Abstract
The genus Amaranthus is one of the few dicotyledonous, non-grass mesophytes that use specialized C4 annuals or short-lived perennials to produce significant amounts of edible small-seeded pseudo cereals. In this study, we characterized the genetic diversity of 120 genotypes of amaranths collected from diverse amaranth-growing regions of Ethiopia using multivariate analysis of yield and yield-related traits. The experiments were carried out at Hawassa University, in the years 2020 and 2021. The experimental design was set up using an alpha lattice design and replicated two times. The collected data were examined for 24 descriptors. Principal component analysis showed that the first six principal components with eigenvalues greater than one contributed 80.41% of the variability. However, the first two principal components explained 52.42% of the total variation. The highest contributing traits in the first component were days to flowering, basal stem diameter, plant height at flowering, plant height at maturity, auxiliary inflorescence length, number of branches, terminal inflorescence lateral length, days to maturity, terminal inflorescence stalk length, leaf number, leaf length, top lateral branch length. The traits with the greatest weight on the second component were leaf area, basal lateral branch length, leaf length, and leaf width, grain filling period, grain sinking filling rate, and grain yield. Therefore, selection based on these traits would be effective for yield improvement in amaranth genotypes. Additionally, the hierarchical clustering grouped all the genotypes into five clusters. The pairwise generalized squared distance (D2) among the five clusters based on Mahalanobis's D2 statistics revealed the maximum and highly significant genetic distance was observed between II and III (277.79), while the minimum inter-cluster distance observed between clusters I and II (39.50). The findings suggest that amaranth genotypes in Ethiopia have a lot of genetic variation, which might be used for future breeding and ought to be conserved.
Keywords: Descriptors, Genotype, Amaranth, Agro-morphological, Characters, Multivariate
1. Introduction
Amaranthus belongs to the Amaranthaceae family consisting of 50–70 species [1]. Amaranthus is a C4 plant that thrives in drought-stressed environments and can withstand adverse abiotic conditions such as excessive salt, acidity, or alkalinity, making it particularly well-suited for subsistence farming [2]. Because of their C4 characteristics, amaranths may grow swiftly. They are accounted for as highly prolific plants as a result [3]. Historical records indicate that the genus includes both domesticated and wild species. Around 8000 years ago, the Mayan civilization of South and Central America was the first to domesticate and produce amaranths [4]. Amaranths are broad-leaved plants with tiny, nutrient-rich grains that are sometimes referred to as pseudocereals. They are one of the few non-grass species that yield a sizable quantity of small-seeded grain [5].
Amaranth was historically a staple of traditional African agriculture and is semi-domesticated in Ethiopia and other nations of East Africa, where it is mostly grown as a vegetable [6]. Today, amaranth is produced as a grain crop in remote locations including the mountains of Ethiopia, the hills of South India, the Himalayas of Nepal, and the plains of Mongolia. It has the best seed quality and the greatest potential for usage as a food component [7,8]. Additionally, Ethiopia has been considered a center for amaranth variety due to the extensive use of regional names and the high level of genetic diversity there. Eleven species have been found in the Ethiopia and Eritrea Flora area [9]. This genus is economically significant, it is regarded as the most important raw resource for food and nutrition, as well as a powerful medicinal plant [10]. These plants exhibit all of the features of a functional food [11], and their nutritional and functional properties, such as lowering blood cholesterol and avoiding gluten, are extremely comparable [12]. As a result, including these grains in one's diet has the potential to enhance one's diet while also providing health advantages. Moreover, amaranth grains have a greater protein content than other typical cereals [13].
Due to their extreme adaptability to various eco-geographical conditions, amaranths also display incredible diversity [14]. Only a few taxonomic characteristics are exclusive to the genus Amaranthus, despite the tremendous morphological diversity between and within species [15,16]. Amaranths have a lot of genetic diversity, which suggests there is a lot of possibility for improvement. It is, therefore, necessary to accomplish initiatives for crop improvement and biodiversity protection in amaranth [17]. The assessment of a crop species' genetic diversity is an excellent place to start when it comes to crop improvement since it provides a framework and a roadmap for choosing parental lines and designing a breeding program. It's crucial to the creation of new crop varieties with desirable characteristics. As a result, determining the genetic diversity of current genotypes is a prerequisite for crop improvement. Unfortunately, genetic research of the genus amaranth is very limited in Ethiopia due to a lack of extensive research, discrimination, and lack of knowledge, and the improvement of this plant is limited. Furthermore, researchers are less interested in this “orphan crop,” as it is known [18]. By establishing a value chain for the production of amaranth-based value-added products, which may help with food security and nutritional quality, the research on amaranth has made a significant contribution to boosting amaranth growth. Even though several countries have reported on the distribution of Amaranth species used like cereal grains and the production of ancient grain products, Ethiopia has yet to conduct similar research.
Various crop species, such as the Arachis and Amaranthus genotypes , have had quantitative variations correctly classified using multivariate statistical methods [[19], [20], [21], [22]]. Although studies on the morphological variety of amaranths [17], and quinoa [23] have been conducted, in-depth agronomic suggestions for yield and yield-related improvement are uncommon. To effectively manage genotypes and make proper use of them in breeding programs, the current study was conducted with the following aims in mind: To optimize yield, it is crucial to assess genetic diversity using multivariate analysis. To enhance leaf and grain yields through selection and subsequent breeding programs, this study set out to evaluate the genetic diversity in amaranth genotypes. The amaranth genus, according to several studies, has a lot of morphological diversity [[24], [25], [26]]. This type of information is critical for selecting better genotypes for breeding efforts. In Ethiopia, no specific study has been conducted to characterize amaranth genotypes so far. The results are helpful in future genetic research to enhance this climate-resilient and highly nutritious neglected crop species using agro-morphological characteristics.
2. Materials and methods
2.1. Experimental site
The experiment was carried out at Hawassa University, which is located at the agricultural experimental site. Geographically, the location is in Ethiopia's Sidama region, around 275 km from the capital, Addis Ababa. The experimental area is located at latitude 7°2′ 54.7503″N and longitude 38°30′ 17.1608″ E, at an elevation of 1709 m above sea level. The soil texture class in the experimental area was clay loam, and the pH ranged from 6–to 6.5. The district's mean monthly minimum and maximum temperatures are 14.1.0 °C and 27.9 °C, respectively. For two growing seasons, the experimental farm received an average of 1379.16 mm of rain throughout the growing seasons.
2.2. Plant material
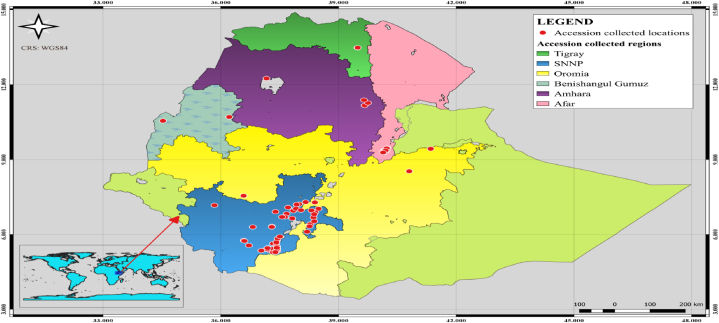
120 amaranth genotypes in total were utilized in this study (Table 1); 34 genotypes came from the Ethiopian Biodiversity Institute (EBI), 2 from the Melkasa research center, 15 from the Were research center (Afar region), 8 from the Sidama, and the remaining 61 genotypes were collected in the SNNP, Oromia, and Tigray regions of Ethiopia in 2019 and characterized for various agro-morphological traits (Fig. 1). One hundred eighteen members have passport information; the other two are released varieties since they don't. To accomplish the aforementioned objective a two-season experiment was run at the agricultural research site of Hawassa University in Ethiopia.
Table 1.
List and location of plant materials included in the study.
| Genotype Name | Easting | Northing | Altitude | |
|---|---|---|---|---|
| KEN-065 | 321813 | 583339 | 1756 | |
| YRC-048 | 411783 | 675273 | 1995 | |
| KAZ-077 | 327268 | 599608 | 1202 | |
| KEN-067 | 321745 | 585148 | 1760 | |
| 211457 | 326440.36 | 589244.33 | 1560 | |
| 242917 | 812954 | 793136 | 2200 | |
| KEN-011 | 299762 | 603070 | 1739 | |
| 241764 | 306077 | 591230 | 1520 | |
| HA-003 | 636126 | 1046188 | 726 | |
| KAZ-076 | 327267 | 599336 | 1202 | |
| 208683 | 758052 | 1043629 | 2270 | |
| KEN-016 | 322071 | 584756 | 1745 | |
| MEK-084 | 552161 | 1490423 | 2155 | |
| HAL-039 | 407697 | 806621 | 1834 | |
| 212892 | 315406 | 622936 | 2200 | |
| 211455 | 328390 | 645017 | 1150 | |
| HAW-041 | 444076 | 776871 | 1750 | |
| KAZ-055 | 326719 | 609804 | 1213 | |
| DRA-053 | 324631 | 614748 | 1290 | |
| ALE-023 | 299964 | 597729 | 1370 | |
| KEN-015 | 320119 | 586057 | 1819 | |
| ALE-069 | 299971 | 597724 | 1376 | |
| DIL-050 | 421120 | 709519 | 1459 | |
| KAZ-057 | 326722 | 609738 | 1214 | |
| KEN-022 | 321811 | 585162 | 1751 | |
| SHIA-007 | 627807 | 1036846 | 735 | |
| 240815 | 354053.54 | 754795.3 | 1950 | |
| 209056 | 312304.71 | 699421.8 | 1660 | |
| ABL-004 | 369796 | 734542 | 1367 | |
| ALE-034 | 295826 | 597501 | 1316 | |
| ALE-024 | 299885 | 597965 | 1368 | |
| SAA-004 | 636126 | 1046188 | 726 | |
| 212582 | 572790 | 1234490 | 1840 | |
| ALE-068 | 299970 | 597736 | 1384 | |
| 212581 | 581861 | 1245570 | 2920 | |
| KAZ-007 | 327352 | 599054 | 1187 | |
| 204645 | 327492.26 | 590316.19 | 1600 | |
| 225717 | 348942 | 740795 | 1440 | |
| KEN-019 | 321816 | 585168 | 1754 | |
| 91003 | 233325 | 835289 | 2040 | |
| 242533 | 665945 | 1167179 | 1250 | |
| KEN-013 | 319707 | 586409 | 1772 | |
| BNT-026 | 247228 | 615060 | 1618 | |
| AC-NL | Released variety | Released variety | 1550–2400 | |
| Madiira II | Released variety | Released variety | 1550–2400 | |
| BKD-027 | 232336 | 637249 | 1427 | |
| 204644 | 334751.47 | 653902.46 | 1200 | |
| 225718 | 348942 | 740795 | 1440 | |
| ALE-070 | 299981 | 597743 | 1329 | |
| OTI-044 | 431913 | 721434 | 1893 | |
| KAZ-008 | 326435 | 604708 | 1204 | |
| MEK-081 | 552793 | 1489902 | 2143 | |
| KAZ-056 | 326684 | 609708 | 1214 | |
| KEN-018 | 321785 | 585144 | 1761 | |
| BA-001 | 628326 | 1026376 | 743 | |
| ALE-074 | 299762 | 603000 | 1655 | |
| 209057 | 341220.34 | 740810 | 1580 | |
| KAZ-059 | 327268 | 599608 | 1202 | |
| KEN-010 | 319325 | 585745 | 1729 | |
| KEN-012 | 319703 | 586470 | 1760 | |
| 215567 | 372960 | 768376 | 2100 | |
| ALE-025 | 300048 | 597628 | 1349 | |
| SHD-001 | 434789 | 761187 | 1874 | |
| ALE-073 | 301337 | 604237 | 1693 | |
| DAL-043 | 429799 | 738442 | 1790 | |
| SA-001/07 | 624955 | 1028024 | 736 | |
| SRA-002 | 632177 | 1031734 | 742 | |
| 219284 | 358268 | 783157 | 1750 | |
| HA-008 | 634804 | 1044976 | 730 | |
| ALE-071 | 299973 | 597729 | 1372 | |
| 240814 | 322866.04 | 764187.65 | 1250 | |
| KAZ-060 | 319338 | 585751 | 1734 | |
| MEK-083 | 552491 | 1490761 | 2151 | |
| BKD-031 | 234460 | 634949 | 1400 | |
| WON-049 | 418632 | 699405 | 1743 | |
| KAZ-058 | 327268 | 599608 | 1202 | |
| KAZ-006 | 327308 | 598524 | 1188 | |
| BKD-029 | 234471 | 634953 | 1409 | |
| WA-003 | 628204 | 1033787 | 741 | |
| YRB-002 | 424075 | 769481 | 1857 | |
| KEN-021 | 321818 | 585166 | 1752 | |
| KAZ-079 | 326433 | 604711 | 1204 | |
| KEN-064 | 321816 | 585168 | 1762 | |
| SHL-040 | 433633 | 805425 | 1662 | |
| KEN-017 | 322084 | 584791 | 1762 | |
| 242532 | 665945 | 1167179 | 1250 | |
| SA-008/07 | 624844 | 1026573 | 740 | |
| 240813 | 323825.31 | 589809.06 | 2750 | |
| 208764 | 698483.15 | 943746.8 | 1850 | |
| MEK-080 | 552552 | 1490147 | 2162 | |
| BA-016 | 629359 | 1027955 | 740 | |
| WA-001 | 627878 | 1033651 | 740 | |
| SA-025/07 | 625039 | 1026746 | 735 | |
| DMG-038 | 380667 | 777365 | 1881 | |
| KAZ-078 | 327269 | 599337 | 1200 | |
| 242531 | 195529 | 1185259 | 1250 | |
| DA-006 | 630310 | 1034519 | 735 | |
| 211456 | 314087.91 | 599272.76 | 1570 | |
| CHU-045 | 431835 | 721533 | 1910 | |
| DUF-003 | 394497 | 770836 | 2113 | |
| ADZ-037 | 389243 | 797508 | 1912 | |
| 212583 | 570917 | 1258445 | 1640 | |
| 208025 | 300580 | 1354876 | 2100 | |
| 225713 | 311940 | 696674 | 1600 | |
| KEN-014 | 319660 | 586850 | 1690 | |
| 91001 | 315299 | 584231 | 1890 | |
| 212893 | 324653 | 628440 | 1380 | |
| ALE-033 | 282131 | 591864 | 589.5 | |
| KEN-020 | 321817 | 585166 | 1753 | |
| K3A-005 | 631670 | 1035541 | 738 | |
| 242530 | 665945 | 1167179 | 1250 | |
| BKD-028 | 234471 | 634953 | 1409 | |
| SA-005/07 | 624847 | 1026721 | 737 | |
| KAZ-009 | 325782 | 604450 | 1213 | |
| 242534 | 665945 | 1167179 | 1250 | |
| 212890 | 382230 | 795996 | 2180 | |
| DAL-042 | 431596 | 753041 | 1780 | |
| MEK-082 | 552972 | 1490517 | 2140 | |
| 225715 | 258450 | 696873 | 1780 | |
| ALE-075 | 299919 | 603075 | 1640 | |
Fig. 1.
