Abstract
Candida albicans SEC4 was cloned by complementing the Saccharomyces cerevisiae sec4-8 mutation, and its deduced protein product (Sec4p) was 63% identical to S. cerevisiae Sec4p. One chromosomal SEC4 allele in C. albicans CAI4 was readily disrupted by homologous gene targeting, but efforts to disrupt the second allele yielded no viable null mutants. Although this suggested that C. albicans SEC4 was essential, it provided no information about this gene’s functions. Therefore, we constructed a mutant sec4 allele encoding an amino acid substitution (Ser-28→Asn) analogous to the Ser-17→Asn substitution in a trans-dominant inhibitor of mammalian Ras protein. GAL1-regulated expression plasmids carrying the mutant sec4 allele (pS28N) had minimal effects in glucose-incubated C. albicans transformants, but six of nine transformants tested grew very slowly in galactose. Incubation of pS28N transformants in galactose also inhibited secretion of aspartyl protease (Sap) and caused 90-nm secretory vesicles to accumulate intracellularly, and plasmid curing restored growth and Sap secretion to wild-type levels. These results imply that C. albicans SEC4 is required for growth and protein secretion and that it functions at a later step in the protein secretion pathway than formation of post-Golgi secretory vesicles. They also demonstrate the feasibility of using inducible dominant-negative alleles to define the functions of essential genes in C. albicans.
The pathway by which proteins are transported between membrane-bound intracellular compartments and then out of the cell is highly conserved in all eukaryotes. This secretion (or vesicular transport) pathway has been studied extensively in Saccharomyces cerevisiae, and several dozen S. cerevisiae genes encoding its key components have been cloned and analyzed in some detail (14, 20, 22). The Candida albicans homologs of three essential S. cerevisiae secretion pathway genes have been described. C. albicans SEC18 (19), SEC14 (18), and SEC4 (5) all complemented the corresponding mutations in S. cerevisiae, and their deduced protein products were 50 to 67% identical to the corresponding proteins in S. cerevisiae. However, the functions of these genes have not yet been studied directly in C. albicans, except that efforts to disrupt both chromosomal alleles of C. albicans SEC14 yielded no viable null mutants (18).
In S. cerevisiae, SEC4 encodes a small Ras-like GTPase (Sec4p) that is required for fusion of post-Golgi secretory vesicles to the plasma membrane (23). Because SEC4 is essential in S. cerevisiae, sec4 null mutants are nonviable. However, temperature-sensitive (23) and dominant-negative (27) mutations in S. cerevisiae SEC4 inhibit growth and protein secretion and cause post-Golgi secretory vesicles to accumulate intracellularly. In the present study, we cloned the C. albicans SEC4 homolog by complementing a temperature-sensitive S. cerevisiae sec4 mutant, and we sought to ascertain this gene’s functions. Gene disruption studies suggested that C. albicans SEC4 was essential, but they provided no specific information about this gene’s functions. Therefore, we used site-directed mutagenesis to construct a mutant sec4 allele analogous to those encoding trans-dominant inhibitors of other ras-like GTPases, and we used this mutant sec4 allele to study the functions of C. albicans SEC4.
MATERIALS AND METHODS
Strains and media.
S. cerevisiae NY28 (MATα ura3-52 sec4-8) (from P. Novick, Yale University) was cultured in YPD (1% yeast extract, 2% peptone, 2% glucose) or minimal glucose (0.67% yeast nitrogen base without amino acids [YNB], 2% glucose). Wild-type C. albicans SC5314 and its derivative CAI4 (Δura3::imm434/Δura3::imm434) (from W. Fonzi, Georgetown University) were cultured in YPD, minimal glucose, or minimal galactose (0.67% YNB, 2% galactose). Aspartyl protease secretion by C. albicans was examined in (i) 0.17% YNB without ammonium sulfate and with 0.4% bovine serum albumin (BSA) and either 2% glucose (glucose-BSA) or 2% galactose (galactose-BSA) or (ii) 0.34% YNB without ammonium sulfate and with 0.2% BSA, 0.2% yeast extract, and either 2% glucose (glucose-BSA-YE) or 2% galactose (galactose-BSA-YE). Uracil auxotrophs were selected on 5-fluoro-orotic acid (FOA) medium (minimal glucose, 0.1 mg of uridine per ml, and 0.7 mg of FOA per ml). Solid media were prepared by adding 2% agar.
Libraries and plasmids.
A library of genomic DNA fragments from C. albicans WO-1 in the Escherichia coli-S. cerevisiae shuttle plasmid pEMBLYe23 was obtained from P. T. Magee (University of Minnesota). Plasmid p5921, which contains a hisG-URA3-hisG selectable marker (7), was obtained from W. Fonzi. Plasmid pGAL2.7 was constructed by (i) ligating the 2.7-kb XbaI restriction fragment containing C. albicans GAL1 from plasmid pCW31 (9) into the XbaI site in the yeast shuttle plasmid YEp352 and (ii) ligating the 2.7-kb SalI-BamHI restriction fragment from this plasmid into SalI- and BamHI-digested plasmid p1041 (10). The autonomously replicating C. albicans plasmid pVEC was obtained from P. T. Magee. pVEC was constructed by ligating the 3.6-kb NruI-SmaI fragment containing C. albicans ARS2 and URA3 from plasmid pRM1 (21) into the NdeI site in pUC18. pSK was from Stratagene (La Jolla, Calif.).
Isolation and properties of C. albicans SEC4.
The plasmid library of C. albicans genomic DNA was introduced into S. cerevisiae NY28 by electroporation, and uracil prototrophs were selected on minimal glucose at 25°C, replica plated, and incubated at 37°C. Plasmids from thermotolerant S. cerevisiae transformants were expanded in E. coli and retested for the ability to complement the sec4-8 mutation in S. cerevisiae NY28. Standard methods were used for restriction mapping, subcloning, Southern hybridizations, DNA amplification by PCR, and DNA sequencing. Chromosomal mapping was performed by B. B. Magee (University of Minnesota), as previously described (4).
Targeted disruption of C. albicans SEC4.
