Abstract
Background
The seasonal human coronaviruses (HCoV) NL63, 229E, OC43, and HKU1 are globally endemic, yet the majority of HCoV infections remain undiagnosed.
Methods
In a cross-sectional study, 2389 serum samples were collected from children and adults in France in 2020. In a longitudinal cohort study, 2520 samples were collected from 898 French individuals followed up between 2020 and 2021. Antibodies to HCoVs were measured using a bead-based multiplex assay.
Results
The rate of waning of anti-HCoV spike immunoglobulin G antibodies was estimated as 0.22–0.47 year−1 for children, and 0.13–0.27 year−1 for adults. Seroreversion was estimated as 0.31–1.37 year−1 in children and 0.19–0.72 year−1 in adults. The estimated seroconversion rate in children was consistent with 20%–39% of children being infected every year with each HCoV.
Conclusions
The high force of infection in children indicates that HCoVs may be responsible for a substantial proportion of fever episodes experienced by children.
Keywords: epidemiology, seasonal human coronavirus, serocatalytic model, serology
After its emergence in 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing coronavirus disease 2019 (COVID-19), led to a global pandemic with far-reaching consequences for human health and medical research [1]. One outcome has been the revitalization of research into other respiratory viruses, most notably the human coronaviruses (HCoVs) responsible for the common cold [2]. Four HCoVs (HCoV-NL63, HCoV-229E, HCoV-OC43, HCoV-HKU1), also referred to as seasonal coronaviruses, are associated with a range of respiratory symptoms, including coughing, sneezing, sore throat, and headaches, as well as more severe symptoms, such as pneumonia and bronchiolitis [3].
Despite the ubiquity of these viruses, surprisingly little is known about HCoV epidemiology owing to a number of practical challenges for surveillance. The large number of cocirculating respiratory pathogens results in very poor specificity of symptomatic surveillance [4]. Routine polymerase chain reaction–based testing of samples collected from patients presenting to hospitals with clinical symptoms can provide important information on temporal trends in HCoV cases, most notably revealing seasonal patterns [5,6]. However, the large proportion of HCoV infections that are asymptomatic or cause minor symptoms make it impossible to estimate the absolute number of infections from polymerase chain reaction testing of clinical cases [4].
Serology, based on the detection of antibodies induced by previous infections, has been frequently used to measure population-level HCoV transmission [7]. As with SARS-CoV-2, the most immunodominant antigens for the 4 HCoVs are spike (S) and nucleocapsid (N). A common serosurveillance method is to measure anti-S or anti-N immunoglobulin (Ig) G antibodies using an enzyme-linked immunosorbent assay, or a comparable immunoassay based on similar principles. Alternatively, live viral neutralization assays have been used to demonstrate the functional activity of immune factors in human serum [8].
Seroepidemiological investigations of HCoVs have typically used 2 categories of study design: cross-sectional studies and longitudinal cohort studies. In cross-sectional studies, blood samples are collected from a target population, from blood donor samples, or from residual samples in hospital clinics. The proportion of samples with antibody levels above a defined cutoff is termed the seroprevalence, providing an estimate of the proportion of a population previously infected. Through analysis of age-stratified seroprevalence, it is also possible to obtain estimates of the force of infection [7,9]. Longitudinal cohort studies involve following up a group of individuals over time, with collection of samples at multiple time points. These studies have been sporadically implemented since the 1970s, most notably for 229E and OC43 [10,11]. By identifying boosts in antibody levels between consecutive samples, longitudinal studies allow for infections and reinfections to be identified [12]. By following up infected individuals during periods when they are not reinfected, longitudinal studies also allow assessment of the duration of immune responses, for example, by measuring the rate of waning of antibody levels [8].
During the COVID-19 pandemic in early 2020, Edridge and colleagues [13] analyzed samples from a longitudinal cohort study of 10 Dutch individuals followed up for up to 35 years, with blood samples collected up to every 3 months. Biobanked samples were tested for anti-N IgG antibodies to the 4 HCoVs, resulting in a detailed investigation of antibody kinetics. The resulting data demonstrated that humoral immunity waned rapidly and that HCoV reinfection could occur within 6 months. This provided critical early insights into the duration of immunity to SARS-CoV-2 and the potential for future reinfection, findings that were subsequently validated [14,15]. Although there are important differences between SARS-CoV-2 and HCoVs [2], improved understanding of HCoV seroepidemiology may yield further insights into the long-term transmission dynamics of SARS-CoV-2 [16].