Map of Ethiopia showing the collection site for the different genotypes of Amaranthus species from different agroecological regions. Maps were generated using QGIS v.3.14.15 Pi, QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Foundation; 2020. http://qgis.osgeo.org.
2.3. Experimental layout and crop management
The experiment was carried out using an imperfect random block design known as an alpha lattice, which involved several investigators [27,28]. Alpha lattice design, as compared to RCBD, is a significant incomplete block design group (IBD) variant that can lower experimental error by removing variability between small blocks, limiting unknown variation within each replication, and so enhancing field trial efficiency [29].
The experimental field was prepared by manually hoeing the ridges, plowing the field when needed, and harrowing it with a tractor. Amaranths were laid out in a setup with two replications, sixteen blocks, and fifteen plots per block. Each unit plot, which had a size of 1.80 m in length, 1.50 m in breadth, and 2.7 m2 in area, was separated from the others by a 0.60 m route between plots, a one m walkway between blocks, and a three m gap between replications. A total of 1,345.2 sq m (38 m*35.4 m) was needed for each season.
The seed was sowed on April 15, 2020, and 2021, at one site in each season, under favorable growth conditions, and throughout the agricultural season. The environment at the experimental location is one growing season in a calendar year. Different genotypes of seeds were continually sowed in two rows with a 0.75 m interval between them. The small seeds were put in seedbeds and covered with finely powdered farmyard manure that had been mixed with sand in a 1:4 ratio. Twice, thinning was carried out at 14 and 22 days after sowing (DAS), with a space of 75 cm between rows and 30 cm between plants. According to Grubben et al. [30] and Shukla et al. [31], the experiment followed standard cultural practices. A single experimental unit (plot) included 36 individuals in two rows, and hand-hoeing was used to control weeds at 2-week intervals following germination and whenever necessary. A total of 12 plants were maintained in each plot.
2.4. Agro-morphological traits and data collection
To characterize the material under study, observations were made on various morphological traits at distinct phonological stages (Table 2). In each plot, the phenotypic characteristics of 10 randomly tagged plants were assessed. Plants were grown to maturity and defined using amaranth descriptors, as recommended by the International Board for Plant Genetic Resources based on taxonomic keys [32]. Characters not on the list that were considered necessary for the characterization were included. In this investigation, a total of 24 quantitative characters were recorded.
Table 2.
List of quantitative agro-morphological traits used along with their phenotype code, description, and phenotype scores.
| Traits | Phenotypic code | Descriptions |
|---|---|---|
| Plant height at flowering (cm) | PHF | At 50% flowering, the height in cm from the ground's surface to the main stem's tip (terminal inflorescence) was measured using a meter tap. |
| Plant height at maturity (cm) | PHM | Using a meter tap, determine a plant's height in centimeters at the 80% maturity stage from the soil's surface to the top of the inflorescence. |
| Leaf length (cm) | LL | A meter tap was used to measure the length of the leaves at the blooming stage for each plant's three typical representative leaf sizes (small, medium, and big). |
| Leaf width (cm) | LW | Three different representative leaf sizes (small, middle, and large) per plant were randomly sampled for measurement of leaf width at the flowering stage by using a meter tap. |
| Stem diameter (mm) | SD | Using a digital caliper, the thickest section of the stem, at the maturity stage, was measured to be 10 cm above the ground. |
| Leaf thickness (mm) | LT | A digital caliper was used to measure the leaf thickness in the center of the leaf at the blooming stage on three distinct representative leaf sizes (small, medium, and big) from each plant. |
| Petiole length (cm) | PL | At the flowering stage, a meter scale was used to measure the distance in centimeters from the base of the stem to the petiole of each leaf for three sample petiole sizes (small, medium, and big) on each plant. measured from the start of the leaf blade to the point where the petiole was inserted. |
| Number of the branch (number) | BN | At the mature growth stage, the number of branches along the main stem was counted. |
| Terminal inflorescence stalk length (cm) | TISL | A meter rule was used to measure the average length of 10 randomly chosen terminal inflorescence stalks from the base to the tip at the maturity stage. |
| Terminal inflorescence laterals length (cm) | TILL | The mean length of 10 randomly selected terminal inflorescence laterals length (cm) was measured at the maturity stage. |
| Number of nodes on the main stem (number) | NN | Many nodes were counted along the main stem at the flowering stage. |
| Grain yield (tone ha−1) | GY | An adjusted moisture content of 12% was used to estimate the grain yield, which was then weighed to calculate the dry matter of the net plot and extrapolated per hectare. |
| Leaf area (cm2) | LA | Three different representative leaf sizes (small, middle, and large) per plant were measured by using a digital leaf area meter. |
| 1000 Seed weight (g) | TSW | Using an analytical balance, a sample of a thousand seeds from each genotype was counted and weighed. |
| Number of leaves (number) | LN | At the flowering stage, the main stem's countable leaves were counted. |
| Days to maturity (days) | DM | Plant maturity occurs when the seed removed from the center of the inflorescence does not change form when crushed between fingers and when the inflorescence turns from green to brown. |
| Days to flowering (days) | DF | From the 90% emergence date until the point where 50% of the plants in a row had ears developed in the terminal inflorescence, it took days for 50% of the plants to bloom. |
| Basal lateral branch length (cm) at maturity | BLBL | Using a meter tap, the maturity stage is measured from the bottom of the stem branches to the top of a branch. |
| Top lateral branch length (cm) at maturity | TLBL | measured using a meter tap at the mature stage from the top of the stem branches to the top of a branch. |
| Axillary inflorescence length (cm) | AIL | At the maturity stage, the mean length of 10 randomly chosen auxiliary inflorescences was measured. |
| Days to emergence (days) | DE | The number of days from planting to 90% of seedlings germinates in each plot was recorded and expressed in days. |
| Leaf yield (tone ha−1) | LY | The separated parts of the leaves were dried for a week (7 days) and weighed to determine the dry matter of the net plot and then extrapolated per hectare. |
| Grain filling period (days) | GFP | The number of days between days for 50% flowering and days to grain maturity. |
| Grain sink filling rate (kg ha−1day −1) | GSFR | GSFR is stated as a ratio of grain yield to the number of days from 50% blooming to seed maturity and is represented in kilograms per hectare per day. |
In each season, data was collected from the field at four phases of growth: germination, vegetative stage, 50% of the plants' developing inflorescences, and plant maturity shortly before and after harvesting. Plant height at flowering and maturity (cm), petiole length (cm), leaf width (cm), leaf thickness (mm), terminal inflorescence stalk length (cm), inflorescence lateral length (cm), number of branches per plant (number), number of leaves (number), number of nodes (number), stem diameter (mm), number of days to emergence (days), number of days to flowering (days), number of days to maturity (days), leaf area (cm2), and 1000-seed weight (g.), leaf yield tone ha−1, grain yield tone ha−1, grain filling period (days), grain filling rate (kg ha−1day−1) are among them. As a result, agro-morphological characteristics were evaluated four times per plot for each replication, for a total of forty observations per plot. Seedling data, vegetative data, inflorescence data, and seed data were among the 24 characteristics evaluated. Except for days to emergence, days to flowering, and days to maturity, which were recorded at the plot level, all ten planted genotypes for each population (two replications) were used to collect quantitative trait data. For the traits, the mean value of 10 plants per plot was measured and recorded. Three different leaf lengths, leaf thickness, leaf width, petiole length, and leaf area (small, medium, and large) were randomly picked per plant for measurement. Leaf length, breadth, thickness, petiole length, and area were computed using the averages of 30 measurements (10 plants per plot for each of the 3 different leaf and petiole sizes). The harvest was done manually, panicles were carefully removed to minimize grain spilling, then the panicles were threshed by a mechanical thresher, seeds were cleaned, and the seeds were held in an electric oven at 100 °C for 48 h to regulate the moisture content to 12%, as advised by Refs. [33,34]. A digital caliper was used to measure the diameter of the stem and the thickness of the leaves (Digimatic Solar DC-S15 m, Mitutoyo, Japan). The LI-3100 AREA METER, an electronic leaf area meter, was used to measure the leaf area (LI, Cor, Inc, Lincoln, Nebraska, USA).
2.5. Data analysis
The mean values of the genotypes of the two seasons were subjected to the procedures of a multivariate analysis. Because the parameters considered in this study did not share a common measurement scale, the quantitative traits for the principal component, hierarchical agglomerative cluster, and inter-cluster and intra-cluster distance analyses (standardized Mahalanobis's D2) were all standardized to a mean of zero and a standard deviation of one [[35], [36], [37]].
