Abstract
The emergence of carbapenem-resistant Klebsiella pneumoniae (CRKP) co-carrying multiple carbapenemases is complicating clinical treatment. This study aimed to investigate the global dissemination trends of CRKP strains that co-carry multiple carbapenemases. The CRKP isolate KP424 co-carrying blaNDM-1 and blaKPC-2, recovered from a stool specimen, was identified by the NG-Test Carba 5 test, and the genome sequence was further determined by using Nanopore MinION and Illumina NovaSeq 6000 technologies. The genome sequences of the CRKP strains carrying multiple carbapenemase genes were further retrieved from the NCBI GenBank database. Thirteen antimicrobial resistance genes, including blaNDM-1 and blaKPC-2, have been identified in KP424, with blaNDM-1 and blaKPC-2 located on different plasmids. In total, 832 genome sequences of CRKP strains co-carrying two carbapenemase genes were retrieved from the NCBI database. Strains carrying both blaNDM and blaOXA-48-like accounted for 665 (79.9 %) of the total strains, ranking first, and those carrying both blaKPC and blaNDM accounted for 103 (12.4 %), ranking second. The prevalence of CRKP strains co-carrying two carbapenemase genes increased significantly over time, from 0.40 % in 2010 to 9.67 % in 2021. The proportion of strains carrying both blaKPC and blaNDM has also increased, from 0.00 % in 2010 to 4.40 % in 2021. The strains carrying both blaKPC and blaNDM had the highest prevalence (66.7 %, 52/78) in China, while those carrying both blaNDM and blaOXA-48-like had the highest prevalence worldwide. Multiple-carbapenemase producers pose a great threat to public health; further research on the mechanisms underlying multiple carbapenemase gene occurrence is required to prevent their global dissemination.
Keywords: Klebsiella pneumoniae, Carbapenemase, NDM, KPC, Whole-genome sequencing
Graphical Abstract
1. Introduction
Klebsiella pneumoniae, an Enterobacterales member, is a clinically significant, gram-negative, opportunistic pathogen that can cause a variety of infections, including respiratory infections, urinary tract infections, surgical site infections, ventilator-associated pneumonia, and bacteraemia. The bactericidal effects of antibiotics on K. pneumoniae are waning gradually as a result of the ongoing rise of carbapenem-resistant K. pneumoniae (CRKP) in clinical settings, making the choice of clinical treatment more challenging.
The main mechanism underlying the carbapenem resistance of CRKP is the production of carbapenemase. K. pneumoniae carbapenemases (KPCs), New Delhi metallo-beta-lactamase (NDM) and oxacillinase-48 (OXA-48) are the three most commonly reported carbapenemases worldwide, and their widespread dissemination poses a global public health threat [1], [2]. The first case of KPC-producing K. pneumoniae was discovered in the United States in 1996 and reported in 2001; such strains have subsequently been identified in many countries [3]. NDM−1 was initially isolated from K. pneumoniae in India in 2009 and subsequently spread quickly worldwide [4]. Throughout much of Europe, Northern Africa, and the Middle East, OXA-48 enzyme has proliferated to become the most prevalent enterobacterial carbapenemase [2]. KPC and OXA-48 are Ambler class A enzymes that are not inhibited by clavulanate or tazobactam but are inhibited by avibactam [5]. NDM is an Ambler class B enzyme, also known as a metallo-beta-lactamase, that is unaffected by inhibitors but susceptible to monobactam lactams such as aztreonam. Compared to nonproducing strains, K. pneumoniae strains producing carbapenemase frequently show higher rates of resistance to other antibiotics, such as aminoglycosides and quinolones, severely limiting clinical treatment options. Ceftazidime/avibactam (CZA) is currently the first-line drug for treating infections caused by KPC or OXA-48 producing K. pneumoniae, but it has no activity against class B carbapenemases [6].
In this study, genomic characterization of a strain carrying both blaKPC and blaNDM was performed. Furthermore, we retrieved the genome sequences of CRKP strains co-carrying two of the five major carbapenemase genes, blaKPC, blaNDM, blaOXA-48-like, blaVIM, and blaIMP, from the NCBI GenBank database reported from 1980 to 2022 in 105 countries. The genomic characteristics and trends of global dissemination of CRKP strains co-carrying two carbapenemase genes were further investigated.
2. Materials and methods
2.1. Clinical isolate
In August 2021, a male patient with severe pneumonia was admitted to a tertiary hospital in Hangzhou, China. The CRKP strain assigned as KP424 was isolated from a stool sample collected from this patient. NG-Test Carba 5 (NG Biotech, France) was used for the detection of common carbapenemases in KP424. The isolate was identified initially using the VITEK MS system (bioMérieux, France) and further confirmed by 16S rRNA Sanger sequencing.