The method of Fonzi and Irwin (7) was used in an effort to disrupt both chromosomal alleles of C. albicans SEC4. The first step was to construct a gene disruption cassette in which the C. albicans SEC4 open reading frame (ORF) was deleted and replaced with the hisG-URA3-hisG selectable marker from plasmid p5921. To accomplish this, Taq DNA polymerase and primers Nok5SacI (5′ GCC GCT GAG CTC TTG GAA CAT TCC TTA TGC AGG C 3′) and CA11 (5′ GAT GAA TTG TTA ATA CTT GAT ATG TTT TCC 3′) were used (i) to amplify 692 bp of 5′ untranslated C. albicans SEC4 DNA (nucleotides 173 to 864 upstream from the start codon) from p412a and (ii) to add a SacI restriction site (underlined). The PCR product was digested with SacI and BglII, and it was ligated into the SacI and BglII sites on the left side of the hisG-URA3-hisG selectable marker in p5921, which yielded pL1. Next, primers Nok3Bam (5′ GAT GCG GGA TCC CCA TGA ACG AAG AAG AAG AAG AAG AG 3′) and Nok3Sal (5′ GGC GTG GTC GAC CAA CCA AAC CAG ATA GTG AGA ATT G 3′) were used (i) to amplify 820 bp of 3′ untranslated C. albicans SEC4 DNA (nucleotides 42 to 861 downstream from the stop codon) from p412a and (ii) to add BamHI and SalI restriction sites (underlined). This PCR product was digested with BamHI and SalI, and it was ligated into the BamHI and SalI sites on the right side of the hisG-URA3-hisG marker in pL1, which yielded pL1R2.
To disrupt the first chromosomal SEC4 allele, pL1R2 was digested with SacI and SalI, and the linearized sec4Δ::hisG-URA3-hisG gene disruption cassette was introduced into C. albicans CAI4 by the lithium acetate method. Uracil prototrophs were selected on minimal glucose, and they were tested for homologous integration of the sec4Δ::hisG-URA3-hisG gene disruption cassette within the SEC4 locus as follows. First, allele-specific PCR with primers G1 (5′ GGT TCT GTC GAA GTC GCG CCG CGC 3′) and 23 (5′ GAG ACT TCT AGA TAG TTC TCG ATG 3′) was used to determine if the sec4Δ::hisG-URA3-hisG gene disruption cassette had integrated homologously within a SEC4 locus. Also, PCR with primers 5 (5′ GTT AGC CAA ACA CGC ATG AAC 3′) and 10 (5′ CCA AAA CTA TTC CAC ATA TCA TTC C 3′) was used to detect changes in the sizes of the chromosomal SEC4 loci. Second, C. albicans genomic DNA was digested with NdeI, ApaI, or both enzymes, and the digests were transferred to nylon membranes. The membranes were hybridized in 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 64°C with a 304-bp 32P-labeled PCR product generated from p412a with primers 21 (5′ GTT TGT TAG GAG GTA TGC TAT TGG G 3′) and 22 (5′ AAT AGT GAG AAA GAA AGA GTG TAG 3′), after which the membranes were washed in 0.3× SSC–0.1% sodium dodecyl sulfate (SDS) at 64°C and analyzed by autoradiography.
To disrupt the second SEC4 allele, selected C. albicans SEC4/sec4Δ::hisG-URA3-hisG mutants were expanded in YPD to permit loss of URA3 by cis recombination between the flanking hisG repeats, uracil auxotrophs were selected on FOA medium, and these strains’ genotypes were determined by PCR and by Southern hybridization as described above. Next, selected C. albicans SEC4/sec4Δ::hisG strains were transformed again with the linearized sec4Δ::hisG-URA3-hisG gene disruption cassette, uracil prototrophs were selected, and these strains’ genotypes were determined by PCR and by Southern hybridization.
Construction of plasmids pS28N and pSEC4.
To overexpress wild-type and mutant SEC4 alleles in C. albicans, it was first necessary to construct an autonomously replicating plasmid containing a regulable promoter. To accomplish this, we used Taq DNA polymerase, M13 sequencing primer, and primer GAL1 (5′ CGG CGG GTC GAC GGA TCC ACT AGT TTA ATT AAG GTA TAA CTC TTT CTT ATA AAA ATC GG 3′) (i) to amplify approximately 1.0 kb of the C. albicans GAL1 promoter from plasmid pGAL2.7 and (ii) to add a convenient BamHI restriction site (underlined). The PCR product was digested with XbaI and BamHI, and it was ligated into XbaI- and BamHI-digested pVEC, which yielded plasmid pYM1.
Next, we constructed a mutant C. albicans sec4 allele encoding arginine instead of serine at position 28 of Sec4p (S28→N), using the Quickchange (Stratagene) site-directed mutagenesis method. First, a 1.03-kb BglII fragment containing the ORF and transcription terminator from C. albicans SEC4 was excised from plasmid p412a and ligated into BamHI-digested pSK, which yielded pSKSEC4. Next, pSKSEC4 was amplified with Pfu DNA polymerase and the mutagenic oligonucleotides S28N5 (5′ CCG GTG TTG GGA AAA ATT GTT TAT TAT TGC GTT TTG 3′) and S28N3 (5′ CAA AAC GCA ATA ATA AAC AAT TTT TCC CAA CAC CGG 3′). This yielded pSKSEC4(S28N), which encodes a mutant Sec4p bearing the S28→N substitution.
The sec4(S28N) allele in pSKSEC4(S28N) was amplified with Pfu DNA polymerase and primers GAL2 (5′ CGG CGG GTC TTA ATT AAA TGA GCG GTA AAG GAA CAT CAT CAA G 3′) and GAL3 (5′ CGG ACG GGA TCC CAA CAA AAT ACC CCA GAT CTA GAG 3′), and the product was ligated into the PacI and BamHI sites in pYM1, which yielded pS28N. Lastly, the ORF and transcription terminator from wild-type C. albicans SEC4 were amplified by the same method and inserted into the PacI and BamHI sites of pYM1, which yielded pSEC4 (Fig. 1). The accuracy of the final DNA constructions was confirmed by DNA sequencing.
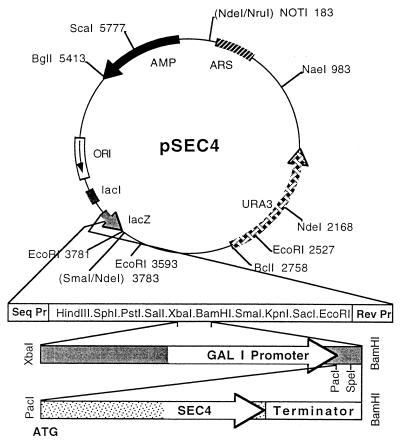
FIG. 1.