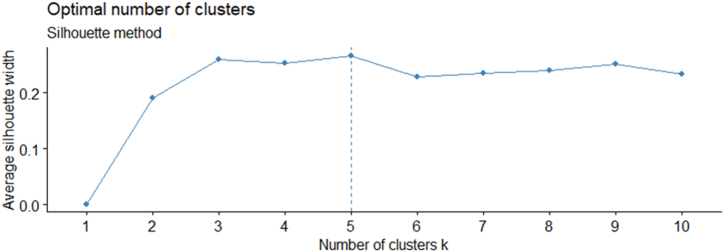
To group the 120 genotypes and to bring out the patterns of similarity and dissimilarity based on 24 quantitative morphological characters, the R version (4.1.2) has been used with various packages to perform cluster-based analysis based on ward D2 hierarchical grouping using Euclidian distance and determine the optimum number of clusters in the data set using the average silhouette method for the dendrogram [38]. The silhouette technique and the NbClust software in R Studio were used to determine the optimum number of clusters [39].
Principal component analysis, eigenvalues, eigenvectors, biplot, and clusters were obtained using FactoMineR and factoextra packages using R studio. At each axis of differentiation, principal component analysis (PCA) displays the importance of the largest contributor to the total variance [40]. PCs with an eigenvalue ≥1.0 were included in the criteria for determining the number of main components (PCs) [41]. In addition, the first PCs that accounted for at least 70% to 80% of the cumulative variance were kept and used to explain the variability [42,43]. Using the “biotools” package of the R-studio, the means of each replication and the overall means of each trait were used to calculate the D2 distance and group the genotypes according to D2 statistics [44]. The important proportion and the cumulative proportion of each variable were determined using the “Singh” statistic from the same package based on the squared generalized Mahalanobis distance [45]. The significance is determined by the generalized Mahalanobis distance squared.
Using the SAS system software package, genetic divergence between clusters was computed based on a correlation matrix and Mahalanobis' D2 statistic [46] as suggested by Ref. [47]. Intercluster distances were calculated based on the standardized Mahalanobis's D2 statistics as:
| D2 ij = (xi – xj)’ cov-1 (xi – xj) |
Where, D2ij = the distance between cases i and j; xi and xj = vectors of the values of the variables for cases i and j; and cov-1 = the pooled within groups variance-covariance matrix.
3. Results
3.1. Principal component analysis (PCA)
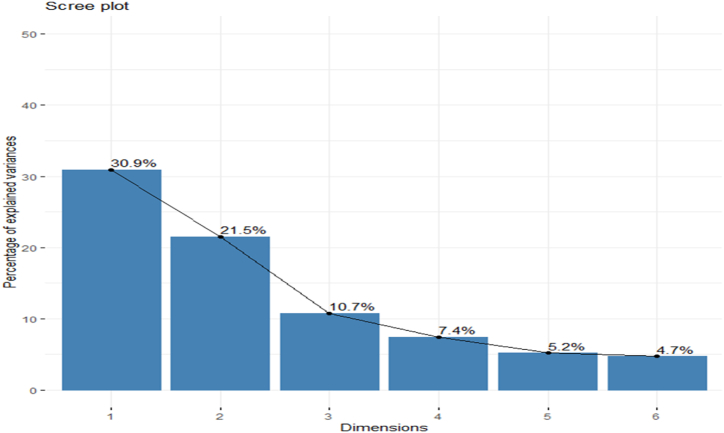
The six primary components of PCs 1 through 6 were generated from the original data. PC1, PC2, PC3, PC4, PC5, and PC6 were maintained from the overall main components, accounting for 80.41% of the total variance, with values of 30.94%, 21.49%, 10.70, 7.44, 5.16, and 4.69%, respectively. In any further data analysis, these main component scores may be used to summarize the original 24 variables, which accounted for the majority of the variance (Table 3).
Table 3.
Eigenvectors and eigenvalues of the first six principal components (PCs) for 24 quantitative characters of 120 genotypes.
| Traits Principal Components | ||||||
|---|---|---|---|---|---|---|
| PC.1 | PC.2 | PC.3 | PC.4 | PC.5 | PC.6 | |
| Eigenvalue | 7.42 | 5.16 | 2.57 | 1.79 | 1.24 | 1.12 |
| Variance. Percent | 30.94 | 21.49 | 10.70 | 7.44 | 5.16 | 4.69 |
| Cumulative. variance. Percent | 30.94 | 52.42 | 63.13 | 70.57 | 75.73 | 80.41 |
| Eigenvectors | ||||||
| Days to emergence [DE] | 0.33 | −0.45 | 0.52 | −0.31 | 0.07 | −0.01 |
| Days to flowering [DF] | 0.86 | 0.31 | 0.04 | −0.03 | 0.06 | −0.22 |
| Plant height at flowering [PHF] | 0.83 | 0.17 | 0.31 | 0.04 | 0.03 | −0.09 |
| Leaf number [LN] | 0.46 | −0.17 | −0.41 | 0.58 | 0.32 | 0.20 |
| Leaf area [LA] | 0.27 | 0.84 | −0.13 | −0.03 | −0.22 | −0.12 |
| Leaf length [LL] | 0.45 | 0.81 | 0.13 | 0.12 | −0.07 | −0.06 |
| Leaf width [LW] | 0.44 | 0.80 | 0.13 | 0.04 | −0.17 | 0.03 |
| Leaf thickness [LT] | −0.21 | 0.54 | 0.12 | 0.41 | 0.01 | 0.05 |
| Petiole length [PL] | 0.43 | 0.06 | 0.3 | 0.29 | −0.43 | 0.47 |
| Leaf yield [LY] | 0.44 | 0.34 | 0.03 | 0.12 | −0.21 | 0.30 |
| Branch number [BN] | 0.77 | −0.33 | 0.35 | 0.14 | 0.11 | 0.04 |
| Basal lateral branch length [BLBL] | 0.15 | −0.83 | 0.29 | −0.05 | 0.07 | 0.25 |
| Top lateral branch length [TLBL] | −0.45 | −0.23 | 0.56 | 0.04 | −0.18 | 0.11 |
| Node number [NN] | 0.42 | −0.25 | −0.45 | 0.57 | 0.30 | 0.25 |
| Days to maturity [DM] | 0.69 | −0.33 | 0.21 | 0.29 | 0.00 | −0.49 |
| Plant height at maturity [PHM] | 0.81 | 0.09 | 0.38 | −0.03 | 0.10 | 0.07 |
| Stem diameter [SD] | 0.84 | 0.30 | 0.18 | −0.10 | 0.10 | −0.05 |
| Auxiliary inflorescence length [AIL] | −0.80 | 0.09 | 0.31 | 0.34 | −0.15 | −0.01 |
| Terminal inflorescence lateral length [TILL] | −0.72 | 0.28 | 0.37 | 0.32 | −0.04 | −0.15 |
| Terminal inflorescence stalk length [TISL] | −0.61 | −0.01 | 0.50 | 0.42 | −0.03 | 0.03 |
| Grain-filling periods [GFP] | 0.15 | −0.67 | 0.23 | 0.38 | −0.05 | −0.43 |
| Grain sink filling rate [GSFR] | −0.38 | 0.62 | 0.29 | −0.14 | 0.54 | 0.14 |
| Thousand seed weight [TSW] | −0.35 | 0.35 | −0.33 | 0.28 | 0.00 | −0.29 |
| Grain yield [GY] | −0.38 | 0.48 | 0.46 | −0.01 | 0.58 | 0.00 |
The first principal component (PC1) accounted for 30.94% of the total variation and was dominated by yield-related traits. The agro-morphological traits that loaded highly for the principal component (PC1) were, DF (0.86), SD (0.84), PHF (0.83), PHM (0.81), AIL (−0.80), BN (0.77), TILL (−0.72), DM (0.69), TISL (−0.61), LN (0.46), LL (0.45), TLBL (−0.45), LW (0.44), LY (0.44), GY (−0.38), GSFR (0.38), and TSW (−0.35). The second principal component (PC2) demonstrated 21.49% of the total variation, which related to traits such as LA (0.84), BLBL (−0.83), LL (0.81), LW (0.80), GFP (−0.67), GSFR (0.62), and GY (0.48). The third principle component explained 10.70% of the total variation and was associated with the TLBL (0.56), DE (0.50), TISL (0.50), GY (0.46), NN (−0.45), the LN (−0.41), PHM (0.38), BN (0.35), and TSW (−0.35). The fourth principal components were selected from 24 components, which accounted for 7.44% of the total variation, for LN (0.58), NN (0.57), TISL (0.42), LT (0.41), GFP (0.38), AIL (0.34), TILL (0.32), DE (-0.31), and DM (0.29). The fifth principal component contributed 5.16% of the total variation and was mainly related to GY (0.58), GSFR (0.54), and PL. The positive and negative loading indicates the presence of positive and negative correlation trends between the components and the variables. Therefore, the above-mentioned traits that load high either positively or negatively contributed more to the diversity, and they were the ones that most distinguished the clusters. (Table 3).
3.2. Scree plot
The Scree plot depicts the relative distances between eigenvalues, which are used to rank the components in order of preference. The first six numbers are regarded to be important (as their eigenvalues are higher or equal to 1). Then the numbers decrease even more substantially, and we decide that these components aren't as important as they once were, and we'll leave them out of subsequent analyses. As shown in Fig. 2, PC1 had 30.94% variability, followed by PC2 with 21.49%, 10.70%, 7.44%, 5.16%, and 4.69%, respectively, with eigenvalues of 7.42, 5.16, 2.56, 1.79, 1.24 and 1.12.
Fig. 2.
Scree plot of principal components (PCs) for 24 quantitative traits in 120 Amaranths genotypes.
3.3. Clustering of genotypes based on regions of the country
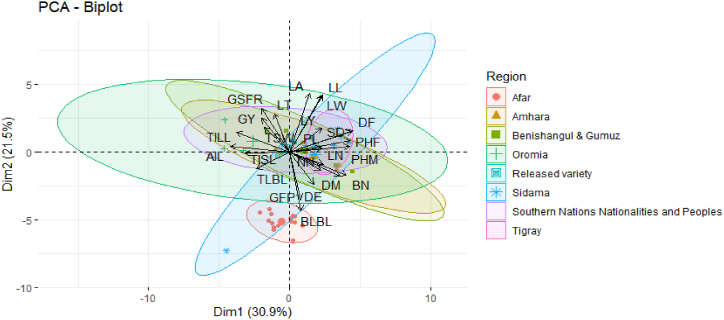
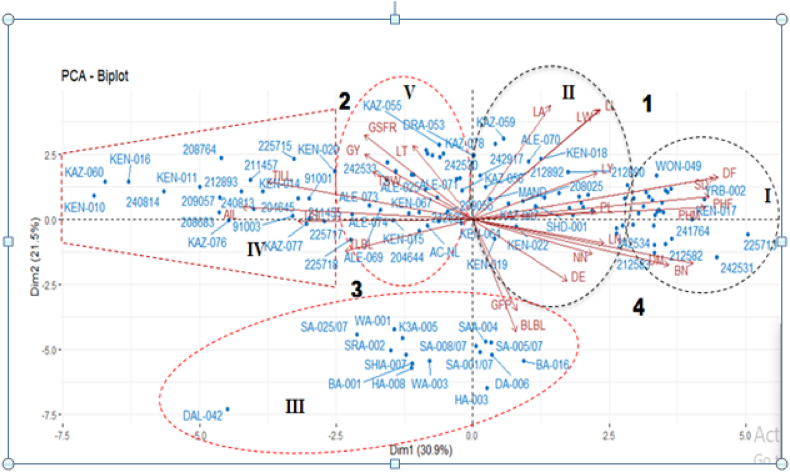
A principal component analysis biplot revealed that dim1 distinguishes two groups of amaranth genotypes: one largely comprised of Tigray and Sidama that had the longest PHM, PHF, PL, DF, and high LN, and several NN, thicker SD when compared to genotypes from Oromia, Amhara, South Nation Nationality People Regional State, and Benishangul and Gumuz. The second group, which included genotypes from Amhara, Benishangul & Gumuz, Oromia, and the South Nation Nationality People's Regional State, had longer TISL, AIL, TILL, GSFR, GY when compared to Tigray, Afar, and the Sidama region (Fig. 3). Dimension 2 also distinguishes between two genotype groups. The longest BLBL, DE, and NN are observed in Afar and Sidama genotypes, whereas the highest LY, LA, LW, GFP, and LL are observed in Amhara, Benishangul & Gumuz, Oromia, and South Nation Nationality People Regional State genotypes.
