Abstract
As a nutrient, heme is important for various cellular processes of organism. Bacteria can obtain heme via heme biosynthesis or/and uptake of exogenous heme from the host. On the other side, absorption of excess heme is cytotoxic to bacteria. Thus, bacteria have developed systems to relieve heme toxicity and contribute to the maintenance of heme homeostasis. In the past decades, the mechanisms underlying heme acquisition and tolerance have been well studied in Gram-positive model bacteria, such as Staphylococcus, Streptococcus and other Gram-positive bacteria. Here, we review the elaborate mechanisms by which these bacteria acquire heme and resist heme toxicity. Since both the heme utilization system and the heme tolerance system contribute to bacterial virulence, this review is not only helpful for a comprehensive understanding of the heme homeostasis mechanism in Gram-positive bacteria but also provides a theoretical basis for the development of antimicrobial agents.
Keywords: Heme utilization, Heme tolerance, Gram-positive bacteria
1. Introduction
As an essential nutrient for most organisms, heme is involved in diverse cellular processes, such as oxygen storage and transportation, electron transfer, aerobic respiration, and gas sensing [[1], [2], [3], [4], [5]]. In vertebrates, free heme is rare because most of it is bound to hemoglobin (Hb), myoglobin (Mb) and hemopexin (Hpx) [3,6]. Infectious bacteria have developed a high-affinity heme utilization system to transport heme from their host to establish infection. Once the bacterial heme utilization system is destroyed, the virulence of the bacteria will be attenuated [2,7,8]. Regarding Gram-positive bacteria, the heme transport system of Staphylococcus aureus was the first identified and is well-studied, which was called the iron-regulated surface determinant (Isd) system [9].
However, excess heme is toxic to bacteria, it causes membrane disruption, membrane protein and lipid oxidation, and DNA damage [2,10]. To overcome heme toxicity, bacteria employ multiple heme tolerance systems to detoxify, including efflux, degradation and sequestration. The mechanisms of heme detoxification were well studied in Gram-positive bacteria, such as S. aureus, Streptococcus agalactiae and Clostridioides difficile. When the heme is overloaded, S. aureus sense the signal through the two-component signal transduction systems (TCS) HssRS, which leads to heme efflux through the heme efflux system HrtAB [11]. Other Gram-positive bacteria, such as S. agalactiae employs porphyrin-regulated efflux system, PefAB and PefRCD, to export heme [12]. C. difficile employs the heme-activated operon HatRT and HsmRA to sense and detoxify heme [7,8]. For degradation, IsdG and IsdI heme oxygenases in S. aureus degrade heme to free iron, staphylobilins and formaldehyde [13].
In recent years, heme detoxification systems have made more progress in Gram-positive bacteria than heme transport system. Herein, we review the mechanisms of heme utilization and detoxification in the classical model of Gram-positive bacteria, S. aureus, and other Gram-positive bacteria.
1.1. Heme acquisition from the host
1.1.1. Heme acquisition mediated by the Isd system of S. aureus
S. aureus is a facultative anaerobe that uses heme during anaerobic nitrate respiration and aerobic respiration [14]. S. aureus could lyse erythrocytes to release Hb through secreted hemolysins, and then binds free Hb and extracts heme [15]. Mazmanian et al. (2003) reported that the high-affinity Isd heme acquisition system of S. aureus is responsible for Hb binding and the passage of heme into the cytoplasm [9]. The Isd system is composed of nine proteins, which are encoded in five Fur-regulated transcriptional units [16]. IsdA, IsdB, IsdC, and IsdH are cell wall-anchored heme-binding surface receptors; IsdDEF is membrane-associated transport system; and IsdG and IsdI are two cytoplasmic heme oxygenases [17].
Four Isd surface receptors contain an N-terminal secretion signal and a C-terminal sortase signal; IsdA, IsdB, and IsdH are anchored by the cell housekeeping sortase A; and IsdC is attached by StrB [9,17]. All the Isd surface proteins reversibly bind heme through the NEAT (near-iron transporter) domain. NEAT domains consist of approximately 120–125 amino acids and share a β-sandwich fold, and it is primarily responsible for binding Hb and heme [17,18]. IsdA and IsdC contain one NEAT domain each, and binds heme with high affinity. IsdB contains two NEAT domains: domain one (N1) binds Hb and domain two (N2) binds heme [17]. IsdH contains three NEAT domains: domain one and two (N1 and N2) are for Hb and Hb-Hp binding, and domain three (N3) is for heme binding [19,20].
The surface receptors IsdB and IsdH are the first step to extract heme molecules from Hb using a tri-domain unit joined by a helical linker domain. The tri-domain unit in IsdB is formed by domains N1 and N2, and that in IsdH is formed by domains N2 and N3 [21,22]. IsdB binds to the β-subunits of the Hb tetramer, leading to Hb dimerization, and IsdB binds to the α-chain of Hb. Then, heme is extracted by IsdB, and the F-helix in the β-subunits is unfolded [23]. IsdH binds the α-globin F helix of Hb, then the F-helix undergoes conformational change that disrupts heme pocket structure and promotes heme release [24]. The IsdH and Hb complex is dynamic. In this complex, the N2 domain binds Hb, and the inter-domain motions within IsdH enable the N3 domain to transiently distort the Hb's pocket resulting in heme exposure [21,25].
Following capture by IsdH and IsdB, heme is unidirectionally passed to IsdA, and subsequently transfers to IsdC [17]. Heme is transferred between NEAT domains through the formation of handclasp complexes [16,26]. Subsequently, heme is transferred to the heme-specific lipoprotein IsdE, and then the permease IsdF transfers heme across the membrane. IsdD was speculated to associated with IsdEF, its function remains to be identified [9]. S. aureus possesses special compartments, functional membrane microdomains [FMMs], for coordinating diverse cellular functions [27]. IsdF is energized by the housekeeping ATPase FhuC, and IsdF interacts with the FMM scaffolding protein flotillin A (FloA), and co-localize on the intact bacterial cells [27]. Isd-dependent heme utilization and proliferation in S. aureus requires FMMs and floA [27].
Cytoplasmic heme can be degraded by heme oxygenases IsdG and IsdI to release free iron or be incorporated into protein as a co-factor [2,17]. Mutations in the Isd system components lead to it being highly deficient in virulence, indicating that heme acquisition is important for S. aureus infection and is a potential target for the development of novel antibiotics [28,29]. Homologues of the Isd system were identified in Bacillus anthracis [30], Bacillus cereus [31], Listeria monocytogenes [32] and Staphylococcus lugdunensis [33].