Plasmid pSEC4 was constructed by inserting into the autonomously replicating C. albicans plasmid pVEC (i) 1.0 kb of genomic DNA immediately 5′ to the C. albicans GAL1 start codon and (ii) the ORF and transcription terminator from C. albicans SEC4. pS28N was identical to pSEC4 except that the sec4(S28N) allele replaced wild-type SEC4.
Effects of pS28N and pSEC4 in C. albicans.
Plasmids pS28N and pSEC4 were introduced into C. albicans CAI4 by the lithium acetate method, and uracil prototrophs were selected on minimal glucose. pS28N-transformed C. albicans was cured of plasmids by culturing for 40 generations in YPD and by plating on FOA medium. Absence of pS28N after curing was verified by PCR, using primers BL3 (5′ CCA ATG CTT AAT CAG TGA GGC AAC 3′) and BL5 (5′ AGT ATT CAA CAT TTC CGT GTC GCC 3′) from the β-lactamase gene. Plasmid copy numbers were estimated by Southern hybridization, using a 0.85-kb probe for β-lactamase DNA that was generated with primers BL3 and BL5 and a 0.75-kb probe for C. albicans ADE2 DNA that was generated with PCR primers CaADE2Spe3 (5′ TTG AGC ACT AGT CAT TTC AAC ACC GAA AAT ACC ACA C 3′) and CaADE2Pac15 (5′ TGG ATG GTT AAT TAA GAA GCA GCA CAT AGA TTG AAT ATC 3′).
Growth rates were determined by measuring the optical density at 600 nm (OD600) at intervals after glucose-grown transformants were washed and resuspended either in minimal glucose or minimal galactose at an OD600 of 0.1, and CFU were determined by plating serial dilutions on minimal glucose.
Secreted aspartyl protease (Sap) expression was induced by growing C. albicans transformants to stationary phase in glucose-BSA or glucose-BSA-YE, after which the cells were washed and resuspended at an OD600 of 10 in (i) glucose-BSA or galactose-BSA or (ii) glucose-BSA-YE or galactose-BSA-YE. The cell suspensions were shaken at 30°C, and cell-free supernatants were obtained after 2 to 24 h and tested (i) for residual BSA by SDS-polyacrylamide gel electrophoresis with Coomassie blue staining and/or (ii) for immunoreactive Sap by Western blotting, using polyclonal rabbit antibodies to Sap (29) (from N. Agabian, University of California at San Francisco).
To determine the effects of pS28N and pSEC4 on the ultrastructural morphology of C. albicans, the transformants were incubated in minimal glucose or minimal galactose for 6.5 h, and the cells were harvested, fixed in 3% glutaraldehyde–0.1 M sodium cacodylate (pH 6.8), washed with 0.1 M sodium cacodylate, and postfixed in 1% OsO4. The cells were then embedded in Epox 812×, and thin sections were stained with Pb citrate-uranyl acetate and examined with a Philips 300 electron microscope.
Nucleotide sequence accession number.
The GenBank accession number for C. albicans SEC4 is AF015306.
RESULTS
Isolation and properties of C. albicans SEC4.
When S. cerevisiae NY28 was transformed with the C. albicans genomic DNA library, plasmids isolated from 10 of 17 thermotolerant transformants contained similar 6.9-kb inserts. One of these plasmids (p412a) again conferred thermotolerance when it was reintroduced into S. cerevisiae NY28. A 1,269-bp portion of the insert from p412a was sequenced in both directions and contained a 630-bp intronless ORF flanked on its 5′ end by four potential TATA elements and on its 3′ end by an S. cerevisiae-type transcription termination sequence (30). This ORF encoded a deduced protein of 210 amino acids and 23 kDa that had 63% identity with S. cerevisiae Sec4p and 45% identity with human Rab3 protein (Rab3p). The deduced protein also contained (i) four consensus guanine nucleotide interaction domains, (ii) a membrane attachment sequence at its C terminus, and (iii) conserved amino acids in positions where single amino acid substitutions cause temperature-sensitive and dominant-negative mutations in S. cerevisiae SEC4 and other Ras-like GTPases (Fig. 2).
FIG. 2.
The deduced Sec4p’s of C. albicans (Ca) and S. cerevisiae (Sc) were 61% identical (:). Conserved GTP-binding (∗) and membrane attachment (+) domains are shown, as are the positions where a G147→D substitution causes a temperature-sensitive mutation (▵) and where S34→N and N133→I substitutions cause dominant-negative mutations (↑) in S. cerevisiae Sec4p.
Targeted disruption of C. albicans SEC4.
Genomic Southern analyses showed that the insert from p412a hybridized to a single-copy gene on SfiI fragment M of C. albicans chromosome R (data not shown). Therefore, we attempted to construct C. albicans sec4 null mutants by homologous gene targeting. The genotypes of 20 uracil prototrophs obtained when C. albicans CAI4 was transformed with the sec4Δ::hisG-URA3-hisG gene disruption cassette were analyzed by allele-specific PCR and by Southern hybridization.
PCR amplification of genomic DNA with primers 23 and G1 (which were derived, respectively, from a part of the C. albicans SEC4 locus that was not included in the gene disruption cassette and from bacterial hisG DNA) (Fig. 3A) generated a product only if the gene disruption cassette integrated homologously within an SEC4 locus (e.g., strains YM5 and YM6) but not from wild-type C. albicans (CAI4) or following ectopic integration (e.g., strain YM7) (Fig. 3B). Also, PCR with primers 5 and 10 (Fig. 3A) generated a 1.2-kb product from wild-type SEC4, no additional product when the large sec4Δ::hisG-URA3-hisG cassette integrated homologously within a SEC4 locus (e.g., strains YM5 and YM6), or a 1.5-kb product when cis recombination between the flanking hisG repeats in the sec4Δ::hisG-URA3-hisG allele generated a smaller sec4Δ::hisG allele (e.g., strains YM5F and YM6F) (Fig. 3B). Overall, analysis by allele-specific PCR indicated (i) that the sec4Δ::hisG-URA3-hisG gene disruption cassette integrated homologously into one wild-type SEC4 allele in 17 of 20 transformants and (ii) that three of three FOA-resistant strains derived from C. albicans SEC4/sec4Δ::hisG-URA3-hisG mutants had the expected SEC4/sec4Δ::hisG genotype.
