Abstract
The bacteriochlorophyll of the purple photosynthetic bacterium Rhodobacter sphaeroides is esterified with phytol. The presence of this alcohol moiety is essential for the correct assembly of the photosynthetic apparatus. Despite this, and the fact that R. sphaeroides is widely used for the study of structure-function relationships in photosynthesis, the molecular genetics of the steps in which the alcohol is added and modified have not previously been investigated in this organism. Sequencing near the center of the photosynthesis gene cluster has now revealed the existence of an open reading frame encoding a putative 394-amino-acid polypeptide displaying strong homology with the products of a number of genes from other photosynthetic organisms, each proposed to be responsible for the reduction of the alcohol moiety of (bacterio)chlorophyll to phytol. An R. sphaeroides transposon mutant in this gene, bchP, possessed a structurally modified photosystem assembled with bacteriochlorophyll esterified with geranylgeraniol, rather than with phytol, implying that the product of this gene was geranylgeranyl-bacteriochlorophyll reductase. This identification was confirmed by the performance of in vitro assays using heterologously expressed protein, providing the first direct demonstration of the activity of a bchP gene product.
The purple photosynthetic bacterium Rhodobacter sphaeroides contains bacteriochlorophyll a (bchla) as its principal photosynthetic pigment. This bchl is esterified with the C20 isoprenoid alcohol, phytol. This phytol moiety constitutes 30% of the total molecular weight and is the principal factor determining the hydrophobic nature of the bchl molecule. Within the bacterial photosynthetic pigment-protein complexes it appears to be crucial for both stability and function, and it is clear that the assembly of these complexes cannot be completed without its attachment (2, 5). The crystallography carried out on the bacterial reaction center and light-harvesting complexes has provided an abundance of structural information (9, 11, 24), such that it is possible to examine the conformation adopted by each phytol chain in atomic detail. In light-harvesting complex 2 (LH2), it appears that the close intertwining of the phytol tails of the B800 and B850 bchls, along with the carotenoid chains, imparts a significant amount of stability to the complex. The tails also play a major role in controlling the orientation of the transition dipoles of the tetrapyrrole rings, a function that is crucial for fast energy transfer. Now that there is structural information on all of the bchl-protein complexes of R. sphaeroides (11, 35), it is appropriate to study the biogenesis of the pigments and protein of the photosynthetic apparatus of this organism.
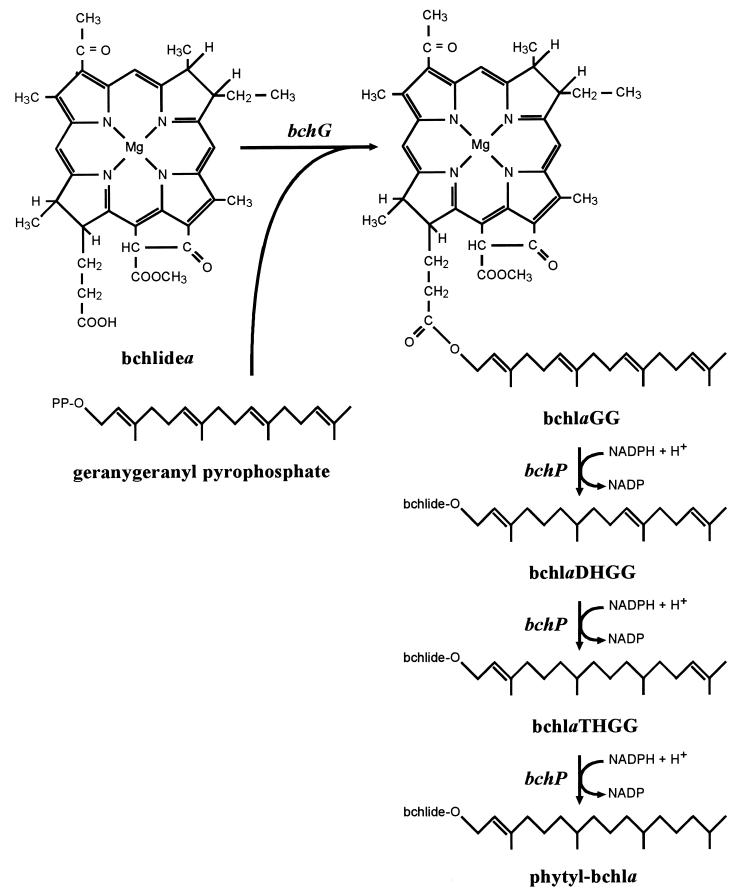
It has long been known that in R. sphaeroides, phytylation is the last stage of bchl biosynthesis (29). More recently, high-performance liquid chromatography (HPLC) analysis of wild-type R. sphaeroides led to the proposal that bacteriochlorophyllide (bchlide) is not, however, directly esterified with phytol (32). Rather, the range of bchla esters detected, and their relative abundances, suggested that bchlide was initially esterified with geranylgeraniol (GG) and that the alcohol moiety then underwent three successive hydrogenation reactions, converting it from GG to dihydrogeranylgeraniol (DHGG), to tetrahydrogeranylgeraniol (THGG), and finally to phytol (Fig. 1). Indeed, mutational analysis has implicated an identical mechanism in the closely related R. capsulatus (5).
FIG. 1.
Proposed pathway for the phytylation of bchlidea in R. sphaeroides and R. capsulatus. The sequence of the hydrogenation reactions is based on that which occurs during chl biosynthesis in greening plants (30). The genes involved are in italics.