1.1.2. Heme acquisition system of Corynebacterium diphtheriae
The human pathogen, C. diphtheriae causes respiratory disease and secretes the potent diphtheria toxin (DT), using heme and Hb from the host as an essential iron source [34,35]. Drazek et al. (2009) showed that the hmu genetic cluster is involved in heme transportation, and its expression is regulated by the diphtheria toxin repressor, DtxR, and iron [36]. The hmu genetic cluster includes ATP binding cassette (ABC) heme transporter HmuTUV and two surface-anchored proteins: HtaA and HtaB. HmuTUV is composed of the heme-binding lipoprotein HmuT, the heme transporter permease protein HmuU and the ATP-binding protein HmuV [35]. HmuT is tethered to the cytoplasmic membrane via an N-terminal lipid region [36]. HtaA and HtaB contain N-terminal signal region and C-terminal transmembrane domain, and HtaA is a membrane-anchored protein and also a secreted protein [36]. HtaA binds heme through two conserved regions: CR1 (conserved region 1) and CR2 [37]. The CR domain contains conserved tyrosine and histidine residues, which is important for heme and hemoproteins binding [38]. The C. diphtheriae CR domain has no similarity with NEAT domains [34]. HtaB contains one CR domain, and it functions as a mediator of heme transportation [39].
Allen et al. (2011) showed that two genetic loci chtA-chtB and chtC-cirA also participate in heme and Hb binding and are regulated by DtxR and iron [39]. ChtA and ChtC are surface-anchored proteins, each contains a single N-terminal CR domain, and ChtB has one CR domain [39]. ChtA has 35% similarity with HtaA, which is only limited to the CR domain, ChtB has 63% similarity with HtaB, and ChtC has 47% similarity with ChtA [39]. This means that C. diphtheriae employ multiple-function membrane proteins to fulfill heme requirements. Homologues of the Hmu system, ChtA, ChtC and ChtB are distributed predominantly in Corynebacteria.
In addition to the heme-binding proteins mentioned above, a unique iron-regulated, Hb- and Hb-Hp binding protein (HbpA) was also identified in C. diphtheriae [40]. HbpA is both located in the membrane and secreted, and exists in a large-molecular-weight aggregate that is important for optimal binding to Hb and Hb-Hp [41]. HbpA does not bind heme and contains a CR domain but can bind Hb and Hb-Hp [40]. Structural analysis of HbpA protein revealed that the C-terminal region is pivotal for the use of heme-iron from Hb-Hp and plays a role in anchoring to the membrane [41]. Homologues of HbpA are only distributed in some of C. diphtheriae strains [40].
The heme acquisition system of C. diphtheriae acquires heme through the following process. At the first step, most Hb and Hb-Hp are bound by secreted or membrane-associated HbpA, and HbpA can promote the binding of Hb-Hp to surface receptors HtaA and ChtA/C [40]. HtaA and ChtA/C bind heme from Hb or Hb-Hp via CR domains and transport heme to HtaB or ChtB [34,41]. Subsequently, heme is transported by HmuTUV across the cell membrane into the cytoplasm and degraded by heme oxygenase HmuO or bound with different apoproteins as an enzymatic co-factor for utilization or storage [34].
1.2. Excess heme is toxic to bacteria
Although heme is essential for the biological function of proteins, it becomes toxic to bacteria when present in excess. For example, when erythrocytes are lysed, infectious bacteria encounter high concentrations of heme. We named this process nutritional intoxication. Excessive free heme causes deleterious effects, such as membrane disordered, oxidative stress damage, lipid oxidation, and DNA damage [2]. Owing to the natural lipophilicity of heme, it tends to gather in the cell membrane. Then, heme and membrane-associated quinone molecules undergo cyclic redox reactions, and generate highly reactive semi-quinones and reduced heme [42]. Both products can react with atmospheric oxygen to produce superoxide radicals, resulting in oxidative stress damage to the cell membrane [42].
Heme metabolites are also toxic to bacteria. Bilirubin is the terminal metabolite in heme catabolism, and can destroy the structure and function of the bacterial cell membrane via direct intercalation, resulting in membrane destabilization [43]. Excess heme results in redundant iron production in two ways. First, heme oxygenases degrade heme and release free iron. Second, iron itself can secede from the middle of the porphyrin ring when oxidation occurs [2,7]. Fenton chemistry reactions and Haber–Weiss reactions are classical chemical reactions that result in the production of ROS components, such as hydroxyl radicals, as the iron cycle moves between the ferrous and ferric states [2]. The hydroxyl and superoxide radicals have strong oxidizing properties, which can lead to DNA damage, membrane disruption, and membrane protein and lipid oxidation, thus resulting in irreversible damage [2,7,10].
Porphyrin is also toxic to bacteria. The reduced porphyrin is a pro-oxidant, whereas the oxidized porphyrin is photosensitive, releasing labile singlet oxygen or hydroxyl radicals under ultraviolet or visible light, which results in different redox reactions [44]. In the early stages of growth in Gram-positive bacteria, porphyrin is in its photo-activated state and can inhibit the growth of bacteria as a result of lipid peroxidation, oxidation of nucleotides in DNA and amino acids in proteins, and protein cross-linking, which leads to bacterial cell damage and death [45].
2. Strategies of Gram-positive bacteria to resist heme toxicity
To overcome heme toxicity, Gram-positive bacteria have evolved and developed various heme detoxification strategies, including efflux, degradation, and sequestration.
2.1. Bacterial heme efflux pump
Bacterial efflux pumps are responsible for the efflux of natural substances produced by the host, such as bile, hormones, and other substances [46,47]. Currently, six families of efflux pumps have been described. These include the primary active transporters from the ABC family, which are dependent on ATP hydrolysis to drive transport [46]. The other five families include the major facilitator superfamily (MFS), resistance-nodulation-cell division (RND) superfamily, small multidrug resistance (SMR) family, multidrug and toxin extrusion (MATE) family, and proteobacterial antimicrobial compound efflux (PACE) family, which are powered by proton motive force (PMF) [46]. In Gram-positive bacteria, the ABC family transporter HrtAB in S. aureus, the MFS family transporter HatT in C. difficile, the MFS family transporter PefA and the ABC family transporter PefCD in S. agalactiae mediate heme efflux [7,12,48,49].
2.2. Heme efflux regulated by the HssRS and HrtAB system of S. aureus
S. aureus can adapt to high concentrations of heme (10 μM) after being pre-exposed to a sub-toxic concentration (2 μM), indicating that S. aureus encode heme detoxification system [50]. Actually, it has been shown that S. aureus employs the heme-sensing TCS (two-component signal transduction systems), HssRS, and the heme-regulated ABC transporter, HrtAB to reduce heme toxicity [51] (Table 1). HssRS is composed of the response regulator, HssR and the histidine kinase protein, HssS. HrtAB consists of the cytoplasmic HrtA and the integral membrane permease protein HrtB [11,51]. When S. aureus is exposed to high concentrations of heme, heme-responsive sensor HssS undergoes auto-phosphorylation at histidine 249 and subsequently transfers its phosphate group to aspartate 52 of HssR. Phosphorylated HssR binds to a direct repeat sequence within the hrtAB promoter and induces the expression of HrtAB, which is responsible for the efflux of excess heme to the extracellular environment [11,51]. Consistently, deletion of hssRS or hrtAB results in high heme sensitivity [48]. HssS-HssR dependent regulation of HrtA and HrtB to alleviate heme pressure is important for S. aureus survival and infection.
Table 1.
