Abstract
Hepatic sinusoids are uniquely organized structures that help maintain a spectrum of hepatic functions. Although several in vitro liver models have been developed to replicate liver sinusoids, most of these platforms require complex, multi-step fabrication methods making it difficult to achieve truly three-dimensional (3D) channel geometries. In this study, a single-step bioprinting technique was demonstrated to simultaneously print a chip platform and develop a perfusable vascularized liver sinusoid in vitro model. The integrated system uses a co-axial-based bioprinting approach to develop a liver sinusoid-like model that consists of a sacrificial core compartment containing a perfusable pre-vascular structure and an alginate-collagen-based shell compartment containing hepatocytes. The lumen-based perfusable 3D liver sinusoid-on-a-chip (LSOC-P) demonstrated significantly better hepatocyte viability, proliferation, and liver-specific gene and protein expression compared to a 3D hepatocyte-based core/shell model with static media and the standard hepatocyte-based 2D sandwich culture system. A drug toxicity evaluation of hepatotoxins highlighted the comparatively higher sensitivity of the LSOC system with a close estimation of the therapeutic range of safe drug concentrations for humans. In conclusion, the current findings indicate that the combinatorial single-step co-axial bioprinting technique is a promising fabrication approach for the development of a perfusable LSOC platform for drug screening applications.
Keywords: Liver-on-a-chip, Bioprinting, Single-step fabrication, Liver sinusoid, Drug screening
Graphical abstract
1. Introduction
Preclinical assessments, such as two-dimensional (2D)-based in vitro cell tests and animal trials, are part of the current drug-discovery process. However, it is well acknowledged in the scientific community that the current testing techniques are inefficient [[1], [2], [3]]. Apart from the significant time and resource requirements of animal model-based studies, they have limited translational efficacy because of the interspecies differences between humans and animals. Additionally, increasing ethical concerns over the use of animals for biological studies have led to a reduction in animal experiments globally [1]. As an alternative, the development of biological in vitro models of human tissues and organs has emerged as a popular and noteworthy avenue for replicating human disease conditions as well as for screening and evaluating potential drug candidates.
The liver is one of the most important organs in the human body [4]. It is a highly complex organ composed of hexagonal functional units called hepatic lobules. These lobules consist of hepatic sinusoids that allow blood to flow from the portal artery and portal vein toward the central vein. Sinusoids are responsible for most of the important functions of the liver, including gas exchange, nutrient absorption, detoxification, and nutrient storage. These functions are performed by highly diversified cells called sinusoidal cells, which include endothelial cells lining the sinusoidal channels flanked by hepatocytes. Because of its major roles in various metabolic functions and the detoxification of endogenous and exogenous compounds, the liver is one of the main organs susceptible to damage because of pharmaceutical drugs [5]. Over the past few years, considerable efforts have been dedicated to establishing in vitro liver systems that can mimic in vivo liver physiology for toxicity screening, thereby helping to comprehend the associated drug metabolism. A 2D liver-on-a-chip with different cell types that imitate liver sinusoids was created using soft lithography and polydimethylsiloxane (PDMS) molding [[6], [7], [8]]. Another technique to fabricate a liver-on-a-chip was developed by applying liver spheroids to premade chip platforms with increased hepatocyte activity [9,10]. However, the organization of cells and extracellular matrix (ECM) components in ordered 2D and 3D microscale structures, which is essential for the creation of living tissue, is challenging because of manufacturing-related limitations [11]. In this regard, bioprinting techniques have greatly enhanced the choices available for recreating the liver microarchitecture.
Bioprinting is based on the additive manufacturing process, which facilitates the creation of patterned cell-laden structures by utilizing various bioinks that are printed following a predefined design. In most of the reported works on bioprinting hepatocytes, the extrusion-based bioprinting technique has been used for developing hepatic models [12,13]. Wang et al. used the extrusion bioprinting technique to create hepatocyte-laden gelatin constructs that exhibited viability and functional characteristics [14]. Since then, many attempts have been made to develop liver models using various types of bioinks, such as chitosan [15], collagen [16], alginate [17], and fibrin [18], as well as extracellular matrix [19], thus indicating the suitability of the bioprinting approach. Recent studies have used combinations of various cell types as bioinks to bioprint co-cultures of hepatocytes and non-parenchymal cells. For example, liver-on-a-chip with a vascular/biliary fluidic channel was developed using cell printing with multiple types of liver cells [20].