In R. sphaeroides there are 11 steps of bchl biosynthesis following the chelation of protoporphyrin IX with magnesium. Most of the loci encoding the enzymes catalyzing these steps have now been identified and sequenced, and all have been found to lie within the 40.7-kb photosynthesis gene cluster, which also contains genes encoding subunits of the reaction center and light-harvesting complex 1 (LH1) and the enzymes of carotenoid biosynthesis (Fig. 2A). However, until now, there has been a paucity of genetic and biochemical data concerning the phytylation stage of bchl biosynthesis in R. sphaeroides. Here, we present a complete characterization of the gene, bchP, responsible for the hydrogenation of the GG moiety to phytol. These data have resulted not only from sequence and mutational analysis but also crucially from in vitro assays, for although mutational analyses and complementation studies can provide much valuable information about the function of gene products, such assays are needed to provide a clear and unambiguous demonstration of their biosynthetic capabilities. In vitro assays have already proved invaluable in the elucidation of the genetics and enzymology of several steps of the bchl biosynthesis pathway. For example, in vitro assays following overexpression of heterologous protein in Escherichia coli allowed the correct assignment of the bchM gene product to the methyltransferase step of bchl biosynthesis in both R. capsulatus (4) and R. sphaeroides (12). Gibson et al. (13) were also able to reconstitute the magnesium-protoporphyrin chelatase activity of R. sphaeroides in vitro, through overexpression of the bchH, bchI, and bchD genes in E. coli, thereby demonstrating a requirement for all three gene products and also for ATP.
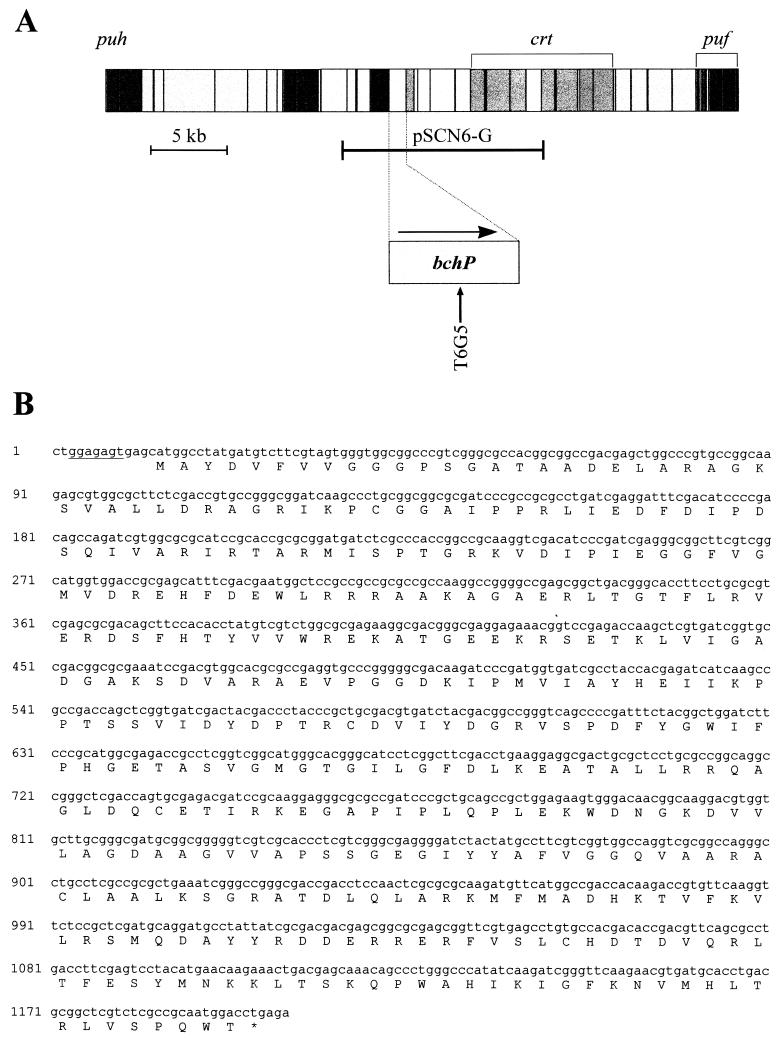
FIG. 2.
Physical map of the R. sphaeroides photosynthesis gene cluster, adapted from reference 8, with an expanded map of the bchP locus indicating its direction of transcription and the position of Tn5 insertion T6G5. Structural genes, and those with putative regulatory or assembly functions, are shown in the darkest grey; the next darkest represents carotenoid biosynthesis genes; bacteriochlorophyll biosynthesis genes are paler again, while very pale grey indicates genes of no known function. For further information on the photosynthesis gene cluster of R. sphaeroides, see reference 27. The longer horizontal bar immediately below the full map represents the insert of pSCN6-G, cloned in the pSUP202 vector. (B) Nucleotide and deduced amino acid sequences of the bchP gene. A potential ribosome-binding sequence is underlined.
In the case of chlorophyll (chl) biosynthesis, the terminal hydrogenation reactions have already been the subject of several in vitro studies. Assays were initially performed with etioplasts or chloroplasts as the sources of enzyme activity, but until recently, the individual enzyme proteins were neither isolated nor characterized, the source of activity being refined no further than a particular plastid fraction, such as thylakoids or chloroplast envelope membranes (for example, references 3 and 34). Now, however, the availability of the sequences of genes proposed to encode enzymes catalyzing these steps has enabled the performance of more precise assays. Keller et al. (20) have recently demonstrated the geranylgeranyl reductase activity of an Arabidopsis thaliana homologue of bchP. Here, we describe the heterologous expression of R. sphaeroides bchP in E. coli and the use of the resulting recombinant protein to catalyze the hydrogenation reaction which generates phytyl-bchla (bchlaP), thereby confirming the role of this gene product as determined by transposon Tn5 mutagenesis. Although a bchP gene has previously been identified and mutated in the closely related R. capsulatus (5), this constitutes the first direct demonstration of the activity of a bchP gene product. Mutation of bchP in R. capsulatus generated a strain with substantial reductions in both its photosynthetic growth rate and its steady-state levels of photopigments (5). Mutation of this gene in R. sphaeroides produced similar phenotypic effects, though on a much reduced scale; these are discussed.
In a parallel study, we have identified and analyzed the open reading frames (ORFs) immediately around bchP in the R. sphaeroides photosynthesis gene cluster (2). These include the gene, bchG, responsible for the initial esterification of bchlide (5, 28).
MATERIALS AND METHODS
DNA sequencing.