Here, we characterize the seroepidemiology of HCoVs in France by combining data from 2 studies with complementary design. The first study is a longitudinal cohort that began recruitment in winter 2020 in a French town north of Paris. Most participants were also enrolled in a previous cross-sectional study that took place in spring 2020 during France's first confinement in response to the COVID-19 pandemic [17]. Overall, these participants were followed up every 6 months for almost 2 years. Notably, the sanitary measures designed to reduce SARS-CoV-2 transmission were also shown to reduce transmission of other respiratory viruses [18]. The availability of samples dating back to spring 2020 provides a rare natural experiment, allowing the duration of immunity to HCoVs to be investigated in the absence of reinfection. In a second study, a cross-sectional cohort was implemented based on residual samples from French hospitals in 2020 [19]. This study collected many samples from children, allowing a detailed assessment of the population-level force of infection. By testing samples from these studies with multiplex serological assays and applying mathematical models to the resulting information, we provide a quantitative description of the seroepidemiology of the 4 HCoVs at endemic equilibrium in France.
METHODS
SeroPed Cross-Sectional Study
From February to August 2020, the SeroPed cross-sectional survey was implemented to evaluate immunity to SARS-CoV-2 and HCoVs in individuals attending French hospitals [19]. Analyzed samples were either anonymous residual serum samples from medical care or samples collected in other clinical studies (INCOVPED NCT04336761) after informed consent. Information on patient age and sex and the date of sampling were collected from medical records or study databases and compiled with the serological results. As the aim of the study was to estimate SARS-CoV-2 prevalence in the general population, samples were not collected from individuals admitted to hospital owing to infection with SARS-CoV-2. In total, we analyzed samples from 2389 individuals 1–100 years of age, with intensive sampling in children <10 years of age (29% [703 of 2389).
COVID-Oise Longitudinal Cohort Study
Scientists at Institut Pasteur initiated the COVID-Oise longitudinal cohort study in the town of Crépy-en-Valois in the Oise Department, the location of the first detected cluster of COVID-19 cases in France [17]. In an initial cross-sectional survey, samples were collected from 2004 individuals in spring 2020 (session 0). Of these individuals, 487 progressed to enroll in the COVID-Oise longitudinal cohort beginning in winter 2020 (session 1), with follow-up sessions in spring 2021 (session 2) and winter 2021 (session 3). A total of 905 individuals have been enrolled during these 3 sessions. In total, epidemiological data and biological specimens were collected on 2582 occasions. After exclusion of samples with insufficient blood volume, 2520 samples from 898 individuals were included in this analysis (Table 1). Participants ranged from 5 years of age to nursing home residents.
Table 1.
Overview of Epidemiological Studies
| Characteristic | SeroPeda | COVID-Oisea |
|---|---|---|
| Study design | Cross-section | Longitudinal cohort |
| Individuals, no. | 2389 | 898 |
| Samples, no. | 2389 | 2520 |
| Samples per individual | ||
| 1 | 2389 | 101 |
| 2 | 0 | 227 |
| 3 | 0 | 315 |
| 4 | 0 | 255 |
| Sex | ||
| Male | 1191 | 570 |
| Female | 1103 | 328 |
| Unknown | 95 | 0 |
| Age, y | ||
| 1–5 | 339 | 0 |
| 5–10 | 364 | 35 |
| >10 | 1686 | 863 |
| Sampling period | Feb–Aug 2020 | Apr 2020–Dec 2021 |
| HCoV serological markers | S-IgG | S-IgG, N-IgG, S-IgA, N-IgA |
Abbreviations: HCoV, human coronavirus; Ig, immunoglobulin; N, nucleocapsid; S, spike.
Values represent no. of individuals unless otherwise specified.
Serological Assays
Serum samples were tested for antibodies to coronaviruses using bead-based multiplex assays [20]. Two versions of the assay were used. The first assay, applied to the SeroPed samples, was a 9-plex assay for measuring IgG antibodies to 5 SARS-CoV-2 antigens (trimeric spike ectodomain of the ancestral variant, receptor-binding domain, spike S2 subunit, nucleocapsid, and membrane-envelope fusion) and the spike ectodomains of the 4 HCoVs (NL63, 229E, OC43, and HKU1). Additional information on the expression of the spike ectodomain proteins can be found in Supplementary File 1. The second assay, applied to the COVID-Oise samples, was a 28-plex assay for measuring IgG and IgA antibodies. In addition to the previously described antigens, this assay included the spike and receptor-binding domain of several SARS-CoV-2 variants (Alpha, Geta, Gamma, and Delta) and the nucleocapsid of the 4 HCoVs. The correlation between measured antibody responses was assessed (Supplementary Figure 1). Plates were read using a Luminex MagPix system, and the median fluorescence intensity was used for analysis. A 5-parameter logistic curve was used to convert median fluorescence intensity to relative antibody units, relative to the standard curve performed on the same plate to account for interassay variation.
