]
- 56.Van de Peer Y, De Wachter R. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput Appl Biosci. 1997;13:227–230. doi: 10.1093/bioinformatics/13.3.227. [DOI] [PubMed] [Google Scholar]
- 57.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees. Comput Appl Biosci. 1997;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 58.Yamamoto K R, Alberts B M, Benzinger R, Lawhorne L, Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970;40:734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]