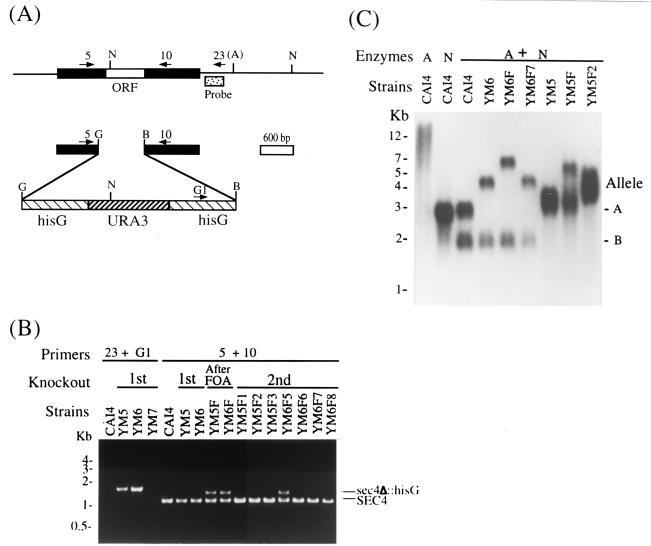
FIG. 3.
Targeted disruption of C. albicans SEC4. (A) Restriction maps of the C. albicans SEC4 locus (top) and the sec4Δ::hisG-URA3-hisG gene disruption cassette (bottom). Primers used for allele-specific PCR and the probe used for Southern hybridizations are shown. Abbreviations: G, BglII; B, BamHI; N, NdeI; A, polymorphic ApaI site. (B) When C. albicans CAI4 was transformed with the linearized sec4Δ::hisG-URA3-hisG gene disruption cassette, PCR with primers 23 and G1 generated a product only if the gene disruption cassette integrated homologously (YM5 and YM6, but not wild-type CAI4 or ectopic integrant YM7). Also, primers 5 and 10 generated a 1.2-kb PCR product from wild-type SEC4, no additional product from the large sec4Δ::hisG-URA3-hisG cassette (YM5 and YM6), and a 1.5-kb product when recombination between flanking hisG repeats generated a smaller sec4Δ::hisG allele (YM5F and YM6F). When YM5F or YM6F (SEC4/sec4Δ::hisG) was transformed again with sec4Δ::hisG-URA3-hisG DNA, homologous (YM5F1, YM5F2, YM5F3, YM6F6, YM6F7, and YM6F8) and ectopic (YM6F5) integrations were observed. However, 20 of 20 transformants analyzed retained a wild-type SEC4 allele. (C) When C. albicans CAI4 genomic DNA was digested with ApaI (A), NdeI (N), or both enzymes and hybridized with the probe shown in panel A, the presence of a polymorphic ApaI restriction site showed that the sec4Δ::hisG-URA3-hisG cassette could replace either of the two SEC4 alleles in C. albicans CAI4 (compare YM6 to YM5). Since URA3 contains an NdeI site, the hybridized fragments from the sec4Δ::hisG alleles in YM6F and YM5F (first knockout after FOA) were larger than those from the sec4Δ::hisG-URA3-hisG alleles in YM5 and YM6. When the sec4Δ::hisG-URA3-hisG cassette was reintroduced into YM6F, it replaced the sec4Δ::hisG allele, but not the remaining wild-type SEC4 allele (see YM6F7). When the gene disruption cassette was reintroduced into YM5F, more complex integration events occurred (see YM5F2), but allele-specific PCR (see YM5F2 in panel B) showed that all transformants retained at least one wild-type SEC4 allele.
The 32P-labeled SEC4 probe (Fig. 3A) hybridized to a single 3-kb band in NdeI digests of C. albicans CAI4 genomic DNA and to two bands (3 and 2 kb) in NdeI and ApaI double digests (Fig. 3C). Thus, the presence of a polymorphic ApaI restriction site allowed us to distinguish between the two SEC4 alleles in C. albicans CAI4. Among the 20 transformants analyzed, the sec4Δ::hisG-URA3-hisG gene disruption cassette (i) replaced the 3-kb band (allele A) in 6 transformants (e.g., strain YM6), (ii) replaced the 2-kb band (allele B) in 12 transformants (e.g., strain YM5), and (iii) did not integrate homologously in 2 transformants (Fig. 3C). Also, since C. albicans URA3 contains an NdeI site (Fig. 3A), the labeled fragments in the sec4Δ::hisG-URA3-hisG alleles (e.g., strains YM5 and YM6) were smaller than the labeled fragments in the sec4Δ::hisG alleles (e.g., strains YM6F and YM5F) (Fig. 3C).
Although these results established that either of the two SEC4 alleles in C. albicans CAI4 could be disrupted by homologous targeting, repeated efforts to disrupt the wild-type SEC4 alleles in several C. albicans SEC4/sec4Δ::hisG mutants yielded no viable sec4 null mutants. When the SEC4/sec4Δ::hisG strains YM6F, YM9F, and YM5F were transformed with the sec4Δ::hisG-URA3-hisG gene disruption cassette, PCR with primers 5 and 10 showed that both homologous (e.g., strains YM5F1, YM5F2, YM5F3, YM6F6, YM6F7, and YM6F8) and ectopic (e.g., strain YM6F5) integrations occurred. However, all 20 transformants analyzed had at least one wild-type SEC4 allele (Fig. 3B). Moreover, genomic Southern analyses showed that the sec4Δ::hisG-URA3-hisG gene disruption cassette either (i) replaced the previously mutagenized sec4Δ::hisG allele (e.g., strain YM6F7), (ii) integrated into the wild-type SEC4 locus without replacing the wild-type gene (e.g., strain YM5F2), or (iii) did not integrate homologously (not shown) (Fig. 3C).
Effects of sec4(S28N) overexpression on growth and viability.