Mixed-Effects Regression Analysis of Longitudinal Data
The imposition of lockdown measures in March 2020 as part of France's response to the COVID-19 crisis resulted in decreased transmission of all respiratory pathogens, including HCoVs [18]. The absence of reinfections allows the kinetics of the HCoV antibody response to be analyzed. Despite the likely absence of reinfections, it is still possible that there were some seasonal coronavirus reinfections. To exclude likely HCoV infections, we analyzed data on 4 biomarkers: S-IgG, N-IgG, S-IgA, and N-IgA. We defined a likely infection to have occurred between consecutive samples when there was an increase in antibody levels by a factor >8 for ≥2 biomarkers. This approach identified 21 likely NL63 infections (18 in children), 17 likely 229E infections (16 in children), 22 likely OC43 infections (18 in children), and 13 likely HKU1 infections (11 in children).
To estimate the rate of antibody waning, linear mixed-effects regression models were applied to longitudinal measures of antibody responses from the COVID-Oise cohort, with removal of data from individuals with likely HCoV infection. For biomarker k to HCoV c, measured in individual i at time tj, the antibody level Ackij can be modeled as follows:
where α denotes the estimated antibody level at session 0, β the rate of antibody waning, and ɛ is a normally distributed error term. Models were implemented in R software using the lme4 library [21].
Classification of Seropositivity
Individuals in the SeroPed study were classified as seropositive for HCoVs based on measured anti-S IgG levels. Because it was not possible to obtain verified negative controls for the HCoVs, we focused on measured antibody levels in children 1–5 years of age to define seropositivity cutoffs. We fitted 2 component gaussian mixture models to the log antibody data using the mclust R package [22]. This model assumes that the first gaussian component corresponds (with lower mean) to measured antibody levels from seronegative individuals and that the second gaussian component corresponds to the measured antibody levels from seropositive individuals. The seropositivity cutoff was defined as the mean of the means of the 2 distributions. For NL63, the data were not consistent with 2 distinct gaussian distributions (Supplementary Figure 2), and we instead defined the cutoff as the median of the measured antibody responses.
Serocatalytic Models
Serocatalytic models describe the dynamics of how individuals seroconvert after infection at rate λ and serorevert at rate ρ owing to antibody waning. The proportion of seropositive individuals in a population is defined to be the seroprevalence P, which varies as a function of age t, according to the following equation:
Assuming that individuals are seronegative at birth (ignoring temporary seropositivity due to maternal immunity), this equation can be solved to give the following:
The second equation assumes that both λ and ρ are constant over time and across all age groups. To account for different epidemiological behaviors between age groups, we can split the populations at age Tc into children with parameters λc and ρc and adults with parameters λa and ρa. Solving the serocatalytic model gives the estimated age-dependent seroprevalence, as follows:
The parameter Tc can be estimated from the data if there is sufficient statistical signal, or it can be predefined by known epidemiological criteria. Here we choose to fix Tc at 10 years. This splits the population into children (aged <10 years), and adults (defined as all individuals >10 years old).
Statistical inference was implemented in a bayesian framework using Markov chain Monte Carlo methods with a Metropolis Hastings algorithm. For the proposal distribution, a multivariate normal covariance matrix was dynamically tuned to obtain a target acceptance rate of 23%, and 200 000 steps were implemented, with the first 20% removed for burn-in. After assessment for convergence, parameters were reported as medians with 95% credible intervals.
Priors for Serocatalytic Models
Minimally informative uniform priors were assumed for the seroconversion rate λ. A common challenge for serocatalytic models is the mutual unidentifiability of λ and the seroreversion rate ρ. To address this, we use knowledge on antibody kinetics from the COVID-Oise longitudinal study to provide prior information for the seroreversion rate in models applied to the SeroPed study. For each quantitative measurement of antibody response in the SeroPed study, we estimated the time to seroreversion using the antibody decay rates with associated uncertainty from the COVID-Oise study. For both adults and children, the distribution of the times to seroreversion were used to define priors on the seroreversion rates.
Ethical Considerations
The majority of serum samples analyzed in the SeroPed study (2404 of 2544) were leftovers from routine medical blood sample processing in French hospitals. Other samples (141 of 2544) were collected for the purpose of a clinical study (INCOVPED, which is registered with ClinicalTrials.gov [NCT04336761]). Personal data processing for this study comply with the requirements of the “reference methodology MR-004” established by the French Data Protection Authority, Commission Nationale Informatique & Libertés (CNIL) regarding data processing in health research (Health Data Hub no. F20210519170759), since specific information for the research was provided to the parents of the INCOVPED study participants. The COVID-Oise study was registered with ClinicalTrials.gov (NCT04644159) and received ethical approval from the Comité de Protection des Personnes Nord Ouest IV. Several COVID-Oise participants participated in the CORSER studies in spring 2020, which were registered with ClinicalTrials.gov (NCT04325646) and approved by the Comité de Protection des Personnes Ile de France III.