2.2. Antimicrobial susceptibility test
Antimicrobial susceptibility testing was carried out in accordance with the Clinical and Laboratory Standards Institute (CLSI) recommendations. The antimicrobial agents cefotaxime, ceftazidime, cefepime, cefoxitin, aztreonam, imipenem, meropenem, ceftazidime/avibactam, gentamicin, amikacin, ciprofloxacin, levofloxacin, sulfamethoxazole/trimethoprim and colistin (Sigma, USA) were used in the test. Antimicrobial susceptibility was determined using breakpoints approved by the CLSI [7]. Standard broth microdilution tests were performed with Mueller–Hinton broth (cation-adjusted; Oxoid Ltd., England) to determine the minimum inhibitory concentration (MIC) of the above antimicrobial agents. Tigecycline (Sigma, USA) was tested using fresh (<12 h) Mueller–Hinton broth. The MIC of cefiderocol (Shionogi, Japan) was determined using iron-depleted cation-adjusted Mueller–Hinton broth (ID-CAMHB) prepared with Chelex® 100 resin (Bio-Rad Laboratories, USA). For quality control, E. coli ATCC 25922 was used. As there are no CLSI breakpoints for tigecycline, the FDA standard was adopted. The EUCAST guidelines were used to interpret the colistin MIC.
2.3. Isolates retrieved from the NCBI GenBank database
CRKP strains co-carrying two of the five major carbapenemase genes were retrieved from the NCBI GenBank database. Strains with assembled genomic data were included in this study, while strains with only raw reads were excluded. A total of 832 genome sequences of CRKP strains were obtained. The strains were reported in 1980–2010 (n = 4, 0.48 %), 2011 (n = 4, 0.48 %), 2012 (n = 6, 0.72 %), 2013 (n = 24, 2.88 %), 2014 (n = 41, 4.93 %), 2015 (n = 71, 8.53 %), 2016 (n = 189, 22.72 %), 2017 (n = 123, 14.78 %), 2018 (n = 130, 15.63 %), 2019 (n = 160, 19.23 %), 2020 (n = 36, 4.33 %), and 2021 to the present (n = 44, 5.29 %).
2.4. DNA extraction and whole-genome sequencing
The genomic DNA of K. pneumoniae KP424 was extracted using the QIAamp DNA Mini Kit (Qiagen, USA). A long-read MinION sequencer (Nanopore, Oxford, UK) and the Illumina NovaSeq 6000 platform (Illumina Inc., San Diego, CA, USA) were used to determine the strain's complete genome sequence. Using Unicycler (v0.4.7) in conservative mode, both long MinION reads and short Illumina reads were subjected to hybrid assembly. Pilon was used to create and correct entire circular contigs with Illumina reads over several rounds until no change was noted [8]. The NCBI Prokaryotic Genome Annotation Pipeline (PGAP) server automatically annotated the entire genome.
2.5. Genomic analysis
Using the BacWGSTdb server, multilocus sequence typing (MLST), virulence gene identification, and plasmid replicon identification for K. pneumoniae KP424 were performed [9], [10]. The ResFinder 4.1 server was used to identify antimicrobial resistance genes (ARGs) of KP424. Capsule and lipopolysaccharide serotypes of KP424 were predicted using Kaptive [11], [12]. Through the use of ISfinder, insertion sequences (ISs) of KP424 were predicted [13]. Plasmid sequences of KP424 were uploaded to the NCBI database, and plasmids that matched the target plasmid were identified. The BLAST Ring Image Generator was used to generate concentrated ring comparisons between the blaNDM- or blaKPC-carrying plasmid and similar plasmids (BRIG) [14]. Genetic analysis of 832 CRKP strains co-carrying two carbapenemase genes was carried out by Kleborate [15].
2.6. Phylogenetic analysis
The BacWGSTdb server was used along with core-genome MLST (cgMLST) methods to conduct the initial analysis of the phylogenetic relationships between KP424 and other K. pneumoniae strains. The phylogenetic tree was also examined using CSI Phylogeny (version 1.4) [16], which is based on a core-genome single-nucleotide polymorphism (SNP) strategy. CSI Phylogeny was applied to call and filter K. pneumoniae KP424 SNPs, perform site validation, and infer phylogeny based on concatenated alignment of the high-quality SNPs. The maximum parsimony algorithm was used to create a phylogenetic tree from the resulting SNPs, which was then visualized on the iTOL webpage [17]. The core SNP alignment of KP424 and other K. pneumoniae strains co-carrying blaNDM and blaKPC carbapenemase genes was generated by Snippy and visualized by the iTOL online webserver [17].
2.7. Nucleotide sequence accession numbers
The whole-genome results of strain KP424 have been deposited in DDBJ/EMBL/GenBank under accession numbers CP109983-CP109991.
3. Results and discussion
The MICs of antimicrobial agents against K. pneumoniae KP424 are presented in Table S1. KP424 is a multidrug-resistant strain that was resistant to most of the antibiotics tested, including cefotaxime, ceftazidime, cefepime, cefoxitin, aztreonam, imipenem, meropenem, ceftazidime/avibactam, gentamicin, amikacin, ciprofloxacin and levofloxacin, but was susceptible to sulfamethoxazole/trimethoprim, tigecycline, colistin and cefiderocol.