Although the gene disruption studies summarized above suggested that C. albicans SEC4 was essential, they provided no specific information about this gene’s functions. Therefore, we examined the effects on growth and viability of C. albicans transformants of GAL1-regulated expression plasmids encoding wild-type Sec4p (pSEC4) or a mutant Sec4p analogous to known trans-dominant inhibitors of several other Ras-like GTPases (1, 2, 8, 10, 21–23) (pS28N). Seven of seven pSEC4 transformants tested grew well (i.e., OD600 of >15 at 24 h) in repressing (minimal glucose) and inducing (minimal galactose) media. Nine of nine pS28N transformants tested also grew well in minimal glucose, but six of these transformants grew slowly in minimal galactose.
That growth inhibition in galactose was mediated by pS28N was verified by plasmid curing experiments. When four galactose-inhibited pS28N transformants were cultured for 40 generations in YPD, uracil protrophs derived from all four transformants retained the galactose-inhibited growth phenotype. In contrast, 15 of 16 FOA-resistant clones derived from the same YPD cultures (4 from each of 4 transformants) grew as well on minimal galactose agar plus uridine as did their parent, C. albicans CAI4. Moreover, PCR analysis showed that the single FOA-resistant clone that retained the galactose-inhibited growth phenotype also contained β-lactamase DNA, which implied that this clone retained a version of pS28N that lacked a functional URA3.
Further analysis of a representative galactose-inhibited pS28N transformant showed that its generation times were 1.7 h in minimal glucose and 18.6 h in minimal galactose, compared to 1.7 h in minimal glucose and 1.9 h in minimal galactose for a pSEC4-transformed control (Fig. 4). Despite its slow growth, the pS28N transformant reached high cell densities after prolonged incubation in minimal galactose (OD600 values of 14 after 3 days and 25 after 6 days). Moreover, when the 6-day culture was diluted to an OD600 of 0.1 in fresh minimal galactose and reincubated, it reached an OD600 of >11 after 24 h, as did a pSEC4-transformed control. One possible explanation for the instability of growth inhibition in galactose was that prolonged incubation in galactose may have selected transformants with low plasmid copy numbers; however, Southern hybridization showed no discernible differences in the ratios of plasmid-derived β-lactamase DNA to chromosomal ADE2 DNA in pS28N transformants grown overnight in minimal glucose or for 6 days in minimal galactose.
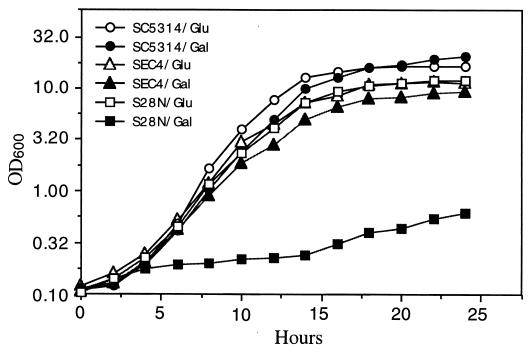
FIG. 4.
Effects of sec4(S28N) overexpression on growth. Wild-type C. albicans (SC5314) and pSEC4-transformed C. albicans CAI4 (SEC4) grew well in minimal glucose (Glu) and minimal galactose (Gal). pS28N-transformed C. albicans CAI4 also grew well in minimal glucose (S28N/Glu), but these transformants grew very slowly when they were transferred to minimal galactose (S28N/Gal).
CFU were enumerated at 0, 2, and 6 h after glucose-grown C. albicans pS28N transformants were washed, resuspended at an OD600 of 0.1 in minimal galactose or minimal glucose, and shaken at 30°C. The CFU per milliliter in minimal glucose and minimal galactose, respectively, were 7.47 × 105 and 7.25 × 105 at 0 h, 5.55 × 105 and 5.47 × 105 at 2 h, and 5.36 × 106 and 5.76 × 105 at 6 h. Thus, overexpression of sec4(S28N) in C. albicans inhibited growth without causing a substantial loss of viability, at least during the period before OD600 values began to rise.
Effects of sec4(S28N) overexpression on protein secretion.
We next examined the abilities of the pSEC4- and pS28N-transformed C. albicans strains to secrete Sap when they were incubated in glucose or galactose medium at high cell densities (OD600 = 10) that permitted minimal cell growth. In one experiment, the cell-free supernatants of pS28N- and pSEC4-transformed C. albicans that had been incubated in glucose-BSA or galactose-BSA for 2 and 7 h contained large amounts of residual BSA and little or no immunoreactive Sap. After 20 h, however, the cell-free supernatants of the galactose-incubated pS28N transformants contained much more residual BSA and much less immunoreactive Sap than the supernatants of the glucose-incubated pS28N transformants or the glucose- or galactose-incubated pSEC4 transformants (Fig. 5). Since the final cell density of the galactose-incubated pS28N transformants was only slightly lower (OD600 = 31) than that in the controls (OD600 = 49 for glucose-incubated pSEC4 transformants, OD600 = 57 for galactose-incubated pSEC4 transformants, and OD600 = 46 for glucose-incubated pS28N transformants), the marked differences in Sap secretion could not be ascribed to differences in final cell densities.
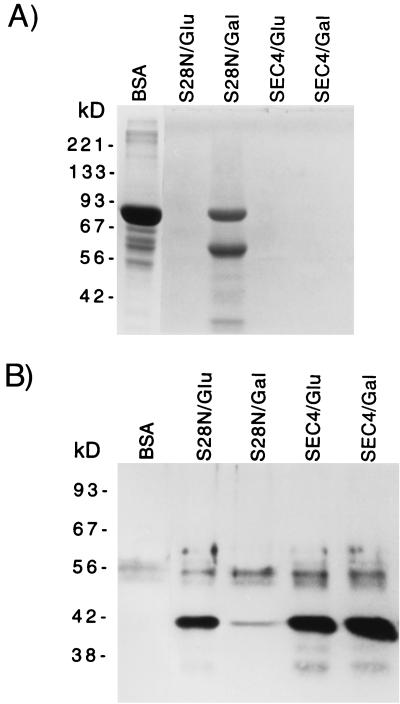
FIG. 5.