Patient Consent Statement
In the SeroPed study, patients or their parents were informed that leftover samples could be used for research studies and had the option to oppose this reuse. Because serum samples were transferred for research completely anonymously and it was not possible to return to individual patients’ files, samples were processed in accordance with existing regulations and guidelines of the French Commission for Data Protection (Commission Nationale de l'Informatique et des Libertés, Health Data Hub no. F20210519170759). For the INCOVPED study, when signing the informed consent form for this study, participants’ parents had been informed and had consented to collected samples being used for other approved research studies. For the COVID-Oise study, all participants or their guardians provided informed consent before the provision of each sample.
RESULTS
Antibody Kinetics in the COVID-Oise Longitudinal Cohort
Between April 2020 and November 2021, there were declines in anti-S IgG levels for the 4 HCoVs, with the most notable decreases occurring between April and December 2020—the period spanning the first and most restrictive lockdown (Figure 1). Data for the anti-N IgG antibody response are presented in Supplementary Figure 3.
Figure 1.
Human coronavirus (HCoV) antibody kinetics in the COVID-Oise study. For the 4 HCoVs, anti–spike (S) immunoglobulin (Ig) G antibody responses were measured in samples collected in April 2020 (session 0), December 2020 (session 1), April 2021 (session 2), and November 2021 (session 3). Antibodies were measured in 2520 samples from 898 unique individuals. Abbreviation: RAU, relative antibody units.
After exclusion of likely infections, the rate of waning of anti-S IgG antibodies over time in children and adults was estimated from the COVID-Oise data (Table 2). For the 4 HCoVs, anti-S IgG antibodies waned more rapidly in children than in adults. The rate of waning in children was in the range 0.22–0.47 year−1, equivalent to a half-life of 1.47–3.15 years. The rate of waning in adults was in the range 0.13–0.27 year−1, equivalent to a half-life of 2.57–5.33 years.
Table 2.
Estimated Antibody Waning and Seroreversion Rates
| HCoV Type | Rate (95% CI), y−1 | |||
|---|---|---|---|---|
| Anti-S IgG Antibody Waning Ratea | Anti-S IgG Seroreversion Ratea | |||
| Children (Aged <10 y) | Adults (Aged >10 y) | Children (Aged <10 y) | Adults (Aged >10 y) | |
| NL63 | 0.47 (.41–.54) | 0.25 (.24–.27) | 1.37 (.15–11.73) | 0.72 (.08–6.32) |
| 229E | 0.38 (.31–.46) | 0.27 (.26–.28) | 0.31 (.05–2.01) | 0.22 (.03–1.41) |
| OC43 | 0.22 (.17–.28) | 0.13 (.12–.15) | 0.32 (.05–1.98) | 0.19 (.03–1.17) |
| HKU1 | 0.39 (.33–.45) | 0.17 (.16–.19) | 0.63 (.09–4.55) | 0.29 (.04–2.10) |
Abbreviations: CI, confidence interval; HCoV, human coronavirus; Ig, immunoglobulin; S, spike.
Antibody waning rates are estimated from linear mixed-effects regression models applied to data from the COVID-Oise longitudinal data. Seroreversion rates are calculated based on antibody waning rates from the COVID-Oise study and seropositivity cutoffs from the SeroPed study.
The antibody waning rate provides a measure of how the concentration of antibodies in blood samples reduces over time, whereas the seroreversion rate measures the rate at which seropositive individuals revert to seronegative as antibody levels drop below the seropositivity cutoff. The distribution of the time to seroreversion is shown in Supplementary Figure 4, and the estimated anti-S IgG seroreversion rates are shown in Table 2.
Age-stratified Anti-S IgG Antibody Levels in the SeroPed Study
In the SeroPed cross-sectional study, there were notable increases with age in anti-S IgG antibody levels to the 4 HCoVs (Figure 2). The dense sampling in the group 1–18 years of age allows for a detailed description of how antibodies are acquired on a population level. Note that samples were not included for children <12 months of age because they may carry maternal antibodies. Above 18 years of age, average antibody levels remain stable, except for anti-S antibodies to 229E, which continued to increase.
Figure 2.
Anti–spike (S) immunoglobulin (Ig) G antibody levels in the SeroPed cross-sectional study. Each point represents a measured antibody response. Horizontal dashed lines denote cutoffs for seropositivity. In total, 2389 samples from unique individuals were included. Abbreviations: HCoV, human coronavirus; RAU, relative antibody units.
