Abstract
Fibroblast growth factors (FGFs) are paracrine or endocrine signaling proteins that, activated by their ligands, elicit a wide range of health and disease-related processes, such as cell proliferation and the epithelial-to-mesenchymal transition. The detailed molecular pathway dynamics that coordinate these responses have remained to be determined. To elucidate these, we stimulated MCF-7 breast cancer cells with either FGF2, FGF3, FGF4, FGF10, or FGF19. Following activation of the receptor, we quantified the kinase activity dynamics of 44 kinases using a targeted mass spectrometry assay. Our system-wide kinase activity data, supplemented with (phospho)proteomics data, reveal ligand-dependent distinct pathway dynamics, elucidate the involvement of not earlier reported kinases such as MARK, and revise some of the pathway effects on biological outcomes. In addition, logic-based dynamic modeling of the kinome dynamics further verifies the biological goodness-of-fit of the predicted models and reveals BRAF-driven activation upon FGF2 treatment and ARAF-driven activation upon FGF4 treatment.
Keywords: Fibroblast, Growth Factors, Kinome, Signaling, Phosphoproteomics, Breast Cancer, Modeling, Targeted MS, SRM, MAPK
Graphical Abstract
Highlights
-
•
Treatment with different FGFs activate distinct signaling pathways in cancer cells.
-
•
A targeted kinome activity assay enables the quantification of kinome dynamics.
-
•
Different FGF stimulations generate disparate kinome dynamics.
-
•
Logic-based dynamic modeling provides biological pathway validation.
-
•
FGF2 treatment induces BRAF activation and FGF4 results in ARAF activation.
In Brief
We provide an extensive overview of FGF-regulated kinome signaling in breast cancer cells, enabled via a custom-made targeted kinome activity assay. We apply logic-based dynamic modeling to verify literature-derived biological pathways. This in-depth comparison between FGF2, FGF3, FGF4, FGF10, and FGF19 signaling revealed differential response and involvement of kinases hitherto undescribed in the FGF context. Moreover, we expanded on the existing FGF-signaling knowledge, for example, by revealing differential activation of ARAF and BRAF for FGF4 and FGF2, respectively.
Fibroblast growth factors (FGFs) and cofactors heparin/heparin-sulfate or beta-klotho induce trans-autophosphorylation upon binding to FGF receptors (FGFRs), thereby activating signaling pathways and regulating diverse biological processes (1, 2, 3, 4, 5). There are 18 FGF ligands known so far that can activate the seven alternatively spliced isoforms of four FGFR genes. Specific combinations of receptor and ligand result in the regulation of a plethora of diverse cellular processes, including cell differentiation, cell proliferation, and epithelial-mesenchymal transition (EMT) (6, 7).
Besides the role of FGFs in health, during development and adult life, dysregulated FGF-FGFR signaling is implicated in various types of cancer, including breast cancer (8, 9, 10, 11). FGF2 is commonly detected in the tumor microenvironment of breast cancer and can induce tumor growth (12, 13). FGF3, FGF4, and FGF19 are located on the 11q13 amplicon, which is amplified in 15 to 20% of breast cancer patients and are all linked to increased tumor progression (14, 15, 16, 17). FGF10 can drive type III EMT in breast cancer, promoting invasiveness (18). These unfavorable effects in breast cancer patients result from diverse and complex FGF-driven cellular signaling (10).
The fine-tuned coordination of the diverse FGF-driven cellular processes is thought to be regulated by the MAPK/ERK pathway, the PI3K pathway, the PLCγ pathway, and the JAK-STAT pathway (19, 20, 21, 22, 23). For example, the MAPK/ERK pathway is thought to drive cell proliferation, and the PI3K pathway is believed to regulate EMT (24, 25). These pathways are highly dependent on multiple kinases that relay signals by adding phosphate groups to proteins or other molecules. Kinase activity is often determined by phosphorylation in the kinase activation loop, which can be measured and quantified using a targeted mass spectrometry–based kinome assay (26, 27). Even though the main pathways involved in FGF signaling are elucidated, molecular mechanistic insights into the regulations of the differential cellular processes are still largely lacking (28, 29).
FGF2, FGF3, FGF4, FGF10, and FGF19 are all associated with breast cancer; however, insights into the differential signaling of these FGFs are lacking. It is unclear what pathways and kinases are regulated by the different FGFs. Also, no mechanistic signaling comparisons are investigated to elucidate the importance of each of the FGFs and their possible roles in breast cancer. Gaining these biological insights is key to understanding the implications of FGF signaling in breast cancer.
Here, we aim to broaden our understanding of FGF signaling by quantifying temporal kinase activation dynamics using a selected reaction monitoring (SRM) assay with broad coverage of kinases that are involved in the FGFR signaling pathway. To verify the biological results from the longitudinal SRM data, we created a dynamic mechanistic model of the signaling pathway using logic-based ordinary differential equations. To explain discrepancies in our developed model, we used modeling-guided analysis of shotgun phosphoproteomics data. Our approach successfully mapped FGF2, FGF3, FGF4, FGF10, and FGF19 signaling in breast cancer cell lines and allowed us to add hitherto unknown involved kinases and signaling dynamics to FGF stimulations.
Experimental Procedures
Cell Culture
MCF-7 (ATCC), BT-474 (ATCC), and EFM-192a (DSMZ) cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Sigma) and 2 mM glutamine. Cells were regularly tested for mycoplasma. All cells were cultured in a humidified incubator equilibrated with 5% CO2 at 37 °C. Experiments were performed after the fifth passage and before the 20th passage to limit cell heterogeneity between experiments.
Sample Preparation for Mass Spectrometry
For mass spectrometry experiments, ∼5 million cells were plated in triplicates in 10 cm plates in regular medium. After 24 h, the medium was changed to serum-starved medium supplemented with 5 μg/ml heparin (Thermo Fisher Scientific). After 24 h, cells were incubated with 50 ng/ml of either FGF2 (Peprotech), FGF3 (KyvoBio), FGF4 (Peprotech), FGF10 (Peprotech), or FGF19 (Peprotech). Cells were washed three times with ice-cold PBS, scraped, and snap-frozen until further sample preparation.
Cell Growth Assay
Triplicate groups of ∼0.1 million cells were plated in 12-well plates again first in regular medium and subsequently in medium with either 5 μg/ml heparin or without. After 24 h, one of the five different FGF ligands was added, and the plate was incubated in an IncuCyte ZOOM at 37 °C/5% CO2 until the end of the experiment. Pictures of each well were taken every hour, of which the percentage plate coverage was determined. Significance between groups was determined using an ANOVA and Tukey’s range test (p < 0.05).
Scratch Wound-Healing Assay
In 12-well plates, triplicates of 3e5 cells were plated in a regular medium; after 24 h, the medium was changed to starved medium supplemented with 5 μg/ml heparin. Subsequently, the cells were verified to be confluent when the scratch assay was performed (30). The scratch assay was analyzed as described before (31). In short, using the ImageJ/Fiji script “Wound Healing Size Tool”, the percentage of wound closure was calculated between t = 24 h and t = 0 (32). Significance between groups was determined using an ANOVA and Tukey’s range test (biological triplicates, p < 0.05).
Spectral Library Generation
Spectral libraries were used to determine peptide fragmentation characteristics and their indexed retention time, which are key for identifying peptides in the tier 2 SRM assay. The custom mix of heavy-labeled peptides (JPT or Thermo Fisher Scientific) was mixed with iRT peptides (Biognosys) and analyzed using an Orbitrap Q-Exactive HF (Thermo Fisher Scientific). An unscheduled parallel reaction monitoring method scanned for the +2 and +3 charged peptides, including all possible methionine oxidations. Peptides were separated using a 2 h gradient; a 120 k resolution was used for the parallel reaction monitoring assay, resulting in a minimum of five spectra per peptide. Raw files were processed with MaxQuant (version 1.6.10.43). As a library, the verified human proteome from UniProtKB (release 09-2019) was used, with the addition of the Biognosys iRT peptides, resulting in 20,370 proteins in total. The digestion mode was set to “specific” using Trypsin/P and a maximal number of miscleavages of 1. Carbamidomethyl cysteine was set as fixed modification, and the variable modifications were set to serine/threonine/tyrosine phosphorylation, methionine oxidation, and isotope labels. The first peptide tolerance was set to 20 ppm, and the main peptide tolerance was set to 4.5 ppm. MS2 tolerance was set to 20 ppm. The search results were filtered using a 1% peptide and protein false discovery rate cut-off; the minimal Andromeda score for modified peptides was set at a threshold of 40 and a minimal delta score of 6. Subsequently, using Skyline (version 20.1.1.83), spectra were manually validated, and when deemed reliable, pseudo-MS2 spectra were generated, which were used as the peptide library.
SRM Assay Development
The SRM assay was developed using previously described methods (27). The assay was developed on a TSQ Altis (Thermo Fisher Scientific). In brief, the 10 most intense fragment ions from the library were used as initial transitions. These transitions were used to optimize multiple parameters, such as retention time and collision energy. The collision energy was optimized per transition using Skyline, with the TSQ Vantage CE formula as starting point (CE = 0.03 m/z + 2.905 for doubly charged precursors and CE = 0.038 m/z + 2.281 for precursor charges of 3 and higher) and optimized using steps of 1 voltage.
Protein Digestion SRM Assay
Snap-frozen protein pellets were lysed, reduced, and alkylated in lysis buffer (1% sodium deoxycholate (SDC), 10 mM tris(2-carboxyethyl)phosphine hydrochloride), 40 mM chloroacetamide, and 100 mM TRIS, pH 8.0 supplemented with phosphatase inhibitor (PhosSTOP, Roche) and protease inhibitor (cOmplete mini EDTA-free, Roche). Cells were heated at 95 °C and sonicated with a Bioruptor Plus (Diagenode) for 15 cycles of 30 s. Bradford protein assay (Bio-Rad Protein Assay Kit I, Bio-Rad) was used to determine the protein amount, after which samples were split into 200 μg aliquots. Proteins were digested overnight at 37 °C with trypsin (1:50 μg/μg) (Sigma-Aldrich) and lysyl endopeptidase (1:75 μg/μg) (Wako). Heavy-labeled phosphopeptides were added to the samples. The SDC was precipitated with 2% formic acid (FA) twice, after which samples were desalted and enriched in an automated fashion using the AssayMap Bravo platform (Agilent Technologies) with corresponding AssayMap C18 (Agilent Technologies) reverse-phase column as previously described (33). The enrichment of phosphorylated peptides, in short, samples were dissolved in 80% ACN/0.1% TFA. Fe3+ IMAC cartridges were used, using 80% ACN/0.1% TFA as washing buffer and 10% Na4OH as elution buffer.
SRM LC-MS/MS Setup
Samples were analyzed on a TSQ Altis (Thermo Fisher Scientific) coupled to an UltiMate 3000 (Thermo Fisher Scientific) and an easy spray analytical column (ES802A, 25 cm, 75 mm ID PepMap RLSC, C18, 100 Å, 2 mm particle size column (Thermo Fisher Scientific)). First, samples were reconstituted in 2% LC-MS grade FA. Samples were loaded on a trap column (Acclaim PepMap 100 C18 HPLC Column 0.3 × 5 mm with 5 μm particles (Thermo Fisher Scientific)) with 2.2% buffer A (0.1% FA) for 3 min and subsequently separated using 0 to 32% buffer B (99.9% ACN, 0.1% FA) in 35 min at 300 nl/min and followed by a 20 min column wash with 80% buffer B at 300 nl/min and 10-min column equilibration at 2.2% B. The TSQ Altis spray voltage was set at 1.9 kV and fragmented at 1.5 mTorr in the second quadrupole. The first quadrupole was set at 0.7 da FWHM, and the third quadrupole was set at 1.2 da FWHM. All transitions were measured with optimized collision energy without scheduling and a cycle time of 1.5 s.