Effects of sec4(S28N) overexpression on protein secretion. (A) pS28N transformants incubated in galactose-BSA at 30°C for 20 h degraded much less extracellular BSA than pS28N transformants incubated in glucose-BSA or pSEC4 transformants incubated in glucose-BSA or galactose-BSA. Reducing SDS-polyacrylamide gel electrophoresis with cell-free supernatants from 0.1 OD600 unit per lane and Coomassie blue staining is shown. (B) Western blotting showed that the galactose-incubated pS28N transformants secreted much less immunoreactive Sap than glucose-incubated pS28N transformants, glucose-incubated pSEC4 transformants, or galactose-incubated pSEC4 transformants (0.5 OD600 unit per lane).
In another experiment, three of four galactose-inhibited pS28N transformants degraded much less extracellular BSA when they were incubated for 16 h in galactose-BSA-YE than when they were incubated in glucose-BSA-YE. In contrast, C. albicans CAI4, C. albicans CAI4 transformed with the control plasmid pSEC4, and two of two non-galactose-inhibited pS28N transformants degraded as much extracellular BSA when they were incubated in galactose-BSA-YE as when they were incubated in glucose-BSA-YE. Moreover, the pS28N transformant that lost the galactose-inhibited growth phenotype after plasmid curing regained the ability to degrade the extracellular BSA in galactose-BSA-YE. In contrast, the pS28N transformant that grew slowly in galactose and contained β-lactamase DNA after curing also did not degrade extracellular BSA in galactose-BSA-YE.
Effects of sec4(S28N) overexpression on ultrastructural morphology.
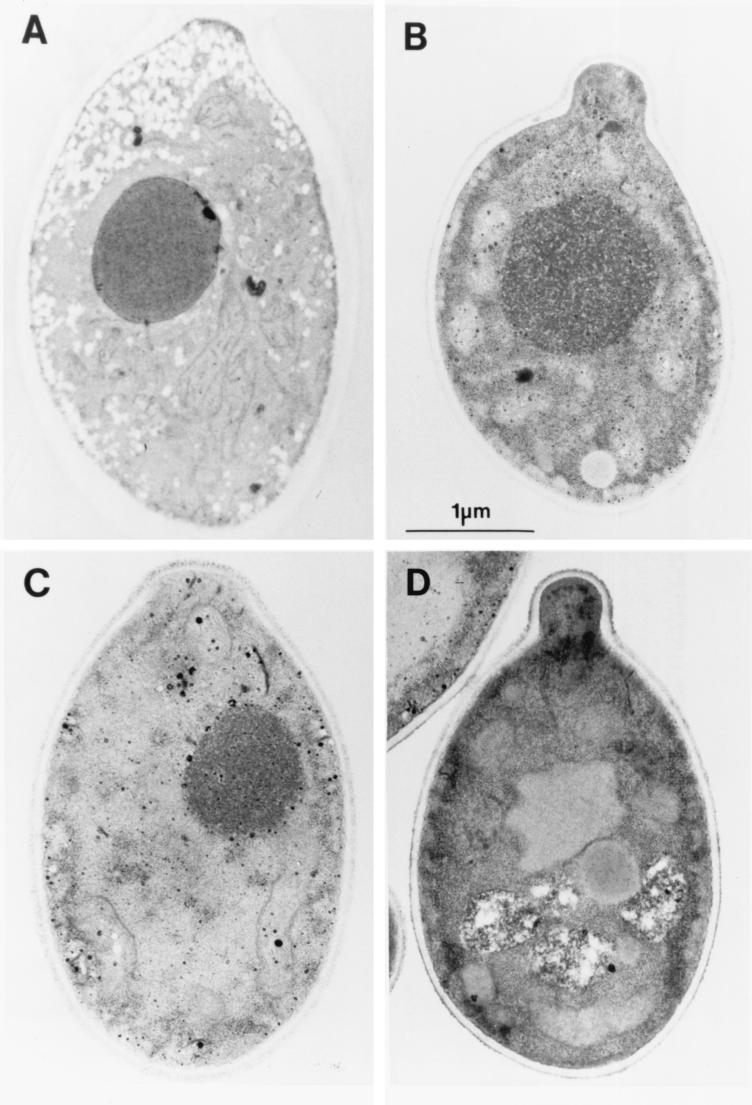
There was marked intracellular accumulation of secretory vesicles when a galactose-inhibited pS28N transformant was incubated in minimal galactose for 6.5 h. These vesicles were approximately 90 nm in diameter (which is similar to the size of the post-Golgi vesicles that accumulate in S. cerevisiae sec4-8 mutants subjected to restrictive temperatures), and they were localized mostly in the region of the bud or adjacent to the plasma membrane. In contrast, there was no intracellular accumulation of transport vesicles in glucose-incubated pS28N transformants or in glucose- or galactose-incubated pSEC4 transformants (Fig. 6).
FIG. 6.
Effects of sec4(S28N) overexpression on ultrastructural morphology. Typical post-Golgi secretory vesicles accumulated in pS28N-transformed C. albicans cells incubated in minimal galactose for 6.5 h (A) but not in pS28N transformants incubated in glucose (B), pSEC4 transformants incubated in galactose (C), or plasmidless C. albicans SC5314 incubated in galactose (D).
DISCUSSION
C. albicans causes more serious infections than any other fungus, but classical genetics are seldom useful for studying this organism because of its diploid genome and its lack of sexual reproduction. Recently, however, powerful molecular methods have been developed for studying C. albicans biology and pathogenesis. For example, many C. albicans genes have been cloned and sequenced, and integrative and episomal DNA transformation systems are well established. Moreover, SfiI macrorestriction maps of each C. albicans chromosome are available (4), and more than 100 genes have been assigned to specific SfiI fragments. Also, a detailed physical map of chromosome 7 has recently been constructed (3), and efforts to sequence the complete C. albicans genome are nearing completion.
Powerful methods for using the rapidly expanding body of C. albicans DNA sequence data to understand the biology of C. albicans and the pathogenesis of C. albicans infections have also been developed. The development of a technically straightforward method for deleting specific genes from a C. albicans ura3Δ strain whose virulence can be restored to wild-type levels by reintroducing URA3 (7) was a major advance, and this and similar methods have been used to ascertain directly the importance of several proposed C. albicans virulence factors (8, 12, 15, 24). Although this experimental approach is extremely useful, one of its major limitations is that disruption of essential genes causes loss of viability. Therefore, many fundamental questions about the biology and virulence of C. albicans cannot be answered simply by cloning and disrupting individual genes. In S. cerevisiae and other model fungi, the functions of essential genes can be studied by generating and analyzing conditional mutants or by overexpressing dominant-negative mutant alleles (6, 13, 17, 27). To our knowledge, the only example in which either of these approaches has been used with medically important fungi was that temperature-sensitive mutations were introduced by homologous gene targeting into the essential NMT genes (encoding myristoyl-coenzyme A:protein N-myristoyl transferase) of Cryptococcus neoformans (16) and C. albicans (28).