The K. pneumoniae KP424 genome consists of nine contigs totalling 6044973 bp. Contig 1 (5460,786 bp) belongs to the chromosome, and the other contigs belong to eight different plasmids (contig 2: 293,179 bp; contig 3: 93,565 bp; contig 4: 86,272 bp; contig 5: 81,442 bp; contig 6: 11,057 bp; contig 7: 10,060 bp; contig 8: 7329 bp; contig 9: 1283 bp). The PGAP server predicted a total of 5639 protein-coding sequences, 89 tRNA genes, and 25 rRNA operons. According to the MLST scheme of K. pneumoniae, KP424 belongs to sequence type 15 (ST15). The KL type of KP424 is predicted to be KL24. The genome contains a variety of IS elements, the majority of which belong to the IS5, IS630, and IS3 families (Table S2).
The thirteen antimicrobial resistance genes that were identified in the KP424 genome are presented in Supplementary Table S3. We identified the β-lactam resistance genes blaSHV-106, blaSHV-28, blaDHA-1, blaNDM-1 and blaKPC-2; the aminoglycoside resistance gene armA; the fosfomycin resistance gene fosA; the macrolide resistance genes mph(E) and msr(E); the quinolone resistance genes qnrB4, oqxA and oqxB; and the sulphonamide resistance gene sul1. The genes oqxA, oqxB, blaSHV-106, blaSHV-28 and fosA were located on the chromosome. The genes mph(E), msr(E), armA, sul1, blaDHA-1 and qnrB4a were located in contig 2; blaNDM-1 was located in contig 4; and blaKPC-2 was located in contig 8. Eight plasmid replicons were also identified in the genome (Table S4): IncFIB and IncHI1B on contig 2, IncFIB and IncFII on contig 4, FIA and IncFII on contig 5, Col on contig 6 and ColRNAI on contig 7.
The blaNDM-1-carrying plasmid (contig 4) was named pNDM-1-KP424. The Basic Local Alignment Search Tool (BLASTN) was used to compare pNDM-1-KP424 to other blaNDM-1-carrying plasmids, and several similar plasmids were found in the NCBI GenBank database (Fig. S1). The most closely related plasmid shared 100 % coverage and 100 % identity with pNDM-1-KP424 (plasmid pRJF866 from K. pneumoniae strain RJF866, accession no. KF732966) [18]. The blaKPC-2-carrying plasmid (contig 8) was named pKPC-2-KP424. Sequence comparisons revealed similarities with many previously reported plasmid sequences, but the plasmids were smaller.
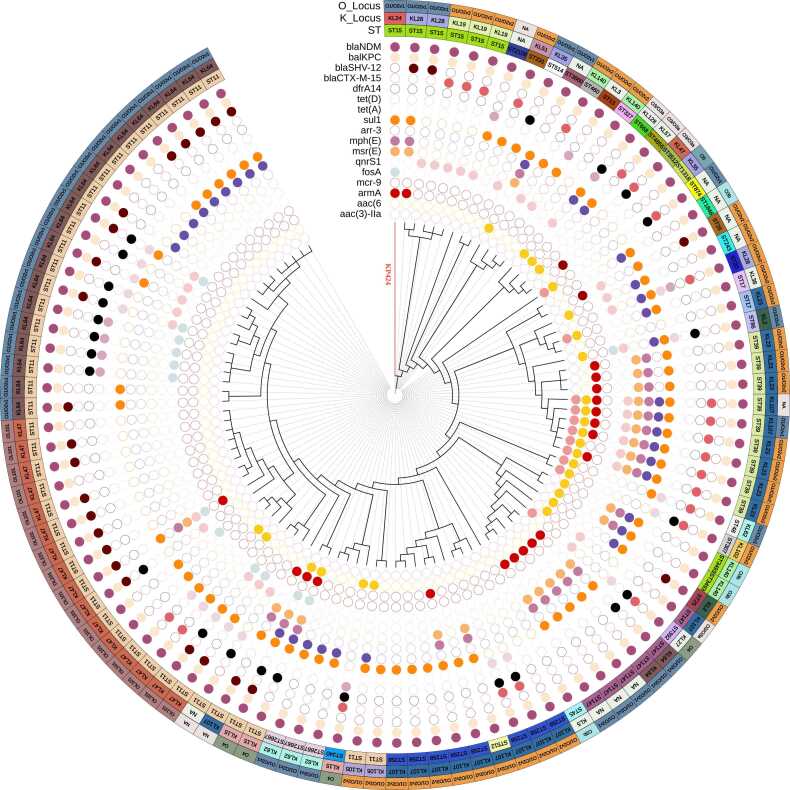
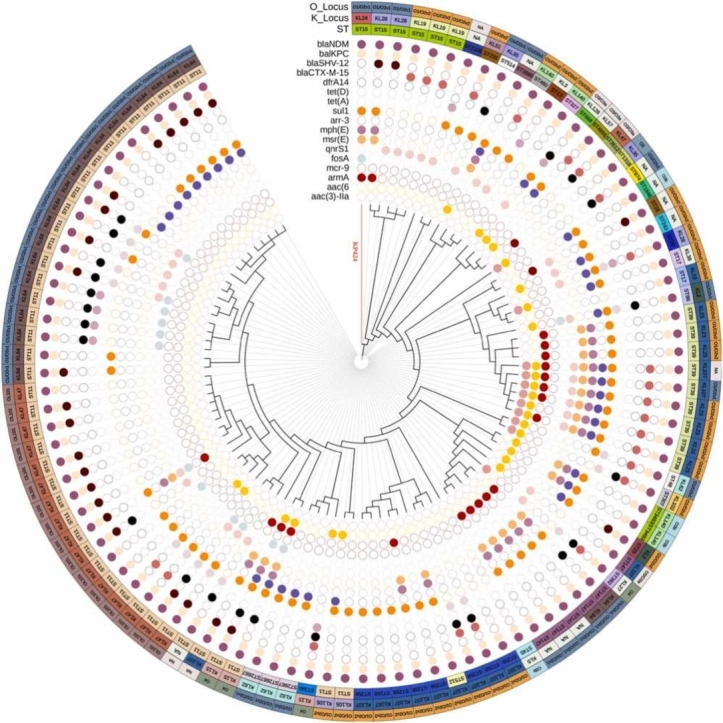
Supplementary Fig. S2 depicts the phylogenetic relationship between KP424 and other ST15 K. pneumoniae strains obtained from the BacWGSTdb using the cgMLST strategy. The database currently contains 281 ST15 K. pneumoniae strains, 25 of which are from China. The phylogenetic relationships between these 25 strains are very close. The strain most similar to KP424 is CL2079, which has 109 different cgMLST loci and carries only the blaKPC-2 gene. This strain was found in a clinical specimen