SRM Data Assessment
All experiments were analyzed using Skyline-Daily (version 20.2.1.404) (34). The quality of the peptides was assessed mainly on the signal similarity between the heavy and the light peptides. The most important aspects were perfect co-elution, peak shape, and relative contributions of each transition between the heavy and the light peptide. A rdotp >0.95 was maintained to indicate the similarity between the heavy and the light peptide. In-house R scripts were used for further data visualization and analysis.
Experiment Design and Statistical Rationale
All treated and untreated cellular groups used in this study entailed n = 3 biological replicates. Samples for phosphoproteomic analyses were enriched for phosphorylated peptides, and all samples were injected separately into the LC-MS/MS system. Each raw file was separately processed using the MaxQuant software. This number was sufficient to evaluate reproducibility and quantitatively compare the various conditions.
Logic-Based Dynamic Modeling
Logic-based dynamic modeling was performed as described earlier (35). In short, first, a prior knowledge network (PKN) was generated using Omnipath and converted to a simple interaction file (36). Normalization was done per kinase across all the FGFs. The average fold change to t = 0 was scaled between 0 to 1 using the 99% interquartile range (biological triplicates) described in Equation 1.
| (1) |
Values <0 or >1 were set to 0 or 1, respectively. The different FGFs were set to 0.75 for their modeling.
The model was trained using the freely available CNORode for all FGFs simultaneously (37). Each kinase can be described using a continuous update function Bi where the activity of a kinase xi is predicted {0,1} using the associated upstream effectors, as shown in Equation 2 (38).
| (2) |
can be interpreted as the kinase responsiveness to upstream effectors where a small value indicates a slower response. Each transfer function is a Hill-type function, as previously described and presented in Equation 3 (39).
| (3) |
The sigmoidal shape curve is determined by parameters n and k. The k parameter can be interpreted as the strength of the interaction where a high k value describes a high signal throughput.
Kinase Dynamic Parameter Estimation
Each kinase is assigned a fixed n value of 3 and a k and τ value determined by the dynamic modeling. CNORode and the MEIGOR toolkit were used, which uses the normalized kinase activity data and the PKN to determine the best k and τ values based on the smallest root-mean-square error (RMSE) (40). The method entails L2 normalization to prevent overfitting, which was set to a value of 10−5. The update function was verified to have achieved optimal performance based on the RMSE response curves. Model goodness-of-fit was determined using Pearson’s r and the RMSE of all measured and predicted time points of all kinases. The biological RMSE was determined using the deviation between the measured values and the mean.
Peptide Work-Up Phosphoproteomics
Peptide work-up was performed identically to the SRM peptide workup except that no heavy-labeled peptides were added after digestion.
Peptide Work-Up Proteomics
Snap-frozen protein pellets were lysed, reduced, and alkylated in lysis buffer (1% SDC, 10 mM tris(2-carboxyethyl)phosphine hydrochloride), 40 mM chloroacetamide, and 100 mM TRIS, pH 8.0 supplemented with protease inhibitor (cOmplete mini EDTA-free, Roche). Cells were heated at 95 °C and sonicated with a Bioruptor Plus (Diagenode) for 15 cycles of 30 s. Bradford protein assay (Bio-Rad Protein Assay Kit I, Bio-Rad) was used to determine the protein amount, after which samples were split into 10 μg aliquots. Proteins were digested overnight at 37 °C with 1:50 trypsin (Sigma-Aldrich) and 1:75 and lysyl endopeptidase (Wako), after which samples were desalted using an Oasis platform, dried down, and stored at −80 until further use.
Data-Dependent Analysis of Phosphoproteomics
Samples were suspended in 2% FA and analyzed on an Exploris (Thermo Fisher Scientific) coupled to an UltiMate 3000 (Thermo Fisher Scientific), fitted with a μ-precolumn (C18 PepMap100, 5 μm, 100 Å, 5 mm × 300 μm; Thermo Fisher Scientific), and an analytical column (120 EC-C18, 2.7 μm, 50 cm × 75 μm; Agilent Poroshell). Peptides are loaded in 9% buffer A (0.1% FA) for 1 min and separated using 9 to 36% buffer B (80% ACN, 0.1% FA) in 97 min at 300 nl/min and followed by a 6 min column wash with 99% buffer B at 300 nl/min and a 10-min column equilibration at 9% B. The mass spectrometer (MS) was operated in data dependent acquisition mode, with the MS1 scans in a range of 375 to 1600 m/z acquired at 60 k, using an automatically set automatic gain control target. MS2 scans were acquired with a 16 s dynamic exclusion at a 30 k resolution, 28% normalized collision energy, and an isolation window of 1.4 m/z.
Raw files were processed via MaxQuant version 1.6.17.0 using the verified human proteome from UniprotKB (release 09-2019) containing 20,369 proteins (41). A maximum of five modifications and two miscleavages were set using fixed carbamidomethyl modification, and the variable modifications oxidized methionine, protein N-terminal acetylation, and serine/threonine/tyrosine phosphorylation. The protein and peptide false discovery rates were set to <0.01 and conducted with match between runs enabled. The first MS1 peptide tolerance was set to 20 ppm, the main MS1 peptide tolerance to 4.5 ppm, and MS2 match tolerance to 20 ppm. No normalization or imputation was applied.
Shotgun Proteomics Analysis
Samples were suspended in 2% FA and analyzed on a Q-Exactive HF (Thermo Fisher Scientific) coupled to an UltiMate 3000 (Thermo Fisher Scientific), fitted with a μ-precolumn (C18 PepMap100, 5 μm, 100 Å, 5 mm × 300 μm; Thermo Fisher Scientific), and an analytical column (120 EC-C18, 2.7 μm, 50 cm × 75 μm; Agilent Poroshell). Peptides are loaded in 9% buffer A (0.1% FA) for 1 min and separated using 9 to 44% buffer B (80%ACN, 0.1% FA) in 155 min at 300 nl/min and followed by a 6 min column wash with 95% buffer B at 300 nl/min and a 10-min column equilibration at 9% B. The MS was operated in data dependent acquisition mode, with the MS1 scans in a range of 375 to 1600 m/z acquired at 60 k, using an automatic gain control target of 3e6. MS2 scans were acquired with a 24 s dynamic exclusion at a 30 k resolution, 27% normalized collision energy, and an isolation window of 1.4 m/z.
Raw files were processed via MaxQuant version 1.6.17.0 using the verified human proteome from UniprotKB (release 09-2019) containing 20,369 proteins (41). A maximum of five modifications and three miscleavages was set using fixed carbamidomethyl modification, and the variable modifications oxidized methionine and protein N-terminal acetylation. The protein and peptide false discovery rates were set to <0.01 and conducted with match between runs enabled. The first MS1 peptide tolerance was set to 20 ppm, the main MS1 peptide tolerance to 4.5 ppm, and MS2 match tolerance to 20 ppm. Further analysis was performed using artMS version 1.12.0 building on MSstats (http://artms.org) (42). MSstats imputation was done using accelerated failure time modeling, and the samples were median normalized after imputation.
FGFR Quantitative Polymerase Chain Reaction Quantification
MCF-7 cells were plated in triplicates. Subsequently, the samples were lysed and prepared for quantitative polymerase chain reaction (qPCR) analysis using the protocol adapted from (43). In short, samples were lysed and isolated using the vendor’s instructions of NucleoSpin RNA plus (Macherey-Nagel) with the addition of a DNase removal step using RNase-Free Dnase (Qiagen). Next, 500 ng of RNA was used to obtain complementary DNA using the vendor’s instructions of AH iScript (Bio-Rad). The qPCR was performed at 95 °C for 10 min, followed by (95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s), a total of 40 times. Normalization was performed using beta-actin and glyceraldehyde-3-phosphate dehydrogenase following (44). The primers used are listed in supplemental Table S2.
RAP1 Activation Assay
RAP1 activity was determined following the supplier’s instructions (Merck, Cat# 17-321). In short, MCF-7 cells were plated in 15 cm plates and incubated for 60 min with FGF2, FGF3, FGF4, FGF10, FGF19, or without FGF and lysed using the provided lysis buffer. After, equal amounts of protein were used for the RAP1 pulldown, including one positive control consisting of MCF-7 cell lysates incubated with GTPγS. Subsequently, a Western blot was conducted using the provided RAP1 antibodies. Linear adjustments were performed using Fiji (32).
Results
Dynamic Kinase Activity Quantification
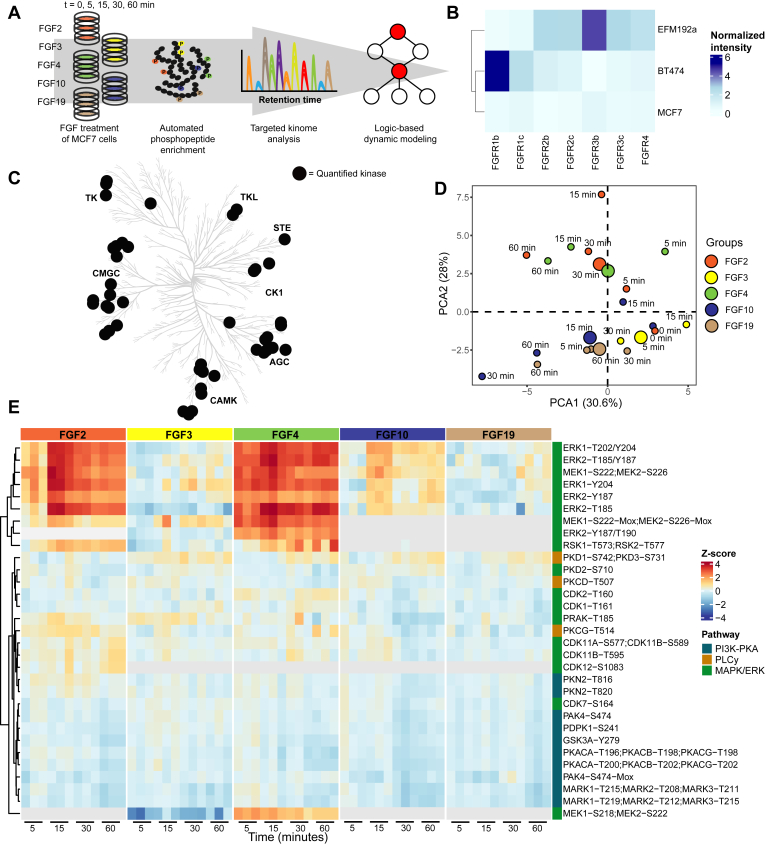
Here, we performed (phospho)proteomics experiments to elucidate the specific effect of different FGF ligands on FGFR activation and downstream signaling. Thereby we focused on FGFR signaling in breast cancer cells induced by either FGF2, FGF3, FGF4, FGF10, or FGF19. To understand FGF signaling, we quantified temporal system-wide kinase activity using a dedicated SRM assay targeting the activation loops of a widespread panel of kinases (Fig. 1A). We performed extensive literature mining of the FGFR signaling pathway to increase coverage of the kinases involved, resulting in the addition of nearly 200 phosphopeptides spanning 50 kinases to the original assay developed by Schmidlin et al. (2019), generating an assay comprising 484 phosphopeptides on 197 kinases (supplemental Table S1) (27).
Fig. 1.
Stimulation with FGF2, FGF3, FGF4, FGF10, or FGF19 results in differential kinome regulation.A, schematic overview of the experimental approach, whereby a targeted kinase activation loop SRM assay was used to monitor system-wide kinase activity upon treatment of MCF-7 cells with distinct FGF ligands. B, qPCR experiments were performed to monitor FGFR expression on three different cell lines. FGFR expression was normalized to beta-actin and glyceraldehyde-3-phosphate dehydrogenase. C, kinome tree with kinases significantly regulated by at least one of the tested FGF ligands represented by black dots (ANOVA p < 0.05, triplicate measurements). D, principal component analysis (PCA) of the kinase activity data at different time points and with the different tested FGFs. Mean values were used for the independent triplicate measurements. E, heatmap through unsupervised hierarchical clustering of all significantly (ANOVA p < 0.05) regulated phosphorylated peptides over all time points and FGFs (with each experiment performed in triplicate). Not identified phosphorylated peptides are represented in gray. FGF, fibroblast growth factor; FGFR, FGF receptor; qPCR, quantitative polymerase chain reaction; SRM, selected reaction monitoring.