Fig. 3.
Principal components analysis biplot showing quantitative variables and individuals on Dimensions 1 & 2. For abbreviated names (codes) of different traits (see Table 2).
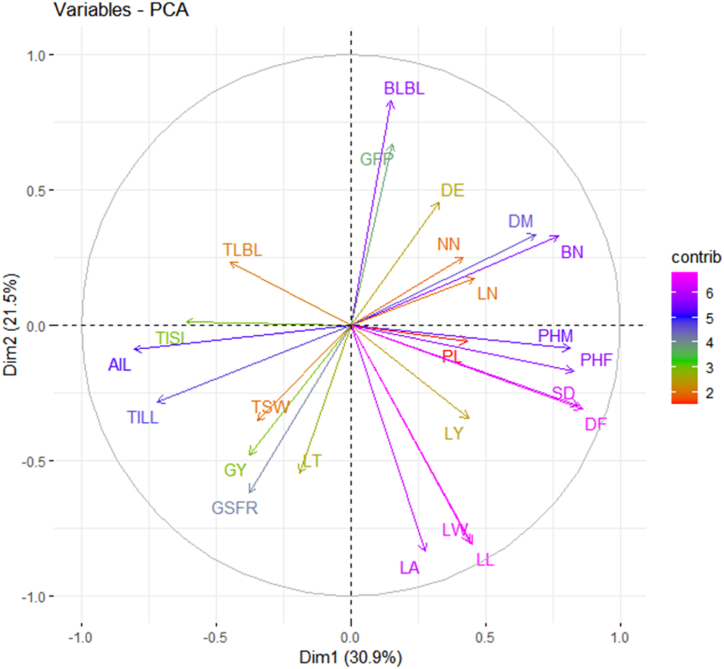
Variable PCA shows that the first two dimensions accounted for 52.42% of the overall variance across amaranth genotypes. Variables that are close to the center of the plot are less important for the first two components. Accordingly, PHM, GSFR, BLBL, LL, DM, DF, LA, AIL, BN, TILL, DE, TLBL, LY, SD, TISL, GSFR, GFP, GY, LW, PHF, and LT were the major variables contributing to the variance of the first two components. NN, LN, PL, and TSW contribute less to principal components 1 and 2. The degree of correlation between the two measured variables is indicated by the angle between the two vectors (Fig. 4). The strongest correlation was found between LL and LW, followed by SD and DF, all of which had a +1 correlation. There was a highly significant correlation between, GY, GSFR, and TSW; LA, LW, LY, and LT; and DM and BN; DF and DM. There was also a positive correlation between LN, NN, DE, and BN. Positive correlations were found between AIL, TISL, GSFR, GY, and TSW. AIL, TISL, GSFR, and GY, as well as TSW, had a negative connection with PHM and PHF. BLBL and GFP had no or very little association with SD; DF, PHM, PL, and TISL. Furthermore, LT is unrelated to SD, DF, PHF, PL, and TISL. There was no correlation between TSW, GY, and GSFR with DE, TISLL with DE, and the BN, as indicated by the angle of around 180° (r = cos180 = −1).
Fig. 4.
Variable correlation plot showing the relationships between 24 quantitative traits in 120 Amaranths genotypes were collected from different regions of Ethiopia. For abbreviated names (codes) of different traits (see Table 2)
3.4. Cluster analysis
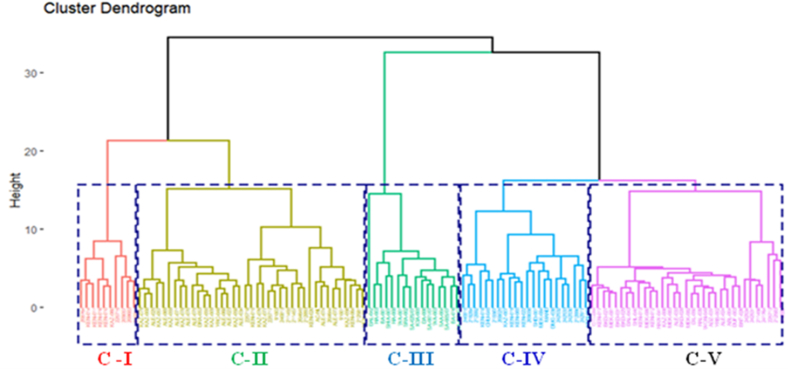
Based on standardized data from two seasons of mean values of 24 quantitative traits, the cluster analysis revealed 120 amaranth genotypes that were typically grouped into five basic clusters based on the level of variation among the genotypes (Fig. 5, Fig. 6). The average values of the morphological characters from each cluster are shown in Table 5.
Fig. 5.
Silhouette width to determine the number of clusters (K) for quantitative data.
Fig. 6.
Cluster analysis showing the relationship among 120 Amaranths genotypes based on 24 quantitative traits; the abbreviated codes on the dendrogram nodes are described in Table 2.
Table 5.
Mean performance of different quantitative traits in each cluster.
| Traits | Clusters |
Grand Mean | ||||
|---|---|---|---|---|---|---|
| I | II | III | IV | V | ||
| Days to emergence (days) | 6.28 | 5.92 | 7.48 | 7.01 | 6.98 | 6.73 |
| Days to flowering (days) | 33.88 | 42.79 | 36.95 | 50.70 | 55.67 | 44.00 |
| Plant height at flowering (cm) | 57.20 | 60.27 | 61.15 | 82.19 | 95.01 | 71.16 |
| Leaf number (number) | 21.59 | 30.26 | 30.41 | 30.1 | 30.86 | 28.64 |
| Leaf area (cm2) | 64.53 | 79.58 | 22.04 | 76.62 | 78.86 | 64.33 |
| Leaf length (cm) | 14.09 | 14.48 | 9.04 | 15.73 | 16.06 | 13.88 |
| Leaf width (cm) | 6.80 | 7.19 | 4.68 | 7.71 | 7.81 | 6.84 |
| Leaf thickness (mm) | 0.40 | 0.39 | 0.35 | 0.39 | 0.37 | 0.38 |
| Petiole length (cm) | 5.75 | 6.05 | 6.44 | 6.29 | 6.56 | 6.22 |
| Leaf yield (t/ha) | 8.89 | 10.04 | 8.71 | 11.69 | 10.92 | 10.05 |
| Branch number (number) | 18.16 | 22.57 | 34.18 | 33.04 | 34.53 | 28.50 |
| Basal lateral branch length (cm) | 39.55 | 22.41 | 134.65 | 45.71 | 54.62 | 59.39 |
| Top lateral branch length (cm) | 33.33 | 18.11 | 25.56 | 20.56 | 18.93 | 23.30 |
| Node number (number) | 20.02 | 27.05 | 27.91 | 26.48 | 27.16 | 25.72 |
| Days to maturity (days) | 82.88 | 85.23 | 101.41 | 102.02 | 106.8 | 95.67 |
| Plant height at maturity (cm) | 166.27 | 169.86 | 188.00 | 232.13 | 263.89 | 204.03 |
| Stem diameter (cm) | 22.79 | 25.29 | 22.92 | 30.29 | 34.92 | 27.24 |
| Auxiliary inflorescence length (cm) | 22.50 | 13.58 | 12.14 | 13.04 | 6.32 | 13.52 |
| Terminal inflorescence lateral length (cm) | 25.58 | 18.78 | 15.82 | 19.11 | 14.76 | 18.81 |
| Terminal inflorescence stalk length (cm) | 38.97 | 29.00 | 30.96 | 34.34 | 21.31 | 30.92 |
| Grain-filling periods (days) | 49.00 | 42.44 | 64.45 | 51.32 | 51.12 | 51.67 |
| Grain sink filling rate (kg ha−1day −1) | 48.45 | 34.59 | 14.08 | 31.44 | 26.15 | 30.94 |
| Thousand seed weight (g) | 0.96 | 1.02 | 0.68 | 0.91 | 0.69 | 0.86 |
| Grain yield (t ha−1) | 2.28 | 1.42 | 0.89 | 1.50 | 1.24 | 1.47 |
The 120 amaranths genotypes were divided into five groups based on quantitative traits by using the hierarchical agglomerative ward D2 dendrogram of the Euclidean distance clustering approach (Fig. 6).
Table 4 shows the number and names of genotypes found in each cluster, as well as their collecting areas. Clusters (II) included 39 genotypes (32.5%) out of a total of 120, followed by V, IV, III, and I which had 33 (27.5%), 22 (18.33%), 16 (13.33%), and 10 (8.3%) genotypes, respectively.
Table 4.
Clustering of 120 Amaranths species. of genotypes into 5 clusters using the mean of 24 agro-morphological quantitative characters.
| Cluster | Number of genotypes | Genotypes comprised | Collection Region |
| I | 10 | 208683, 240814, 209057 225715, 208764, KEN-014, KEN-010, KAZ-060, KEN-011, KEN-016 | South Nation Nationality People's Regional State and Oromia. |
| II | 39 | ALE-074, KAZ-078, KAZ-057, KEN-020, KAZ-076,204645,ALE-068, KAZ-059, KAZ-055, 240813, KAZ-008, KAZ-009, ALE-023, 225718, 211457, ALE-073, KAZ-077, KEN-015, 212581, 91001, KAZ-058, KEN-064, KEN-021, 225717, AC-NL, KAZ-056, 211455, KAZ-079, ALE-034, 204644, ALE-025, DRA-053, 91003, 212893, ALE-071, ALE-070, YRC-048, ALE-075, ALE-069 | South Nation Nationality, Amhara, Released Varieties, and Oromia. |
| III | 16 | SAA-004, HA-003, DA-006, BA-016, SRA-002, K3A-005, SA-025/07, SA-008/07, SA-001/07, SHIA-007, SA-005/07, DAL-042, HA-008, BA-001, WA-001, WA-003 | Afar and Sidama |
| IV | 22 | 212890, CHU-045, 215567, 209056, 242533, KEN-065, 219284, 242917, 242530, 242532, SHD-001, 212892, 208025, MEK-084, KEN-022, MAND, KEN-067, 240815, KAZ-007, KEN-019, DMG-038, KEN-018 | South Nation NationalityPeople RegionalState, Sidama, Benishangul & Gumuz, Amhara, Tigray, and Released varieties. |
| V | 33 | MEK-081, BKD-031, OTI-044, KEN-012, WON-049,242534, 211456, 241764, MEK-082, 242531, ALE-033, DIL-050, KAZ-006, BKD-027, BKD-029, ALE-024, DAL-043, HAW-041, BKD-028, MEK-083, 212583, KEN-017, HAL-039, MEK-080, SHL-040, ABL-004, KEN-013, DUF-003, 225713, 212582, ADZ-037, YRB-002,BNT-026 | Southern Nations Nationalities and Peoples, Released varieties, Tigray, Amhara, Oromia, Sidama, Benishangul & Gumuz. |
3.5. Performance of genotypes under different clusters
The cluster mean values for 24 traits are presented in Table 5. Surprisingly, Cluster II had the highest mean values genotypes for LA and TSW. This group of genotypes has the smallest mean value for BLBL, DE, GFP, and TLBL were primarily collected from different regions of Ethiopia, including South Nation Nationality People Regional State, (n = 36, 92.31%), Amhara (n = 1, 2.56%), Released varieties (n = 1, 2.56%), and Oromia (n = 1, 2.56%). Clusters (V) had 33 genotypes and accounting for 27.50% of total genotypes, are dominated by genotypes from South Nation Nationality People Regional State (n = 19, 57.58%), Sidama (n = 5, 15.15%), Tigray (n = 4, 12.12%), Benshangul-Gumz (n = 2, 6.06%), Amhara (n = 2, 6.06%), and Oromia (n = 1, 3.03%). In comparison to the other four clusters, genotypes in this cluster had the greatest mean values for DF to DM, LL, LN, LW, BN, PHF, PHM, PL, and SD. Their mean values for AIL, TILL, and TISL were all the lowest. Cluster IV encompassed 22 genotypes, contributing to 18.33% of all genotypes (n = 14, 63.64%) from South Nation Nationality People Regional State and (n = 3, 13.64%) from Benshangul-Gum, Sidama (n = 2, 9.09%), Amhara (n = 1, 4.55%), released varieties (n = 14.55%), Tigray region (n = 1, 4.55%). In comparison to the other four clusters, genotypes in this cluster had the greatest mean values for LY. AIL, TLBL, TILL, GY, GFP, and TISL all had intermediate mean values (Table 5). Interestingly, cluster III is dominated by Afar genotypes accounting for 13.33% of total genotypes (n = 15, or 93.75%) and other genotypes from Sidama (n = 1, or 6.25%). This group of genotypes had the greatest mean values for the BLBL, DE, GFP, and NN. GSFP, GY, TSW, LA, LL, LW, LT, and LY had the lowest mean values. Cluster I comprises 10 genotypes, accounting for 8.33% of total genotypes, collected from South Nation Nationality People Regional State (n = 8, 80.00%), and Oromia (n = 2, 20%). Genotypes in this cluster were characterized by the highest mean value for LT, TLBL, TISL, TILL, AIL, and GSFR. As a result, genotypes from clusters I and IV had the greatest mean values of grain yield-related characteristics and leaf yield-related features, which is desirable for breeding (Table 5).