The heme efflux pumps and its regulators.
| Bacteria | Transporters | Regulators | signaling | References |
|---|---|---|---|---|
| S. aureus, B. anthracis, B. thuringiensis, S. agalactiae, L. monocytogenes | HrtAB | HssRS | heme | [[52], [53], [54], [55],86] |
| C. diphtheriae | HrtAB | ChrSA | heme | [56] |
| L. lactis | HrtAB | HrtR | heme | [57] |
| E. faecalis | HrtAB | FhtR | heme | [58] |
| S. agalactiae | PefAB/PefCD | PefR | heme | [12] |
| C. difficile | HatT | HatR | heme | [7] |
The homologues of HssRS and HrtAB have been characterized in other Gram-positive bacteria, including B. anthracis [52], Bacillus thuringiensis [53], Streptococcus agalactiae [54], and L. monocytogenes [55], indicating that the heme detoxification strategies of HssRS and HrtAB systems are generally conserved in these bacteria (Table 1). However, although the homologues of HrtAB have been described in C. diphtheriae [56], Lactococcus lactis [57], and Enterococcus faecalis [58], no TCS HssRS homologues identified in these bacteria, suggesting that they have a unique transcriptional regulator for activating HrtAB function [58].
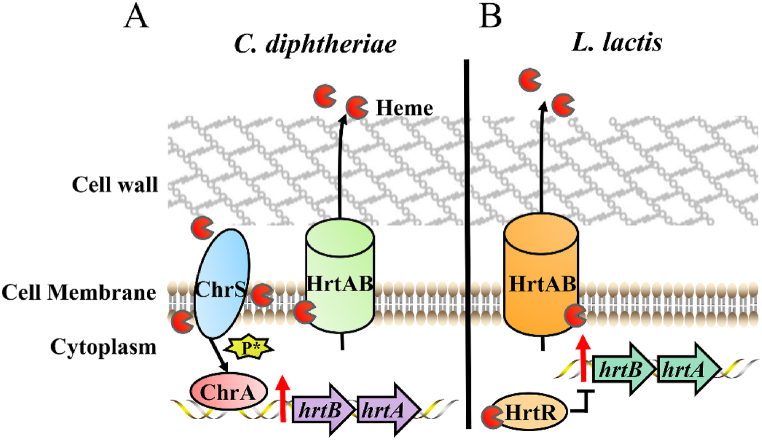
C. diphtheriae employs the TCS consisting of cytoplasmic membrane sensor ChrS and response regulator ChrA to mediate the transcription of hrtAB [56] (Fig. 1A) (Table 1). ChrS senses heme and undergoes auto-phosphorylation [59,60]. The signal is transferred to ChrA, and then phosphorylated ChrA binds to the target promoter regions, activating the transcription of hrtAB [56] (Fig. 1A). ChrA consists of an N-terminal regulatory domain and a C-terminal DNA-binding region, and the linker region of the N-terminal could create a dimerization interface binding DNA when receiving a heme-sensing signal [61]. Deletion of chrAB failed to trigger the expression of hrtAB under heme pressure, and mutations in hrtAB cause high heme sensitivity and the accumulation of cytoplasmic heme [56,62].
Fig. 1.
Heme efflux systems of C. diphtheriae and L. lactis [56,57]. (A) In C. diphtheriae, cytoplasmic membrane ChrS senses heme and autophosphorylation occurs. The phosphate group is transferred to ChrA such that phosphorylation ensues. ChrA binds the DNA sequence within the promoter of hrtAB, thereby activating the expression of hrtA and hrtB and resulting in the extrusion of heme via the HrtAB efflux pump. P* represents the phosphate group. (B) In L. lactis, the regulator HrtR binds to heme and alleviates the inhibition on the hrtAB promoter, leading to the extrusion of heme by HrtAB.
Recently, the crystal structures of the C. diphtheriae HrtAB and HrtAB-heme complex was resolved [63]. The entire HrtAB comprises two copies of HrtA-HrtB pair, HrtA contains a nucleotide-binding domain, and HrtB contains a transmembrane domain and an extracytoplasmic domain [63]. The HrtB dimer contains a Glu219 residue for the heme-binding site, which is located on the surface of the outer leaflet of the membrane. The heme-binding site captures heme from the membrane, and the heme is placed in the rearranged transmembrane helix bundle. The HrtA dimerization is driven by ATP-binding, and the heme-binding site is squeezed to extrude the bound heme from the cytoplasmic membrane [63]. The reduction of heme in the cytoplasmic membrane leads to a corresponding decrease in heme in the cytoplasm, as heme is evenly distributed between the cytoplasm and the membrane [64,65].
L. lactis is generally used for industrial fermentation and requires heme to activate aerobic respiration. HrtA and HrtB from L. lactis shares 50% and 37% homology with HrtA and HrtB from S. aureus, respectively. The heme-regulated transport regulator, HrtR, belonging to TetR family, is responsible for heme sensing and is adjacent to hrtAB [57,66] (Fig. 1B) (Table 1). The expression of hrtA and hrtB is repressed by HrtR through direct binding to the 15-nt palindromic sequence within the hrtAB promoter [57]. HrtR binds heme at the histidine residues His72 and His149, resulting in a coil-to-helix transition of the α4 helix in the heme-sensing domain and structural changes in HrtR, further leading to HrtR dissociating from the binding site of the hrtAB promoter [57,66]. Therefore, the expression of the HrtAB transporter alleviates heme pressure. Consistently, a hrtRAB mutant of L. lactis exhibited deficient growth when plated in a solid medium containing excess heme [57]. The homologues of HrtR have also been identified in other food and commensal bacteria, such as Lactobacillus amylolyticus and Leuconostoc citreum [57].
E. faecalis colonizes the human intestinal flora and is a threat to immunocompromised patients leading to severe diseases [67]. Heme is beneficial for E. faecalis as it facilitates the activation of respiration and the expression of heme-dependent catalase A (katA) [58]. HrtA and HrtB from E. faecalis share 24% and 45% homology with HrtA and HrtB from S. aureus, respectively [58]. Neither hssRS nor hrtR homologues have been identified in E. faecalis. The E. faecalis heme transport regulator, FhtR belongs to the TetR family and is located upstream of hrtAB [58] (Table 1). FhtR binds the two distinct 14-nt inverted repeat regions within the hrtAB promoter and inhibits hrtAB expression [58]. FhtR binds internalized heme at a pivotal tyrosine residue Tyr132, and FhtR-heme complexes dissociated from the hrtAB operon, resulting in HrtAB-mediated heme exportation [58]. The homologues of FhtR primarily distributed in enterococci, vagococci, and carnobacteri [58].
In summary, considering the contribution of HrtAB to heme toxicity and its widespread conservation in Gram-positive bacteria, targeting HrtAB as a designed drug to inhibit its function would be beneficial for limiting bacterial infections.