To select an appropriate cell line, this SRM assay was initially used to monitor the system-wide kinome activity response of a set of breast cancer cell lines, namely MCF-7, BT-474, and EFM-192a cells, upon FGF2 and FGF4 stimulation as these bind the majority of FGFR spliceoforms. From these data, we concluded that MCF-7 cells especially displayed a broad kinome response after stimulation (supplemental Fig. S1). We reasoned this would be explained by FGFR expression; however, surprisingly, qPCR quantification of FGFR expression in the panel of tested cells showed that the MCF-7 cells exhibited an overall low expression of FGFRs (Fig. 1B and supplemental Fig. S2 and supplemental Table S2). This highlights that FGFR expression alone does not solely determine the extent of downstream signaling. Due to the observed broad kinome response, we did proceed with the MCF-7 cells, which were incubated with either FGF2, FGF3, FGF4, FGF10, or FGF19 and the cofactor heparin for 0, 5, 15, 30, and 60 min (45, 46). Using the kinase activation loop SRM assays, we quantified kinase activity profiles of 46 phosphorylated sites spanning 44 kinases (supplemental Table S3). Of these, 35 kinases displayed significant regulation over time (ANOVA p < 0.05) upon stimulation with at least one of the five tested FGF ligands. Each of the tested ligands resulted in differential regulation of kinases across most kinase families (Fig. 1C) that were primarily members of the MAPK/ERK, PKA, and/or PLCγ pathways (Fig. 1, D and E) (1, 47).
Fine-Tuned Activation of the MAPK/ERK Signaling Pathway
As the MAPK/ERK pathway is known to be involved in FGF signaling, we first compared the kinase activity profiles acquired with the SRM assays of kinases involved in this pathway. FGF-stimulated MAPK/ERK activation is commonly regarded to be directed via the RAS-RAF-MEK-ERK signaling cascade (25, 48, 49, 50, 51, 52, 53). In MCF-7 cells, only FGF2, FGF4, and FGF10 treatments significantly activated several of the kinases in the MAPK/ERK pathway (Fig. 2A).
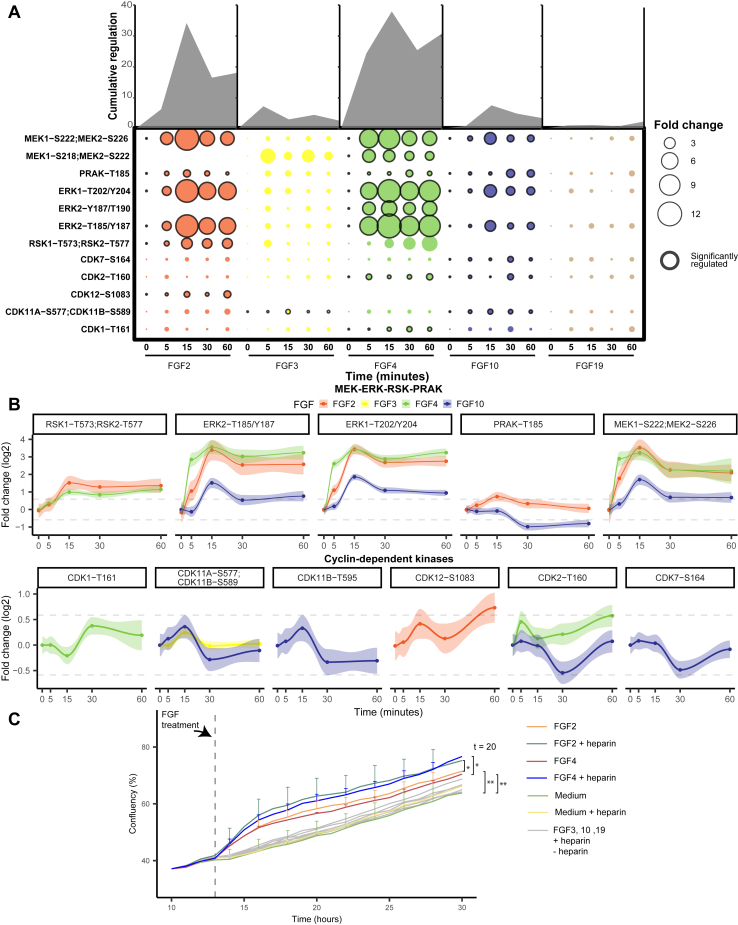
Fig. 2.
Regulation of kinases and cyclins implicated in the MAPK/ERK pathway.A, measured phosphorylated peptides involved in the MAPK/ERK pathway that show significant regulation (ANOVA p < 0.05, independent triplicate measurements) following incubation with at least one of the tested FGFs. Black-edged circles represent significant phosphosites. The area of the circle represents cumulative regulation in fold change of the measured phosphopeptides measured in triplicate over five subsequent time points. B, The dynamic phosphorylation of the sites is color-coded by the FGF ligand used, and only plots are depicted when the ligand changed the phosphorylation at that site significantly. Gray lines represent a 1.5 fold change, and 90% confidence intervals are presented per phosphopeptide. C, influence of incubation with the FGF ligand and heparin on cellular growth. Growth curves of MCF-7 cells, incubated with 50 ng/ml of each of the tested FGF ligands with or without 5 μg/ml of heparin. The confluency percentage was taken as a readout to analyze cell growth and plotted (data was acquired in triplicate showing the SDs per time point and ligand used). FGF, fibroblast growth factor.
Investigating the kinases involved in the MAPK/ERK pathway after either FGF2, FGF4, or FGF10 treatment showed rapid and high regulation of especially the main signaling hub of the MAPK/ERK pathway, namely MEK (MEK1 and MEK2) and ERK (ERK1 and ERK2) (Fig. 2B) (54). FGF2 and FGF4 treatment resulted in an >10-fold increase of MEK- and ERK-activating phosphorylation and FGF10 resulted in an >2-fold increase. Notably, MEK and ERK activation was about 10-fold higher than the other kinases in the MAPK/ERK pathway, supporting their central role as signaling hub (Fig. 2B). MEK and ERK dynamics per FGF treatment showed high correlation, displaying direct regulation of ERK as the target of MEK. However, MEK and ERK signaling dynamics showed a lower correlation between different FGF stimulations, also in the case of strong activation via FGF2 and FGF4. FGF4 treatment resulted in fast activation peaking at 15 min and showing an additional increase after the 30-min time point. FGF2 treatment on the other hand showed slightly slower activation with also the maximum at 15 min that plateaus from 30 min onwards (Fig. 2B). This suggests differential MAPK/ERK pathway regulation.
Even though FGF2, FGF4, and FGF10 all activated MEK and ERK, all ligands resulted in unique downstream activation, which could either be the result of different activation mechanisms and feedback loops or due to different activation dynamics of the same pathway (55, 56). After FGF2 and FGF4 treatment, but not FGF10, MEK and ERK dynamics highly correlated with RSK1 and RSK2 dynamics, which are regulators of cell proliferation and cell survival (Fig. 2B) (57, 58, 59). FGF2 incubation resulted in the activation of CDK12 and a transient 1.5-fold increase in activating phosphorylation of PRAK, of which the role in the context of FGF has remained elusive (60, 61). FGF4 incubation resulted in the activation of CDK1 and CDK2. Interestingly, cyclin dependent kinase (CDK) activation dynamics are relatively modest, with a maximum increase in activating phosphorylation of 60% (Fig. 2B). Uniquely, FGF10 treatment did not activate kinases downstream of ERK but inactivated CDK2, CDK7, CDK11a, CDK11b, and PRAK. Inactivation of these kinases occurred concurrently after 30 min, which may originate from a negative feedback loop (62). FGF10 may initiate this feedback loop by recycling its receptor FGFR2b to the cell membrane, or FGFR2b intracellular transport may expose the receptor to the substrates responsible for the feedback loop (63). Notably, only FGF10 showed sustained PRAK inactivation, which has been associated with decreased tumor progression (64, 65).
FGF3 and FGF19 have been described to activate the MAPK/ERK pathway in a subset of cell lines through FGFR4 activation (66, 67, 68, 69). In contrast, in our dataset, we did not observe any activation of the MAPK/ERK pathway after FGF3 and FGF19 stimulations, although, in our proteome profiles of MCF-7 cells after 24 h of incubation with different FGFs, we did clearly identify the FGFR4 receptor.
In the context of FGF stimulation, MAPK/ERK pathway activation is considered to drive cell growth and increase tumor progression (52, 70, 71). To verify whether cell growth was indeed induced in our experimental conditions, we monitored cell growth after FGF stimulations using an IncuCyte ZOOM. Only after stimulation with FGF2 and FGF4, we temporarily observed significantly increased cell growth (two-tailed t test, p < 0.05) (Fig. 2C). This finding was in line with the high MAPK/ERK pathway activation quantified in FGF2- and FGF4-stimulated cells. FGF10 stimulation did not substantially increase cell growth even though the MAPK/ERK pathway was activated. This suggests that a signaling threshold must be reached to activate proliferation or that alternative signaling is required for cell growth. Notably, adding heparin significantly increased the proliferation rate of FGF2- and FGF4-treated MCF-7 cells, while only adding heparin did not increase cell proliferation (Fig. 2C).
Consistent Downregulation of the PKA Pathway
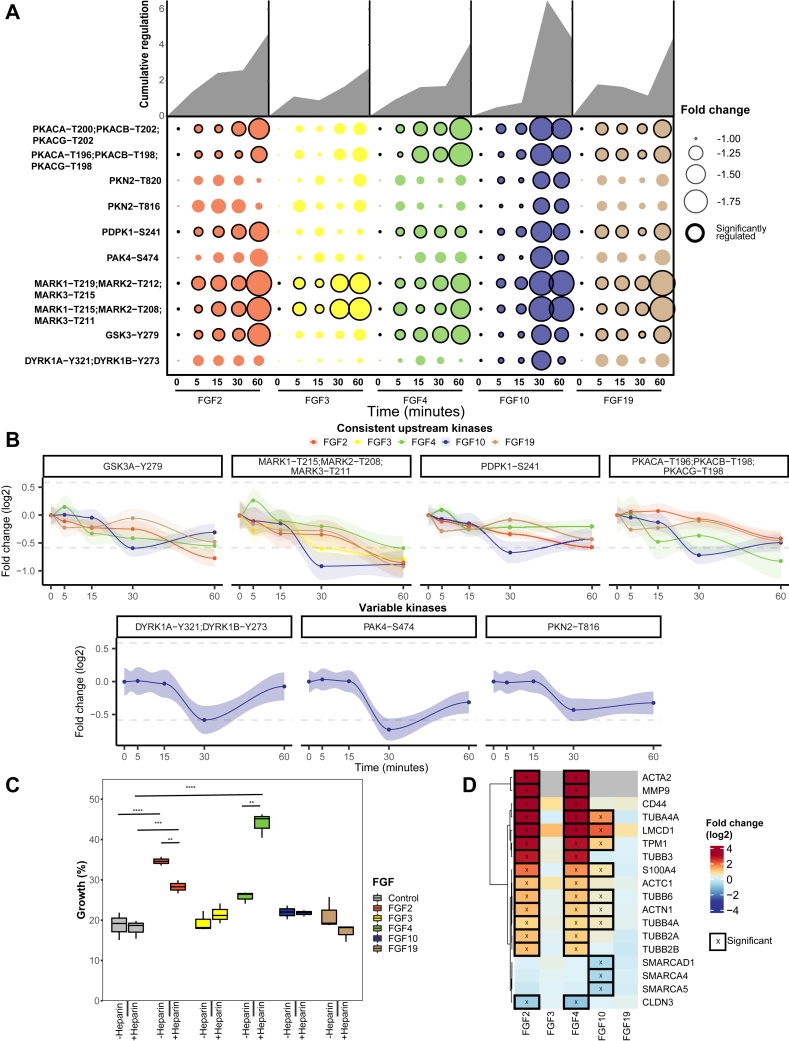
Next, we examined the PKA pathway. In our analysis, incubation with each of the tested FGFs, except FGF3, resulted in the significant inactivation of the PKA pathway (Fig. 3A). We quantified the change in phosphorylation of the upstream regulator PDPK1, which directly regulates PKA activity by phosphorylating Thr-197 (72). All measured kinases involved in the PKA pathway highly correlated with PDPK1 dynamics for all FGF stimulations in our dataset.