from Jiangsu, China, in 2012. Using NCBI Pathogen Detection, a total of seven ST15 K. pneumoniae strains carrying both blaNDM-1 and blaKPC-2 could be identified in the database (Table S5). A phylogenetic tree was constructed for KP424 and the seven strains using a core-genome SNP strategy (Fig. S3). Five of the eight strains originated in China, two in Turkey, and one in Vietnam. They are classified into four serotypes: KL19, KL28, KL24, and KL112. They all contain fosA, oqxA, oqxB, blaSHV-28, blaSHV-106, blaNDM and blaKPC genes. Except for three strains from Jinan, China, which are closely related, all of the strains are distantly related. Fig. 1 depicts the phylogenetic relationships of KP424 and 103 other strains carrying both blaNDM and blaKPC in terms of STs and K-Locus, O-Locus, and antibiotic resistance genes. Multiple antibiotic resistance genes are present in these strains, and the phylogenetic relationships of strains with the same ST are relatively close. Interestingly, only KP424 belonged to KL24, while all ST39 strains belonged to KL23, and all ST258 strains belonged to KL107.
Fig. 1.
Phylogenetic analyses of KP424 and 103 K. pneumoniae strains co-carrying blaNDM and blaKPC. The three outer circles indicate the O_Locus, K_Locus and sequence types, with different colours representing different types and accompanied by text notes. Cells of various colours indicate the presence of various antimicrobial resistance genes in the inner 17 circles, while blank cells indicate the absence of the gene. The branches of KP424 are highlighted in red in the figure.
From 1980 to 2022, 832 genome sequences of CRKP strains co-carrying two of the five major carbapenemase genes from 105 countries were retrieved from the NCBI GenBank database. The strains that co-carried blaNDM and blaOXA-48-like genes accounted for 665 (79.9 %) of these, ranking first, and strains that co-carried blaKPC and blaNDM genes accounted for 103 (12.4 %), ranking second. In recent decades, the prevalence of CRKP co-carrying two carbapenemase genes showed a clear upwards trend over time, from 0.40 % before 2010–9.67 % in 2021 (Table 1). The frequency of strains carrying both blaKPC and blaNDM also increased, from 0.00 % before 2010–4.40 % in 2021. Furthermore, we investigated if the two carbapenemases were located on the same plasmid. However, only 49 of the 832 were complete genomes, while the remaining 783 strains were draft genomes. Carbapenemases were all found in separate plasmids in the 49 complete genomes. In the 783 draft genomes, the two carbapenemase genes are located in different contigs in the majority of strains, with the exception of six strains that have both carbapenemase genes on the same contig.
Table 1.
Prevalence of CRKP carrying two carbapenemases from 1980 to 2022.