Serocatalytic Models Applied to Age-Stratified Seroprevalence Data
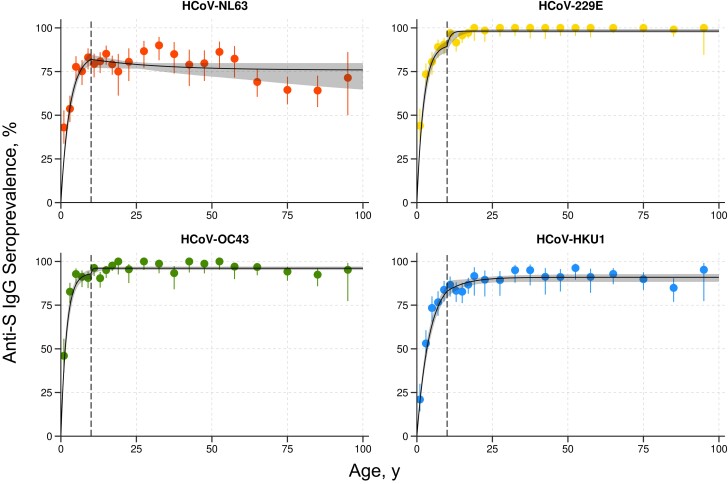
Continuous measurements of antibody titers were converted to age-stratified levels of seroprevalence (Figure 3). Serocatalytic models with separate parameters for adults and children were fit to this seroprevalence data. The model captured the age-dependent variation in the data. Estimated parameter values are shown in Table 3. For the seroconversion rate in children, it was possible to obtain precise estimates with narrow credible intervals. The seroconversion and seroreversion rates in adults had wide credible intervals, in most cases consistent with the uniform prior distribution. This indicates that the force of infection in adults is not identifiable from cross-sectional data. The estimated seroconversion rate in children ranged from λc = 0.22–0.50 year−1, which is equivalent to of children becoming infected every year.
Figure 3.
Serocatalytic models fitted to age-stratified seroprevalence data. Points denote measured seroprevalence, with vertical bars representing 95% confidence intervals. Solid black line denote the posterior median model fit, with shaded regions representing 95% credible intervals. Vertical dashed lines denote the separation between children (aged ≤10 years) and adults (aged >10 years). Abbreviations: HCoV, human coronavirus; Ig, immunoglobulin; S, spike.
Table 3.
Estimated Seroconversion and Seroreversion Rates
| HCoV Type | Rate, Posterior Median (95% CrI), y−1 | |||
|---|---|---|---|---|
| Seroconversion Rate λ | Seroreversion Rate ρ | |||
| Children (Aged <10 y) | Adults (Aged >10 y) | Children (Aged <10 y) | Adults (Aged >10 y) | |
| NL63 | 0.29 (.24–.34) | 0.03 (.01–4.47) | 0.052 (.020–.081) | 0.010 (.004–1.23) |
| 229E | 0.40 (.33–.48) | 0.96 (.23–4.87) | 0.042 (.020– .070) | 0.018 (.002–.113) |
| OC43 | 0.50 (.42–.59) | 2.50 (.27–4.86) | 0.035 (.018– .059) | 0.100 (.009–.213) |
| HKU1 | 0.22 (.19–.26) | 0.13 (.06– .53) | 0.021 (.004–.048) | 0.013 (.005–.064) |
Abbreviations: CrI, credible interval; HCoV, human coronavirus.
The estimated seroreversion rate in children was ρc = 0.021–0.052 year−1, equivalent to a seroreversion half-life of log2/ρc = 13–33 years. Notably, this is substantially higher than the prior estimates (Table 2). For adults, seroreversion rates were ρa = 0.010–0.101 year−1. Analysis of the posterior distribution did not provide evidence of difference in seroreversion rates between children and adults.
DISCUSSION
We investigate HCoV seroepidemiology by combining data from a cross-sectional study and a longitudinal cohort. The differing designs allow the strengths and weaknesses of each study to complement each other. The longitudinal cohort allows for an analysis of antibody kinetics, but the sharp reduction in transmission of respiratory viruses after confinement in early 2020 prevents estimation of an endemic force of infection. Children <5 years old were not enrolled in the cohort, preventing analysis of an immunonaive population. In contrast, the cross-sectional study included many samples from children <5 years of age. Because the cross-sectional study includes only 1 sample per individual, we cannot identify when individuals seroconverted. Rather than identifying individual infection events, cross-sectional studies provide estimates of force of infection through age-stratified increases in seroprevalence, most notably in young children.
We estimated seroconversion rates in children in the range of 0.22–0.50 year−1, equivalent to 20%–39% of previously uninfected children becoming infected every year. Summing across the 4 HCoVs , this equates to 1.17 infections per year. This estimate would increase in the event of reinfection and decrease if there was cross-protective immunity between HCoVs. In addition, children will be exposed to other respiratory viruses that were not included in our assay, such as respiratory syncytial virus, influenza, adenoviruses, and rhinoviruses [23]. Switching from population-level data to individual anecdotes, these figures may be familiar to parents with young children in daycare, who typically observe multiple episodes of fever and other symptoms consistent with respiratory virus infection. These infections are seldom diagnosed and are often handled by parents in the absence of interactions with formal health systems, except for a minority of cases that result in severe symptoms. It is arguable that the collective experience of parents of young children can be a better source of information on transmission levels of nonsevere respiratory viruses than official epidemiological records.