Fig. 3.
Regulation of kinases implicated in the PKA pathway.A, all significantly regulated phosphopeptides of the PKA pathway are represented as the mean across triplicates (p < 0.05 ANOVA). Cumulative absolute regulation in fold change is represented in the area plot to show overall pathway regulation. B, regulation of significant (ANOVA p < 0.05) changing phosphopeptides is plotted from the PKA pathway. Gray lines represent a 1.5 fold-change, and 90% confidence intervals are presented per phosphopeptide. C, MCF-7 cells were subjected to a scratch wound assay, and after 24 h, percentage closure of the scratch was measured. The boxplots represent triplicate measurements of FGF-stimulated cells with or without 5 μg/μl heparin. A two-sided t test was used to validate the significance. D, proteins associated with EMT were extracted from the shotgun dataset. Significantly regulated proteins are displayed using an asterisk (FDR <0.05). EMT, epithelial-to-mesenchymal transition; FGF, fibroblast growth factor.
PDPK1 can also initiate PI3K pathway activation. Thus, we examined changes in substrate phosphorylation of the central regulators of the PI3K pathway, namely AKT1 and AKT2, using shotgun phosphoproteomics analysis of FGF-stimulated MCF-7 cells. All FGF treatments resulted in minimal changes in AKT1 and AKT2 substrate phosphorylation (supplemental Fig. S4). Only FGF2 and FGF4 treatments increased the phosphorylation of two and four substrates, respectively. However, these phosphorylated residues are also downstream targets of the MAPK/ERK pathway. Therefore, minimal changes in the PI3K pathway activity were observed.
All tested FGFs, except for FGF3, resulted in the inactivation of PKA pathway kinases PKA, GSK3A, and MARK kinases (Fig. 3B). Inactivation was consistent but modest. The most significant decrease was a 2-fold decrease on two phosphorylated sites in the activation loop of MARK1, MARK2, and MARK3, respectively (supplemental Fig. S3). Notably, no relation has been described between MARK kinases and FGF signaling up to this day. MARK kinases control cell polarity by regulating microtubules, and reduced MARK kinase activity has been linked to EMT, which is in line with the EMT-inducing effects of FGFs (73, 74).
Besides the similar PKA pathway dynamics by FGF2, FGF4, FGF10, and FGF19 quantified by the targeted kinome assay, solely FGF10 stimulation led to a decrease in phosphorylation of the downstream kinases PAK4, DYRK1A, DYRK1B, and PKN (Fig. 2B). This reveals broad FGF10-induced negative regulatory mechanisms. Notably, the inhibited PAK4, which plays a role in cell adhesion, can be regulated via ERK, the PI3K pathway, and direct binding by PKA (75, 76, 77, 78). The observed kinase activity dynamics of PAK4 strongly correlated with the rest of the PKA pathway while opposing the MAPK/ERK pathway activity dynamics. This observation suggests that PAK4 is linked to the PKA pathway, not the MAPK/ERK pathway.
All herein quantified kinases in the PKA pathway are implicated in EMT, a key process in regulating tumor metastasis (79, 80). FGF2, FGF4, FGF10, and FGF19 have all been described to induce EMT. To investigate whether the tested FGFs induced EMT, we next performed a wound-healing assay that assays cell migration capabilities, which is a key process in EMT (81, 82). These assays revealed that only FGF2 and FGF4 showed a significant increase in wound-healing capacity of 15 and 5% compared to unstimulated MCF-7 cells, respectively (Fig. 3C). Interestingly, with the addition of heparin, this dampened to 10% for FGF2 and increased to 40% for FGF4, revealing a modest role for heparin in regulating EMT. To find further support for FGF-induced EMT, we extracted proteins from the EMTome database associated with EMT, specifically focusing on proteins that directly trigger EMT or are key markers for EMT (83). In their proteomic profiles (supplemental Tables S4 and S5), FGF2 and FGF4 stimulations showed an identical profile of 15 EMT-associated proteins significantly regulated after 24 h (Fig. 3D), supporting an EMT-like phenotype downstream of FGF2 and FGF4. FGF10 stimulation resulted in less pronounced expression changes in 7 of the 15 observed EMT proteins, in part confirming the role of FGF10 in inducing EMT, whereas FGF3 and FGF19 showed no significant expression changes in EMT-related proteins (84). This is further supported by gene set enrichment analysis of the hallmarks of EMT as provided by MSigDB, which in our proteome data are only significantly upregulated after FGF2 and FGF4 treatment (supplemental Fig. S5) (85). Altogether, these findings show that FGF2, FGF4, FGF10, FGF19 all inactivated numerous kinases involved in EMT. However, only FGF2 and FGF4 treatment resulted in increased wound-healing capacities and an EMT-like phenotype on proteome level, FGF10 treatment only resulted in a more EMT-like phenotype on proteome level, and FGF19 did not show either. Further mechanisms must thus be regulated to induce EMT.
Undistinguished PLCγ Signaling Along the FGF–FGFR Axis
Next, we explored the measured activity profiles of the kinases within the PLCγ pathway. The PLCγ pathway is relatively understudied in the context of FGFR stimulation and regulates specialized functions (86, 87, 88, 89, 90, 91). In our current study, PKD1, PKD2, PKD3, PKCδ, and PKCγ showed significant regulation when incubated with at least one of the tested FGFs (supplemental Fig. S6). Kinase activation dynamics were nonlinear, hinting at the presence of multiple feedback loops (62). Moreover, kinases in the PLCγ pathway showed a relatively low correlation in their activation dynamics, and all tested FGFs showed distinct kinase regulation (supplemental Fig. S6).
Indeed, only FGF2 transiently activated PKCα/β/γ by activating phosphorylation Thr-514, yet no other PLCγ pathway kinases were regulated (92). FGF4, FGF10, and FGF19 all activated PKD1 and PKD3, whereas FGF4 and FGF10 also showed the inactivation of PKCδ and PKD2 or only PKCδ, respectively.
Distinct FGF Ligands Induce Distinct and Diverse Temporal Dynamics in Phospho-Signaling
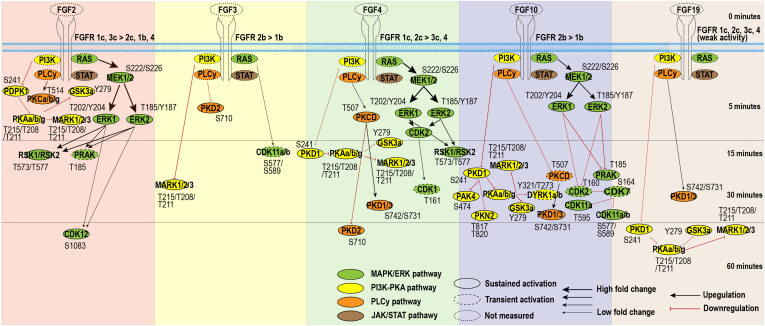
Not only does FGF specificity to the various FGFRs determine the biological outcome, but also the affinity for the various FGFRs is crucial. In receptor tyrosine kinases biology, it is known that ligands with high affinity to the receptor can lead to fast, transient activation, while lower affinity ligands, binding to the same receptor, lead to a slower sustained activation, resulting in a different biological outcome (93, 94). To evaluate whether each FGF differentially regulated signaling dynamics, OmniPath was used to construct biological networks in which kinases are ordered based on the initial time point when regulation was observed (Fig. 4) (36).
Fig. 4.
Temporal kinome dynamics following FGF treatment. MCF-7 cells were treated with either FGF2, FGF3, FGF4, FGF10, or FGF19, together with heparin. The resulting temporal kinome dynamics were quantified using the targeted activation loop assay and normalized to the t = 0 time point. The presented kinome signaling dynamics are separated per FGF used for stimulation, and kinases are ordered based on significant initial activation (ANOVA + Tukey’s range test, p < 0.05, biological triplicates). Black and red arrows indicate whether measured kinase activity increased or decreased over time, respectively. The fold change compared to the t = 0 time point is represented by the thickness of the arrows. MARK1/2/3 phosphorylated site T215/T208/T211 is ambiguous to T219/T212/T215. PKAa/b/g phosphorylation site T196/T198/T198 is ambiguous to T200/T202/T202. FGF, fibroblast growth factor.
Indeed, in our data, each of the tested FGFs did lead to distinct timing of initial pathway regulation. FGF2 induced a fast initial activation within 5 min of all measured pathways. This is expected as FGF2 binds to most FGFRs with high affinity (21). Although FGF4 binds the same FGFR subset as FGF2, except for FGFR1b, it does so with different affinity. As a potential consequence, and in contrast to FGF2, FGF4 stimulation inactivated the PKA pathway in our experiments only after 15 min (Fig. 4). FGF10 stimulation activated the MAPK/ERK pathway within 5 min, similar to FGF2 and FGF4. However, this was followed by a strong downregulation after 30 min of more downstream targets. Last, FGF3 and FGF19 resulted in relatively slow (and modest) activation only 30 min after stimulation (Fig. 4).
Logic-Based Dynamic Modeling Validates the Known FGF Pathways but also Identifies Putative New Players
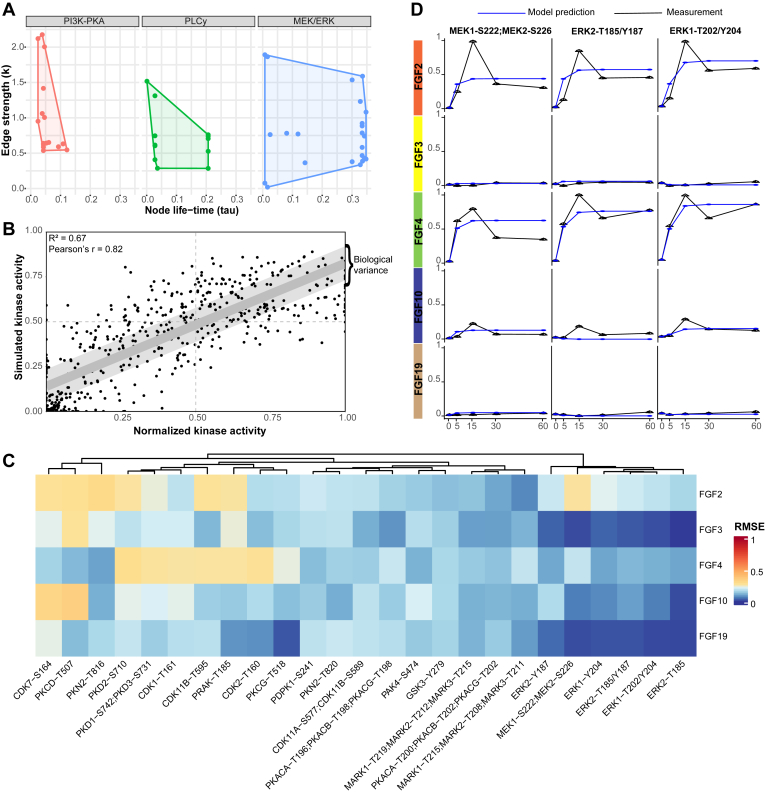
Pathway models such as in Figure 5 are based on existing knowledge and are thus inherently biased towards well-characterized pathways. Therefore, validation of the biological model is needed to identify either missing or inaccurate connections between kinases or missing signaling nodes. To verify our biological model, predict signaling dynamics between kinases, and find possible gaps, we used a dynamic mechanistic model based on logic-based ordinary differential equations (95). First, a PKN was built using information available via OmniPath using only kinases quantified in all FGF stimulations (Fig. 4) (36). Next, the logistic-based ordinal differentiations were calculated using the quantitative longitudinal kinase activity data of all FGF stimulations together. For each node, a speed factor (τ) was calculated to represent the responsiveness of a kinase’s activation to upstream kinases activation (38). Low values indicate a slow transfer of activation from kinases’ upstream activators. For each node, also an edge-specific transmission parameter (k) was calculated, which represents the quantitative signal that is transferred between kinases (38). High values of the nonlinear k parameters indicate that relatively little quantitative signal is transferred via the edge. To evaluate the quality of the predicted τ and k values, Pearson’s r and the RMSE of all the quantitative kinome values in the model were assessed and compared to the RMSE between biological replicates (Fig. 5, A and B). The RMSE of the model (0.18) is almost as low as the RMSE observed between the biological replicates (0.1). The model thus successfully predicts most of the kinase activity, with a small error likely due to unknown entries in the PKN.