DNA was sequenced by using an ABI 373A sequencer in conjunction with an ABI DyeDeoxy reagent kit (Perkin-Elmer). Sequence analysis was performed with Lasergene software (DNAStar) and the Genetics Computer Group package (10, 11a) on the UNIX computer of the SEQNET facility at Daresbury, United Kingdom. The polypeptide sequence was also analyzed by using Block Searcher on the World Wide Web (14).
Transposon Tn5 mutagenesis.
An R. sphaeroides genomic library constructed in mobilizable vector pSUP202 (15, 33) was screened with restriction fragments of the R. sphaeroides photosynthesis gene cluster encompassing the newly sequenced bchP gene. This resulted in the isolation of a clone, pSCN6-G, containing 13.5 kb of genomic DNA, including bchP (Fig. 2A). Random insertions of transposon Tn5 into pSCN6-G were generated by the method of Simon et al. (33). The sites of Tn5 insertion were determined by restriction analysis, after which selected insertions were transferred into the genome of wild-type R. sphaeroides NCIB 8253 by conjugation and homologous recombination (8, 33).
Growth of R. sphaeroides.
R. sphaeroides strains were routinely grown semiaerobically in the dark at 34°C in M22+ medium (17). Antibiotic concentrations were 20 μg/ml for neomycin and 1 μg/ml for tetracycline. For growth under photosynthetic conditions, anaerobic cultures were placed under low light intensity (3 W/m2) at room temperature. To overcome problems of phototoxicity, tetracycline was replaced with doxycycline, at a concentration of 0.25 μg/ml, in photosynthetically grown cultures. Growth of cultures was monitored by measurements of absorbance at 680 nm.
Complementation of R. sphaeroides mutants.
The oligonucleotides 5′-GATCTAGAGGAGACGACCATATGGCCTATGATGTCTTCGTAGT-3′ and 5′-TCCTAAGCTTGGATCCTCAGGTCCATTGCGGCGAGAC-3′ were used to amplify the R. sphaeroides bchP gene by PCR and introduce XbaI and HindIII sites which enabled cloning into pRKSK1 (16), generating plasmid pSK1bchP. Vector pRKSK1 provided gene expression under the transcriptional control of the R. sphaeroides puc operon promoter.
Plasmid pSK1bchP was transformed into E. coli S17-1 (33), permitting conjugative transfer into R. sphaeroides Tn5 mutant T6G5. The parental plasmid pRKSK1 was independently introduced as a negative control.
Whole-cell absorbance spectroscopy.
Absorbance spectra of dark, semiaerobically grown cultures at identical cell densities were obtained on a Guided Wave model 260 spectrophotometer.
Absorbance spectra of acetone-methanol extracts.
Pigments were extracted from dark, semiaerobically grown cultures of R. sphaeroides by using acetone-methanol, (7:2 [vol/vol]). Absorbance spectra were recorded between 350 and 950 nm on a Beckman DU640 spectrophotometer.
HPLC of bchls.
Bchls present in acetone-methanol extracts were separated by HPLC on a Merck LiChrospher 100 RP-18 column (250 by 4 mm, inside diameter). The flow rate was 1 ml/min, with an initial composition of 64% methanol–16% acetone–20% water, changing over the course of 10 min to 80% methanol–20% acetone. This buffer composition was maintained for 15 min, during which time the bchla esters were eluted. Elution of bchls was monitored with a Waters 996 photodiode array detector, scanning from 350 to 800 nm every 2 s. The chromatograms at 365 nm were derived from the accumulated absorbance scans, using the Millennium software (Waters). With this system, bchlaGG eluted at approximately 18.4 min, bchlaDHGG eluted at approximately 19 min, bchlaTHGG eluted at approximately 19.8 min, and bchlaP eluted at approximately 20.7 min. These pigments were identified by direct comparison with Rhodospirillum rubrum, which has bchlaGG as its principal bchl pigment (19), and to wild-type R. sphaeroides, which possesses mainly bchlaP but also has quantities of the three less reduced esters (32).
HPLC of carotenoids.
Carotenoid pigments were initially extracted into acetone-methanol (2:1 [vol/vol]) and subsequently transferred into ether by the method of Britton and Riesen (6). The samples were then evaporated to dryness under N2, and the pigments were redissolved in ethyl acetate-acetonitrile-water (10:9:1 [vol/vol]). Carotenoids were separated by HPLC on a Spherisorb ODS2 column (250 by 4.6 mm, inside diameter), using a protocol provided by A. Young (Liverpool John Moores University, Liverpool, United Kingdom). The flow rate was 1 ml/min, with an initial solvent composition of 20% ethyl acetate–72% acetonitrile–8% water, changing over the course of 6 min to 60% ethyl acetate–36% acetonitrile–4% water. This solvent composition was maintained for 15 min, during which time the carotenoids eluted. Elution of carotenoids was monitored with a Waters 996 photodiode array detector, scanning from 270 to 600 nm every 2 s.
Heterologous expression of bchP in E. coli.
In addition to XbaI and HindIII sites, the pair of primers used to amplify bchP for homologous expression in R. sphaeroides contained restriction sites for NdeI and BamHI. These permitted cloning of the PCR product into vector pET9a (Novagen) for heterologous expression in E. coli. The resulting plasmid, pET9a-bchP, as well as the parental plasmid pET9a, was transformed into E. coli BL21(DE3)pLysS (Novagen).
Strains of BL21(DE3)pLysS containing pET9a derivatives were grown at 25°C in 400 ml of Luria-Bertani (LB) medium containing neomycin (20 μg/ml) and chloramphenicol (34 μg/ml) until the optical density at 600 nm (OD600) of the cultures had reached 0.6 to 1. Protein synthesis was induced by the addition of isopropyl-β-d-thiogalactopyranoside to the cultures to a final concentration of 0.4 mM. The cells were harvested after approximately 18 h, and the cell pellets were washed. To assess protein production and solubility, the cells from a 1-ml sample of culture were resuspended to an OD600 of 5 in assay buffer (see below) and disrupted by sonication (three bursts of 20 s each) on ice. Soluble and insoluble fractions were then isolated by centrifugation at 21,000 × g for 15 min at 4°C, and the fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Samples were separated on a 10% acrylamide gel by the method of Laemmli (23) and stained with Coomassie blue.