The absence of reinfection in the COVID-Oise cohort allowed assessment of the duration of HCoV antibody responses. We estimated that anti-S IgG antibodies had a half-life of 1.47–3.15 years in children, and 2.57–5.33 years in adults, consistent with previous reports for other coronaviruses [8,22–27]. Based on these data, we assumed prior estimates for seroreversion rates of 0.3–1.4 year−1 in children and 0.2–0.7 year−1 in adults, consistent with estimates of 0.9–3.8 year−1 published by Rees et al [7]. However, after we fit serocatalytic models to the cross-sectional SeroPed data, our posterior median estimates for the half-life of seroreversion were in the range 13–33 years in children and 7–68 years in adults, substantially longer than prior estimates. This discrepancy may be due to model identifiability—many parameter combinations can accurately reconstruct the age-stratified seroprevalence (Figure 3). Furthermore, age-stratified seroprevalence from a cross-sectional study is not an optimal study design for evaluating the duration of an antibody response—the statistical inference procedure favors accurate reconstruction of age-stratified seroprevalence over biologically relevant prior information on seroreversion rates.
An alternative explanation is model misspecification. We assumed exponential decay of both seropositivity and antibody levels, although it is known that waning antibody levels are better described by a biphasic exponential function [28]. In addition, our model does not allow for the potential role of slower seroreversion due to boosting of antibody levels with new infections. A further limitation of our model is that we do not capture the potential association between antibody levels and the prevention of infection or symptoms. Although preexisting immunity has been shown to be associated with protection from HCoV infection [8], there are no available data on a serological correlate of protection.
There are several limitations across the multiple components of our analysis, from epidemiological study design to serological assays and statistical models. For the COVID-Oise study, 4 biomarkers were measured (anti-S IgG, anti-N IgG, anti-S IgA, anti-N IgA) in each sample, whereas only anti-S IgG was measured in the SeroPed cross-section. It is possible that application of serocatalytic models to biomarkers other than anti-S IgG antibodies would yield substantially different results. An ideal approach would integrate information from several biomarkers, but the development of serocatalytic models for multiplex data remains an open challenge. An additional limitation of our study was that we did not use viral neutralization assays. Often considered the reference standard for serological studies, neutralization assays measure the level of functional protection of immune components in human serum against a target virus [10,11]. Validating our immunoassay against neutralization assays would substantially strengthen our findings. A further limitation is the challenge of obtaining verified negative control samples for globally endemic viruses that routinely infect young children. We addressed this challenge by fitting gaussian mixture models to samples from young children to identify positive and negative subpopulations, but this was not possible for all HCoVs.
In the early stages of the COVID-19 pandemic, findings from seroepidemiological studies of HCoVs yielded many useful insights into the future behavior of SARS-CoV-2, for example, that humoral immunity would wane over time [8] and that there would likely be frequent reinfection [13]. Our seroepidemiological analysis provides a description of the long-term endemicity of the HCoVs NL63, 229E, OC43 and HKU1 in France. The good fit of the serocatalytic model is consistent with these viruses being at endemic equilibrium for a long time owing to a balance between increased immunity from new infections, waning antibody levels, and the introduction of newly susceptible children. This differs drastically from the immune landscape of SARS-CoV-2, which has been circulating for <3 years, in contrast to HCoVs, which have been circulating for most individuals’ lifetimes. The long-term patterns of endemic equilibrium of the HCoVs provide one possibility of the future epidemiology of SARS-CoV-2. However, widespread vaccination against COVID-19 will substantially alter the SARS-CoV-2 immune landscape and possibly the endemic equilibrium of HCoVs if there is significant cross-protective immunity.
Supplementary Material
Acknowledgments
The authors thank the participants who agreed to participate into the different studies and the medical and paramedical teams who were involved in sample and data collection. They thank the teams of Crépy-en-Valois town hall and the director and technical services of the local hospital for their help in implementing the COVID-Oise study. They also thank the Investigation Clinique et Accès aux Ressources Biologiques technical team for management and distribution of the samples.
Contributor Information
Alix De Thoisy, Infectious Disease Epidemiology and Analytics G5 Unit, Department of Global Health, Institut Pasteur, Université Paris Cité, Paris, France.
Tom Woudenberg, Infectious Disease Epidemiology and Analytics G5 Unit, Department of Global Health, Institut Pasteur, Université Paris Cité, Paris, France.
Stéphane Pelleau, Infectious Disease Epidemiology and Analytics G5 Unit, Department of Global Health, Institut Pasteur, Université Paris Cité, Paris, France.
Françoise Donnadieu, Infectious Disease Epidemiology and Analytics G5 Unit, Department of Global Health, Institut Pasteur, Université Paris Cité, Paris, France.
Laura Garcia, Infectious Disease Epidemiology and Analytics G5 Unit, Department of Global Health, Institut Pasteur, Université Paris Cité, Paris, France.
Laurie Pinaud, Epidemiology of Emerging Diseases Unit, Department of Global Health, Institut Pasteur, Université Paris Cité, Paris, France.