2.3. Heme efflux mediated by the Pef system of Streptococcus agalactiae
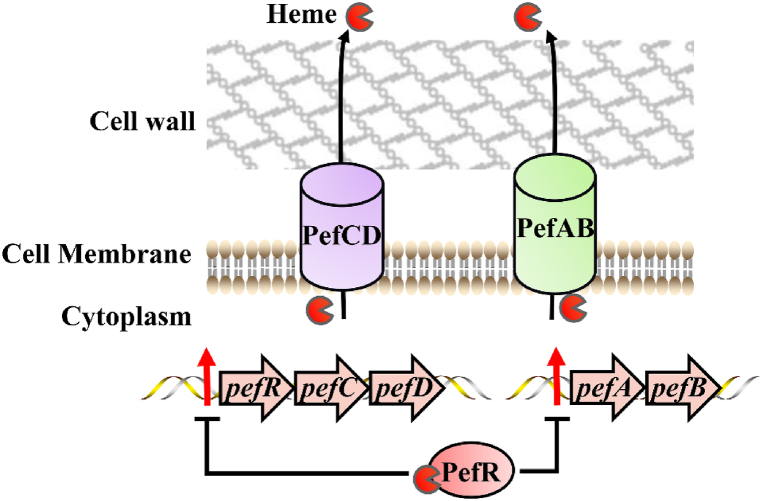
The opportunistic pathogen S. agalactiae infects the human gastrointestinal tract, female genitourinary tracts, and bovine mammary glands, causing septicemia and meningitis [12,54]. S. agalactiae cannot synthesize heme and requires exogenous heme for respiration via the activation of the terminal oxidase cytochrome bd quinol oxidase (CydAB) [54]. Heme-regulated detoxification systems HrtAB and HssRS have been identified in S. agalactiae [54]. Furthermore, it was demonstrated that another porphyrin-regulated efflux system, PefAB and PefRCD, is also important for heme detoxification [12] (Fig. 2) (Table 1). PefAB is composed of the MFS transport protein, PefA and hypothetical protein, PefB. PefRCD is composed of the MarR-superfamily protein PefR and the ABC multidrug transporter PefCD [12]. PefR represses the expression of pefAB and pefRCD operons by binding to the DNA inverted repeat motifs (IR) within the promoter regions. When heme is in excess, the PefR-heme complex is formed. PefR then dissociates from the promoter regions, and PefAB and PefCD expel excess heme [12] (Fig. 2).
Fig. 2.
Pef system of S. agalactiae [12]. PefR binds heme and thus derepresses the pefAB and pefRCD operon. Consequently, PefAB and PefCD are able to efflux heme and protect S. agalactiae from heme damage.
The pefR mutant strain displays a reduced respiration rate, due to the over-expression of pefAB and pefCD, indicating that PefR is important for restricting the expression of Pef efflux pumps. This allows maintenance of intracellular heme homeostasis and ensures that respiration and proliferation function optimally in S. agalactiae [12]. Compared with the HrtAB/HssRS system, the Pef system is activated under lower heme conditions (0.1–0.5 μM) [12]. This distinction allows S. agalactiae to rapidly adapt to intracellular heme levels under different cellular physiological conditions [12,54]. In summary, S. agalactiae relies on the HrtAB/HssRS and PefAB/PefRCD systems to synergistically maintain heme homeostasis.
The homologues of PefAB are distributed in Enterococcus faecalis, Streptococcus uberis, and Lactobacillus gasseri, and the homologues of PefRCD are primarily distributed in Streptococcus genus, including Streptococcus dysgalactiae, Streptococcus thermophilus, and S. uberis [12].
2.4. Heme efflux mediated by the HatRT system of Clostridioides difficile
C. difficile is a spore-forming obligate anaerobe that infects the colon and causes severe damage to the intestinal epithelial layer, thereby causing inflammation and bleeding through the release of the bacterial toxins, TcdA and TcdB. The release of toxins leads to high levels of heme from lysed erythrocytes, causing heme toxicity [7]. C. difficile does not encode homologues of HrtAB, PefAB, or PefRCD. Compared to untreated cultures of C. difficile, the expression of an operon comprising two genes was upregulated by 5- to 11-fold the concentrations of heme-treated cultures (50 μM) in C. difficile [7]. These genes were shown to encode a TetR family transcriptional regulator and an MFS family multidrug transporter referred to as heme activated transporter regulator (HatR) and heme activated transporter (HatT), respectively [7] (Fig. 3) (Table 1).
Fig. 3.
HatRT and HsmRA systems of C. difficile [8]. HatR binds heme and derepresses the hatRT operon, which allows HatT to extrude heme. Similarly, HsmR senses heme and activates the expression of the hsmRA operon, and then HsmA binds heme and resists oxidative stress.
Compared to the wild-type strain, hatT and hatR mutant strains were more sensitive to heme, and the sensitivity of the hatT mutant strain was even more prominent when exposed to excess heme [7]. Despite the constitutive expression of hatT, the hatR mutant strain was still sensitive to heme, indicating that HatR may function as a heme-binding protein where the histidine residue His99 was identified to be crucial for heme binding [7].
In general, in the presence of excess heme, HatR binds heme directly to form the HatR-heme complex and dissociates from the hatRT operon [7]. Subsequently, hatT transcription is activated and excess heme is pumped out [7] (Fig. 3). The HatRT system is essential for protecting C. difficile from damage caused by heme toxicity, for growth and for maintaining heme homeostasis.
2.5. Heme degradation and sequestration
2.5.1. Heme degradation mediated by HO-1 and IsdG-like heme oxygenases
Heme oxygenases (HOs) are mainly divided into two families in Gram-positive bacteria: the canonical HO-1 heme oxygenases and the non-canonical IsdG-like heme oxygenases (Fig. 4). The first HO-1 heme oxygenases is HO-like family protein (HmuO) in C. diphtheriae, which has significant homology with the human heme oxygenase protein HO-1 (33% identity and 70% similarity) [68]. The HO-1 heme oxygenases are a monomeric helical protein with an α-helical fold that similar to mammal HO-1, and the fold forms a heme-binding pocket containing two ordered water molecules [69,70]. HmuO oxidatively cleaves the α-carbon of heme to produce α-biliverdin, carbon monoxide (CO), and free iron [34,70] (Fig. 4A). The hmuO gene is activated by two paralogous TCSs, ChrS-ChrA and HrrS-HrrA in a heme- and Hb-dependent manner and is repressed by DtxR under iron-replete conditions [71,72]. Under heme accumulation conditions, the sensor kinases ChrS and HrrS detect heme and activate the phosphorylation of the response regulators ChrA and HrrA. The phosphorylated response regulators directly bind upstream of the hmuO promoter, thereby activating expression [72].
Fig. 4.
The heme degradation systems [72,73,79]. (A) In C. diphtheriae, the sensors ChrS and HrrS sense heme and transfer signals to ChrA and HrrA. ChrA and HrrA bind the hmuO promoter, and the expression of hmuO is activated. HmuO degrades heme to Fe2+, CO, and biliverdin. (B) In S. aureus, heme oxygenases IsdG and IsdI degrade heme to Fe2+, formaidehyde, and staphylobilin. (C) In M. tuberculosis, MhuD degrades heme to Fe2+ and mycobilin.