3.6. Intra and inter-cluster distances
The intra and inter-cluster distances of all clusters have been presented in (Table 6). The intra-cluster D2 values ranged from 2.23 (cluster II) to 4.95 (cluster I). The intra-cluster distance was observed highest in cluster I (4.95) followed by cluster III (4.01), cluster IV (3.38), and cluster V (2.63), and was recorded as the lowest in cluster II (2.23). However, the inter-cluster D2 values varied between clusters II and III (277.79). The pairwise generalized squared distance (D2) among the five clusters based on Mahalanobis's D2 statistics revealed the maximum and highly significant genetic distance was observed between II and III (277.79), followed by clusters I and III (275.14), cluster III and V (226.87), clusters III and IV (221.48), cluster I to V (86.07), cluster II and V (52.40), cluster II to IV (49.95), cluster II to IV (27.69), cluster IV and V (21.49), and the minimum inter-cluster distance observed between cluster I and II (39.50). As previously stated, the fundamental reason for variety in a cluster's composition is large inter-cluster distance, which may be leveraged in a hybridization program. The most divergent clusters were observed between clusters II and III. On average, Cluster III is the most distant group among the five clusters.
Table 6.
Pairwise generalized square distance (D2) between (below diagonal) intercluster and (within diagonal or Boldface) intracluster distance based on the standardized mean values of 120 Amaranths species genotypes.
| From CLS | I | II | III | IV | V | Mean |
|---|---|---|---|---|---|---|
| I | 4.95 | 112.67 | ||||
| II | 39.50** | 2.23 | 99.35 | |||
| III | 275.14** | 277.79** | 4.01 | 158.26 | ||
| IV | 49.95** | 27.69ns | 221.48** | 3.38 | 131.50 | |
| V | 86.07** | 52.40** | 226.87** | 21.49ns | 2.63 | 97.46 |
* = significant at P < 0.05, and ** = highly significant at P < 0.01; NS = non-significant at P > 0.05. Chi-Square (X2) = 24, DF = 36.415 at P < 0.05 and 42.980 at P < 0.01.
Fig. 7 shows the biplot loadings for the first two PCs of the 24 agro-morphological parameters in the amaranth genotypes. The findings showed that depending on the length, direction, and proximity of the vector lines, the analyzed traits could be divided into five groups. In the current experiment, principal component biplot loadings analysis and cluster analysis based on Euclidian distances both revealed comparable patterns that appear to support one another. In general, the PCA biplot pattern in the first two PCs reflected the findings of the cluster analysis. Fig. 7 shows clearly that the features representing the DF, DM, PHF, BN, and SD had a significant impact on the genotypes of cluster I, which correspond to quadrants 1 and 4, as compared to this, eight traits LA, LL, LW, LN, PL, NN, LY, and DE that correspond to quadrants 1 and 2 significantly influenced cluster II genotypes. One of two characters, the BLBL of cluster III, which belongs to the fourth quadrant, was influenced by the genotypes. The AIL, TILL, and TISL of the investigated features had a significant impact on the genotypes of cluster IV, which belongs to the second quadrant. GSFR, GY, and TLBL three of the five traits, all had a significant impact on the genotypes of cluster V, which correspond to the second and third quadrants (Fig. 7).
Fig. 7.
Genotype by trait PCA biplots of the first two principal components (PC1 and PC2) combining the score plots of the 120 amaranth genotypes showing their grouping pattern and the corresponding loading plots based on 24 quantitative traits. For abbreviated names (codes) of different traits (see Table 2) above.
3.7. The relative contribution of different characters to genetic divergence
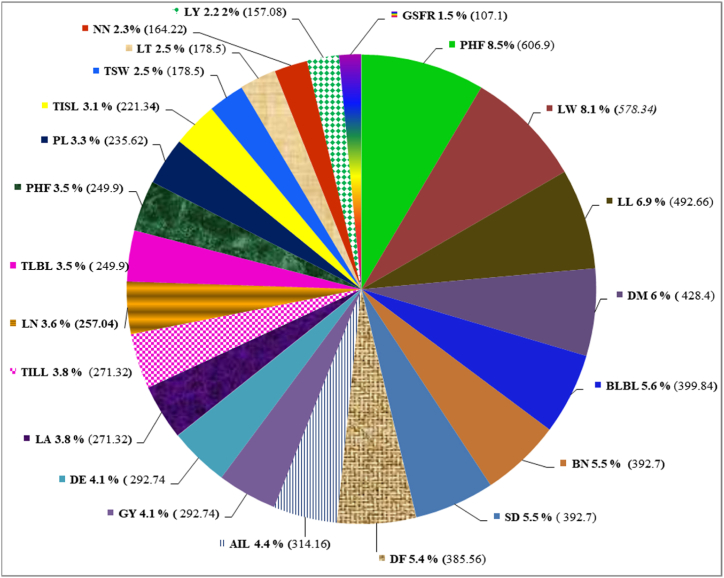
By estimating the relative contribution of distinct traits to genetic divergence, the D2 analysis's effectiveness is increased. Fig. 8 shows the results of the genetic contribution to diversity. The top five characteristics among all the characters were plant height at maturity (8.5%), leaf width (8.10), leaf length (6.90%), days to maturity (6.0%), and basal lateral branch length, which together accounted for the majority of the overall divergence (5.60%. These were the main contributors to diversity. While the amount of divergence caused by grain sink filling rate (1.5%), leaf yield (2.20%), node number (2.3%), leaf thickness (2.50%), thousand seed weight (2.50%), and other factors are insignificant.
Fig. 8.
Percent contribution and sometimes ranked first of different characters towards diversity in amaranth genotypes. For abbreviated names (codes) of different traits (see Table 2) above. Note number under the parenthesis indicates the number of times ranked first.
4. Discussion
4.1. Principal component analysis
Without a constant supply of biologically diverse plants, modern plant breeding would not be conceivable, much alone successful [48]. The availability of genetic resources and the evaluation of their diversity are both markers of breeding program success for any crop, including amaranth [49,50]. The initial stage in plant breeding is to determine genetic diversity in agro morphological variability and characterization in plant breeding. Diversities in morphology result from differences in the crop's genetic makeup [51,52] as well as the likely effect of planting places and times. Plant breeders may need to gather germplasm from underutilized crops to generate improved cultivars that will feed fast-growing populations [53]. Through trait-assisted selection, trait variation aids in the selection of the best lines for improvement [[54], [55], [56]].
Principal component analysis (PCA) is the re-validation technique of cluster analysis [57]. PCA was also helpful in detecting the correlations between traits, as well as recognizing the independent principle components that are effective on plant attributes and assessing diversity [58]. Moreover, PCA is a well-known dimension reduction approach for condensing a big collection of variables into a small set that retains the majority of the information from the large set [59]. It reflects the importance of the largest contributor to the total variation at each axis of differentiation [60]. According to Chahal et al. [61], characters with greater absolute values close to unity in the first principal component influence cluster more than those with lower absolute values close to zero. As a result, rather than a little contribution from each character, the current study found that the diversification of genotypes into separate clusters was due to the rather substantial contribution of a few traits. Therefore, characteristics with higher coefficients, typically 0.6 or more, on the PC axis are generally considered more important [62]. The positive and negative loading indicates the presence of positive and negative correlation trends between the components and the variables. Therefore, the above-mentioned traits that load high either positively or negatively contributed more to the diversity, and they were the ones that most distinguished the clusters.
The results of multivariate analysis for 24 attributes revealed that the first six PCs accounted for the majority of the variability. All 24 quantitative traits were determined as essential agro morphological makeup, contributing to total trait variability and hence being appropriate traits for amaranth selection and improvement, according to the PCA. In this study, the first component 30.94% (PC1) accounted for a greater proportion of the variation than the second component 21.49% (PC2). Similar results were reported by Showemimo et al. [63] of total variations at 39.87% (PC1) and 18.64% (PC2) in Amaranths species. Several studies also support these findings [26, 64]. In addition, PC1 is the most powerful criterion for selection for yield improvement [64]. Days to flowering, basal stem diameter, plant height at maturity, plant height at flowering, auxiliary inflorescence length, number of branches, leaf length, leaf width, terminal inflorescence lateral length, leaf yield, grain yield, grain sink filling rate, and thousand seed weight were all investigated in this study have a greater impact on PC1. A similar result was identified by Refs. [65,66]. The substantial relationship between PC1 and grain yield contributing parameters, as well as PC2 and leaf yield contributing traits, suggests a reasonable selection direction. Several researchers [63,67] supported our findings. Plant height, days to maturity, days to flowering, and length of inflorescence were also recognized as key direct drivers of grain output investigations, This is in agreement with the findings of Kandel et al. [68] in amaranths.
Another widely used method for displaying graphical groupings of genotypes for examining interrelationships between agro-morphological features in various crops is the biplot [26, 69]. According to Naidoo et al. [69], traits that were exhibited towards the center of the biplot had a smaller impact on the overall variation than those that were placed farther out. In the biplot analysis, the length of the vector lines and their proximity to the axis are often related to the size of the vector loadings. Furthermore, the loading variables in the PCA biplot indicated how strongly each trait affects a PC and how they are connected. For traits with longer and closer vector lines to the X-axis in the first two PCs, as opposed to traits with shorter and longer vector lines with a narrow and wide distance of the vector trait from the axis, respectively, For instance, traits of PHM, PHF, SD, and DF had longer vector lines which are closer to the X-axis as compared to BLBL which also had longer vector lines but the line was far from the X-axis. This indicates the contribution of PHM, PHF, SD, and DF for the total variation was higher in the first PC as compared to BLBL, and the contribution of these traits to the total variation in the first PC is noticeably higher. However, BLBL had a longer vector line which is very close to the Y-axis indicating that the contribution of this trait for the second PC was higher as compared to other traits.