|
∼2010 (n = 1001)a |
2011 (n = 735) |
2012 (n = 1032) |
2013 (n = 1969) |
2014 (n = 2766) |
2015 (n = 2290) |
2016 (n = 2904) |
2017 (n = 2855) |
2018 (n = 2874) |
2019 (n = 1591) |
2020 (n = 544) |
2021∼ (n = 455) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blaNDM and blaOXA-48-like | 0 | 1 | 3 | 16 | 31 | 60 | 180 | 101 | 101 | 126 | 22 | 24 |
| blaNDM and blaKPC | 0 | 0 | 0 | 1 | 0 | 4 | 6 | 18 | 20 | 24 | 10 | 20 |
| blaKPC and blaOXA-48-like | 0 | 0 | 0 | 2 | 3 | 7 | 1 | 2 | 0 | 9 | 2 | 0 |
| blaKPC and blaVIM | 4 | 2 | 3 | 4 | 3 | 0 | 0 | 1 | 3 | 0 | 2 | 0 |
| blaNDM and blaIMP | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 2 | 1 | 0 | 0 |
| blaNDM and blaVIM | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 0 | 0 |
| blaVIM and blaOXA-48-like | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| blaKPC and blaIMP | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| blaVIM and blaIMP | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| blaIMP and blaOXA-48-like | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 4 | 4 | 6 | 24 | 41 | 71 | 189 | 123 | 130 | 160 | 36 | 44 |
| Prevalence of CRKP carrying blaNDM and blaKPC | 0.00 % | 0.00 % | 0.00 % | 0.05 % | 0.00 % | 0.17 % | 0.21 % | 0.63 % | 0.70 % | 1.51 % | 1.84 % | 4.40 % |
| Prevalence of CRKP carrying two carbapenemases | 0.40 % | 0.54 % | 0.58 % | 1.21 % | 1.48 % | 3.10 % | 6.51 % | 4.31 % | 4.52 % | 10.06 % | 6.61 % | 9.67 % |
Number of CRKP genome sequences retrieved from the National Center for Biotechnology Information GenBank database by the year.
In terms of geographical distribution, Thailand (284 strains), the United States (82 strains), and China (78 strains) had the greatest prevalence of two carbapenemase-producing strains (Table 2). All strains screened from Thailand carried both blaNDM and blaOXA-48-like (284 strains, 100 %). The strains that carried both blaNDM and blaOXA-48-like were predominant in the United States (54 strains, 65.9 %), followed by those carrying both blaNDM and blaKPC (26 strains, 31.7 %). The strains that carried both blaNDM and blaKPC predominated among the 78 strains from China. The strains that carried both blaNDM and blaOXA-48-like formed the bulk of strains recovered from the United Kingdom (61 strains, 93.8 %). The strains from other Asian and European nations were likewise dominated by strains that carried both blaNDM and blaOXA-48-like, which accounted for 91.7 % and 78.0 %, respectively. The most prevalent sequence types discovered were ST16 (34.9 %), ST147 (10.2 %), ST14 (9.6 %), and ST11 (9.4 %), with ST16, ST147, and ST14 dominated by strains that carried both blaNDM and blaOXA-48-like and ST11 by strains that carried both blaNDM and blaKPC. KL51 (33.8 %), KL64 (11.9 %), and KL2 (7.7 %) were the three most prevalent K loci, whereas O3b (34.7 %), O1/O2v1 (31.0 %), and O1/O2v2 (27.7 %) dominated the O loci.
Table 2.
Characteristics of strains carrying two carbapenemases.
| Total (n = 832) |
blaNDM and blaOXA-48-like (n = 665) |
blaNDM and blaKPC (n = 103) |
blaKPC and blaOXA-48-like (n = 26) |
blaKPC and blaVIM (n = 22) |
blaNDM and blaIMP (n = 7) |
blaNDM and blaVIM (n = 4) |
blaVIM and blaOXA-48-like (n = 3) |
blaKPC and blaIMP (n = 2) |
blaVIM and blaIMP (n = 0) |
blaIMP and blaOXA-48-like (n = 0) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| MLST | |||||||||||
| ST11 | 78 | 17(2.6 %) | 42(40.8 %) | 13(50.0 %) | 2(9.1 %) | 0 | 2(50.0 %) | 1(33.3 %) | 1(50.0 %) | 0 | 0 |
| ST16 | 290 | 290(43.6 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST14 | 80 | 80(12.0 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST39 | 24 | 7(1.1 %) | 9(8.7 %) | 8(30.8 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST258 | 9 | 0 | 8(7.8 %) | 1(3.8 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST147 | 85 | 69(10.4 %) | 6(5.8 %) | 1(3.8 %) | 8(36.4 %) | 0 | 0 | 1(33.3 %) | 0 | 0 | 0 |
| ST15 | 21 | 16(2.4 %) | 5(4.9 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST2667 | 3 | 0 | 3(2.9 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST307 | 59 | 56(8.4 %) | 1(1.0 %) | 1(3.8 %) | 1(4.5 %) | 0 | 0 | 0 | 0 | 0 | 0 |