The IsdG-like heme oxygenases degrade heme without CO production, which is distinct from HO-1 protein, and have no similarity to the HO-like family proteins. The IsdG-like heme oxygenases include IsdG/I in S. aureus and MhuD in Mycobacterium tuberculosis [13,70,73]. IsdG and IsdI share 64% identity, can degrade heme to release free iron, linear tetrapyrrole staphylobilins (5-oxo-δ-bilirubin and 15-oxo-β-bilirubin), and formaldehyde in the presence of NADPH cytochrome P450 oxidoreductase or ascorbate [74,75] (Fig. 4B). MhuD degrades heme to mycobilin isomers and free iron, and has 45% similarity with IsdG/I [73] (Fig. 4C).
Structural analysis of IsdG-like heme oxygenases revealed that these proteins have a ferredoxin-like fold and a β-barrel at the dimeric interface, and form homodimers of subunits containing only one polar amino acid, which is distinct from HO-like family proteins [76]. After being bound by IsdG-like proteins, heme undergoes significant heme ruffling [77]. Each dimer of IsdG and IsdI contains heme-binding pockets, and the residues responsible for heme binding are conserved between them, including His, Trp and Asn residues [76,77]. MhuD of M. tuberculosis can bind one or two molecules of heme at the same active site, whereas IsdG/I just binds a single heme at an active site [73]. Both diheme-MhuD and monoheme-MhuD complexes are enzymatically active and are able to degrade heme [78]. IsdG-like proteins have also been identified in S. epidermidis [79], B. anthracis [80], and L. monocytogenes [81].
2.5.2. Heme sequestration mediated by the HsmRA system
In C. difficile, the hatRT mutation did not influence colonization or persistence, indicating that C. difficile possesses another mechanism to resist heme toxicity [7,8]. Knippel et al. (2020) identified the heme-sensing membrane protein HsmRA, which is regulated by heme and responsible for heme sequestration and oxidative stress resistance [8]. HsmRA is composed of the MarR family transcriptional regulator HsmR, and the membrane protein HsmA. Excessive heme is bound by HsmR through a conserved histidine residue, His50, and then induces hsmRA transcription. HsmA reduces free heme concentrations through sequestration; moreover, heme-HsmA complexes could provide protection from oxidative stress [8,82] (Fig. 3). Consistently, hsmR or hsmA mutant strains are increasingly sensitive to excess heme when compared to the wild-type strain [8]. Homologues of HsmR are widely distributed in Clostridia, with a homology of 53%–78% [8]. Homologues of HsmA are widely distributed in Clostridia, Bacillus, Lactobacillus, Enterococcus, Bacteroides, and Geobacter, with a homology of 37%–74%, indicating that HsmA is a conserved strategy for heme sequestration across several bacterial genera [8].
2.5.3. Heme homeostasis mediated by HrrSA of Corynebacteria
The TCS HrrSA system, which is highly conserved among almost all corynebacterial species, including C. diphtheriae and Corynebacterium glutamicum, plays a crucial role in maintaining the homeostasis of heme when there is an excess of exogenous or endogenous heme [60,83,84]. The similarity between HrrSA in C. glutamicum and HrrSA in C. diphtheriae is 87% [84].
The regulatory functions of HrrSA encompass a wide range of activities. Under excessive heme conditions, HrrSA inhibits heme synthesis by suppressing the expression of heme synthesis enzymes such as GtrR, UroD, and CpfC [72,83,84]. Additionally, HrrSA activates bacterial respiration by upregulating the expression of genes involved in the respiratory chain, including ctaE-qcrCAB, ctaD, ctaF, and cydAB [84]. HrrSA also activates the expression of the gene katG, which encodes catalase, to counteract oxidative stress induced by heme [84]. Moreover, HrrSA stimulates the expression of genes related to cell membrane remodeling, such as murA and aftC [84]. Furthermore, HrrSA can activate the expression of other transcriptional regulators, including ramA, ramB, and amtR [84]. In summary, the HrrSA system coordinates the expression of genes involved in heme biosynthesis, respiration, oxidative stress response, and cell membrane remodeling to maintain heme homeostasis. The regulatory network of the HrrSA system is highly complex, and its conservation among corynebacterial species suggests its potential as a target for drug development.
3. Conclusion and perspective
Gram-positive bacteria employ finely regulated heme uptake and tolerance systems to adapt to varying heme levels in different environments. The mechanisms of heme uptake include the secretion of haemophore, active uptake and transport processes involving various receptors and associated uptake systems. Recently, it was shown that Gram-positive bacterium, Dietzia sp. DQ12-45-1b, releases membrane vesicles (MVs) to participate in extracellular heme capture [85]. Whether haemophore is secreted as MVs requires further investigation. On the other hand, when exposed to excessive heme, tolerance systems play a vital role in protecting bacteria from the toxic effects of heme, including mechanisms such as heme oxygenases, heme-binding proteins, and heme efflux pumps. Among six different efflux systems, only two efflux systems have been found to be involved in heme efflux. Other types of heme efflux systems may be identified in the future. Taken together, these heme acquisition and tolerance systems provide valuable insights into the adaptation and survival strategies of Gram-positive bacteria in diverse environments. Understanding the physiological and pathological implications of these systems may lead to novel therapeutic approaches and the development of drugs targeting heme-related processes in bacterial infections.
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Data availability statement
No data was used for the research described in the article.
Additional information
No additional information is available for this paper.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 32072825 and Grant No. 32273003, http://www.nsfc.gov.cn/), the Natural Science Foundation of Sichuan Province (2022NSFSC0007), the earmarked fund for China Agriculture Research System (CARS-42-17), and the Sichuan Veterinary Medicine and Drug Innovation Group of the China Agricultural Research System (SCCXTD-2020-18).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Contributor Information
Mafeng Liu, Email: liumafengra@163.com.
Anchun Cheng, Email: chenganchun@vip.163.com.