Fig. 5.
Logic-based dynamic modeling reveals unknowns in FGF-induced signaling.A, logic-based dynamic modeling was used to predict a speed factor (τ) and a transmission parameter (k) for the kinases across the PKA, PLCγ, and MAPK/ERK pathways. These represent the signal transduction speed and the quantitative signal transferred between kinases, respectively. B, the root-mean-squared error (RMSE) of the predicted values by logic-based dynamic modeling and the measured values by the targeted kinome loops assay. The values were normalized using the 99% interquartile range. The light gray area represents the biological variation in the measurements. The dots represented are the mean values of the replicates and all time points. C, mean RMSE values for the measured versus predicted kinase activity values. The modeling was performed using identical networks, meaning downstream kinase-kinase relations constitute the same predictive k and tau values. Therefore, predictive downstream errors may indicate differential regulation between FGFs. D, line plots of the measured and predicted kinase activity using the function with the lowest error across all FGF stimulations. Before the logic-based dynamic modeling, the average of the quantified kinome values was taken (biological triplicates) and normalized using the 99% interquartile range. The blue line represents the model prediction, and the black line represents the quantified kinase activity using the targeted kinome assay. FGF, fibroblast growth factor; RMSE, root-mean-square error.
To explore these unknowns in the PKN, the RMSE of individual phosphopeptides was evaluated (Fig. 5C). High RMSE suggests that the model is insufficient to predict a kinase activation state, which results from missing or erroneous connections between nodes in the network. Therefore, a high predictive error can be used to find novel biological connections or nodes. The model showed no highly contradictive prediction errors for single kinases (RMSE error >0.5), which occurs when activation of one kinase leads to activation of the next kinase, but inactivation is measured. However, some kinases showed errors that were higher than the biological variance.
Kinases with a relatively high error are part of the PLCγ and MAPK/ERK pathways. Error in kinases regulated by the PLCγ pathway is expected due to the low pathway coverage (Fig. 4). Surprising, however, is the substantial error in MEK activity prediction after FGF2 stimulation (Fig. 5C). The model failed to predict the fast activation of MEK and ERK and did not incorporate the oscillatory patterns typical for feedback loops (Fig. 5D). Further, following MEK-ERK signaling downstream, all measured CDKs, including CDK1, CDK2, CDK7, CDK11a, and CDK11b, show a relatively high predictive error. This suggests differential MEK-ERK signaling to their downstream effectors. We, therefore, hypothesized that the error in MEK-ERK-CDK signaling is indicative of unknown links between kinases or missing nodes in the current model. We will focus on this more in the next section.
Modeling-Guided Analysis Unveils Differential FGF Signaling
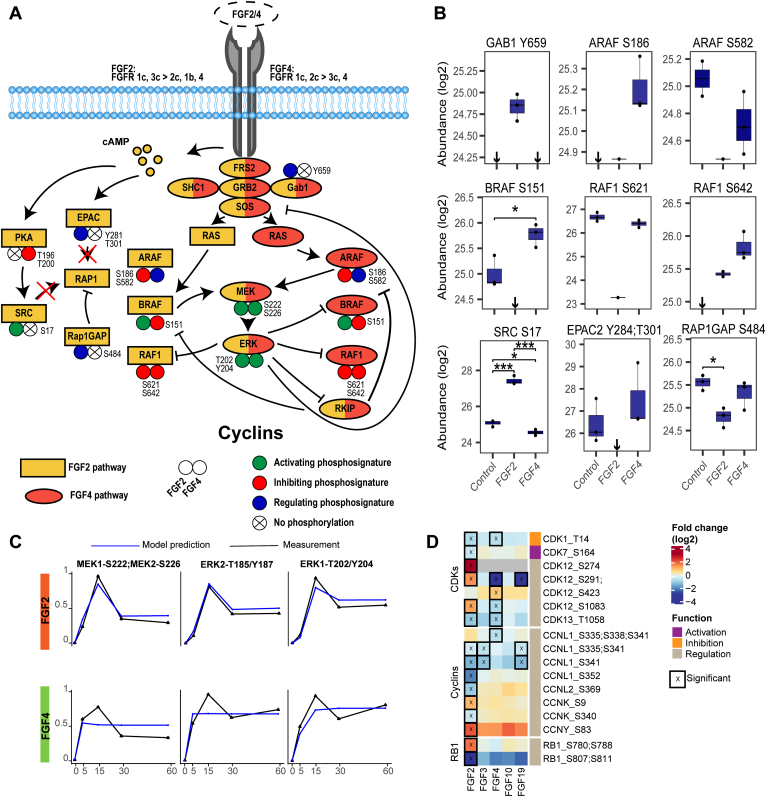
With the aim to explain the predictive modeling errors for MEK, ERK, and CDKs, we expanded the model using manually curated literature mining, shotgun phosphoproteomics analysis of FGF-stimulated MCF-7 cells (supplemental Table S6), and our targeted kinome data. Significantly regulated proteins were used to construct a more refined pathway (Fig. 6A) (96, 97, 98).
Fig. 6.
Regulation of RAF family kinases modulates ERK signaling.A, mapping of phosphorylations of proteins involved in ERK activation shows tight regulation of the RAF kinase family members. The regulation does occur in the RAP1 activation pathway of ERK, yet no RAP1 activation was measured, suggesting this does not contribute to ERK activation. B, quantified peptide abundances corresponding to (A). Significance is depicted using ∗ (p < 0.05) or ∗∗∗ (p < 0.001) using a two-sided t test (biological triplicates). If all values are below the detection limit, this is shown using a ↓. Abundances are acquired using shotgun phosphoproteomics after 60-min stimulation with the different FGFs. C, line plots of the measured kinase activity and the predicted kinase activity using the function with the lowest error across all FGF stimulations. The PKN used is the updated biological pathways, also presented in (A). The average of biological triplicates was taken and normalized using the 99% interquartile range. Model predictions are shown in blue, and quantified kinase activity is shown in black. D, phosphorylation of cell cycle–regulating proteins from the phosphoproteomics data. Significant regulated sites are displayed (two-sided t test, p < 0.05, biological triplicates). Scores represent log2 fold changes. FGF, fibroblast growth factor; PKN, prior knowledge network.
A potential missing link came from the phosphoproteomics data that suggested a RAP1 activation signature exclusively for FGF2. RAP1 is an alternative activator of MEK-ERK, whose activators include EPAC2 and proto-oncogene tyrosine-protein kinase (SRC), and its main negative regulator is RAP1gap (Fig. 6A) (99, 100, 101). Uniquely, FGF2 treatment abolished the signal of Tyr-284 and Thr-301 phosphorylation of EPAC2, which is important for EPAC2 membrane localization. Moreover, FGF2 treatment increased activating phosphorylation Ser-17 of SRC about 4-fold and resulted in a 1.6-fold increase in regulating phosphorylation Ser-484 on RAP1gap (102, 103, 104, 105, 106, 107). These phosphorylations highlight possible RAP1 activation. Therefore, we conducted a RAP1 activity assay. However, this RAP1 activation assay showed no significant RAP1 activation in all tested ligands (supplemental Fig. S7). From these data, we concluded that although pathways commonly involved in RAP1 activation were regulated, RAP1 was not activated and thus was not the cause of differential MEK-ERK dynamics.
Next, we compared FGF2- and FGF4-induced signaling along a more detailed RAS-RAF-MEK-ERK signaling axis (Fig. 6, A and B). FGF2 and FGF4 treatment resulted in fine-tuned and distinct regulation along this signaling axis, especially of the RAF family members (serine/threonine-protein kinase Raf, A-Raf, B-Raf (ARAF, BRAF, RAF1)) that coordinate MEK-ERK activation (108). Only FGF2 treatment enabled BRAF activity by abolishing the signal of the inhibitory ERK target site Ser-151 on BRAF (Fig. 6B) (109). Moreover, FGF2 treatment resulted in reduced activity of ARAF following significant downregulation of Ser-582 phosphorylation, which is needed for 14-3-3 binding to increase the activity of ARAF (110). FGF2 also resulted in a reduced active state of RAF1 implied by an 8-fold lower signal of Ser-621 phosphorylation, necessary for 14-3-3 activation, and by the negative feedback phosphorylation of Ser-642 by ERK on RAF1 (Fig. 6B) (111, 112, 113). Contrarily, FGF4 stimulation showed an activating signature for ARAF, indicated by the phosphorylation of the regulatory site Ser-186 on ARAF (114). Further, FGF4 stimulation resulted in inhibitory phosphorylation on BRAF and RAF1, with a two-fold increase in Ser-151 phosphorylation on BRAF and a strong increase in Ser-642 phosphorylation on RAF1, which was absent in the control (Fig. 6B). In conclusion, the RAF family members showed differential regulation as FGF2 treatment indicated BRAF-driven activation, while FGF4 treatment indicated ARAF-driven activation.
To further validate these signaling differences, we again applied logic-based dynamic modeling using the data from the targeted kinome assay. In the updated model, FGF2 signaling was directed via BRAF and FGF4 via ARAF. Moreover, to model the negative feedback loops, one negative feedback loop between ERK and the FGF activation of ARAF and BRAF was added, as well as a negative feedback loop from ERK to RKIP and from RKIP to ARAF and BRAF activation of MEK (115). The updated pathway showed improved modeling accuracy (Fig. 6C). Especially the FGF2 signaling prediction now has high accuracy that follows the measured feedback loops, giving confidence to the predicted biological pathway. Prediction of FGF4 signaling dynamics was also improved over the initial model, with more accurately modeled activation dynamics, however, is not optimal yet (Fig. 6C). Indeed, the updated model supports the two different modes of ERK activation downstream of FGF2 and FGF4, yet alternative regulators need to be identified to fully explain FGF signaling dynamics.
Following ERK activation further downstream, we set out to use the shotgun phosphoproteomics data to confirm predictive errors for the CDKs and validate differential regulation downstream of ERK. Cumulatively, 17 different phosphorylated sites on proteins that regulate the cell cycle were quantified, including CDKs, cyclins that regulate CDK activity, and RB1, which are all central to cell cycle progression (Fig. 6D) (116, 117). FGF3, FGF10, and FGF19 showed little CDK regulation in our model, in line with the targeted kinome data and the modeling results. FGF2 and FGF4 showed distinct activation patterns of CDKs (Fig. 6D), agreeing with the targeted kinome data and the modeling error. These distinct activation patterns confirm the predictive error of the dynamic model and show that FGF2 and FGF4 regulate cell cycle progression differently.
Discussion
By investigating the FGF-induced dynamic kinome regulation using a targeted kinome assay, we quantified and compared the signaling responses of FGF2, FGF3, FGF4, FGF10, and FGF19. All FGF stimulations resulted in a unique biological response in MCF-7 cells, with FGF2 and FGF4 having the broadest kinome response, FGF10 having a moderate response, and FGF3 and FGF19 showing a modest response. We find complex activation mechanisms that initiate FGF signaling as biological responses upon FGF stimulation vary between cell lines, do not correlate with FGFR expression level, and are influenced by heparin.