Geranylgeranyl-bchl reductase assays.
The assay procedure and buffer composition used were based primarily on those used by Oster et al. (28) for bchl synthetase assays, with some modifications in line with the bacterial magnesium-protoporphyrin chelatase assays of Gibson et al. (13). Harvested E. coli cell pellets were resuspended to an OD600 of 50 in assay buffer consisting of 50 mM HEPES (pH 8.0), 120 mM potassium acetate, 10 mM magnesium acetate, 0.3 M glycerol, 0.1% (vol/vol) Triton X-100, and 10 mM dithiothreitol, and the cells were ruptured by sonication as described above. To 30 μl of E. coli extract (containing the equivalent of 1.5 OD600 units of sonicated cells) in an Eppendorf tube on ice were then added phenylmethylsulfonyl fluoride to a final concentration of 0.5 mM, NADPH to 0.5 mM, ATP to 5 mM, and 30 μl of French-pressed cells of R. sphaeroides bchP mutant T6G5 containing approximately 5 nmol of bchlaGG. The volume was made up to 300 μl with assay buffer. Incubations were carried out in the dark for 90 min at 30°C. The pigments were then extracted with acetone-methanol 7:2 (vol/vol), and 50 μl was used for HPLC analysis.
RESULTS
Nucleotide and deduced amino acid sequences of the bchP gene of the R. sphaeroides photosynthesis gene cluster.
The complete nucleotide sequence of a previously unknown ORF near the center of the R. sphaeroides photosynthesis gene cluster was obtained (Fig. 2) (26). The putative 394-amino-acid gene product has a predicted molecular weight of 43,449 and conforms to a compiled codon usage table for R. sphaeroides photosynthesis genes. Analysis using the Kyte-Doolittle hydropathy scale (22) and the Klein algorithm (21) suggested that the polypeptide is largely hydrophilic and has no regions buried in the membrane bilayer.
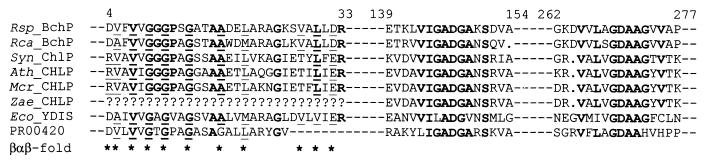
The predicted R. sphaeroides gene product has 73.7% identity with the gene product of R. capsulatus bchP, a gene likewise present within the respective photosynthesis gene cluster (7); therefore, we have designated the R. sphaeroides gene bchP. The R. sphaeroides bchP gene product is also 38.4% identical to the product of the chlP gene of Synechocystis sp. strain PCC 6803 (EMBL accession no. X97972). This gene partially complements an R. sphaeroides bchP mutant, implying functional homology (1). Further substantial homologies exist with the products of several plant genes: 37.7% identity with a geranylgeranyl reductase enzyme in Arabidopsis thaliana (EMBL accession no. Y14044) (20) and similar levels of homology with closely related sequences in Mesembryanthemum crystallinum (EMBL accession no. AF069318) and Zantedeschia aethiopica (EMBL accession no. AF055296); the ability of the A. thaliana enzyme to reduce both chlaGG and GG-pyrophosphate (GGPP) to their phytylated equivalents has been demonstrated in vitro (20). Among other, weaker homologues, the majority of which have no assigned function, is E. coli YDIS (Swissprot accession no. P77337), a probable electron transfer flavoprotein-quinone oxidoreductase. All of these proteins have regions displaying strong sequence similarities with three of six elements of an aromatic ring hydroxylase (flavoprotein monooxygenase) signature (block PR00420) (14) (Fig. 3). The first of these conserved domains is found in many eukaryotic and prokaryotic proteins known to bind flavin adenine dinucleotide (FAD) and/or NAD(P)H cofactors and aligns with an amino acid fingerprint predicting the occurrence of an ADP-binding βαβ fold (36); the function of the other two is unknown.
FIG. 3.
Partial alignment of R. sphaeroides bchP deduced amino acid sequence (Rsp_BchP) with sequences of other putative gene products. Fully conserved residues are in bold type. Sequences (and, in parentheses, accession numbers) are as follows: Rca_BchP, R. capsulatus BchP (P26172); Syn_ChlP, Synechocystis sp. strain PCC 6803 ChlP (X97972); Ath_CHLP, A. thaliana CHL P (Y14044); Mcr_CHLP, M. crystallinum CHL P (AF069318); Zae_CHLP, Z. aethiopica CHL P partial sequence (AF055296); Eco_YDIS, E. coli YDIS (P77337); PR00420, segments representing blocks of the flavoprotein monooxygenase signature. Asterisks below the alignment mark the positions of significant amino acids within a putative ADP-binding βαβ fold (36); residues which match the proposed fingerprint are underlined. Numbers above the alignment are amino acid positions within R. sphaeroides BchP.
Analysis of the transposon Tn5 mutant T6G5. (i) Southern blot analysis.
A novel bchP mutant of wild-type R. sphaeroides was generated by transposon Tn5 mutagenesis. Southern blot analysis confirmed that the location of the Tn5 insertion in this mutant strain, T6G5, was identical to that in pSCN6-G, the plasmid in which the insertion originated (data not shown). The transposon insertion site was subsequently more precisely determined by sequencing from the left inverted repeat of Tn5 in the mutated plasmid. Insertion T6G5 was found to reside in codon 199 of bchP.
(ii) Photosynthetic growth analysis.
Insertion T6G5 led to a reduction in the maximal growth rate under illumination of approximately 27% in comparison with the wild type, although the final cell density was not significantly affected. The doubling times were approximately 22 and 16 h for T6G5 and the wild type, respectively.
(iii) Analysis by absorbance spectroscopy.