References
- 1.Krüger A., Keppel M., Sharma V., Frunzke J. The diversity of heme sensor systems - heme-responsive transcriptional regulation mediated by transient heme protein interactions. FEMS Microbiol. Rev. 2022;46 doi: 10.1093/femsre/fuac002. [DOI] [PubMed] [Google Scholar]
- 2.Choby J.E., Skaar E.P. Heme synthesis and acquisition in bacterial pathogens. J. Mol. Biol. 2016;428:3408–3428. doi: 10.1016/j.jmb.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimizu T., Lengalova A., Martínek V., Martínková M. Heme: emergent roles of heme in signal transduction, functional regulation and as catalytic centres. Chem. Soc. Rev. 2019;48:5624–5657. doi: 10.1039/c9cs00268e. [DOI] [PubMed] [Google Scholar]
- 4.Mayfield J.A., Dehner C.A., DuBois J.L. Recent advances in bacterial heme protein biochemistry. Curr. Opin. Chem. Biol. 2011;15:260–266. doi: 10.1016/j.cbpa.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plate L., Marletta M.A. Nitric oxide-sensing H-NOX proteins govern bacterial communal behavior. Trends Biochem. Sci. 2013;38:566–575. doi: 10.1016/j.tibs.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaer D.J., Buehler P.W., Alayash A.I., Belcher J.D., Vercellotti G.M. Hemolysis and free hemoglobin revisited: exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood. 2013;121:1276–1284. doi: 10.1182/blood-2012-11-451229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knippel R.J., Zackular J.P., Moore J.L., Celis A.I., Weiss A., Washington M.K., DuBois J.L., Caprioli R.M., Skaar E.P. Heme sensing and detoxification by HatRT contributes to pathogenesis during Clostridium difficile infection. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knippel R.J., Wexler A.G., Miller J.M., Beavers W.N., Weiss A., de Crécy-Lagard V., Edmonds K.A., Giedroc D.P., Skaar E.P. Clostridioides difficile senses and hijacks host heme for incorporation into an oxidative stress defense system. Cell Host Microbe. 2020;28:411–421.e6. doi: 10.1016/j.chom.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazmanian S.K., Skaar E.P., Gaspar A.H., Humayun M., Gornicki P., Jelenska J., Joachmiak A., Missiakas D.M., Schneewind O. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 10.Anzaldi L.L., Skaar E.P. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect. Immun. 2010;78:4977–4989. doi: 10.1128/IAI.00613-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stauff D.L., Torres V.J., Skaar E.P. Signaling and DNA-binding activities of the Staphylococcus aureus HssR-HssS two-component system required for heme sensing. J. Biol. Chem. 2007;282:26111–26121. doi: 10.1074/jbc.M703797200. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez A., Lechardeur D., Derré-Bobillot A., Couvé E., Gaudu P., Gruss A. Two coregulated efflux transporters modulate intracellular heme and protoporphyrin IX availability in Streptococcus agalactiae. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skaar E.P., Gaspar A.H., Schneewind O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 2004;279:436–443. doi: 10.1074/jbc.M307952200. [DOI] [PubMed] [Google Scholar]
- 14.Choby J.E., Skaar E.P. Staphylococcus aureus coproporphyrinogen III oxidase is required for aerobic and anaerobic heme synthesis. mSphere. 2019;4 doi: 10.1128/mSphere.00235-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spaan A.N., Reyes-Robles T., Badiou C., Cochet S., Boguslawski K.M., Yoong P., Day C.J., de Haas C.J., van Kessel K.P., Vandenesch F., Jennings M.P., Le Van Kim C., Colin Y., van Strijp J.A., Henry T., Torres V.J. Staphylococcus aureus targets the duffy antigen receptor for chemokines (DARC) to lyse erythrocytes. Cell Host Microbe. 2015;18:363–370. doi: 10.1016/j.chom.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abe R., Caaveiro J.M., Kozuka-Hata H., Oyama M., Tsumoto K. Mapping ultra-weak protein-protein interactions between heme transporters of Staphylococcus aureus. J. Biol. Chem. 2012;287:16477–16487. doi: 10.1074/jbc.M112.346700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conroy B.S., Grigg J.C., Kolesnikov M., Morales L.D., Murphy M.E.P. Staphylococcus aureus heme and siderophore-iron acquisition pathways. Biometals. 2019;32:409–424. doi: 10.1007/s10534-019-00188-2. [DOI] [PubMed] [Google Scholar]
- 18.Andrade M.A., Ciccarelli F.D., Perez-Iratxeta C., Bork P. NEAT: a domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-research0047. Research0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis-Guardiola K., Mahoney B.J., Clubb R.T. NEAr transporter (NEAT) domains: unique surface displayed heme chaperones that enable gram-positive bacteria to capture heme-iron from hemoglobin. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.607679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sæderup K.L., Stødkilde K., Graversen J.H., Dickson C.F., Etzerodt A., Hansen S.W., Fago A., Gell D., Andersen C.B., Moestrup S.K. The Staphylococcus aureus protein IsdH inhibits host hemoglobin scavenging to promote heme acquisition by the pathogen. J. Biol. Chem. 2016;291:23989–23998. doi: 10.1074/jbc.M116.755934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clayton J., Ellis-Guardiola K., Mahoney B.J., Soule J., Liu W., Clubb R.T., Wereszczynski J. Directed inter-domain motions enable the IsdH Staphylococcus aureus receptor to rapidly extract heme from human hemoglobin. J. Mol. Biol. 2022;434 doi: 10.1016/j.jmb.2022.167623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowden C.F.M., Chan A.C.K., Li E.J.W., Arrieta A.L., Eltis L.D., Murphy M.E.P. Structure-function analyses reveal key features in Staphylococcus aureus IsdB-associated unfolding of the heme-binding pocket of human hemoglobin. J. Biol. Chem. 2018;293:177–190. doi: 10.1074/jbc.M117.806562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Bei O., Marchetti M., Ronda L., Gianquinto E., Lazzarato L., Chirgadze D.Y., Hardwick S.W., Cooper L.R., Spyrakis F., Luisi B.F., Campanini B., Bettati S. Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2116708119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickson C.F., Jacques D.A., Clubb R.T., Guss J.M., Gell D.A. The structure of haemoglobin bound to the haemoglobin receptor IsdH from Staphylococcus aureus shows disruption of the native α-globin haem pocket. Acta Crystallogr. D Biol. Crystallogr. 2015;71:1295–1306. doi: 10.1107/S1399004715005817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis-Guardiola K., Clayton J., Pham C., Mahoney B.J., Wereszczynski J., Clubb R.T. The Staphylococcus aureus IsdH receptor forms a dynamic complex with human hemoglobin that triggers heme release via two distinct hot spots. J. Mol. Biol. 2020;432:1064–1082. doi: 10.1016/j.jmb.2019.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaudin C.F., Grigg J.C., Arrieta A.L., Murphy M.E. Unique heme-iron coordination by the hemoglobin receptor IsdB of Staphylococcus aureus. Biochemistry. 2011;50:5443–5452. doi: 10.1021/bi200369p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adolf L.A., Müller-Jochim A., Kricks L., Puls J.S., Lopez D., Grein F., Heilbronner S. Functional membrane microdomains and the hydroxamate siderophore transporter ATPase FhuC govern Isd-dependent heme acquisition in Staphylococcus aureus. Elife. 2023;12 doi: 10.7554/eLife.85304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pishchany G., McCoy A.L., Torres V.J., Krause J.C., Crowe J.E., Jr., Fabry M.E., Skaar E.P. Specificity for human hemoglobin enhances Staphylococcus aureus infection. Cell Host Microbe. 2010;8:544–550. doi: 10.1016/j.chom.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visai L., Yanagisawa N., Josefsson E., Tarkowski A., Pezzali I., Rooijakkers S.H.M., Foster T.J., Speziale P. Immune evasion by Staphylococcus aureus conferred by iron-regulated surface determinant protein IsdH. Microbiology (Read.) 2009;155:667–679. doi: 10.1099/mic.0.025684-0. [DOI] [PubMed] [Google Scholar]