| ST231 | 19 | 16(2.4 %) | 1(1.0 %) | 2(7.7 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST101 | 29 | 29(4.4 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST383 | 22 | 20(3.0 %) | 0 | 0 | 2(9.1 %) | 0 | 0 | 0 | 0 | 0 | 0 |
| other | 113 | 65(9.8 %) | 28(27.2 %) | 0 | 9(40.9 %) | 7(100.0 %) | 2(50.0 %) | 1(33.3 %) | 1(50.0 %) | 0 | 0 |
| K locus | |||||||||||
| KL64 | 99 | 63(9.5 %) | 22(21.4 %) | 2(7.7 %) | 8(36.4 %) | 1(14.3 %) | 1(25.0 %) | 1(33.3 %) | 1(50.0 %) | 0 | 0 |
| KL47 | 20 | 0 | 17(16.5 %) | 3(11.5 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| KL23 | 19 | 5(0.8 %) | 8(7.8 %) | 6(23.1 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| KL107 | 20 | 5(0.8 %) | 13(12.6 %) | 0 | 2(9.1 %) | 0 | 0 | 0 | 0 | 0 | 0 |
| KL106 | 1 | 0 | 0 | 1(3.8 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| KL105 | 4 | 0 | 2(1.9 %) | 0 | 0 | 0 | 2(50.0 %) | 0 | 0 | 0 | 0 |
| KL102 | 58 | 56(8.4 %) | 1(1.0 %) | 1(3.8 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| KL17 | 22 | 22(3.3 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| KL30 | 20 | 18(2.7 %) | 0 | 0 | 2(9.1 %) | 0 | 0 | 0 | 0 | 0 | 0 |
| KL51 | 281 | 279(42.0 %) | 1(1.0 %) | 1(3.8 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| KL2 | 64 | 62(9.3 %) | 2(1.9 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| other | 224 | 155(23.3 %) | 37(35.9 %) | 12(46.2 %) | 10(45.5 %) | 6(85.7 %) | 1(25.0 %) | 2(66.7 %) | 1(50.0 %) | 0 | 0 |
| O locus | |||||||||||
| O1/O2v2 | 172 | 121(18.2 %) | 32(31.1 %) | 10(38.5 %) | 5(22.7 %) | 0 | 2(50.0 %) | 1(33.3 %) | 1(5.0 %) | 0 | 0 |
| O1/O2v1 | 258 | 199(29.9 %) | 36(35.0 %) | 6(23.1 %) | 11(50.0 %) | 1(14.3 %) | 2(50.0 %) | 2(66.7 %) | 1(5.0 %) | 0 | 0 |
| O3b | 289 | 275(41.4 %) | 4(3.9 %) | 6(23.1 %) | 3(13.6 %) | 1(14.3 %) | 0 | 0 | 0 | 0 | 0 |
| O4 | 25 | 18(2.7 %) | 4(3.9 %) | 0 | 2(9.1 %) | 1(14.3 %) | 0 | 0 | 0 | 0 | 0 |
| OL101 | 33 | 14(2.1 %) | 16(15.5 %) | 3(11.5 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| O5 | 5 | 1(0.2 %) | 1(1.0 %) | 0 | 1(4.5 %) | 2(28.6 %) | 0 | 0 | 0 | 0 | 0 |
| other | 50 | 37(5.6 %) | 10(9.7 %) | 1(3.8 %) | 0 | 2(28.6 %) | 0 | 0 | 0 | 0 | 0 |
| Country | |||||||||||
| USA | 82 | 54(8.1 %) | 26(25.2 %) | 2(7.7 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| China | 78 | 7(1.1 %) | 52(50.5 %) | 9(34.6 %) | 2(9.1 %) | 6(85.7 %) | 0 | 0 | 2(100.0 %) | 0 | 0 |
| Thailand | 284 | 284(42.7 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| United Kingdom | 65 | 61(9.2 %) | 1(1.0 %) | 1(3.8 %) | 2(9.1 %) | 0 | 0 | 0 | 0 | 0 | 0 |
| Africa | 3 | 3(0.5 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| other Asian countries | 109 | 100(15.0 %) | 5(4.9 %) | 1(3.8 %) | 0 | 1(14.3 %) | 1(25.0 %) | 1(33.3 %) | 0 | 0 | 0 |
| other European countries | 182 | 142(21.4 %) | 11(10.7 %) | 13(50.0 %) | 13(59.1 %) | 0 | 1(25.0 %) | 2(66.7 %) | 0 | 0 | 0 |
| other North American countries | 10 | 9(1.4 %) | 0 | 0 | 1(4.5 %) | 0 | 0 | 0 | 0 | 0 | 0 |
| Oceania | 5 | 5(0.8 %) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| South America | 14 | 0 | 8(7.8 %) | 0 | 4(18.2 %) | 0 | 2(50.0 %) | 0 | 0 | 0 | 0 |
The geographic distribution of two common carbapenemase combinations is depicted in Fig. 2. K. pneumoniae strains co-carrying blaNDM and blaOXA-48-like were most common in Thailand (42.7 %), Germany (10.2 %), and the United Kingdom (9.2 %). ST16 predominated in Thailand (89.9 %, 254/283), ST147 in the United States (21.0 %, 13/62) and ST14 in the United Kingdom (40.0 %, 30/75). K. pneumoniae strains co-carrying blaKPC and blaNDM were mainly predominant in China (50.5 %), the United States (25.2 %), and Russia (5.8 %). ST11 was most common in China (95.2 %, 40/42), ST258 in the United States (100 %, 8/8) and ST39 in Russia (66.7 %, 6/9) for strains co-carrying blaKPC and blaNDM.
Fig. 2.
Geographic distribution of two common carbapenemase combinations. a) The main geographical distribution of K. pneumoniae strains co-carrying blaNDM and blaOXA-48-like. b) The main geographical distribution of K. pneumoniae strains co-carrying blaKPC and blaNDM. The pie charts depict the proportion of ST types in each country. The names of the countries and the total number of strains are also labelled within the pie chart. The sequence types are differentiated and annotated by colour.