Looking at the FGFs in a breast cancer context, FGF-stimulated cells activate biological pathways that can contribute to the hallmarks of cancer (7, 118). The MAPK/ERK pathway is thought to drive cell proliferation, and the PI3K pathway is believed to regulate EMT (51). However, we find that simply activating these pathways does not per se lead to cell proliferation or EMT, respectively. Importantly, this irregularity between kinome or pathway activation and predicted biological outcome emphasizes the complexity of these processes and their incomplete understanding. FGF2 and FGF4 increased cell proliferation and EMT in MCF-7 cells. However, FGF3, FGF10, and FGF19 are reported to regulate cell proliferation and EMT but were not able to regulate these processes in our system. Additional signaling factors may be needed to sensitize or costimulate the cells for a more pronounced biological response (15, 66, 119).
The quantification of dynamic kinase responses instead of single time points is highly advantageous for understanding FGF-stimulated signaling because these dynamics expose unknown signaling routes and improve the reliability of the predicted signaling network. Often, biological networks are deduced from literature without proper validation. For this purpose, logic-based dynamic modeling provides a suitable solution. Logic-based dynamic modeling of the FGF stimulations resulted in an overall low network error implying feasible network predictions. Mainly the PLCγ pathway showed higher predictive errors due to a higher sparsity of the network, partly due to limited insights into PLCγ signaling in the FGF context. This highlights the importance of further studying PLCγ signaling to understand its functions in FGF signaling (120).
The dynamic modeling highlighted differential and fine-tuned regulation of the MAPK/ERK pathway. Regulating phosphorylations of the RAF kinases indicate that FGF2 stimulation is directed via BRAF, while FGF4 stimulation is directed via ARAF. Literature on RAF kinase family regulation by FGFs is limited; however, understanding RAF regulation is essential because different RAF kinases perform different biological functions (121, 122). Moreover, understanding RAF signaling provides targeted insights that can be exploited to successfully deploy RAF-specific inhibitors in various diseases, such as cancer (123). For example, Metzner et al. show that FGF-driven melanoma is, in some cases, sensitive to the BRAF inhibitor RG7204 (124).
To conclude, this study highlights the differential signaling of FGFs and adopts existing logic-based dynamic modeling techniques to direct, strengthen, and increase the discovered biological knowledge.
Data availability
The mass spectrometry proteomics data are available via ProteomeXchange with identifier PXD038836:
(Username: reviewer_pxd038836@ebi.ac.uk; Password: CCMgJt8J)
The proteomics spectra can be viewed with MS-Viewer (https://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer) using the identifier: zm2mbcdjk1
The untargeted phosphoproteomics data are available via ProteomeXchange with identifier PXD038808:
(Username:reviewer_pxd038808@ebi.ac.uk; Password: mYXS1PfR)
The phosphoproteomics spectra can be viewed with MS-Viewer (https://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer) using the identifier: qyxvxuqbss
The targeted kinome data are available via ProteomeXchange with identifier PXD039005:
(Username: panorama+reviewer164@proteinms.net; Password: AIheltWY)
The used R scripts are available at: https://github.com/TVeth/FGF_Veth_2023
Supplemental data
This article contains supplemental data.
Conflicts of interest
The authors declare that they have no conflict of interest with the contents of this article.
Acknowledgments
We thank Mara Diks and Suzan Thijssen (Utrecht University) for their guidance with the qPCR experiments. We thank Jennifer Haworth and Joseph Parsons (CF team) for providing qPCR sequences. This work has been supported by EPIC-XS, project number 823839, funded by the Horizon 2020 programme of the European Union and the NWO funded Netherlands Proteomics Centre through the National Road Map for Large-scale Infrastructures program X-Omics, Project 184.034.019. Research in CF lab is supported by the Wellcome Trust (107636/Z/15/Z and 107636/Z/15/A), the Biotechnology and Biological Sciences Research Council (BB/R015864/1), and Medical Research Council (MR/T016043/1).
Author contributions
T. S. V. and M. A. conceptualization; T. S. V. and C. F. methodology; T. S. V. data curation; T. S. V. formal analysis; T. S. V. investigation; T. S. V. resources; T. S. V. software; T. S. V. visualization; T. S. V. and M. A. writing–original draft; T. S. F., C. F., A. J. R. H., and M. A. writing–review and editing; A. J. R. H. and M. A. supervision; A. J. R. H. and M. A. funding acquisition.
Supplementary Data
References
- 1.Ornitz D.M., Itoh N. New developments in the biology of fibroblast growth factors. Wires Mech. Dis. 2022;14 doi: 10.1002/wsbm.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarabipour S., Hristova K. Mechanism of FGF receptor dimerization and activation. Nat. Commun. 2016;7 doi: 10.1038/ncomms10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su N., Jin M., Chen L. Role of FGF/FGFR signaling in skeletal development and homeostasis: learning from mouse models. Bone Res. 2014;2 doi: 10.1038/boneres.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuro-o M. The Klotho proteins in health and disease. Nat. Rev. Nephrol. 2019;15:27–44. doi: 10.1038/s41581-018-0078-3. [DOI] [PubMed] [Google Scholar]
- 5.Spivak-Kroizman T., Lemmon M.A., Dikic I., Ladbury J.E., Pinchasi D., Huang J., et al. Heparin-induced oligomerization of FGF molecules is responsible for FGF receptor dimerization, activation, and cell proliferation. Cell. 1994;79:1015–1024. doi: 10.1016/0092-8674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 6.Chen L. Roles of FGF signaling in skeletal development and human genetic diseases. Front. Biosci. 2005;10:1961. doi: 10.2741/1671. [DOI] [PubMed] [Google Scholar]
- 7.Xie Y., Su N., Yang J., Tan Q., Huang S., Jin M., et al. FGF/FGFR signaling in health and disease. Signal Transduct. Target. Ther. 2020;5:181. doi: 10.1038/s41392-020-00222-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francavilla C., O’Brien C.S. Fibroblast growth factor receptor signalling dysregulation and targeting in breast cancer. Open Biol. 2022;12 doi: 10.1098/rsob.210373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korc M., Friesel R. The role of fibroblast growth factors in tumor growth. Curr. Cancer Drug Targets. 2009;9:639–651. doi: 10.2174/156800909789057006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Presta M., Chiodelli P., Giacomini A., Rusnati M., Ronca R. Fibroblast growth factors (FGFs) in cancer: FGF traps as a new therapeutic approach. Pharmacol. Ther. 2017;179:171–187. doi: 10.1016/j.pharmthera.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y., Wu C., Lu G., Hu Z., Chen Q., Du X. FGF/FGFR signaling pathway involved resistance in various cancer types. J. Cancer. 2020;11:2000–2007. doi: 10.7150/jca.40531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giulianelli S., Riggio M., Guillardoy T., Pérez Piñero C., Gorostiaga M.A., Sequeira G., et al. FGF2 induces breast cancer growth through ligand-independent activation and recruitment of ERα and PRBΔ4 isoform to MYC regulatory sequences. Int. J. Cancer. 2019;145:1874–1888. doi: 10.1002/ijc.32252. [DOI] [PubMed] [Google Scholar]
- 13.Sharpe R., Pearson A., Herrera-Abreu M.T., Johnson D., Mackay A., Welti J.C., et al. FGFR signaling promotes the growth of triple-negative and basal-like breast cancer cell lines both In Vitro and In Vivo. Clin. Cancer Res. 2011;17:5275–5286. doi: 10.1158/1078-0432.CCR-10-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsson E., Waltersson M.A., Bostner J., Pérez-Tenorio G., Olsson B., Hallbeck A.-L., et al. High-resolution genomic analysis of the 11q13 amplicon in breast cancers identifies synergy with 8p12 amplification, involving the mTOR targets S6K2 and 4EBP1. Genes. Chromosomes Cancer. 2011;50:775–787. doi: 10.1002/gcc.20900. [DOI] [PubMed] [Google Scholar]
- 15.Wang W., Chen T., Li H., Chen Y., Wu Z., Feng T., et al. Screening a novel FGF3 antagonist peptide with anti-tumor effects on breast cancer from a phage display library. Mol. Med. Rep. 2015;12:7051–7058. doi: 10.3892/mmr.2015.4248. [DOI] [PubMed] [Google Scholar]
- 16.Zhang C., Yang T., Jiang H. miR-511 inhibits proliferation and metastasis of breast cancer cells by targeting FGF4. J. Gene Med. 2020;22 doi: 10.1002/jgm.3168. [DOI] [PubMed] [Google Scholar]
- 17.Zhao X., Xu F., Dominguez N.P., Xiong Y., Xiong Z., Peng H., et al. FGFR4 provides the conduit to facilitate FGF19 signaling in breast cancer progression. Mol. Carcinog. 2018;57:1616–1625. doi: 10.1002/mc.22884. [DOI] [PubMed] [Google Scholar]
- 18.Abolhassani A., Riazi G.H., Azizi E., Amanpour S., Muhammadnejad S., Haddadi M., et al. FGF10: type III epithelial mesenchymal transition and invasion in breast cancer cell lines. J. Cancer. 2014;5:537–547. doi: 10.7150/jca.7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dailey L., Ambrosetti D., Mansukhani A., Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson H.R., Smith M.P., Francavilla C. Fibroblast Growth Factor Receptors (FGFRs) and noncanonical partners in cancer signaling. Cells. 2021;10:1201. doi: 10.3390/cells10051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ornitz D.M., Xu J., Colvin J.S., McEwen D.G., MacArthur C.A., Coulier F., et al. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 22.Touat M., Ileana E., Postel-Vinay S., André F., Soria J.-C. Targeting FGFR signaling in cancer. Clin. Cancer Res. 2015;21:2684–2694. doi: 10.1158/1078-0432.CCR-14-2329. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X., Ibrahimi O.A., Olsen S.K., Umemori H., Mohammadi M., Ornitz D.M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh M., Nakagama H. FGF Receptors: cancer biology and therapeutics: cancer biology and therapeutics on FGF receptors. Med. Res. Rev. 2014;34:280–300. doi: 10.1002/med.21288. [DOI] [PubMed] [Google Scholar]
- 25.Tomita H., Tanaka K., Hirata A., Okada H., Imai H., Shirakami Y., et al. Inhibition of FGF10-ERK signal activation suppresses intraductal papillary neoplasm of the bile duct and its associated carcinomas. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108772. [DOI] [PubMed] [Google Scholar]
- 26.Nolen B., Taylor S., Ghosh G. Regulation of protein kinases: controlling activity through activation segment conformation. Mol. Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 27.Schmidlin T., Debets D.O., van Gelder C.A.G.H., Stecker K.E., Rontogianni S., van den Eshof B.L., et al. High-throughput assessment of kinome-wide activation states. Cell Syst. 2019;9:366–374.e5. doi: 10.1016/j.cels.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gurzu S., Kobori L., Fodor D., Jung I. Epithelial mesenchymal and endothelial mesenchymal transitions in hepatocellular carcinoma: a review. Biomed Res. Int. 2019;2019 doi: 10.1155/2019/2962580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramos C., Becerril C., Montaño M., García-De-Alba C., Ramírez R., Checa M., et al. FGF-1 reverts epithelial-mesenchymal transition induced by TGF-β1 through MAPK/ERK kinase pathway. Am. J. Physiol. Lung Cell. Mol. Physiol. 2010;299:L222–L231. doi: 10.1152/ajplung.00070.2010. [DOI] [PubMed] [Google Scholar]