- 30.Maresso A.W., Garufi G., Schneewind O. Bacillus anthracis secretes proteins that mediate heme acquisition from hemoglobin. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abi-Khalil E., Segond D., Terpstra T., André-Leroux G., Kallassy M., Lereclus D., Bou-Abdallah F., Nielsen-Leroux C. Heme interplay between IlsA and IsdC: two structurally different surface proteins from Bacillus cereus. Biochim. Biophys. Acta. 2015;1850:1930–1941. doi: 10.1016/j.bbagen.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Malmirchegini G.R., Sjodt M., Shnitkind S., Sawaya M.R., Rosinski J., Newton S.M., Klebba P.E., Clubb R.T. Novel mechanism of hemin capture by Hbp2, the hemoglobin-binding hemophore from Listeria monocytogenes. J. Biol. Chem. 2014;289:34886–34899. doi: 10.1074/jbc.M114.583013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heilbronner S., Monk I.R., Brozyna J.R., Heinrichs D.E., Skaar E.P., Peschel A., Foster T.J. Competing for iron: duplication and amplification of the isd locus in Staphylococcus lugdunensis HKU09-01 provides a competitive advantage to overcome nutritional limitation. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen C.E., Schmitt M.P. Utilization of host iron sources by Corynebacterium diphtheriae: multiple hemoglobin-binding proteins are essential for the use of iron from the hemoglobin-haptoglobin complex. J. Bacteriol. 2015;197:553–562. doi: 10.1128/JB.02413-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drazek E.S., Hammack C.A., Schmitt M.P. Corynebacterium diphtheriae genes required for acquisition of iron from haemin and haemoglobin are homologous to ABC haemin transporters. Mol. Microbiol. 2000;36:68–84. doi: 10.1046/j.1365-2958.2000.01818.x. [DOI] [PubMed] [Google Scholar]
- 36.Allen C.E., Schmitt M.P. HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae. J. Bacteriol. 2009;191:2638–2648. doi: 10.1128/JB.01784-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen C.E., Schmitt M.P. Novel hemin binding domains in the Corynebacterium diphtheriae HtaA protein interact with hemoglobin and are critical for heme iron utilization by HtaA. J. Bacteriol. 2011;193:5374–5385. doi: 10.1128/JB.05508-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uluisik R.C., Akbas N., Lukat-Rodgers G.S., Adrian S.A., Allen C.E., Schmitt M.P., Rodgers K.R., Dixon D.W. Characterization of the second conserved domain in the heme uptake protein HtaA from Corynebacterium diphtheriae. J. Inorg. Biochem. 2017;167:124–133. doi: 10.1016/j.jinorgbio.2016.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen C.E., Burgos J.M., Schmitt M.P. Analysis of novel iron-regulated, surface-anchored hemin-binding proteins in Corynebacterium diphtheriae. J. Bacteriol. 2013;195:2852–2863. doi: 10.1128/JB.00244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyman L.R., Peng E.D., Schmitt M.P. Corynebacterium diphtheriae iron-regulated surface protein HbpA is involved in the utilization of the hemoglobin-haptoglobin complex as an iron source. J. Bacteriol. 2018;200 doi: 10.1128/JB.00676-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyman L.R., Peng E.D., Schmitt M.P. The Corynebacterium diphtheriae HbpA hemoglobin-binding protein contains a domain that is critical for hemoprotein binding, cellular localization, and function. J. Bacteriol. 2021;203 doi: 10.1128/JB.00196-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wakeman C.A., Hammer N.D., Stauff D.L., Attia A.S., Anzaldi L.L., Dikalov S.I., Calcutt M.W., Skaar E.P. Menaquinone biosynthesis potentiates haem toxicity in Staphylococcus aureus. Mol. Microbiol. 2012;86:1376–1392. doi: 10.1111/mmi.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nobles C.L., Green S.I., Maresso A.W. A product of heme catabolism modulates bacterial function and survival. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Celis A.I., Choby J.E., Kentro J., Skaar E.P., DuBois J.L. Control of metabolite flux during the final steps of heme b biosynthesis in gram-positive bacteria. Biochemistry. 2019;58:5259–5270. doi: 10.1021/acs.biochem.9b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y., Qin R., Zaat S.A.J., Breukink E., Heger M. Antibacterial photodynamic therapy: overview of a promising approach to fight antibiotic-resistant bacterial infections. J. Clin. Transl. Res. 2015;1:140–167. [PMC free article] [PubMed] [Google Scholar]
- 46.Du D., Wang-Kan X., Neuberger A., van Veen H.W., Pos K.M., Piddock L.J.V., Luisi B.F. Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol. 2018;16:523–539. doi: 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- 47.Guo F., Wang M., Huang M., Jiang Y., Gao Q., Zhu D., Wang M., Jia R., Chen S., Zhao X., Yang Q., Wu Y., Zhang S., Huang J., Tian B., Ou X., Mao S., Sun D., Cheng A., Liu M. Manganese efflux achieved by MetA and MetB affects oxidative stress resistance and iron homeostasis in riemerella anatipestifer. Appl. Environ. Microbiol. 2023;89 doi: 10.1128/aem.01835-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stauff D.L., Skaar E.P. The heme sensor system of Staphylococcus aureus. Contrib. Microbiol. 2009;16:120–135. doi: 10.1159/000219376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donegan R.K. The role of host heme in bacterial infection. Biol. Chem. 2022;403:1017–1029. doi: 10.1515/hsz-2022-0192. [DOI] [PubMed] [Google Scholar]
- 50.Torres V.J., Stauff D.L., Pishchany G., Bezbradica J.S., Gordy L.E., Iturregui J., Anderson K.L., Dunman P.M., Joyce S., Skaar E.P. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe. 2007;1:109–119. doi: 10.1016/j.chom.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stauff D.L., Bagaley D., Torres V.J., Joyce R., Anderson K.L., Kuechenmeister L., Dunman P.M., Skaar E.P. Staphylococcus aureus HrtA is an ATPase required for protection against heme toxicity and prevention of a transcriptional heme stress response. J. Bacteriol. 2008;190:3588–3596. doi: 10.1128/JB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stauff D.L., Skaar E.P. Bacillus anthracis HssRS signalling to HrtAB regulates haem resistance during infection. Mol. Microbiol. 2009;72:763–778. doi: 10.1111/j.1365-2958.2009.06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt R.M., Carter M.M., Chu M.L., Latario C.J., Stadler S.K., Stauff D.L. Heme sensing in Bacillus thuringiensis: a supplementary HssRS-regulated heme resistance system. FEMS Microbiol. Lett. 2016;363:fnw076. doi: 10.1093/femsle/fnw076. [DOI] [PubMed] [Google Scholar]