In recent years, there has been an increase in reports of K. pneumoniae isolates containing two carbapenemases [19], [20], [21], [22]. In addition to K. pneumoniae, there is an increasing number of strains that produce multiple carbapenemases in other enterobacterial species [23]. Multiple-carbapenemase-producing bacteria exhibit an increase in antimicrobial resistance, giving them an advantage over antimicrobial treatments. In some cases, these strains have resulted in hospital transmission and outbreaks [24]. In this study, we present a complete genome sequence analysis of an ST15 K. pneumoniae strain that carries both blaNDM and blaKPC. ST15 K. pneumoniae strains carrying both blaKPC and blaNDM are uncommon. This strain carries multiple drug resistance genes that can be transmitted horizontally between strains via plasmids.
Of the 832 strains carrying two carbapenemase genes retrieved from the NCBI GenBank database, a total of 779 strains carried the blaNDM gene, and most of the strains co-producing two carbapenemases carried the blaNDM gene. Multiple-carbapenemase-producing isolates that express metallo-β-lactamases are resistant to β-lactam/β-lactamase inhibitor combinations such as ceftazidime/avibactam. Reportedly, during treatment, KPC-2-producing K. pneumoniae acquired a blaNDM-5-carrying plasmid, leading to resistance to ceftazidime/avibactam [6]. The acquisition of the blaNDM gene by blaKPC- or blaOXA-48-like-carrying strains can help them better resist additional antimicrobial agents, i.e., ceftazidime/avibactam [25]. Cefiderocol, a new siderophore antibiotic, provides potent broad-spectrum antimicrobial protection against gram-negative pathogens such as Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacterales, including carbapenem-resistant isolates [26]. Cefiderocol is an important complement for treatments against current multidrug-resistant, clinically refractory gram-negative infections. Cefiderocol showed effective activity against most Enterobacterales that co-produced two carbapenemases [23]. However, cefiderocol resistance has been reported in isolates of Enterobacterales co-producing two carbapenemases, and the presence of multiple copies of NDM affects the antibacterial effect of cefiderocol [23], [27]. Therefore, it is important to test cefiderocol on carbapenem-resistant Enterobacterales. Over the last two decades, the OXA-48 enzyme has spread to become one of the most prevalent carbapenemases in Europe, Northern Africa, the Mediterranean, and the Middle East. Since OXA-48-like enzymes frequently only result in low-level resistance to carbapenems, it is possible that the prevalence of OXA-48 is underestimated. This would also indicate that the prevalence of double carbapenemase producers may also be underestimated.
4. Conclusions
Over the last ten years, the proportion of CRKP strains carrying multiple carbapenemase genes has increased significantly. CRKP strains carrying both blaNDM and blaOXA-48-like have already shown a global proliferation trend, whereas those carrying both blaNDM and blaKPC are more commonly recovered in China. blaNDM was found to be more prevalent in multiple-carbapenemase-producing strains. Multiple-carbapenemase producers represent a greater threat to public health because they are more difficult to control and may serve as a reservoir of carbapenemase genes for other bacterial pathogens. To prevent their global expansion, further studies on the mechanisms underlying the co-occurrence of multiple carbapenemase genes are warranted.
CRediT authorship contribution statement
FH and JX designed the experiments. HG, YYW and JFW performed the experiments and were the major contributors in writing the manuscript. LRL and JX analyzed the data. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82172314), Zhejiang Provincial Medical and Health Science and Technology plan (2022KY022, 2023KY484 and 2021KY827), and Research Foundation of Zhejiang Provincial Department of Education (Y202045219).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.07.013.
Contributor Information
Juan Xu, Email: 2020000275@hmc.edu.cn.
Fang He, Email: hefang@hmc.edu.cn.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Xu J., Zhao Z., Ge Y., He F. Unravelling the genome sequence of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae ST11 isolated from a bloodstream infection. J Glob Antimicrob Resist. 2020;20:339–341. doi: 10.1016/j.jgar.2020.01.021. [DOI] [PubMed] [Google Scholar]
- 2.Boyd S.E., Holmes A., Peck R., Livermore D.M., Hope W. OXA-48-Like β-Lactamases: global epidemiology, treatment options, and development pipeline. Antimicrob Agents Chemother. 2022;66 doi: 10.1128/aac.00216-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yigit H., Queenan A.M., Anderson G.J., Domenech-Sanchez A., Biddle J.W., Steward C.D., et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yong D., Toleman M.A., Giske C.G., Cho H.S., Sundman K., Lee K., et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush K., Bradford P.A. Epidemiology of β-lactamase-producing pathogens. Clin Microbiol Rev. 2020:33. doi: 10.1128/CMR.00047-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J., Zhang S., Zhao Z., Chen M., Cao Y., Li B. Acquisition of a stable and transferable bla(NDM-5)-positive plasmid with low fitness cost leading to ceftazidime/avibactam resistance in KPC-2-producing Klebsiella pneumoniae during treatment. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.658070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CLSI. Clinical and Laboratory Standards Institute (CLSI) M100, 2021. Performance Standards for Antimicrobial Susceptibility Testing, 31th ed. 2021.