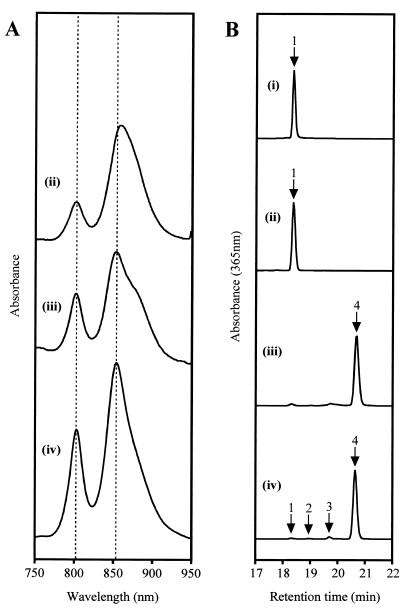
Whole-cell absorbance spectroscopy of a nonphotosynthetic culture of the T6G5 mutant revealed significant alterations in its content of photosynthetic pigment-protein complexes (Fig. 4A). The mutant displayed a dramatic reduction, amounting to approximately one-third, in the overall level of absorbance by the light-harvesting complexes in comparison to wild-type R. sphaeroides. Most apparent was a massive reduction in the B800 peak of LH2 to approximately 30% of its normal height. The B850 peak was lowered by only about 34% but was also red-shifted by approximately 6 nm relative to the wild-type peak (from 853.5 to 859.5 nm), although the underlying effect of the LH1 B875 bchls was impossible to assess.
FIG. 4.
Spectral analyses of R. sphaeroides strains. (A) Whole-cell absorbance spectra balanced according to cell density; (B) HPLC traces of acetone-methanol extracts. R. sphaeroides strains: (ii), bchP mutant T6G5; (iii), T6G5[pSK1bchP]; (iv), wild-type. HPLC trace (i) represents a Rhodospirillum rubrum cell extract. Labeled peaks on HPLC traces: 1, bchlaGG; 2, bchlaDHGG; 3, bchlaTHGG; 4, bchlaP.
(iv) Porphyrin analysis.
Absorbance spectroscopy of the acetone-methanol extract of cells of T6G5 produced a spectrum almost identical to that of wild-type R. sphaeroides (data not shown), implying the presence of either bchla or bchlidea, and presumably the former, since the mutant retained photosynthetic ability. HPLC analysis demonstrated that T6G5 was completely lacking in bchlaP, the principal bchl pigment of wild-type R. sphaeroides; bchlaP had a retention time of approximately 20.7 min under the conditions used (Fig. 4B). Instead it possessed a different bchla species with a shorter retention time, of approximately 18.4 min, which is also found as a minor constituent in wild-type R. sphaeroides cells, particularly those undergoing rapid growth and pigment biosynthesis [Fig. 4B, trace (iv)] (32). The retention time was also identical to that of the principal bchl pigment of Rhodospirillum rubrum [Fig. 4B, trace (i)]. In both species, previous studies have identified this pigment as bchlaGG (19, 32).
(v) Carotenoid analysis.
The carotenoid pigments of T6G5 were also analyzed by HPLC to determine whether these too had been affected by the transposon insertion. Analyses were performed on non-photosynthetically-grown cells; if unaltered in carotenoid biosynthesis or accumulation, these cells were expected to contain principally spheroidenone. This pigment, which had a major absorbance peak at 485.5 nm in the solvents used, eluted after approximately 13.8 min. It was found to be by far the most abundant carotenoid in T6G5.
Complementation analysis of transposon mutants. (i) Porphyrin analysis.
Introduction of plasmid pSK1bchP into T6G5 provided expression of bchP under the transcriptional control of the puc operon promoter. HPLC analysis revealed that complete complementation of the lesion in pigment biosynthesis had been achieved: virtually all of the bchla in stationary-phase cells was now found to be bchlaP, with levels of the other bchla esters being almost identical to those in wild-type cells at the same growth stage (Fig. 4B). The presence alone of vector pRKSK1 had no detectable effect on the pigment composition, with only bchlaGG being found.
(ii) Photosynthetic growth analysis.
The doubling time of T6G5 containing pSK1bchP was approximately 22 h under illumination, compared with 33 h for the same mutant containing only the insert-free plasmid pRKSK1. Therefore, expression of bchP increased the maximal photosynthetic growth rate by approximately 50%.
(iii) Whole-cell absorbance spectroscopy.
The introduction of plasmid pSK1bchP into T6G5 produced an absorbance spectrum very similar to that of wild-type R. sphaeroides. The red shift of T6G5 was fully reversed, and the B800/B850 ratio returned closer to the wild-type level [Fig. 4A, trace (iii)]. The overall absorbance due to LH2 was also increased relative to that of LH1, but an enhanced contribution to the spectrum by LH1 was still made apparent by a very obvious shoulder on the B850 peak, which may have exaggerated the height of this peak relative to that of the B800 peak. In the presence of pRKSK1, or a derivative of this vector, the overall levels of complexes appeared to be lowered proportionately. The reason for this is not known; the shape of the T6G5 spectrum was not, however, altered significantly by the presence of pRKSK1 alone.
Heterologous expression of bchP in E. coli.
The R. sphaeroides bchP gene was cloned into vector pET9a for overexpression studies in E. coli, since the assignment of function to this gene is otherwise circumstantial, relying as it does on the phenotype of the mutant. Protein production from the pET9a construct was induced in E. coli BL21(DE3)pLysS by the addition of isopropyl-β-d-thiogalactopyranoside. The cells were subsequently harvested and fractionated by sonication and centrifugation, and soluble and insoluble fractions were analyzed by SDS-PAGE. In the cells carrying pET9a-bchP, an additional, inducible protein of approximately the correct size (43.4 kDa) was found to be present. At least 50% of this protein appeared to reside in the soluble fraction, although a significant proportion remained in the pellet.
Activity of BchP in vitro.