In general, the cosine of the angle between a vector and an axis indicates the significance of the contribution of the corresponding trait to the PC. As a result, the PHM in the first PC and BLBL in the second PC had a notable contribution to the total variation due to the small cosine of the angle between the traits and the respective PC. The narrower the angle produced by two vectors, the better the relationships. The origin of a PCA biplot shows the average value of all characteristics [62], whereas the measure of an angle between the vectors of two traits indicates their correlation relative to their variance among genotypes. An angle of zero signifies a complete or perfect positive correlation (+1), an angle of = 90 indicates no association, inferring independence or the absence of a relationship between the two traits, an angle >90 indicates a negative correlation and an angle of 180 indicates a lack of correlation or perfect negative correlation (−1) [70,71]. Among various quantitative traits, significant and positive relationships were observed. As a result, a positive correlation between the traits suggested that improving one trait would also improve the other required trait. As a result, the biplot loadings revealed that there was a significant positive association between leaf area, leaf breadth, leaf length, and leaf yield. Since leaf area is a function of leaf breadth and length, this was predicted. Across amaranth genotypes, Gerrano et al. [72], Hailu et al. [73], Akin-Idowu et al. [65], and Mwakha et al. [74] found comparable results.
In addition, TSW and GY, and BN show a strong association. Furthermore, there is a significant link between BN and DM. This is consistent with Yoshihira et al. [75] on soybean. Furthermore, it was discovered that economically important grain yield and grain yield parameters including TISL, TILL, AIL, GSFR, and TSW are positively correlated. This demonstrated that improving inflorescence-related features leads to an increase in the number of grains and grain yield contributing to production. A similar result was observed in wheat by Sokoto et al. [76]; Hailu et al. [73]; Guo et al. [77], and Espitia-Rangel [78] in amaranths. The DM is comprised of the vegetative stage and the GFP. As a result, they should be associated with it. DM was correlated with the vegetative phase and GFP in this study. The rate of grain filling and the length of the filling period should both influence cereal production [79]. In the present study, grain yield and yield-related features were shown to be negatively correlated with grain filling periods but positively correlated with grain filling rates. GY did not correlate with DF, DM, or GFP, as found in, [80]. The lack of a correlation between grain yield and vegetative and grain filling periods, in particular, suggests that in our situation, these different growth duration traits would not be yield-limiting factors in Amaranthus genotypes and that breeding efforts should be directed toward other grain yield-related characters. In addition, early genotypes are more likely than delayed genotypes to have shorter vegetative and grain-filling periods. Days to maturity are comprised of vegetative and grain-filling stages, thus they should be interconnected, as shown in our research. Others have identified consistency in the correlations between grain filling periods, days to maturity, and grain yield. Others, on the other hand, have observed disparities in the correlations between grain filling time, days to maturity, and grain yield.
DF, DM, and BN per plant all showed a negative relationship with grain yield-related traits such as TISL, TILL, AIL, GY, and TSW per plant. This is in agreement with the findings of Hailu et al. [73]; Kigel et al. [81] in amaranths., meaning that genotypes with late flowering, matured, and a limited number of branches per plant had poor seed yield and vice versa. The grain yield contributing characters were negatively correlated to the leaf yield parameters, indicating that genotypes with high leaf yield also had lower grain production. The result was in line with [82]. Similarly, characteristics associated with vegetable yield contributing characters, such as LN, BN, PH, and LA, are also positively correlated. A similar observation was recorded by Sagar et al. [83] and Dinssa et al. [82]. This signifies that the amount of vegetables produced has increased. Tejaswini et al. [84], found a positive correlation between LW, LL, and PH, as well as a negative relationship between vegetable yield traits and the number of branches. In the current study also, DM, DF, and PH were positively correlated. A comparable finding was reported by Yadav et al. [85], and Nyasulu et al. [21]. Because the leaf affects the interception of solar radiation, resource allocation, water uptake method, and water stress tolerance, the leaf area is an important characteristic in determining the plant's production [86]. By enlarging the leaf area, the plant's ability to capture solar radiation can be improved. Valencia et al. [87] reported that a reduced leaf area encourages flowering to begin early, resulting in a positive relationship between leaf area and days to flowering. The current investigation found a similar positive association between leaf area and flowering time. Flowering occurs later in the season, indicating that more resources are spent to plant development, resulting in more photosynthesis available for seed formation. As a result, a plant with delayed blooming and a large leaf will collect more carbon, resulting in a larger seed and a higher yield when harvested [88].
4.2. Clustering of genotypes
Cluster analysis is another method for determining the pattern and degree of correlations between or among genotypes. The 120 amaranth genotypes were grouped into five clusters based on the mean performance of quantitative agro-morphological traits. One of the most effective ways to make use of genetic diversity for yield enhancement is to cluster a large number of genotypes according to how well they perform on average. Genotypes obtained from the same region of the country more or less clustered together in some cases, as was observed in Cluster III with genotypes SAA-004, HA-003, DA-006, BA-016, SRA-002, K3A-005, SA-025/07, SA-008/07, SA 001/07, SHIA-007, SA-005/07, HA-008, BA-001, WA-001, and WA-003, which are from the Afar region. This might indicate that some traits are more restricted to specific geographical locations and that genotypes are more suited to the region's agro ecological circumstances than others. Akin-Idowu et al. [65], for example, made comparable observations. The clustering pattern of distinct genotypes, on the other hand, did not reflect their geographical distribution and was somewhat fairly random. In this study, the genotypes from South Nation Nationality People's Regional State, Amhara, Sidama, Tigray, Benishangul, and Gumuz region, and released varieties of Ethiopia, for example, showed some regional consistency. Clusters IV and V were created to group them. Shanmugam et al. [89], reported that the falling of materials of the same origin into separate clusters was an indication of the broad genetic base of the genotypes belonging to that origin. The current results were consistent with the findings of Akin-Idowu et al. [65]. This shows that genotypes from the same origin seen in distinct clusters are indicative of the origin's wide genetic foundation. This is in agreement with the findings of Kemal et al. [90] in Safflower; Gebeyehu et al. [91] in Guizotia abyssinica.
4.3. Performance of genotypes under different clusters
The highest grain yield-producing clusters were typically found in the first clusters, and the highest leaf yield-producing clusters were typically found in the fourth cluster. LT, TLBL, TISL, TILL, AIL, and GSFR all exhibited high values in the first cluster. In terms of LY, PHF, PHM, LL, LW, SD, TISL, TISL, and GY, genotypes classified in the fourth cluster performed better on average. The best-performing genotypes in terms of the aforementioned traits were likewise found in this cluster, and the mean values of these traits were also higher than the overall grand mean of the corresponding trait aggregated across all clusters. The GY produced by the genotypes in the fourth cluster was high even though grain yield-related parameters, particularly GSFR, TISL, LT, and TILL, were high in this cluster. This could be a result of the smaller seed size (TSW) that the genotypes in this cluster generate, which may have a major impact on the GY per unit area. In terms of DF, LY, LL, LW, LA, PHM, PHF, LT, BN, DM, GFP, SD, TISL, TILL, GSFP, and GY, the genotypes classified in the fourth cluster had higher mean performances, top-performing genotypes, and higher mean values than the grand mean. However, the genotypes included in the fourth cluster had the intermediate mean values of PL, LN, TLBL, BLBL, AIL, and TSW. This result indicated that high GY-producing genotypes had shorter plant height at flowering and plant height at maturity performance. Different scholars reported that modern cultivars (high grain yielders) had short plant heights as compared to landraces which produce low grain yield [92,93].
Moreover, high cluster means for yield and its components should be given significant consideration when choosing genotypes for inclusion in the hybridization program based on genetic diversity [94]. Furthermore, the germplasm from these clusters might be utilized to generate high-yielding varieties or hybridization programs to create materials with desirable traits [95]. Hybridization of genotypes from distinct clusters with high cluster means for quality attributes will generate Amaranthus genotypes that outperform their parents. Furthermore, the greatest inter-cluster variability demonstrated by the genotypes categorized into the various clusters would provide immense potential for Amaranthus genotypes. improvement through hybridization programs involving genotypes from various clusters. A multivariate investigation of bread wheat found similar results [96]. These results suggest that the genotypes grouped in different clusters may be used as potential parental lines for hybridization programs to develop desirable genotypes, as genetic diversity can be better exploited and the chances of finding better segregants increased. In the future, amaranth tree clusters will be used as parents for hybridization or other population enhancement procedures. The cluster that contributed the most to the divergence was given priority when choosing the kind of cluster for selection and the parents for hybridization.
4.4. Genetic divergence
The standard Mahalanobis (D2) statistic is a powerful tool for quantifying the genotypic level variation and may be an effective tool in estimating the genetic diversity in genotypes. Due to their similar performance for the examined variables, the genotypes grouped within a cluster showed restricted genetic variation, as evidenced by the observed intra-cluster genetic distance being smaller than the inter-cluster genetic distance. This finding is also consistent with those of Ahammed et al. [97], and Kumar [98], who found that in amaranth genotypes, the intra-cluster genetic distance was typically smaller than the intercluster genetic distance. Although collection regions were far apart, C II and C III, clusters I and III, had the largest distances between groups, but C I and cluster II had the smallest distance between clusters, which should be expected in terms of spatial distribution. In comparison to genotypes in other clusters, the genotypes in these clusters were quite near to each other.
A broad Mahalanobis distance (D) between the genotypes of two dissimilar clusters could maximize transgressive segregation [99]. Crossing genotypes from those clusters, according to Rahman et al. [100] may not result in a greater heterotic value in F1 and a restricted range of variability in the segregating F2 population. To preserve a somewhat diverse genetic foundation, such analysis was intended to prevent selecting parents from genetically homogenous groups. Spatial dispersion may enhance the accumulation of competing alleles between the two populations, enhancing genetic dissimilarity between them. Clusters II and III, followed by clusters I and III, were the most distant of the five clusters on average. Because crosses between genetically divergent lines will generate heterotic segregates or a broad spectrum of variability in subsequent generations, and thus could be exploited in a hybridization program, the results revealed that a large inter-cluster distance was the major driving force of variation in a cluster's makeup and may be used as parents for hybridization programs to develop desirable varieties [101,102]. This suggests that there are significant genetic differences across these clusters; by choosing genotypes according to the intended use of the crop, yield enhancements may be possible. When parents chosen from these genetically distant clusters are crossed, it is thought that the biggest genetic divergence between them will result in the greatest amount of genetic recombination and differences. However, based on the specific purposes of crossing or hybridizing, the choice of parents from these distant clusters should be made following the peculiar traits of each cluster and the performance of each genotype within each cluster [103]. Crosses involving parents from the clusters with the greatest distances are predicted to have the highest levels of genetic recombination and diversity in the following generation [104]. Based on both group performance and the performance of individual genotypes for specific traits, parents with one or more of the desired traits can be chosen from each cluster [105]. Kumar's findings are consistent with the present findings [98]. Accordingly, suitable parents for heterosis breeding can be genotypes belonging to clusters C I, C III, C IV, and C V.
4.5. The relative contribution of different characters to genetic divergence
The contribution of individual characters toward divergence was estimated by Sneha et al. [106]. Out of the 23 traits examined, PHM (8.5%), LW (8.10), LL (6.90%), DM (6.0%), and BLBL (5.6%) contributed the largest contributions to variety. These traits were responsible for expressing maximum variety between the clusters, and so should be given considerable weightage during selection. The remaining characters contributed little to divergence. A similar result was observed in Wheat (Triticum aestivum L.) genotypes [107].