We chose to study C. albicans SEC4 because (i) Ras-like GTPases are highly conserved even among distantly related eukaryotes and (ii) single amino acid substitutions in conserved domains of S. cerevisiae Sec4p and related Ras-like GTPases cause temperature-sensitive and dominant-negative mutations. The first step was to clone and sequence a C. albicans gene that complemented the temperature-sensitive sec4-8 mutation in S. cerevisiae, and we found that the gene of interest was highly homologous to S. cerevisiae SEC4 and identical to the C. albicans SEC4 gene recently reported by Clement et al. (5). The first chromosomal SEC4 allele was readily disrupted in C. albicans CIA4, but efforts to disrupt the wild-type SEC4 allele in three C. albicans SEC4/sec4Δ::hisG strains yielded no viable null mutants. One possible explanation for the inability to disrupt the second chromosomal allele of any gene may be that there is insufficient similarity between the gene disruption cassette and the second allele to facilitate homologous recombination. In the case of C. albicans SEC4, however, the presence of a polymorphic ApaI restriction site allowed us to show that the sec4Δ::hisG-URA3-hisG gene disruption cassette we used was capable of replacing either of the two SEC4 alleles in C. albicans CAI4. Southern analyses also demonstrated homologous integration of the gene disruption cassette within the wild-type SEC4 locus of a SEC4/sec4Δ::hisG-URA3-hisG strain, but allele-specific PCR showed that all of the resulting transformants retained a wild-type SEC4 allele. Since these results suggested that SEC4 is essential in C. albicans, it was clear that other approaches would be needed to define this gene’s functions.
Small Ras-like GTPases in the Rab/SEC4/YPT1 family contain conserved domains required for guanine nucleotide binding, GTP-GDP exchange, and GTP hydrolysis. The S17→N substitution in mammalian Ras protein alters GTP-GDP exchange-dependent signalling and results in dominant oncogenic transformation. Similarly, S. cerevisiae Ypt1p, mammalian Rab1p, and mammalian Rab3p with S→N substitutions at analogous positions are potent trans-dominant inhibitors of protein transport (1, 2, 11, 13, 25–27). Therefore, we reasoned that it might be possible to define the functions of C. albicans SEC4 by (i) introducing a mutation encoding the S28→N substitution and (ii) overexpressing the resulting sec4(S28N) allele in wild-type C. albicans. We found that autonomously replicating GAL1-regulated plasmids expressing sec4(S28N) (pS28N) had no discernible effects on growth or Sap secretion in C. albicans transformants incubated in glucose, but these plasmids markedly inhibited growth and Sap secretion in galactose-incubated transformants. In addition, typical post-Golgi secretory vesicles accumulated intracellularly in galactose-incubated pS28N transformants but not in glucose-incubated controls. We concluded from these results that C. albicans SEC4 (like S. cerevisiae SEC4) (i) is required for growth and protein secretion and (ii) functions at a later step in the protein secretion pathway than the formation of post-Golgi secretory vesicles.
Two unexpected findings were that sec4(S28N) overexpression did not inhibit growth and protein secretion in a minority of galactose-incubated transformants and that the growth inhibition phenotype was lost after prolonged incubation in galactose. Plasmid curing studies clearly established that inhibition of growth and protein secretion in galactose was mediated by pS28N, and Southern hybridization studies did not support the hypothesis that variable growth phenotypes were caused by marked differences in plasmid copy number. Therefore, the most likely explanation for the instability of the galactose inhibition phenotype is that the deleterious effects of GAL1-regulated sec4(S28N) overexpression selected for clones in which (i) a loss-of-function mutation occurred in one or more key elements in pS28N [e.g., the sec4(S28N) coding sequence, the GAL1 promoter, or ARS2) or (ii) the S28N mutation itself was lost, either by reversion or gene conversion.
The fact that the sec4 mutant phenotype could be induced despite the presence of two wild-type SEC4 alleles implies that sec4(S28N) functioned as a dominant-negative mutant allele. To our knowledge, C. albicans SEC4 is the first essential gene of any medically important fungus whose functions have been elucidated by constructing and overexpressing a dominant-negative mutant allele. It should be noted that construction of the sec4(S28N) allele required prior knowledge of the sequences of trans-dominant inhibitors of several other Ras-like GTPases. Thus, dominant-negative alleles may be most useful for ascertaining the functions of highly conserved genes required for fundamental cellular processes such as protein secretion, cell cycle control, and signal transduction. Also, further studies will be needed to determine why the sec4 mutant phenotype was unstable and to develop methods for maintaining the stability of dominant-negative alleles in C. albicans. Despite these limitations, the general approach used in this study may prove to be useful for studying the functions of additional essential genes in organisms like C. albicans, whose diploid genomes and lack of sexual reproduction preclude the use of more traditional genetic methods.
ACKNOWLEDGMENTS
We thank P. T. Magee and B. B. Magee (University of Minnesota) for the chromosomal mapping studies, the C. albicans genomic DNA library in pEMBLYe23, and plasmid pVEC; Peter Novick (Yale) for S. cerevisiae NY28; William Fonzi (Georgetown) for C. albicans CAI4 and SC5314 and plasmid p5921; Nina Agabian (University of California, San Francisco) for antibodies to C. albicans Sap; Theodore White (University of Washington) for suggestions about the secretion assays; Lillemor Wallmark for the electron microscopy; and Zimei Zhang for technical assistance.
This work was supported by grants from the Department of Veterans’ Affairs and the National Institute of Allergy and Infectious Diseases (R01 AI-36684).