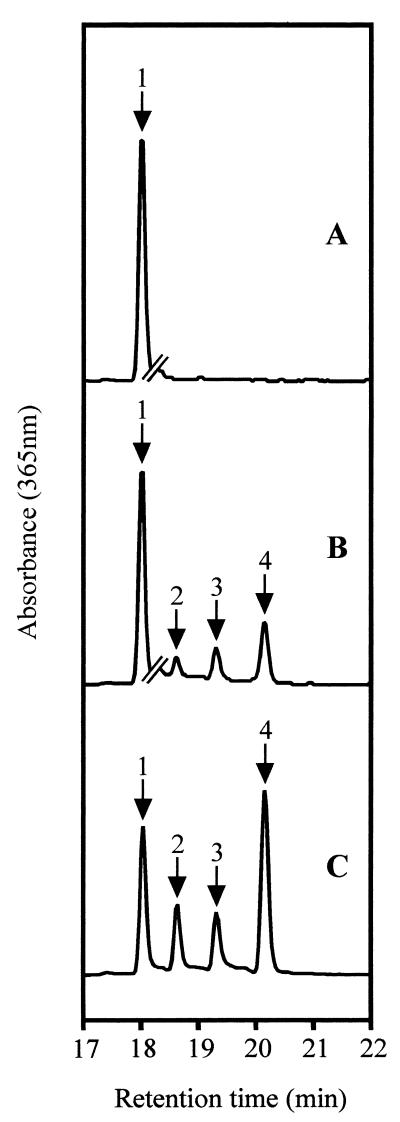
Extracts of induced E. coli cells carrying pET expression plasmids were incubated with a cell extract of R. sphaeroides bchP insertion mutant T6G5, which initially contained only bchlaGG. The resulting bchl pigments were analyzed by HPLC (Fig. 5). When the E. coli extract was of a culture containing the parental plasmid pET9a, no conversion of the bchlaGG was observed. However, when a pET9a-bchP extract was used, 11.7% of the bchl was hydrogenated: 2.1% to bchlaDHGG, 3.2% to bchlaTHGG, and 6.3% to bchlaP.
FIG. 5.
HPLC analysis of geranylgeranyl-bchl reductase assays. Chromatograms are of acetone-methanol extracts of incubations containing porphyrin substrates and E. coli extracts as follows: bchlaGG plus pET9a control (trace A) and bchlaGG plus pET9a-bchP (trace B). Trace C represents a mixture of bchlidea esters. The breaks in traces A and B indicate a change in vertical scale: to the right of the breaks, the height of these traces is expanded by a factor of 4. Labeled peaks: 1, bchlaGG; 2, bchlaDHGG; 3, bchlaTHGG; 4, bchlaP.
DISCUSSION
This paper presents the analysis of a previously unknown ORF in the photosynthesis gene cluster of the purple nonsulfur bacterium R. sphaeroides. This ORF, bchP, is situated less than 2 kb from the bchG locus, responsible for the esterification of bchlide (2, 28), and is likewise now demonstrated to have a role in the final stage of bchl biosynthesis in this organism. We describe the results of sequencing and Tn5 mutagenesis of the bchP gene and also the establishment of an in vitro assay demonstrating the activity of the bchP gene product. In vitro assays of a bchP gene product have not previously been reported.
Sequence analysis of bchP.
The bchP gene encodes a putative soluble 394-amino-acid polypeptide with a predicted molecular mass of 43.4 kDa. It has 73.7% identity with R. capsulatus BchP (5) and also shows significant homology throughout its length to the products of a cyanobacterial gene and of several plant genes. The cyanobacterial gene, chlP of Synechocystis sp. strain PCC 6803, has been shown to be capable of complementing a bchP mutant of R. sphaeroides (1), while a homologous A. thaliana gene product has been demonstrated by in vitro assays to be a geranylgeranyl reductase with the ability to catalyze both the reduction of chlaGG to chlaP and the reduction of GGPP to phytyl pyrophosphate (20). In the N-terminal region of each polypeptide is a section, represented by amino acids 4 to 32 of R. sphaeroides BchP, which fits an amino acid sequence fingerprint for ADP-binding βαβ folds at 10 of 11 positions (36). Such a score is considered sufficient for a prediction that one of these structures is likely to be formed. An ADP-binding βαβ fold binds the ADP moiety of FAD, NADH, or NADPH and may function as a nucleation center for the folding of an entire dinucleotide binding domain (31). Interestingly, the two BchP polypeptides each have an aspartate residue at the final position of the βαβ-fold fingerprint. This acidic residue would be likely to interact very unfavorably with the negatively charged 2′-phosphate of NADPH and is replaced with a nonacidic residue in some NADPH-binding βαβ folds (25, 36). This suggests that NADH, rather than NADPH, may function as the cofactor for BchP in vivo. However, the plant (and cyanobacterial) homologues each also have an acidic residue at this position, yet work on chl biosynthesis in plants has provided evidence that the cofactor for the equivalent reaction is NADPH (3, 34), and this was the electron donor supplied both in the in vitro assays using the A. thaliana geranylgeranyl reductase (20) and to R. sphaeroides BchP itself in the in vitro assays reported in this paper.
Functional analysis of bchP.