- 30.Liang C.-C., Park A.Y., Guan J.-L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 31.Suarez-Arnedo A., Torres Figueroa F., Clavijo C., Arbeláez P., Cruz J.C., Muñoz-Camargo C. An image J plugin for the high throughput image analysis of in vitro scratch wound healing assays. PLoS One. 2020;15 doi: 10.1371/journal.pone.0232565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Post H., Penning R., Fitzpatrick M.A., Garrigues L.B., Wu W., MacGillavry H.D., et al. Robust, sensitive, and automated phosphopeptide enrichment optimized for low sample amounts applied to primary hippocampal neurons. J. Proteome Res. 2017;16:728–737. doi: 10.1021/acs.jproteome.6b00753. [DOI] [PubMed] [Google Scholar]
- 34.Pino L.K., Searle B.C., Bollinger J.G., Nunn B., MacLean B., MacCoss M.J. The skyline ecosystem: informatics for quantitative mass spectrometry proteomics. Mass Spectrom. Rev. 2020;39:229–244. doi: 10.1002/mas.21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tognetti M., Gabor A., Yang M., Cappelletti V., Windhager J., Rueda O.M., et al. Deciphering the signaling network of breast cancer improves drug sensitivity prediction. Cell Syst. 2021;12:401–418.e12. doi: 10.1016/j.cels.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Türei D., Korcsmáros T., Saez-Rodriguez J. OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods. 2016;13:966–967. doi: 10.1038/nmeth.4077. [DOI] [PubMed] [Google Scholar]
- 37.Terfve C., Cokelaer T., Henriques D., MacNamara A., Goncalves E., Morris M.K., et al. CellNOptR: a flexible toolkit to train protein signaling networks to data using multiple logic formalisms. BMC Syst. Biol. 2012;6:133. doi: 10.1186/1752-0509-6-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wittmann D.M., Krumsiek J., Saez-Rodriguez J., Lauffenburger D.A., Klamt S., Theis F.J. Transforming boolean models to continuous models: methodology and application to T-cell receptor signaling. BMC Syst. Biol. 2009;3:98. doi: 10.1186/1752-0509-3-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eduati F., Doldàn-Martelli V., Klinger B., Cokelaer T., Sieber A., Kogera F., et al. Drug resistance mechanisms in colorectal cancer dissected with cell type–specific dynamic logic models. Cancer Res. 2017;77:3364–3375. doi: 10.1158/0008-5472.CAN-17-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egea J.A., Henriques D., Cokelaer T., Villaverde A.F., MacNamara A., Danciu D.-P., et al. MEIGO: an open-source software suite based on metaheuristics for global optimization in systems biology and bioinformatics. BMC Bioinformatics. 2014;15:136. doi: 10.1186/1471-2105-15-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 42.Choi M., Chang C.-Y., Clough T., Broudy D., Killeen T., MacLean B., et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–2526. doi: 10.1093/bioinformatics/btu305. [DOI] [PubMed] [Google Scholar]
- 43.Korsten S.G.P.J., Peracic L., van Groeningen L.M.B., Diks M.A.P., Vromans H., Garssen J., et al. Butyrate prevents induction of CXCL10 and Non-Canonical IRF9 expression by activated human intestinal epithelial cells via HDAC inhibition. Int. J. Mol. Sci. 2022;23:3980. doi: 10.3390/ijms23073980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor S.C., Nadeau K., Abbasi M., Lachance C., Nguyen M., Fenrich J. The Ultimate qPCR experiment: producing publication quality, reproducible data the first time. Trends Biotechnol. 2019;37:761–774. doi: 10.1016/j.tibtech.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Eswarakumar V.P., Lax I., Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Wolf I., Levanon-Cohen S., Bose S., Ligumsky H., Sredni B., Kanety H., et al. Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene. 2008;27:7094–7105. doi: 10.1038/onc.2008.292. [DOI] [PubMed] [Google Scholar]
- 47.Ornitz D.M., Itoh N. The fibroblast growth factor signaling pathway. Wires Dev. Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azami T., Waku T., Matsumoto K., Jeon H., Muratani M., Kawashima A., et al. Klf5 maintains the balance of primitive endoderm versus epiblast specification during mouse embryonic development by suppression of Fgf4. Development. 2017;144:3706–3718. doi: 10.1242/dev.150755. [DOI] [PubMed] [Google Scholar]
- 49.Bockorny B., Rusan M., Chen W., Liao R.G., Li Y., Piccioni F., et al. RAS–MAPK reactivation facilitates acquired resistance in FGFR1 -amplified lung cancer and underlies a rationale for upfront FGFR–MEK blockade. Mol. Cancer Ther. 2018;17:1526–1539. doi: 10.1158/1535-7163.MCT-17-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho K.-W., Cai J., Kim H.-Y., Hosoya A., Ohshima H., Choi K.-Y., et al. ERK activation is involved in tooth development via FGF10 signaling. J. Exp. Zoolog. B Mol. Dev. Evol. 2009;312:901–911. doi: 10.1002/jez.b.21309. [DOI] [PubMed] [Google Scholar]
- 51.Kunath T., Saba-El-Leil M.K., Almousailleakh M., Wray J., Meloche S., Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 52.Lovicu F.J., McAvoy J.W. FGF-induced ERK activation in the lens. Development. 2001;128:10. doi: 10.1242/dev.128.24.5075. [DOI] [PubMed] [Google Scholar]
- 53.Shalaby A.A., Presneau N., Idowu B.D., Thompson L., Briggs T.R., Tirabosco R., et al. Analysis of the fibroblastic growth factor receptor-RAS/RAF/MEK/ERK-ETS2/brachyury signalling pathway in chordomas. Mod. Pathol. 2009;22:996–1005. doi: 10.1038/modpathol.2009.63. [DOI] [PubMed] [Google Scholar]
- 54.Lavoie H., Gagnon J., Therrien M. ERK signalling: a master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 2020;21:607–632. doi: 10.1038/s41580-020-0255-7. [DOI] [PubMed] [Google Scholar]
- 55.Aoki K., Kumagai Y., Sakurai A., Komatsu N., Fujita Y., Shionyu C., et al. Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol. Cell. 2013;52:529–540. doi: 10.1016/j.molcel.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 56.Raina D., Fabris F., Morelli L.G., Schröter C. Intermittent ERK oscillations downstream of FGF in mouse embryonic stem cells. Development. 2022;149:dev199710. doi: 10.1242/dev.199710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anjum R., Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 58.Houles T., Roux P.P. Defining the role of the RSK isoforms in cancer. Semin. Cancer Biol. 2018;48:53–61. doi: 10.1016/j.semcancer.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 59.Romeo Y., Zhang X., Roux P.P. Regulation and function of the RSK family of protein kinases. Biochem. J. 2012;441:553–569. doi: 10.1042/BJ20110289. [DOI] [PubMed] [Google Scholar]
- 60.Maher P. p38 Mitogen-activated protein kinase activation is required for fibroblast growth factor-2-stimulated cell proliferation but not differentiation. J. Biol. Chem. 1999;274:17491–17498. doi: 10.1074/jbc.274.25.17491. [DOI] [PubMed] [Google Scholar]
- 61.New L. PRAK, a novel protein kinase regulated by the p38 MAP kinase. EMBO J. 1998;17:3372–3384. doi: 10.1093/emboj/17.12.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuo C.-C., Savage N.S., Chen H., Wu C.-F., Zyla T.R., Lew D.J. Inhibitory GEF phosphorylation provides negative feedback in the yeast polarity circuit. Curr. Biol. 2014;24:753–759. doi: 10.1016/j.cub.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith M.P., Ferguson H.R., Ferguson J., Zindy E., Kowalczyk K.M., Kedward T., et al. Reciprocal priming between receptor tyrosine kinases at recycling endosomes orchestrates cellular signalling outputs. EMBO J. 2021;40 doi: 10.15252/embj.2020107182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun P., Yoshizuka N., New L., Moser B.A., Li Y., Liao R., et al. PRAK is essential for ras-Induced senescence and tumor suppression. Cell. 2007;128:295–308. doi: 10.1016/j.cell.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y., Wang W., Wu H., Zhou Y., Qin X., Wang Y., et al. The essential role of PRAK in tumor metastasis and its therapeutic potential. Nat. Commun. 2021;12:1736. doi: 10.1038/s41467-021-21993-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Desnoyers L.R., Pai R., Ferrando R.E., Hötzel K., Le T., Ross J., et al. Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene. 2008;27:85–97. doi: 10.1038/sj.onc.1210623. [DOI] [PubMed] [Google Scholar]
- 67.Shi W., Peyrot S.M., Munro E., Levine M. FGF3 in the floor plate directs notochord convergent extension in the Ciona tadpole. Development. 2009;136:23–28. doi: 10.1242/dev.029157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shinya M., Koshida S., Sawada A., Kuroiwa A., Takeda H. Fgf signalling through MAPK cascade is required for development of the subpallial telencephalon in zebrafish embryos. Development. 2001;128:4153–4164. doi: 10.1242/dev.128.21.4153. [DOI] [PubMed] [Google Scholar]
- 69.Teng Y., Guo B., Mu X., Liu S. KIF26B promotes cell proliferation and migration through the FGF2/ERK signaling pathway in breast cancer. Biomed. Pharmacother. 2018;108:766–773. doi: 10.1016/j.biopha.2018.09.036. [DOI] [PubMed] [Google Scholar]
- 70.Koledova Z., Sumbal J., Rabata A., de La Bourdonnaye G., Chaloupkova R., Hrdlickova B., et al. Fibroblast growth factor 2 protein stability provides decreased dependence on heparin for induction of FGFR signaling and alters ERK signaling dynamics. Front. Cell Dev. Biol. 2019;7:331. doi: 10.3389/fcell.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun Y., Fan X., Zhang Q., Shi X., Xu G., Zou C. Cancer-associated fibroblasts secrete FGF-1 to promote ovarian proliferation, migration, and invasion through the activation of FGF-1/FGFR4 signaling. Tumor Biol. 2017;39 doi: 10.1177/1010428317712592. [DOI] [PubMed] [Google Scholar]
- 72.Cauthron R.D., Carter K.B., Liauw S., Steinberg R.A. Physiological phosphorylation of protein kinase A at Thr-197 Is by a protein kinase a kinase. Mol. Cell. Biol. 1998;18:1416–1423. doi: 10.1128/mcb.18.3.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Drewes G., Ebneth A., Preuss U., Mandelkow E.-M., Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 74.Sonntag T., Moresco J.J., Vaughan J.M., Matsumura S., Yates J.R., Montminy M. Analysis of a cAMP regulated coactivator family reveals an alternative phosphorylation motif for AMPK family members. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qu J., Cammarano M.S., Shi Q., Ha K.C., de Lanerolle P., Minden A. Activated PAK4 regulates cell adhesion and anchorage-independent growth. Mol. Cell. Biol. 2001;21:3523–3533. doi: 10.1128/MCB.21.10.3523-3533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramos-Alvarez I., Jensen R.T. P21-activated kinase 4 in pancreatic acinar cells is activated by numerous gastrointestinal hormones/neurotransmitters and growth factors by novel signaling, and its activation stimulates secretory/growth cascades. Am. J. Physiol. Gastrointest. Liver Physiol. 2018;315:G302–G317. doi: 10.1152/ajpgi.00005.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thillai K., Lam H., Sarker D., Wells C.M. Deciphering the link between PI3K and PAK: an opportunity to target key pathways in pancreatic cancer? Oncotarget. 2017;8:14173–14191. doi: 10.18632/oncotarget.13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Won S.-Y., Park J.-J., Shin E.-Y., Kim E.-G. PAK4 signaling in health and disease: defining the PAK4–CREB axis. Exp. Mol. Med. 2019;51:1–9. doi: 10.1038/s12276-018-0204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Katoh M., Katoh M. Cross-talk of WNT and FGF signaling pathways at GSK3ß to regulate ß-catenin and SNAIL signaling cascades. Cancer Biol. Ther. 2006;5:1059–1064. doi: 10.4161/cbt.5.9.3151. [DOI] [PubMed] [Google Scholar]