The study of agro-morphological based genetic diversity across genotypes from various phytogeography areas is significant because a species' capacity to adjust to changing conditions is essentially subordinate to the amount of variety accessible in its gene pool. We looked examined the agro-morphology diversity in Amaranthus genotypes from a variety of perspectives. According to Sarker et al. [108], genetic variability in amaranths is impacted by both genetic and environmental variability. As a result, the use of molecular markers is essential to supplement morphological data and to increase the precise degree of genetic diversity. As a result, the current characterization information makes strides in the utilization of amaranth genotypes in breeding programs and guarantees viable genotype collection administration. As a result, further molecular inquiry about the subject is required.
5. Conclusion
The expression of significant genotypic heterogeneity as well as environmental factors may help to explain the current investigation of the phenotypic variability shown in 120 amaranth genotypes in Ethiopia. The first two principal components had a high contribution to the total variation in amaranth genotypes. Among the studied traits, PHM, BLBL, LL, DM, DF, LA, AIL, BN, TILL, DE, TLBL, SD, TISL, GY, GSFR, GFP, TSW, LW, PHF, LT, and LY all had a high contribution to the total variation in amaranth genotypes. Therefore, the selection of these traits could be effective for yield improvement in amaranth genotypes. Moreover, the occurrence of remarkable variation among amaranth genotypes for measured traits indicates the potential of amaranth genotypes in future breeding programs for improving LY, GY, and yield-related traits. Similar to how PCA emphasized the agro-morphological aspects contributing to the components, the biplot further splits the genotypes and associated attributes into two primary principal component axes. Cluster analysis grouped the genotypes into five clusters, based on the performances of measured traits. High-leaf yield-producing genotypes were found in the fourth cluster, but high-grain yield-producing genotypes were identified in the first cluster. The genetic divergence also showed that the clusters' variability existed. The second and third clusters had the greatest inter-cluster genetic distance, followed by the first and third clusters, and the first and second clusters had the smallest inter-cluster genetic distance. The morphological and agronomic distinctions revealed in this study might be used as pre-breeding tools to get Ethiopia's amaranth genotypes breeding program off to a good start. To detect alleged discrepancies, additional studies, such as the use of molecular marker research, may be required. Finally, phenotypic diversity data can be coupled with genetic markers to drive an efficient hybrid program.
Author contribution statement
Mekonnen Yeshitila: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Andargachew Gedebo; Bizuayehu Tesfaye; Hewan Demissie; Temesgen Magule Olango: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Data availability statement
Data will be made available on request.
Additional information
No additional information is available for this paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
I owe a great deal of gratitude and thanks to my friend Gezahagn Kebede from the Ethiopian Holetta Agricultural Research Center for his assistance during my studies. The Norwegian Embassy in Addis Ababa and the Dilla College of Education in Dilla, Ethiopia have generously provided financial support, which is sincerely acknowledged.
References
- 1.Costea M., et al. Stem morphology and anatomy in Amaranthus L.(Amaranthaceae), taxonomic significance. J. Torrey Bot. Soc. 2001:254–281. [Google Scholar]
- 2.Maughan P., et al. Development, characterization, and linkage mapping of single nucleotide polymorphisms in the grain amaranths (Amaranthus sp.) Plant Genome. 2011;4 [Google Scholar]
- 3.Andini R., et al. Amaranthus genetic resources in Indonesia: morphological and protein content assessment in comparison with worldwide amaranths. Genet. Resour. Crop Evol. 2013;60:2115–2128. [Google Scholar]
- 4.Rastogi A., et al. Amaranth: a new millennium crop of nutraceutical values. Crit. Rev. Food Sci. Nutr. 2013;53:109–125. doi: 10.1080/10408398.2010.517876. [DOI] [PubMed] [Google Scholar]
- 5.Santra D., et al. Book Amaranth Part 2—Sustainability, Processing, and Applications of Amaranth. Elsevier; 2017. Amaranth Part 2—sustainability, processing, and applications of amaranth; pp. 257–264. [Google Scholar]
- 6.Alemayehu F.R., et al. The potential for utilizing the seed crop amaranth (Amaranthus spp.) in East Africa as an alternative crop to support food security and climate change mitigation. J. Agron. Crop Sci. 2015;201:321–329. [Google Scholar]
- 7.Brink M., et al. PROTA Foundation Wageningen; The Netherlands: 2006. Plant Resources of Tropical Africa 1: Cereals and Pulses. [Google Scholar]
- 8.Sokolova D., et al. Comparative characteristics of the amino acid composition in amaranth accessions from the VIR Collection. Turk. J. Agric. For. 2021;45:68–78. [Google Scholar]
- 9.Demissew S. The Ethiopian Flora project. Lessons learned, in Proceedings of the Fourth Global Botanic Gardens Congress. 2010:13–18. Dublin. [Google Scholar]
- 10.Shodiev D., et al. Useful properties of the amaranth plant. ResearchJet Journal of Analysis and Inventions. 2021;2:55–58. [Google Scholar]
- 11.Ishimoto E.Y., et al. Quinoa (Chenopodium quinoa willd quinoa (Chenopodium quinoa willd) as functional food. Revista de Atenção à Saúde. 2010;8 [Google Scholar]
- 12.Almeida S.G., et al. Amaranto (Amaranthus ssp) e quinoa (Chenopodium quinoa) alimentos alternativos para doentes celíacos. Ensaios Ciência: Ciências Biológicas, Agrárias e da Saúde. 2009;13:77–92. [Google Scholar]
- 13.Gamel T.H., et al. Nutritional study of raw and popped seed proteins of Amaranthus caudatus L and Amaranthuscruentus L. J. Sci. Food Agric. 2004;84:1153–1158. [Google Scholar]
- 14.Lee J.-R., et al. Characterization of microsatellite loci developed for Amaranthus hypochondriacus and their cross-amplifications in wild species. Conserv. Genet. 2008;9:243–246. [Google Scholar]
- 15.Sammour R.H., et al. Electrophoretic variations in Amaranthus, bot. Bull. Acad. Sin. 1993;34:37–42. [Google Scholar]
- 16.Juan R., et al. Electrophoretic characterization of Amaranthus L. seed proteins and its systematic implications. Bot. J. Linn. Soc. 2007;155:57–63. [Google Scholar]
- 17.Shah L.R., et al. Morphological characterization of Amaranthus spp. under temperate environment using NBPGR descriptor. J. Pharmacogn. Phytochem. 2018;7:2716–2718. [Google Scholar]
- 18.Jonah P., et al. Variation in pod yield characters and heritability estimates in some cultivars of Bambara groundnut (Vigna subterranean (L.) Verdc. Acad. J. Plant Sci. 2012;5:50–55. [Google Scholar]
- 19.Chandran K., et al. Morphological characterization of Arachis species of section Arachis. Plant Genet. Resour. Newsl. 2000;121 [Google Scholar]
- 20.Ahammed A., et al. Multivariate analysis in stem amaranth (Amaranthus tricolor) Bangladesh J. Plant Breed Genet. 2013;26:11–17. [Google Scholar]
- 21.Nyasulu M., et al. Agromophological characterisation of amaranth accessions from Malawi. Am. J. Plant Sci. 2021;12:1528–1542. [Google Scholar]
- 22.Dhakal A., et al. Multivariate analysis of phenotypic diversity of rice (Oryza sativa L.) landraces from Lamjung and Tanahun Districts, Nepal. Int. J. Agron. 2020;2020 doi: 10.1155/2020/1589150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Harty E.H., et al. Morphological and molecular characterization of quinoa genotypes. Agriculture. 2021;11:286. [Google Scholar]
- 24.Molin W.T., et al. Morphological characterization of Amaranthus palmeri x A. spinosus hybrids. Am. J. Plant Sci. 2017;8:1499–1510. [Google Scholar]
- 25.Srivastava R., et al. Assessment of morphological diversity of selected Amaranthus species. J. Glob. Biosci. 2015;4:3044–3048. [Google Scholar]
- 26.Oyetunde O.A., et al. Genetic diversity and trait profiles of some Amaranthus genotypes. Adv. Hortic. Sci. 2021;35:277. [Google Scholar]
- 27.Masood M.A., et al. Improvement in the precision of agricultural field experiments through design and analysis. Pak. J. Life Soci. Sci. 2008;6:89–91. [Google Scholar]
- 28.Ghareeb Z., et al. Evaluating the precision of faba bean field experiments. Egyp. J. Agr. Sci. 2015;66:288–296. [Google Scholar]
- 29.Kashif M., et al. Efficiency of alpha lattice design in rice field trials in Pakistan. J. Sci. Res. 2011;3:91. 91. [Google Scholar]
- 30.G. Grubben, et al., Genetic Resources of Amaranth-A Global Plan of Action, IBPGR1981.