Laura Tondeur, Epidemiology of Emerging Diseases Unit, Department of Global Health, Institut Pasteur, Université Paris Cité, Paris, France.
Annalisa Meola, Structural Virology Unit, Department of Virology and CNRS UMR 3569, Institut Pasteur, Université Paris Cité, Paris, France.
Laurence Arowas, Investigation Clinique et Accès aux Ressources Biologiques (ICAReB), Center for Translational Research, Institut Pasteur, Paris, France.
Nathalie Clement, Coordination Clinique du CRT, Center for Translational Research, Institut Pasteur, Paris, France.
Marija Backovic, Structural Virology Unit, Department of Virology and CNRS UMR 3569, Institut Pasteur, Université Paris Cité, Paris, France.
Marie-Noëlle Ungeheuer, Investigation Clinique et Accès aux Ressources Biologiques (ICAReB), Center for Translational Research, Institut Pasteur, Paris, France.
Arnaud Fontanet, Epidemiology of Emerging Diseases Unit, Department of Global Health, Institut Pasteur, Université Paris Cité, Paris, France; PACRI Unit, Conservatoire National des Arts et Métiers, Paris, France.
Michael White, Infectious Disease Epidemiology and Analytics G5 Unit, Department of Global Health, Institut Pasteur, Université Paris Cité, Paris, France.
COVID-Oise and SeroPed study teams:
Tom Woudenberg, Stéphane Pelleau, Laurie Pinaud, Laura Tondeur, Marie-Noëlle Ungeheuer, Arnaud Fontanet, Michael White, Sandrine Fernandes Pellerin, Raphaël Guiheneuf, Catherine Delamare, Karl Stefic and Julien Marlet, Etienne Brochot, Sandrine Castelain, Olivier Augereau, Jean Sibilia, François Dubos, Christéle Gras-Le Guen, Marianne Coste-Burel, Berthe-Marie Imbert-Marcille, Cyril Schweitzer, Amélie Gatin, Aline Joulié, Hervé Haas, Aymeric Cantais, Frederique Bertholon, Marie-France Chinazzo-Vigouroux, Cécile Duru, and Aymar Davy Koffi
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Study teams. The COVID-Oise and SeroPed study teams include the following members: Tom Woudenberg, Stéphane Pelleau, Laurie Pinaud, Laura Tondeur, Marie-Noëlle Ungeheuer, Arnaud Fontanet, Michael White, and Sandrine Fernandes Pellerin (Institut Pasteur, Paris France); Raphaël Guiheneuf (Centre Hospitalier Simone Veil de Beauvais, Beauvais, France); Catherine Delamare (CHR Metz Thionville, Metz, France); Karl Stefic and Julien Marlet (Service de Bactériologie-Virologie, Hôpital Bretonneau, CHRU de Tours, Tours, France); Etienne Brochot and Sandrine Castelain, (Service de Virologie, CHU Amiens Picardie, UR 4294 AGIR UPJV, Amiens, France); Olivier Augereau (Service de Microbiologie, Hôpitaux Civils de Colmar, Colmar, France); Jean Sibilia (Laboratoire de Virologie, CHU de Strasbourg, Strasbourg, France); François Dubos (Université de Lille, CHU Lille, Urgences Pédiatriques et Maladies Infectieuses, Lille, France); Christéle Gras-Le Guen (Urgences Pédiatrique et Pédiatrie Générale Hopital Mère Enfant CHU de Nantes, Nantes, France); Marianne Coste-Burel and Berthe-Marie Imbert-Marcille (Service de Virologie CHU Nantes, Nantes, France); Cyril Schweitzer (Hôpital d'Enfants, CHRU de Nancy, Vandoeuvre-Les-Nancy, France); Amélie Gatin (Pediatric Emergency Unit, Hôpital d’Enfants, CHRU Nancy); Aline Joulié and Hervé Haas (Urgences Pédiatriques et Pédiatrie Générale, Hôpitaux Pédiatriques CHU Lenval, Nice); Aymeric Cantais and Frederique Bertholon (Pediatric Emergency Department, Hospital University of St Etienne, France); Marie-France Chinazzo-Vigouroux (Urgences Pédiatriques Hopital Clocheville, CHRU de Tours, Tours, France); and Cécile Duru and Aymar Davy Koffi (Hôpital de Crépy-en-Valois, Crépy-en-Valois, France)
Data availability. All data produced in the present study are available on request to the authors.
Financial support. This work was supported by the Fondation pour la Recherche Médicale (CorPopImm to M. W.); the European Research Council (MultiSeroSurv 852373 to M. W.); the French government’s “Integrative Biology of Emerging Infectious Diseases” (Investissement d'Avenir grant ANR-10-LABX-62-IBEID) and INCEPTION programs (Investissement d’Avenir grant ANR-16-CONV-0005); and the “URGENCE COVID-19” fundraising campaign of Institut Pasteur (TooLab project awarded to M.B.). The COVID-Oise cohort is funded by Alliance Tous Unis Contre le Virus, Institut Pasteur, AP-HP, and Fondation de France.