Random transposon Tn5 mutagenesis was used to generate an insertion mutant of the bchP locus. In this mutant, T6G5, both the total level of complexed pigment and the maximal photosynthetic growth rate were reduced to approximately two-thirds of the wild-type levels. HPLC analysis showed that the bchl pigment was no longer phytylated but was instead esterified with GG, suggesting that there was a lesion in bchl biosynthesis at the stage of alcohol hydrogenation. These phenotypic effects are identical to those identified by Bollivar et al. (5) for a bchP mutant of R. capsulatus, although in this case the reductions in photosynthetic growth rate and photopigment levels were much more severe, each being in the order of sixfold. In R. sphaeroides, the functional assignment was further supported by complementation analysis of T6G5: when bchP was expressed under the transcriptional control of the puc operon promoter, bchlaP again comprised the vast majority of the bchl present. Finally, R. sphaeroides bchP was heterologously expressed in E. coli, resulting in the accumulation of detectable amounts of the putative enzyme protein. An E. coli extract containing this protein was then used successfully to synthesize reduced bchl esters from a cell extract of R. sphaeroides bchP mutant T6G5, which had previously only contained bchlaGG. It is concluded that bchP encodes the enzyme geranylgeranyl-bchl reductase, responsible for catalyzing the terminal hydrogenation steps of bchl biosynthesis in R. sphaeroides. It is still unclear, however, whether these hydrogenation steps occur before or after the esterification reaction, since the presence of unesterified GGPP and bchlide, and of the esterifying enzyme, within the cell extract from the bchP mutant cannot be ruled out; therefore, the occurrence of hydrogenation at the level of unattached GGPP, followed by esterification of the reduced products, cannot be ruled out either. It may be the case that the R. sphaeroides enzyme has the ability, like its A. thaliana homologue, to catalyze the reduction of both the free pyrophosphate and the alcohol moiety of a porphyrin molecule (20). The apparent ability of the esterifying enzyme to utilize GGPP as a substrate suggests that hydrogenation is likely to be at least possible after esterification. The introduction of a workable assay system for these enzymes should provide the opportunity for such uncertainties to be eliminated by reduction in the amount of contaminating material present. It should also permit the cofactor requirements to be established. It is interesting that partially hydrogenated intermediates are detectable in R. sphaeroides, as this suggests that the enzyme and substrate may frequently dissociate between hydrogenation reactions.
Assembly of the R. sphaeroides photosystem with bchlaGG.
The bchP mutant, T6G5, shows that R. sphaeroides is clearly able to utilize bchlaGG instead of bchlaP in the formation of its photosynthetic complexes. However, it also shows that bchlaGG is an inferior pigment for this purpose in R. sphaeroides, since a mutant containing only this pigment possessed a significantly lowered photosynthetic growth rate. This corresponded with a reduction in the level of photopigments, suggesting that it was the formation or stability of the complexes rather than their electron transfer capability, which was affected. This agrees with the findings of Bollivar et al. (5), who found that reaction center function and energy transfer from light-harvesting to reaction center complexes were both unimpaired in a bchP mutant of R. capsulatus. This is despite the fact that the phenotype of the mutant in R. capsulatus was considerably more severe than that in R. sphaeroides.
The whole-cell absorbance spectrum of T6G5 would appear to show a predominance of LH1 over LH2. This indicates that either the assembly or the stability of LH2 is impaired more than that of LH1 (since the supply of complex components, including bchl, should be unchanged). As stated in the introduction, crystallographic analysis of LH2 (24) has suggested that close intertwining of the phytol tails of the B800 and B850 bchls, along with the carotenoid chains, helps to stabilize the complex. GG is more polar than phytol and is predicted to be less flexible due to its three additional, unreduced C-C double bonds. Clearly, this greater rigidity could severely disrupt the packing of the pigment molecules within the LH2 complex. In particular, the phytol chains of the nine B850 bchls coordinated to β-apoproteins are bent to curve round those of the nine B800 bchls and pass parallel to their bacteriochlorin rings at a distance of just 4.0 Å. It seems likely that the change to GG has excluded a number of the B800 bchls from the structure. This, in turn, could account in part for a decrease in the overall abundance of LH2, through a decrease in its stability. LH1 may be less susceptible to change in the nature of the alcohol moiety of its bchls as a consequence of having only a single ring of bchl molecules (18).
ACKNOWLEDGMENT
This work was supported by a grant from the BBSRC (United Kingdom).
REFERENCES
- 1.Addlesee H A, Gibson L C D, Jensen P E, Hunter C N. Cloning, sequencing and functional assignment of the chlorophyll biosynthesis gene, chlP, of Synechocystis sp. PCC 6803. FEBS Lett. 1996;389:126–130. doi: 10.1016/0014-5793(96)00549-2. [DOI] [PubMed] [Google Scholar]
- 2.Addlesee, H. A., L. Fiedor, and C. N. Hunter. Unpublished data.
- 3.Benz J, Wolf C, Rüdiger W. Chlorophyll biosynthesis: hydrogenation of geranylgeraniol. Plant Sci Lett. 1980;19:225–230. [Google Scholar]
- 4.Bollivar D W, Jiang Z-Y, Bauer C E, Beale S I. Heterologous expression of the bchM gene product from Rhodobacter capsulatus and demonstration that it encodes S-adenosyl-l-methionine:Mg-protoporphyrin IX methyltransferase. J Bacteriol. 1994;176:5290–5296. doi: 10.1128/jb.176.17.5290-5296.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bollivar D W, Wang S, Allen J P, Bauer C E. Molecular genetic analysis of terminal steps in bacteriochlorophyll a biosynthesis: characterization of a Rhodobacter capsulatus strain that synthesizes geranylgeraniol-esterified bacteriochlorophyll a. Biochemistry. 1994;33:12763–12768. doi: 10.1021/bi00209a006. [DOI] [PubMed] [Google Scholar]