REFERENCES
- 1.Barbacid M. Ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 2.Burstein E S, Brondyk W H, Macara I G. Amino acid residues in the Ras-like GTPase Rab3A that specify sensitivity to factors that regulate the GTP/GDP cycling of Rab3A. J Biol Chem. 1992;267:22715–22718. [PubMed] [Google Scholar]
- 3.Chibana H, Magee B B, Grindle S, Ran Y, Scherer S, Magee P T. A physical map of chromosome 7 of Candida albicans. Genetics. 1998;149:1739–1752. doi: 10.1093/genetics/149.4.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu W S, Magee B B, Magee P T. Construction of an SfiI macrorestriction map of the Candida albicans genome. J Bacteriol. 1993;175:6637–6651. doi: 10.1128/jb.175.20.6637-6651.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clement M, Fournier H, Repentigny L D, Belhumeur P. Isolation and characterization of the Candida albicans SEC4 gene. Yeast. 1998;14:675–680. doi: 10.1002/(SICI)1097-0061(199805)14:7<675::AID-YEA252>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 6.Dunsmuir P, Hynes M J. Temperature sensitive mutants affecting the activity and regulation of the acetamidase of Aspergillus nidulans. Mol Gen Gen. 1973;123:333–346. doi: 10.1007/BF00433650. [DOI] [PubMed] [Google Scholar]
- 7.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gale C A, Bendel C M, McClellan M, Hauser M, Becker J M, Berman J, Hostetter M K. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science. 1998;279:1355–1358. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- 9.Gorman J A, Chan W, Gorman J W. Repeated use of GAL1 for gene disruption in Candida albicans. Genetics. 1991;129:19–24. doi: 10.1093/genetics/129.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goshorn A K, Grindle S M, Scherer S. Gene isolation by complementation in Candida albicans and applications to physical and genetic mapping. Infect Immun. 1992;60:876–884. doi: 10.1128/iai.60.3.876-884.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holz R W, Brondyk W H, Senter R A, Kuizon L, Macara I G. Evidence for the involvement of Rab3A in Ca(2+)-dependent exocytosis from adrenal chromaffin cells. J Biol Chem. 1994;269:10229–10234. [PubMed] [Google Scholar]
- 12.Hube B, Sanglard D, Odds F C, Hess D, Monod M, Schafer W, Brown A J, Gow N A. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect Immun. 1997;65:3529–3538. doi: 10.1128/iai.65.9.3529-3538.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones S, Litt R J, Richardson C J, Segev N. Requirement of nucleotide exchange factor for Ypt1 GTPase mediated protein transport. J Cell Biol. 1995;130:1051–1061. doi: 10.1083/jcb.130.5.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazar T, Gotte M, Gallwitz D. Vesicular transport: how many Ypt/Rab-GTPases make a eukaryotic cell? Trends Biochem Sci. 1997;22:468–472. doi: 10.1016/s0968-0004(97)01150-x. [DOI] [PubMed] [Google Scholar]
- 15.Leberer E, Ziegelbauer K, Schmidt A, Harcus D, Dignard D, Ash J, Johnson L, Thomas D Y. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr Biol. 1997;7:539–546. doi: 10.1016/s0960-9822(06)00252-1. [DOI] [PubMed] [Google Scholar]
- 16.Lodge J K, Jackson-Machelski E, Toffaletti D L, Perfect J R, Gordon J I. Targeted gene replacement demonstrates that myristoyl-CoA:protein N-myristoyltransferase is essential for viability of Cryptococcus neoformans. Proc Natl Acad Sci USA. 1994;91:12008–12012. doi: 10.1073/pnas.91.25.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacDonald D W, Cove D J. Studies of temperature-sensitive cnx mutants in the fungus Aspergillus nidulans. Biochem J. 1972;127:19. doi: 10.1042/bj1270019pa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monteoliva L, Sanchez M, Pla J, Gil C, Nombela C. Cloning of Candida albicans SEC14 gene homologue coding for a putative essential function. Yeast. 1996;11:1097–1105. doi: 10.1002/(SICI)1097-0061(19960915)12:11%3C1097::AID-YEA990%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 19.Nieto A, Sanz P, Sentandreu R, del Castillo Agudo L. Cloning and characterization of the SEC18 gene from Candida albicans. Yeast. 1993;8:875–887. doi: 10.1002/yea.320090808. [DOI] [PubMed] [Google Scholar]
- 20.Novick P, Field C, Sheckman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 21.Pla J, Perez-Diaz R M, Navarro-Garcia F, Sanchez M, Nombela C. Cloning of the Candida albicans HIS1 gene by direct complementation of a C. albicans histidine auxotroph using an improved double-ARS shuttle vector. Gene. 1995;165:115–120. doi: 10.1016/0378-1119(95)00492-o. [DOI] [PubMed] [Google Scholar]
- 22.Pryer N K, Wuestehube L J, Scheckman R. Vesicle-mediated protein sorting. Annu Rev Biochem. 1992;61:471–516. doi: 10.1146/annurev.bi.61.070192.002351. [DOI] [PubMed] [Google Scholar]
- 23.Salminen A, Novick P J. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- 24.Sanglard D, Hube B, Monod M, Odds F C, Gow N A. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect Immun. 1997;65:3539–3546. doi: 10.1128/iai.65.9.3539-3546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tisdale E J, Bourne J R, Khosravi-Far R, Der C J, Balch W E. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol. 1992;119:749–761. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valencia A, Chardin P, Wittinghofer A, Sander C. The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry. 1991;30:4638–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- 27.Walworth N C, Goud B, Kabcenell A K, Novick P J. Mutational analysis of SEC4 suggests a cyclical mechanism for the regulation of vesicular traffic. EMBO J. 1989;8:1685–1693. doi: 10.1002/j.1460-2075.1989.tb03560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinberg R A, McWherter C A, Freeman S K, Wood D C, Gordon J I, Lee S C. Genetic studies reveal that myristoylCoA:protein N-myristoyltransferase is an essential enzyme in Candida albicans. Mol Microbiol. 1995;16:241–250. doi: 10.1111/j.1365-2958.1995.tb02296.x. [DOI] [PubMed] [Google Scholar]
- 29.White T C, Miyasaki S H, Agabian N. Three distinct secreted aspartyl proteinases in Candida albicans. J Bacteriol. 1993;175:6126–6133. doi: 10.1128/jb.175.19.6126-6133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaret K S, Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982;28:563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]