- 54.Joubert L., Dagieu J.B., Fernandez A., Derré-Bobillot A., Borezée-Durant E., Fleurot I., Gruss A., Lechardeur D. Visualization of the role of host heme on the virulence of the heme auxotroph Streptococcus agalactiae. Sci. Rep. 2017;7 doi: 10.1038/srep40435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dos Santos P.T., Larsen P.T., Menendez-Gil P., Lillebæk E.M.S., Kallipolitis B.H. Listeria monocytogenes relies on the heme-regulated transporter hrtAB to resist heme toxicity and uses heme as a signal to induce transcription of lmo1634, encoding Listeria adhesion protein. Front. Microbiol. 2018;9:3090. doi: 10.3389/fmicb.2018.03090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bibb L.A., Schmitt M.P. The ABC transporter HrtAB confers resistance to hemin toxicity and is regulated in a hemin-dependent manner by the ChrAS two-component system in Corynebacterium diphtheriae. J. Bacteriol. 2010;192:4606–4617. doi: 10.1128/JB.00525-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lechardeur D., Cesselin B., Liebl U., Vos M.H., Fernandez A., Brun C., Gruss A., Gaudu P. Discovery of intracellular heme-binding protein HrtR, which controls heme efflux by the conserved HrtB-HrtA transporter in Lactococcus lactis. J. Biol. Chem. 2012;287:4752–4758. doi: 10.1074/jbc.M111.297531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saillant V., Lipuma D., Ostyn E., Joubert L., Boussac A., Guerin H., Brandelet G., Arnoux P., Lechardeur D. A novel Enterococcus faecalis heme transport regulator (FhtR) senses host heme to control its intracellular homeostasis. mBio. 2021;12 doi: 10.1128/mBio.03392-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ito Y., Nakagawa S., Komagata A., Ikeda-Saito M., Shiro Y., Nakamura H. Heme-dependent autophosphorylation of a heme sensor kinase, ChrS, from Corynebacterium diphtheriae reconstituted in proteoliposomes. FEBS Lett. 2009;583:2244–2248. doi: 10.1016/j.febslet.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 60.Keppel M., Davoudi E., Gätgens C., Frunzke J. Membrane topology and heme binding of the histidine kinases HrrS and ChrS in Corynebacterium glutamicum. Front. Microbiol. 2018;9:183. doi: 10.3389/fmicb.2018.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doi A., Nakamura H., Shiro Y., Sugimoto H. Structure of the response regulator ChrA in the haem-sensing two-component system of Corynebacterium diphtheriae. Acta Crystallogr. F Struct. Biol. Commun. 2015;71:966–971. doi: 10.1107/S2053230X15009838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wakeman C.A., Stauff D.L., Zhang Y., Skaar E.P. Differential activation of Staphylococcus aureus heme detoxification machinery by heme analogues. J. Bacteriol. 2014;196:1335–1342. doi: 10.1128/JB.01067-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakamura H., Hisano T., Rahman M.M., Tosha T., Shirouzu M., Shiro Y. Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2123385119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Light W.R., 3rd, Olson J.S. The effects of lipid composition on the rate and extent of heme binding to membranes. J. Biol. Chem. 1990;265:15632–15637. [PubMed] [Google Scholar]
- 65.Cannon J.B., Kuo F.S., Pasternack R.F., Wong N.M., Muller-Eberhard U. Kinetics of the interaction of hemin liposomes with heme binding proteins. Biochemistry. 1984;23:3715–3721. doi: 10.1021/bi00311a022. [DOI] [PubMed] [Google Scholar]
- 66.Sawai H., Yamanaka M., Sugimoto H., Shiro Y., Aono S. Structural basis for the transcriptional regulation of heme homeostasis in Lactococcus lactis. J. Biol. Chem. 2012;287:30755–30768. doi: 10.1074/jbc.M112.370916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arias C.A., Murray B.E. The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmitt M.P. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 1997;179:838–845. doi: 10.1128/jb.179.3.838-845.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen H., Moreau Y., Derat E., Shaik S. Quantum mechanical/molecular mechanical study of mechanisms of heme degradation by the enzyme heme oxygenase: the strategic function of the water cluster. J. Am. Chem. Soc. 2008;130:1953–1965. doi: 10.1021/ja076679p. [DOI] [PubMed] [Google Scholar]
- 70.Wilks A., Heinzl G. Heme oxygenation and the widening paradigm of heme degradation. Arch. Biochem. Biophys. 2014;544:87–95. doi: 10.1016/j.abb.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kunkle C.A., Schmitt M.P. Comparative analysis of hmuO function and expression in Corynebacterium species. J. Bacteriol. 2007;189:3650–3654. doi: 10.1128/JB.00056-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bibb L.A., Kunkle C.A., Schmitt M.P. The ChrA-ChrS and HrrA-HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect. Immun. 2007;75:2421–2431. doi: 10.1128/IAI.01821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nambu S., Matsui T., Goulding C.W., Takahashi S., Ikeda-Saito M. A new way to degrade heme: the Mycobacterium tuberculosis enzyme MhuD catalyzes heme degradation without generating CO. J. Biol. Chem. 2013;288:10101–10109. doi: 10.1074/jbc.M112.448399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reniere M.L., Ukpabi G.N., Harry S.R., Stec D.F., Krull R., Wright D.W., Bachmann B.O., Murphy M.E., Skaar E.P. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol. Microbiol. 2010;75:1529–1538. doi: 10.1111/j.1365-2958.2010.07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reniere M.L., Skaar E.P. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol. Microbiol. 2008;69:1304–1315. doi: 10.1111/j.1365-2958.2008.06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu R., Skaar E.P., Zhang R., Joachimiak G., Gornicki P., Schneewind O., Joachimiak A. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J. Biol. Chem. 2005;280:2840–2846. doi: 10.1074/jbc.M409526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee W.C., Reniere M.L., Skaar E.P., Murphy M.E. Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J. Biol. Chem. 2008;283:30957–30963. doi: 10.1074/jbc.M709486200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Snyder S.N., Mak P.J. Structure-function characterization of the mono- and diheme forms of MhuD, a noncanonical heme oxygenase from Mycobacterium tuberculosis. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2021.101475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsui T., Nambu S., Ono Y., Goulding C.W., Tsumoto K., Ikeda-Saito M. Heme degradation by Staphylococcus aureus IsdG and IsdI liberates formaldehyde rather than carbon monoxide. Biochemistry. 2013;52:3025–3027. doi: 10.1021/bi400382p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Skaar E.P., Gaspar A.H., Schneewind O. Bacillus anthracis IsdG, a heme-degrading monooxygenase. J. Bacteriol. 2006;188:1071–1080. doi: 10.1128/JB.188.3.1071-1080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Skaar E.P., Schneewind O. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microb. Infect. 2004;6:390–397. doi: 10.1016/j.micinf.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 82.Péchiné S., Collignon A. Immune responses induced by Clostridium difficile. Anaerobe. 2016;41:68–78. doi: 10.1016/j.anaerobe.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 83.Burgos J.M., Schmitt M.P. The ChrSA and HrrSA two-component systems are required for transcriptional regulation of the hemA promoter in Corynebacterium diphtheriae. J. Bacteriol. 2016;198:2419–2430. doi: 10.1128/JB.00339-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Keppel M., Hünnefeld M., Filipchyk A., Viets U., Davoudi C.F., Krüger A., Mack C., Pfeifer E., Polen T., Baumgart M., Bott M., Frunzke J. HrrSA orchestrates a systemic response to heme and determines prioritization of terminal cytochrome oxidase expression. Nucleic Acids Res. 2020;48:6547–6562. doi: 10.1093/nar/gkaa415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang M., Nie Y., Wu X.L. Extracellular heme recycling and sharing across species by novel mycomembrane vesicles of a Gram-positive bacterium. ISME J. 2021;15:605–617. doi: 10.1038/s41396-020-00800-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stauff D.L., Torres V.J., Skaar E.P. Signaling and DNA-binding activities of the Staphylococcus aureus HssR-HssS two-component system required for heme sensing. J. Biol. Chem. 2007;282:26111–26121. doi: 10.1074/jbc.M703797200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.