- 6.Britton G, Riesen R. Worked examples of isolation and analysis, example 3: bacteria. In: Britton G, Liaaen-Jensen S, Pfander H, editors. Carotenoids. 1A. Isolation and analysis. Basel, Switzerland: Birkhäuser; 1995. pp. 227–238. [Google Scholar]
- 7.Burke D H, Alberti M, Armstrong G A, Hearst J E. The complete nucleotide sequence of the 46 kb photosynthesis gene cluster of Rhodobacter capsulatus. GenBank accession no. Z11165. 1991. [Google Scholar]
- 8.Coomber S A, Chaudri M, Connor A, Britton G, Hunter C N. Localized transposon Tn5 mutagenesis of the photosynthetic gene cluster of Rhodobacter sphaeroides. Mol Microbiol. 1990;4:977–989. doi: 10.1111/j.1365-2958.1990.tb00670.x. [DOI] [PubMed] [Google Scholar]
- 9.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3Å resolution. Nature. 1985;318:618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- 10.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ermler U, Fritzsch G, Buchanan S K, Michel H. Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.65 Å resolution: cofactors and protein-cofactor interactions. Structure. 1994;2:925–936. doi: 10.1016/s0969-2126(94)00094-8. [DOI] [PubMed] [Google Scholar]
- 11a.Genetics Computer Group. Program manual for the Wisconsin Package, version 8. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 12.Gibson L C D, Hunter C N. The bacteriochlorophyll biosynthesis gene, bchM, of Rhodobacter sphaeroides encodes S-adenosyl-l-methionine:Mg protoporphyrin methyltransferase. FEBS Lett. 1994;352:127–130. doi: 10.1016/0014-5793(94)00934-1. [DOI] [PubMed] [Google Scholar]
- 13.Gibson L C D, Willows R D, Kannangara C G, von Wettstein D, Hunter C N. Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: reconstitution of activity by combining the products of the bchH, -I and -D genes expressed in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:1941–1944. doi: 10.1073/pnas.92.6.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henikoff S, Henikoff J G. Protein family classification based on searching a database of blocks. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- 15.Hunter C N, Coomber S A. Cloning and oxygen-regulated expression of the bacteriochlorophyll biosynthesis genes bch E, B, A and C of Rhodobacter sphaeroides. J Gen Microbiol. 1988;134:1491–1497. [Google Scholar]
- 16.Hunter C N, Hundle B S, Hearst J E, Lang H P, Gardiner A T, Takaichi S, Cogdell R J. Introduction of new carotenoids into the bacterial photosynthetic apparatus by combining the carotenoid biosynthetic pathways of Erwinia herbicola and Rhodobacter sphaeroides. J Bacteriol. 1994;176:3692–3697. doi: 10.1128/jb.176.12.3692-3697.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter C N, Turner G. Transfer of genes coding for apoproteins of reaction centre and light-harvesting LH1 complexes to Rhodobacter sphaeroides. J Gen Microbiol. 1988;134:1471–1480. [Google Scholar]
- 18.Karrasch S, Bullough P A, Ghosh R. The 8.5Å projection map of the light-harvesting complex I from Rhodospirillum rubrum reveals a ring composed of 16 subunits. EMBO J. 1995;14:631–638. doi: 10.1002/j.1460-2075.1995.tb07041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katz J J, Strain H H, Harkness A L, Studier M H, Svec W A, Janson T R, Cope B T. Esterifying alcohols in the chlorophylls of purple photosynthetic bacteria. A new chlorophyll, bacteriochlorophyll(gg), all-trans-geranylgeranyl bacteriochlorophyllide a. J Am Chem Soc. 1972;94:7938–7939. doi: 10.1021/ja00777a054. [DOI] [PubMed] [Google Scholar]
- 20.Keller Y, Bouvier F, D’Harlingue A, Camara B. Metabolic compartmentation of plastid prenyllipid biosynthesis: evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur J Biochem. 1998;251:413–417. doi: 10.1046/j.1432-1327.1998.2510413.x. [DOI] [PubMed] [Google Scholar]
- 21.Klein P, Kanehisa M, Delisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 22.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.McDermott G, Prince S M, Freer A A, Hawthornthwaite-Lawless A M, Papiz M Z, Cogdell R J, Isaacs N W. Crystal structure of an integral membrane light-harvesting complex from photosynthetic bacteria. Nature. 1995;374:517–521. [Google Scholar]
- 25.McKie J H, Douglas K T. Evidence for gene duplication forming similar binding folds for NAD(P)H and FAD in pyridine nucleotide-dependent flavoenzymes. FEBS Lett. 1991;279:5–8. doi: 10.1016/0014-5793(91)80236-v. [DOI] [PubMed] [Google Scholar]
- 26.Naylor G W. The complete nucleotide sequence of the Rhodobacter sphaeroides photosynthesis gene cluster. EMBL accession no. AJ010302. 1999. [Google Scholar]
- 27.Naylor, G. W., H. A. Addlesee, L. C. D. Gibson, and C. N. Hunter. The photosynthesis gene cluster of Rhodobacter sphaeroides. Photosyn. Res., in press.
- 28.Oster U, Bauer C E, Rüdiger W. Characterization of chlorophyll a and bacteriochlorophyll a synthases by heterologous expression in Escherichia coli. J Biol Chem. 1997;272:9671–9676. doi: 10.1074/jbc.272.15.9671. [DOI] [PubMed] [Google Scholar]
- 29.Richards W R, Lascelles J. The biosynthesis of bacteriochlorophyll. The characterization of latter stage intermediates from mutants of Rhodopseudomonas spheroides. Biochemistry. 1969;8:3473–3482. doi: 10.1021/bi00836a051. [DOI] [PubMed] [Google Scholar]
- 30.Schoch S, Schäfer W. Tetrahydrogeranylgeraniol, a precursor of phytol in the biosynthesis of chlorophyll a: localization of the double bonds. Z Naturforsch. 1978;33c:408–412. [Google Scholar]
- 31.Schulz G E, Schirmer R H. Principles of protein structure. New York, N.Y: Springer Verlag; 1979. pp. 128–129. [Google Scholar]
- 32.Shioi Y, Sasa T. Terminal steps of bacteriochlorophyll a phytol formation in purple photosynthetic bacteria. J Bacteriol. 1984;158:340–343. doi: 10.1128/jb.158.1.340-343.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon R, Preifer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 34.Soll J, Schultz G, Rüdiger W, Benz J. Hydrogenation of geranylgeraniol: two pathways exist in spinach chloroplasts. Plant Physiol. 1983;71:849–854. doi: 10.1104/pp.71.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walz T, Jamieson S J, Bowers C M, Bullough P A, Hunter C N. Projection structures of three photosynthetic complexes from Rhodobacter sphaeroides: LH2 at 6 Å, LH1 and RC-LH1 at 25 Å. J Mol Biol. 1998;282:833–845. doi: 10.1006/jmbi.1998.2050. [DOI] [PubMed] [Google Scholar]
- 36.Wierenga R K, Terpstra P, Hol W G J. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]