- 31.Shukla S., et al. Mineral profile and variability in vegetable amaranth (Amaranthus tricolor) Plant Foods Hum. Nutr. 2006;61:21–26. doi: 10.1007/s11130-006-0004-x. [DOI] [PubMed] [Google Scholar]
- 32.Grubben G., et al. Food and Agriculture Organisation; Rome: 1981. Genetic Resources of Amaranths, International Board for Plant Genetic Resources. [Google Scholar]
- 33.Biru A. 1978. Agronomy Research Manual. [Google Scholar]
- 34.de Jesus Souza F.F., et al. Physiological quality of quinoa seeds submitted to different storage conditions. Afr. J. Agric. Res. 2016;11:1299–1308. [Google Scholar]
- 35.Manly B. Chapman &, Hall Ltd; London: 1986. Multivariate Statistical Methods: a Primer. [Google Scholar]
- 36.Sneath P.H., et al. 1973. Numerical Taxonomy: the Principles and Practice of Numerical Classification. [Google Scholar]
- 37.Mohammadi S.A., et al. Analysis of genetic diversity in crop plants—salient statistical tools and considerations. Crop Sci. 2003;43:1235–1248. [Google Scholar]
- 38.Maechler M. vol. 2. R package version; 2019. (Finding Groups in Data'': Cluster Analysis Extended Rousseeuw et). [Google Scholar]
- 39.Charrad M., Boiteau V., Niknafs Azam, et al. NbClust:: an R package for determining the relevant number of clusters in a data set. J. Stat. Software. 2014;61:1–36. [Google Scholar]
- 40.Menzir A. Phenotypic variability, divergence analysis and heritability of characters in sesame (Sesamum indicum L.) genotypes. Nat. Sci. 2012;10:117–126. [Google Scholar]
- 41.Kaiser H.F. The application of electronic computers to factor analysis. Educ. Psychol. Meas. 1960;20:141–151. [Google Scholar]
- 42.MacCallum R.C., et al. Sample size in factor analysis. Psychol. Methods. 1999;4:84. [Google Scholar]
- 43.Stevens J.P. Routledge; 2012. Applied Multivariate Statistics for the Social Sciences. [Google Scholar]
- 44.Da Silva A.R., et al. Package ‘biotools’. 2017 https://CRAN.R-project.org/package=∼biotools Avaliable online at: accessed December 27, 2020. [Google Scholar]
- 45.Singh . 1981. The Relative Importance of Characters Affecting Genetic Divergence. [Google Scholar]
- 46.Mahalanobis P.C. 1936. On the Generalized Distance in Statistics. [Google Scholar]
- 47.Rao C.R. 1952. Advanced Statistical Methods in Biometric Research. [Google Scholar]
- 48.Duvick D.N. 2007. Breeding of Plants. [Google Scholar]
- 49.Hoisington D., et al. Plant genetic resources: what can they contribute toward increased crop productivity? Proc. Natl. Acad. Sci. USA. 1999;96:5937–5943. doi: 10.1073/pnas.96.11.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choudhary P., et al. Genetic study and selection indices for grain yield of mungbean. Leg. Res. Int. J. 2017;40:836–841. [Google Scholar]
- 51.Manimekalai M., et al. Genetic diversity in the barnyard millet (Echinochola frumentacea) germplasms revealed by morphological traits and simple sequence repeat markers. Curr. Plant Biol. 2018;14:71–78. [Google Scholar]
- 52.Soe I., et al. Genetic diversity analyses of rice germplasm using morphological traits. J. Plant Breed Crop Sci. 2019;11:128–136. [Google Scholar]
- 53.Nelson R.L. Managing self-pollinated germplasm collections to maximize utilization. Plant Genetic Res. 2011;9:123–133. [Google Scholar]
- 54.Moradi Y., et al. Morphological and pomological characterizations of cornelian cherry (Cornus mas L.) to select the superior accessions. Sci. Hortic. 2019;249:208–218. [Google Scholar]
- 55.Dewi, et al. Evaluation of SSR and important agronomical characters of promising mutant lines of soybean. Biodiversitas J. Biolog. Diver. 2020;21 [Google Scholar]
- 56.Fasahat P., et al. Principles and utilization of combining ability in plant breeding. Biometr. Biosta. Int. J. 2016;4:1–24. [Google Scholar]
- 57.Khan M.M.H., et al. Genetic analysis and selection of Bambara groundnut (Vigna subterranea [L.] Verdc.) landraces for high yield revealed by qualitative and quantitative traits. Sci. Rep. 2021;11:1–21. doi: 10.1038/s41598-021-87039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beheshtizadeh H., et al. Principal component analysis and determination of the selection criteria in bread wheat (Triticum aestivum L.) genotypes. Intl. J. Agric. Crop Sci. 2013;5:2024. [Google Scholar]
- 59.Sharma, et al. Multistream video fusion using local principal components analysis. Infrared Tech. App. 1998;XXIV:717–725. [Google Scholar]
- 60.Jolliffe I.T. 2002. Principal Components in Regression Analysis, Principal Component Analysis; pp. 167–198. [Google Scholar]
- 61.Chahal G., et al. Alpha Science Int'l Ltd.; 2002. Principles and Procedures of Plant Breeding: Biotechnological and Conventional Approaches. [Google Scholar]
- 62.DeLacy I., et al. Louisiana State University; Baton Rouge, LA, USA: 1990. Pattern Analysis for the Analysis of Regional Variety Trials, Genotype-By-Environment Interaction and Plant Breeding; pp. 301–334. [Google Scholar]
- 63.Showemimo F., et al. Traits selection criteria for genetic improvement of grain and leafy Amaranth (Amaranthus spp) using Principal Component Analysis. Egyp. J. Agr. Res. 2021;99:170–179. [Google Scholar]
- 64.Adéoti K., et al. Agromorphological characterization of Sesamum radiatum (Schum. and Thonn.), a neglected and underutilized species of traditional leafy vegetable of great importance in Benin. Afr. J. Agric. Res. 2012;7:3569–3578. [Google Scholar]
- 65.Akin-Idowu P.E., et al. Characterization of grain amaranth (Amaranthus spp.) germplasm in southwest Nigeria using morphological, nutritional, and random amplified polymorphic DNA (RAPD) analysis. Resources. 2016;5:6. [Google Scholar]
- 66.Shankar R., et al. Diversity analysis of fleshy leaf type Amaranthus for semi-arid ecosystems. Int. J. Plant Breed. 2012;6:27–33. [Google Scholar]
- 67.Subaşı I. Agro-morphological characterization and some seed characteristics of wild crambe (brassicaceae) species in Turkey. Sustainability. 2022;14:287. [Google Scholar]
- 68.Kandel M., et al. Evaluation and identification of stable and high yielding genotypes for varietal development in amaranthus (Amaranthus hypochondriacus L.) under hilly region of Nepal. J. Agr. Food Res. 2021;5 [Google Scholar]
- 69.Naidoo S.I., et al. Morpho-agronomical characterization of local and international sweet potato germplasm from the South African collection. S. Afr. J. Plant Soil. 2020;37:308–320. [Google Scholar]
- 70.Feltaous Y. Genetic diversity among some Egyptian bread wheat cultivars based on morphological characters and SSR markers. Assiut J. Agric. Sci. 2019;50:35–50. [Google Scholar]
- 71.Malik R., et al. Genetic diversity of agro-morphological characters in Indian wheat varieties using GT biplot. Aust. J. Crop. Sci. 2014;8:1266–1271. [Google Scholar]
- 72.Gerrano A., et al. Vol. 1035. 2013. Agro-morphological variability of Amaranthus genotypes in South Africa; pp. 183–187. (VI International Symposium on the Taxonomy of Cultivated Plants). [Google Scholar]
- 73.Hailu A.F., et al. Estimation of association characters in Amaranths germplasm accessions (Amaranthus spp.) under Mizan and Tepi conditions, South West Ethiopia. Int. J. Res. 2015;2:1–25. [Google Scholar]
- 74.Mwakha F.A., et al. Agro-morphological characterization of Kenyan Slender Leaf (Crotalaria brevidens and C. ochroleuca) accessions. Int. J. Agron. 2020;2020 [Google Scholar]
- 75.Yoshihira T., et al. Effects of maturity group and stem growth habit on the branching plasticity of soybean cultivars grown at various planting densities. Plant Prod. Sci. 2020;23:385–396. [Google Scholar]
- 76.Sokoto M., et al. Correlation analysis of some growth, yield, yield components and grain quality of wheat (Triticum aestivum L.) Niger. J. Basic Appl. Sci. 2012;20:349–356. [Google Scholar]
- 77.Guo Z., et al. Manipulation and prediction of spike morphology traits for the improvement of grain yield in wheat. Sci. Rep. 2018;8:1–10. doi: 10.1038/s41598-018-31977-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Espitia-Rangel E. Book Breeding of Grain Amaranth. CRC Press; 2018. Breeding of grain amaranth; pp. 23–38. [Google Scholar]
- 79.Daynard T., et al. Duration of the grain filling period and its relation to grain yield in corn, Zea mays L. 1. Crop Sci. 1971;11:45–48. [Google Scholar]
- 80.Nass H., et al. Grain filling period and grain yield relationships in spring wheat. Can. J. Plant Sci. 1975;55:673–678. [Google Scholar]
- 81.Kigel J., et al. Development and ecophysiology of amaranths. Amaranth: Biology Chem. Tech. 1994:39–73. [Google Scholar]
- 82.Dinssa F., et al. Effect of leaf harvest on grain yield and nutrient content of diverse amaranth entries. Sci. Hortic. 2018;236:146–157. [Google Scholar]
- 83.Sagar K., et al. Genotypic correlation coefficients among growth, yield and quality parameters in Amaranthus genotypes (Amaranthus tricolor L.) Int. J. Curr. Microb. App. Sci. 2018;7:2708–2714. [Google Scholar]
- 84.Tejaswini N., et al. Correlation and path coefficient analysis in vegetable amaranth (Amaranthus tricolor L.) genotypes. Intl. J. Curr. Microb. Appl. Sci. 2017;6:2977–2996. [Google Scholar]
- 85.Yadav R., et al. Analysis of variability parameters for morphological and agronomic traits in grain amaranth (amaranthus sp) genotypes. Bioscan. 2014;9:1661–1665. [Google Scholar]
- 86.Weraduwage S.M., et al. The relationship between leaf area growth and biomass accumulation in Arabidopsis thaliana. Front. Plant Sci. 2015;6:167. doi: 10.3389/fpls.2015.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Valencia E., et al. Plant size and leaf area influence phenological and reproductive responses to warming in semiarid Mediterranean species, Perspectives in plant ecology. Evolu. System. 2016;21:31–40. doi: 10.1016/j.ppees.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sharma, et al. Study of correlation, path coefficient and linkage of flower color and hairiness with yield controlling quantitative traits in segregating population of cluster bean. Curr. Plant Biol. 2021;26 [Google Scholar]
- 89.Shanmugam A., et al. Vigna radiata (L.) Wilczek); 1982. Genetic Diversity for Quantitative Characters in Green Gram. [Google Scholar]
- 90.Kemal A., et al. Genetic diversity of Safflower (Carthamus tinctorius L.) genotypes at Wollo, Ethiopia using agro-morphological traits. Trop. Plant Res. 2019;6:157–165. [Google Scholar]
- 91.Gebeyehu A., et al. vol. 11. Agronomy; 2021. p. 1479. (Characterization of Oilseed Crop Noug (Guizotia Abyssinica) Using Agro-Morphological Traits). [Google Scholar]
- 92.Reynolds M., et al. Raising yield potential in wheat. J. Exp. Bot. 2009;60:1899–1918. doi: 10.1093/jxb/erp016. [DOI] [PubMed] [Google Scholar]
- 93.Feng T., et al. vol. 11. Agronomy; 2021. p. 663. (Reduced Vegetative Growth Increases Grain Yield in Spring Wheat Genotypes in the Dryland Farming Region of North-West China). [Google Scholar]
- 94.Kumar, et al. Multivariate analysis and clustering of Cuphea procumbens inbred lines. Genetika. 2006;38:23–30. [Google Scholar]
- 95.Ajmal S.U., et al. Multivariate analysis of genetic divergence in wheat (Triticum aestivum) germplasm, Pak. J. Bot., Le. 2013;45:1643–1648. [Google Scholar]
- 96.Ahmad H.M., et al. Multivariate analysis of some metric traits in bread wheat (Triticum aestivum L.) Eur. J. Biotech. Biosci. 2014;1:22–26. [Google Scholar]
- 97.Ahammed A., et al. 2013. Characterization and Evaluation of Stem Amaranth Genotypes in the Summer Season. [Google Scholar]
- 98.Kumar Y. 2019. Genetic Divergence Analysis in amaranthus (Amaranthus spp.) Genotypes for Yield and its Component Characters. [Google Scholar]
- 99.Ayana A., et al. Multivariate analysis of morphological variation in sorghum (Sorghum bicolor (L.) Moench) germplasm from Ethiopia and Eritrea. Genet. Resour. Crop Evol. 1999;46:273–284. [Google Scholar]
- 100.Rahman M.M., et al. Genetic variability, correlation and path coefficient analysis of some physiological traits of transplanted Aman rice (Oryza sativa L.) Middle East J. Sci. Res. 2012;11:563–566. [Google Scholar]
- 101.Afroz S., et al. Multivariate analysis approach to select parents for hybridization aiming at yield improvement in cucumber (Cucumis sativus L.) J. Environ. Sci. Nat. Res. 2013;6:33–36. [Google Scholar]
- 102.Kabir M., et al. Triticum aestivum L.); 2013. Selection of Parents for Hybridization in Wheat. [Google Scholar]
- 103.Singh . Options Méditerranéennes-Série-Séminaries-no; 1990. Prospects of Developing New Genetic Material and Breeding Methodologies for Chickpea Improvement, Present Status and Future Prospects of Chickpea Crop Production and Improvement in the Mediterranean Countries; pp. 43–50. [Google Scholar]
- 104.Sawarkar A., et al. Genetic divergence within some genotypes of tossa jute (Corchorus olitorius L.) Environ. Ecol. 2015;33:749–752. [Google Scholar]
- 105.Singh B. Book Plant Breeding: Principles & Methods. Kalyani publishers; 2016. Plant breeding: principles & methods. [Google Scholar]
- 106.Sneha M., et al. An appraisal of genetic divergence in some indigenous collections of mungbean (Vigna radiata (L.) Wileczek) Electron. J. Plant Breed. 2020;11:620–625. [Google Scholar]
- 107.Mecha B., et al. Genetic diversity based on multivariate analysis for yield and its contributing characters in bread wheat (Triticum aestivum L.) genotypes. Agr. Res. Tech. 2017;8:1–10. [Google Scholar]
- 108.Sarker U., et al. Color attributes, betacyanin, and carotenoid profiles, bioactive components, and radical quenching capacity in selected Amaranthus gangeticus leafy vegetables. Sci. Rep. 2021;11:1–14. doi: 10.1038/s41598-021-91157-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.