References
- 1. World Health Organization. WHO coronavirus (COVID-19) dashboard. Available at: https://covid19.who.int. Accessed 12 June 2022.
- 2. Huang AT, Garcia-Carreras B, Hitchings MDT, et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nature Commun 2020; 11:4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vabret A, Mourez T, Gouarin S, Petitjean J, Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin Inf Dis 2003; 8:985–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Al-Tawfiq JA, Zumla A, Gautret P, et al. Surveillance for emerging respiratory viruses. Lancet Inf Dis 2014; 14:992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gaunt ER, Hardie A, Claas ECJ, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol 2010; 48:2940–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aldridge RW, Lewer D, Beale S, et al. Seasonality and immunity to laboratory-confirmed seasonal coronaviruses (HCoV-NL63, HCoV-OC43, and HCoV-229E): results from the flu watch cohort study. Wellcome Open Res 2020; 5:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rees EM, Waterlow NR, Lowe R, Kucharski AJ; CMMID COVID-19 Working Group . Estimating the duration of seropositivity of human seasonal coronaviruses using seroprevalence studies. Wellcome Open Res. 2021; 6:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Callow KA, Parry HF, Sergeant M, Tyrrell DAJ. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Inf 1990; 105:435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kolehmainen P, Heroum J, Jalkanen P, et al. Serological follow-up study indicates high seasonal coronavirus infection and reinfection rates in early childhood. Mircobiol Spectrum 2022; 10:e0196721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaye HS, Marsh HB, Dowdle WR. Seroepidemiologic survey of coronavirus (strain OC43) related infections in a children's population. Am J Epidemiol 1971; 94:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaye HS, Dowdle WR. Seroepidemiologic survey of coronavirus (strain 229E) infections in a population of children. Am J Epidemiol 1975; 101:238–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wenzel RP, Hendley JO, Davies JA, Gwaltney JM. Coronavirus infections in military recruits: three-year study with coronavirus strains OC43 and 229. Am Rev Respir Dis 1974; 109:621–4. [DOI] [PubMed] [Google Scholar]
- 13. Edridge AWD, Kaczorowska J, Hoste ACR, et al. Seasonal coronavirus protective immunity is short-lasting. Nature Med 2020; 26:1691–3. [DOI] [PubMed] [Google Scholar]
- 14. Nordström P, Ballin M, Nordström A. Risk of SARS-CoV-2 reinfection and COVID-19 hospitalisation in individuals with natural and hybrid immunity: a retrospective, total population cohort study in Sweden. Lancet Inf Dis 2022; 22:781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dan J, Mateus J, Kato Y, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021; 371:eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kissler SM, Tedijanto C, Goldstein E, Grad YH, Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science 2020; 368:860–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fontanet A, Tondeur L, Grant R, et al. SARS-CoV-2 infection in schools in a northern French city: a retrospective serological cohort study in an area of high transmission, France, January to April 2020. Eurosurveillance 2021; 26:2001695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Launay T, Souty C, Vilcu AM, et al. Common communicable diseases in the general population in France during the COVID-19 pandemic. PLoS One 2021; 16:e025839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Woudenberg T, Pelleau S, Anna F, et al. Humoral immunity to SARS-CoV-2 and seasonal coronaviruses in children and adults in north-eastern France. EBioMed 2021; 70:103495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rosado JJ, Pelleau S, Cockram C, et al. Multiplex assays for the identification of serological signatures of SARS-CoV-2 infection: an antibody-based diagnostic and machine learning study. Lancet Microbe 2021; 2:e60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bates D, Machler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Software 2015; 67:1–48. [Google Scholar]
- 22. Scrucca L, Fop M, Murphy TB, Raftery AE. Mclust 5: clustering, classification and density estimation using gaussian finite mixture models. R Journal 2016; 8:289–317. [PMC free article] [PubMed] [Google Scholar]
- 23. Pappas DE, Hendley JO. The common cold. Principles Practice Ped Inf Dis 2008; e1:203–6. [Google Scholar]
- 24. Wu LP, Wang NC, Chang YH, et al. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis 2007; 13:1562–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mo H, Zeng G, Ren X, et al. Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology 2006; 11:49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cao WC, Liu W, Zhang PH, Zhang F, Richardus JH. Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med 2007; 357:1162–3. [DOI] [PubMed] [Google Scholar]
- 27. Liu W, Fontanet A, Zhang PH, et al. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J Inf Dis 2006; 193:792–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pelleau S, Woudenberg T, Rosado J, et al. Kinetics of the severe acute respiratory syndrome coronavirus 2 antibody response and serological estimation of time since infection. J Inf Dis 2021; 224:1489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.