- 80.Zhou B.P., Deng J., Xia W., Xu J., Li Y.M., Gunduz M., et al. Dual regulation of Snail by GSK-3β-mediated phosphorylation in control of epithelial–mesenchymal transition. Nat. Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 81.Grada A., Otero-Vinas M., Prieto-Castrillo F., Obagi Z., Falanga V. Research techniques made simple: analysis of collective cell migration using the wound healing assay. J. Invest. Dermatol. 2017;137:e11–e16. doi: 10.1016/j.jid.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 82.Yan C., Grimm W.A., Garner W.L., Qin L., Travis T., Tan N., et al. Epithelial to mesenchymal transition in human skin wound healing is induced by tumor necrosis factor-α through bone morphogenic protein-2. Am. J. Pathol. 2010;176:2247–2258. doi: 10.2353/ajpath.2010.090048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vasaikar S.V., Deshmukh A.P., den Hollander P., Addanki S., Kuburich N.A., Kudaravalli S., et al. EMTome: a resource for pan-cancer analysis of epithelial-mesenchymal transition genes and signatures. Br. J. Cancer. 2021;124:259–269. doi: 10.1038/s41416-020-01178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat. Rev. Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 85.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim H.-J., Kim J.-H., Bae S.-C., Choi J.-Y., Kim H.-J., Ryoo H.-M. The protein kinase C pathway plays a central role in the fibroblast growth factor-stimulated expression and transactivation activity of Runx2. J. Biol. Chem. 2003;278:319–326. doi: 10.1074/jbc.M203750200. [DOI] [PubMed] [Google Scholar]
- 87.Lima F., Niger C., Hebert C., Stains J.P. Connexin43 potentiates osteoblast responsiveness to fibroblast growth factor 2 via a protein kinase C-Delta/Runx2–dependent mechanism. Mol. Biol. Cell. 2009;20:2697–2708. doi: 10.1091/mbc.E08-10-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mohammadi M., Jaye M., Rubinstein M., Schlessinger J. A tyrosine-phosphorylated carboxy-terminal peptide of the fibroblast growth factor receptor (Flg) is a binding site for the SH2 domain of phospholipase C-gamma 1. Mol. Cell. Biol. 1991;11:5068–5078. doi: 10.1128/mcb.11.10.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Niger C., Hebert C., Stains J.P. Interaction of connexin43 and protein kinase C-delta during FGF2 signaling. BMC Biochem. 2010;11:14. doi: 10.1186/1471-2091-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ranieri D., Nanni M., Persechino F., Torrisi M.R., Belleudi F. Role of PKCε in the epithelial-mesenchymal transition induced by FGFR2 isoform switch. Cell Commun. Signal. 2020;18:76. doi: 10.1186/s12964-020-00582-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Szybowska P., Kostas M., Wesche J., Haugsten E.M., Wiedlocha A. Negative regulation of FGFR (Fibroblast Growth Factor Receptor) signaling. Cells. 2021;10:1342. doi: 10.3390/cells10061342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kelher M.R., McLaughlin N.J.D., Banerjee A., Elzi D.J., Gamboni F., Khan S.Y., et al. LysoPCs induce Hck- and PKCδ-mediated activation of PKCγ causing p47 phox phosphorylation and membrane translocation in neutrophils. J. Leukoc. Biol. 2017;101:261–273. doi: 10.1189/jlb.3A0813-420RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang Z., Tan Y., Gu J., Liu Y., Song L., Niu J., et al. Uncoupling the mitogenic and metabolic functions of FGF1 by Tuning FGF1-FGF receptor dimer stability. Cell Rep. 2017;20:1717–1728. doi: 10.1016/j.celrep.2017.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kiyatkin A., van Alderwerelt van Rosenburgh I.K., Klein D.E., Lemmon M.A. Kinetics of receptor tyrosine kinase activation define ERK signaling dynamics. Sci. Signal. 2020;13:eaaz5267. doi: 10.1126/scisignal.aaz5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morris M.K., Saez-Rodriguez J., Sorger P.K., Lauffenburger D.A. Logic-based models for the analysis of cell signaling networks. Biochemistry. 2010;49:3216–3224. doi: 10.1021/bi902202q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gotoh N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 2008;99:1319–1325. doi: 10.1111/j.1349-7006.2008.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hadari Y.R., Kouhara H., Lax I., Schlessinger J. Binding of Shp2 tyrosine phosphatase to FRS2 Is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol. Cell. Biol. 1998;18:3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang W., Klaman L.D., Chen B., Araki T., Harada H., Thomas S.M., et al. An Shp2/SFK/Ras/Erk signaling pathway controls trophoblast stem cell survival. Dev. Cell. 2006;10:317–327. doi: 10.1016/j.devcel.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 99.Looi C.-K., Hii L.-W., Ngai S.C., Leong C.-O., Mai C.-W. The Role of Ras-Associated Protein 1 (Rap1) in Cancer: bad Actor or Good Player? Biomedicines. 2020;8:334. doi: 10.3390/biomedicines8090334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schmitt J.M., Stork P.J.S. PKA Phosphorylation of Src Mediates cAMP’s inhibition of cell growth via Rap1. Mol. Cell. 2002;9:85–94. doi: 10.1016/s1097-2765(01)00432-4. [DOI] [PubMed] [Google Scholar]
- 101.Stokman G., Qin Y., Booij T.H., Ramaiahgari S., Lacombe M., Dolman M.E.M., et al. Epac-rap signaling reduces oxidative stress in the tubular epithelium. J. Am. Soc. Nephrol. 2014;25:1474–1485. doi: 10.1681/ASN.2013070679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Daumke O., Weyand M., Chakrabarti P.P., Vetter I.R., Wittinghofer A. The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature. 2004;429:197–201. doi: 10.1038/nature02505. [DOI] [PubMed] [Google Scholar]
- 103.de Rooij J., Rehmann H., van Triest M., Cool R.H., Wittinghofer A., Bos J.L. Mechanism of regulation of the Epac Family of cAMP-dependent RapGEFs. J. Biol. Chem. 2000;275:20829–20836. doi: 10.1074/jbc.M001113200. [DOI] [PubMed] [Google Scholar]
- 104.Fukuyama T., Ogita H., Kawakatsu T., Fukuhara T., Yamada T., Sato T., et al. Involvement of the c-Src-Crk-C3G-Rap1 Signaling in the Nectin-induced Activation of Cdc42 and formation of adherens junctions. J. Biol. Chem. 2005;280:815–825. doi: 10.1074/jbc.M411099200. [DOI] [PubMed] [Google Scholar]
- 105.Fukuyama T., Ogita H., Kawakatsu T., Inagaki M., Takai Y. Activation of Rac by cadherin through the c-Src–Rap1–phosphatidylinositol 3-kinase–Vav2 pathway. Oncogene. 2006;25:8–19. doi: 10.1038/sj.onc.1209010. [DOI] [PubMed] [Google Scholar]
- 106.Rehmann H., Prakash B., Wolf E., Rueppel A., de Rooij J., Bos J.L., et al. Structure and regulation of the cAMP-binding domains of Epac2. Nat. Struct. Biol. 2003;10:26–32. doi: 10.1038/nsb878. [DOI] [PubMed] [Google Scholar]
- 107.Rehmann H., Das J., Knipscheer P., Wittinghofer A., Bos J.L. Structure of the cyclic-AMP-responsive exchange factor Epac2 in its auto-inhibited state. Nature. 2006;439:625–628. doi: 10.1038/nature04468. [DOI] [PubMed] [Google Scholar]
- 108.Maurer G., Tarkowski B., Baccarini M. Raf kinases in cancer–roles and therapeutic opportunities. Oncogene. 2011;30:3477–3488. doi: 10.1038/onc.2011.160. [DOI] [PubMed] [Google Scholar]
- 109.Marquette A., André J., Bagot M., Bensussan A., Dumaz N. ERK and PDE4 cooperate to induce RAF isoform switching in melanoma. Nat. Struct. Mol. Biol. 2011;18:584–591. doi: 10.1038/nsmb.2022. [DOI] [PubMed] [Google Scholar]
- 110.Baljuls A., Schmitz W., Mueller T., Zahedi R.P., Sickmann A., Hekman M., et al. Positive regulation of A-RAF by phosphorylation of isoform-specific hinge segment and identification of novel phosphorylation sites. J. Biol. Chem. 2008;283:27239–27254. doi: 10.1074/jbc.M801782200. [DOI] [PubMed] [Google Scholar]
- 111.Dhillon A.S., Yip Y.Y., Grindlay G.J., Pakay J.L., Dangers M., Hillmann M., et al. The C-terminus of Raf-1 acts as a 14-3-3-dependent activation switch. Cell. Signal. 2009;21:1645–1651. doi: 10.1016/j.cellsig.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 112.Zang M., Gong J., Luo L., Zhou J., Xiang X., Huang W., et al. Characterization of Ser338 Phosphorylation for Raf-1 Activation. J. Biol. Chem. 2008;283:31429–31437. doi: 10.1074/jbc.M802855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dougherty M.K., Müller J., Ritt D.A., Zhou M., Zhou X.Z., Copeland T.D., et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol. Cell. 2005;17:215–224. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 114.Stuart S.A., Houel S., Lee T., Wang N., Old W.M., Ahn N.G. A Phosphoproteomic comparison of B-RAFV600E and MKK1/2 inhibitors in melanoma cells. Mol. Cell. Proteomics. 2015;14:1599–1615. doi: 10.1074/mcp.M114.047233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shin S.-Y., Rath O., Choo S.-M., Fee F., McFerran B., Kolch W., et al. Positive- and negative-feedback regulations coordinate the dynamic behavior of the Ras-Raf-MEK-ERK signal transduction pathway. J. Cell Sci. 2009;122:425–435. doi: 10.1242/jcs.036319. [DOI] [PubMed] [Google Scholar]
- 116.Loog M., Morgan D.O. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 117.Valverde J., Altelaar M., Dubra G., van den Toorn H.W.P., van Mierlo G., Vermeulen M., et al. A CDK-mediated phosphorylation switch of disordered protein condensation. Res. Square. 2022 doi: 10.21203/rs.3.rs-1370895/v1. [preprint] [DOI] [Google Scholar]
- 118.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 119.Watson J., Francavilla C. Regulation of FGF10 signaling in development and disease. Front. Genet. 2018;9:500. doi: 10.3389/fgene.2018.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brewer J.R., Mazot P., Soriano P. Genetic insights into the mechanisms of Fgf signaling. Genes Dev. 2016;30:751–771. doi: 10.1101/gad.277137.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dumaz N. Mechanism of RAF isoform switching induced by oncogenic RAS in melanoma. Small GTPases. 2011;2:289–292. doi: 10.4161/sgtp.2.5.17814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wellbrock C., Karasarides M., Marais R. The RAF proteins take centre stage. Nat. Rev. Mol. Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 123.Saini K.S., Loi S., de Azambuja E., Metzger-Filho O., Saini M.L., Ignatiadis M., et al. Targeting the PI3K/AKT/mTOR and Raf/MEK/ERK pathways in the treatment of breast cancer. Cancer Treat. Rev. 2013;39:935–946. doi: 10.1016/j.ctrv.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 124.Metzner T., Bedeir A., Held G., Peter-Vörösmarty B., Ghassemi S., Heinzle C., et al. Fibroblast growth factor receptors as therapeutic targets in human melanoma: synergism with BRAF Inhibition. J. Invest. Dermatol. 2011;131:2087–2095. doi: 10.1038/jid.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data are available via ProteomeXchange with identifier PXD038836:
(Username: reviewer_pxd038836@ebi.ac.uk; Password: CCMgJt8J)
The proteomics spectra can be viewed with MS-Viewer (https://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer) using the identifier: zm2mbcdjk1
The untargeted phosphoproteomics data are available via ProteomeXchange with identifier PXD038808:
(Username:reviewer_pxd038808@ebi.ac.uk; Password: mYXS1PfR)
The phosphoproteomics spectra can be viewed with MS-Viewer (https://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer) using the identifier: qyxvxuqbss
The targeted kinome data are available via ProteomeXchange with identifier PXD039005:
(Username: panorama+reviewer164@proteinms.net; Password: AIheltWY)
The used R scripts are available at: https://github.com/TVeth/FGF_Veth_2023