- 8.Ruan Z., Wu J., Chen H., Draz M.S., Xu J., He F. Hybrid genome assembly and annotation of a pandrug-resistant Klebsiella pneumoniae strain using nanopore and illumina sequencing. Infect Drug Resist. 2020;13:199–206. doi: 10.2147/IDR.S240404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng Y., Zou S., Chen H., Yu Y., Ruan Z. BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 2021;49:D644–d50. doi: 10.1093/nar/gkaa821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruan Z., Feng Y. BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 2016;44:D682–D687. doi: 10.1093/nar/gkv1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyres K.L., Wick R.R., Gorrie C., Jenney A., Follador R., Thomson N.R., et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016;2 doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam M.M.C., Wick R.R., Judd L.M., Holt K.E., Wyres K.L. Kaptive 2.0: updated capsule and lipopolysaccharide locus typing for the Klebsiella pneumoniae species complex. Microb Genom. 2022;8 doi: 10.1099/mgen.0.000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kichenaradja P., Siguier P., Pérochon J., Chandler M. ISbrowser: an extension of ISfinder for visualizing insertion sequences in prokaryotic genomes. Nucleic Acids Res. 2010;38:D62–D68. doi: 10.1093/nar/gkp947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alikhan N.F., Petty N.K., Ben Zakour N.L., Beatson S.A. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genom. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam M.M.C., Wick R.R., Watts S.C., Cerdeira L.T., Wyres K.L., Holt K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12:4188. doi: 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaas R.S., Leekitcharoenphon P., Aarestrup F.M., Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PloS One. 2014;9 doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J., Sun L., Ding B., Yang Y., Xu X., Liu W., et al. Outbreak of NDM-1-producing Klebsiella pneumoniae ST76 and ST37 isolates in neonates. Eur J Clin Microbiol Infect Dis: Publ Eur Soc Clin Microbiol. 2016;35:611–618. doi: 10.1007/s10096-016-2578-z. [DOI] [PubMed] [Google Scholar]
- 19.Wei D.D., Wan L.G., Liu Y. Draft genome sequence of an NDM-1- and KPC-2-coproducing hypervirulent carbapenem-resistant Klebsiella pneumoniae strain isolated from burn wound infections. Genome Announc. 2018:6. doi: 10.1128/genomeA.00192-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Long D., Xiang T.X., Du F.L., Wei D.D., Wan L.G., et al. Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J Antimicrob Chemother. 2019;74:1233–1240. doi: 10.1093/jac/dkz023. [DOI] [PubMed] [Google Scholar]
- 21.Bes T., Nagano D., Martins R., Marchi A.P., Perdigão-Neto L., Higashino H., et al. Bloodstream Infections caused by Klebsiella pneumoniae and Serratia marcescens isolates co-harboring NDM-1 and KPC-2. Ann Clin Microbiol Antimicrob. 2021;20:57. doi: 10.1186/s12941-021-00464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumarasamy K., Kalyanasundaram A. Emergence of Klebsiella pneumoniae isolate co-producing NDM-1 with KPC-2 from India. J Antimicrob Chemother. 2012;67:243–244. doi: 10.1093/jac/dkr431. [DOI] [PubMed] [Google Scholar]
- 23.Bianco G., Boattini M., Comini S., Casale R., Iannaccone M., Cavallo R., et al. Occurrence of multi-carbapenemases producers among carbapenemase-producing Enterobacterales and in vitro activity of combinations including cefiderocol, ceftazidime-avibactam, meropenem-vaborbactam, and aztreonam in the COVID-19 era. Eur J Clin Microbiol Infect Dis: Publ Eur Soc Clin Microbiol. 2022;41:573–580. doi: 10.1007/s10096-022-04408-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raro O.H.F., da Silva R.M.C., Filho E.M.R., Sukiennik T.C.T., Stadnik C., Dias C.A.G., et al. Carbapenemase-producing Klebsiella pneumoniae from transplanted patients in Brazil: phylogeny, resistome, virulome and mobile genetic elements harboring bla (KPC-) (2) or bla (NDM-) (1) Front Microbiol. 2020;11:1563. doi: 10.3389/fmicb.2020.01563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soriano A., Carmeli Y., Omrani A.S., Moore L.S.P., Tawadrous M., Irani P. Ceftazidime-avibactam for the treatment of serious gram-negative infections with limited treatment options: a systematic literature review. Infect Dis Ther. 2021;10:1989–2034. doi: 10.1007/s40121-021-00507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito A., Sato T., Ota M., Takemura M., Nishikawa T., Toba S., et al. In vitro antibacterial properties of cefiderocol, a novel siderophore cephalosporin, against gram-negative bacteria. Antimicrob Agents Chemother. 2018:62. doi: 10.1128/AAC.01454-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simner P.J., Mostafa H.H., Bergman Y., Ante M., Tekle T., Adebayo A., et al. Progressive Development of Cefiderocol Resistance in Escherichia coli During Therapy Is Associated with Increased blaNDM-5 Copy Number and Gene Expression. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material