During the development of in vitro models, imitating the degree of complexity of liver sinusoid architecture is critical for achieving homeostasis of resident hepatocytes and sinusoid-specific functions. Researchers have tried to achieve this tissue complexity in the 3D model by using patterned co-cultures of hepatocytes with different non-parenchymal cells through successive printing of different bioinks. A few recent studies have used co-axial nozzle-based bioprinting that can simultaneously extrude two different cell-laden bioinks to form a core/shell co-culture construct. Genderen et al. used a co-axial system to develop an alginate/gelatin-based coiled perfusable microfiber to replicate kidney proximal convoluted tubules [21]. In a recent article, Singh et al. developed cell-printed glomerular-on-a-chip that recapitulates the human glomerular filtration barrier for the screening of nephrotoxic drugs [22]. This in vitro model was able to generate functional transporters and polarized monolayers. A pump-driven perfusable in vitro free-standing vascular model was constructed by Gao et al. using a co-axial cell-printing technique [23]. Upon maturation, this vascular model exhibited representative vascular functions along with directional angiogenesis and inflammatory responses. The same group also showed that coaxially-bioprinted veins could aid in ischemic limb repair in a rodent model. In their study, only cell-laden constructs were capable of limb salvage and were further improved by the inclusion of a statin drug [24]. A 3D in vitro liver model was co-axially developed by Taymor et al. to create a co-culture model with fibroblasts (murine NIH 3T3) loaded in the core and hepatocytes (human HepG2) enclosed in the shell [25]. However, not many such attempts have yet been made to develop an in vitro liver sinusoid model, which closely mimics the complex 3D physiological architecture of a liver sinusoid with a perfusable vascularized core. In this study, we have attempted to fabricate a 3D lumen-based perfusable liver sinusoid-on-a-chip (LSOC) in a single step that can imitate liver sinusoid structural and functional aspects.
In addition, in most studies related to the development of organ-on-a-chip models, separate steps are required to fabricate the chip platform and develop the hydrogel-based in vitro model. This process requires several preparatory stages and is, therefore, rather labor-intensive. Furthermore, the preparative stages and assembly of the chip platform with the 3D in vitro model increase the chances of contamination. Thus, by using a single-step bioprinting technique, the fabrication process can be minimized. This study revealed that the developed LSOC device exhibited increased hepatocyte growth and cluster formation with enhanced liver functionality. On treatment with various concentrations of hepatotoxic drugs, the LSOC demonstrated a noteworthy toxicity prediction along with a close estimation of therapeutic ranges of safe drug concentrations. The specificity of toxicity prediction of the LSOC was further verified by investigating various liver-specific functional aspects of the model (e.g., the expression of liver proteins and enzymes). Consequently, our results indicate that the single-step bioprinted perfusable LSOC device can be an effective potential platform for screening hepatotoxic drugs.
2. Materials and methods
2.1. Material preparation
Polyethylene vinyl acetate (PEVA, Polysciences Inc., Warrington, PA, USA) was used as the structural material for printing the chip platform on transparent sterilized glass slide plates. Sodium alginate (medium viscosity, Sigma), gelatin (porcine skin, type A, Sigma), and collagen type I (Dalim Tissen, Korea) were used as bioinks for cell printing. A 30 mg/ml gelatin bioink was prepared by dissolving gelatin (w/v) in endothelial growth medium (EGM, Lonza). Alginate bioink (40 mg/ml) was prepared by dissolving sodium alginate (w/v) in sterilized water overnight with constant stirring. Collagen foam was dissolved in 17.5 mM acetic acid to obtain a 25 mg/ml collagen solution. The final collagen solution with a concentration of 15 mg/ml was obtained after pH adjustment.
2.2. Cell maintenance
Human hepatocellular carcinoma (HepG2) cells were purchased from ATCC (USA). These HepG2 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Corning, USA) with 10% (v/v) fetal bovine serum (FBS, Corning, USA) and 1% (v/v) penicillin/streptomycin (P/S, Corning, USA) at 37 °C in a humidified 5% CO2 atmosphere. HUVECs were purchased from Lonza (Basal, Switzerland), cultured in complete EGM (EGM-2 BulletKit, Lonza), and maintained at 37 °C in a humidified 5% CO2 atmosphere. The medium was changed every alternate day.
2.3. Preparation of collagen-alginate composite bioink
The polymeric formulation of the collagen-alginate (CA) composite bioink was prepared by mixing different ratios of collagen (15 mg/ml) and alginate (40 mg/ml), namely C:A 1:1, 2:1 and 1:2, thoroughly to achieve a uniform distribution. The mixing was carried out using a magnetic stirrer for 15 min at 4 °C under sterilized conditions.
2.4. Physicochemical characterization of CA bioink
The rheological characteristics of the CA composite bioinks prepared with different C:A ratios were evaluated using a rheometer (Anton Paar). The viscosity values of the prepared composite bioinks were determined by performing a steady sweep analysis. The viscosity of the hydrogel was measured at 15 °C, which is the approximate temperature of the 3D cell printing process. The composite bioinks were crosslinked with 2% calcium chloride solution for 60 min at 37 °C. A dynamic frequency sweep analysis was conducted to measure the frequency-dependent storage (G′) and loss (G″) moduli of the crosslinked bioinks. The angular frequencies of the dynamic sweeps were 1–100 rad/s under a constant shear strain of 0.5% at 37 °C.
The porosity values of the crosslinked CA bioinks were measured using the absolute ethanol absorption technique [26]. The weight of the empty beaker was noted as W1. Approximately 1 g of CA bioink was placed in the beaker, and 10 ml of absolute ethanol was poured in slowly. The weight (W2) of the beaker containing the ethanol and bioink was noted. Furthermore, the weight of the beaker was measured after removing the CA bioink from the beaker (W3). It was assumed that the void spaces in the hydrogel were filled with absolute ethanol. Therefore, the volume of ethanol in the hydrogel pore was considered the pore volume. The porosity of the porous gel (P) was calculated using the following equations:
| (1) |
| (2) |
| (3) |
| (4) |
where ρh is the density of the CA bioink (g/cm3), ρe is the density of the absolute ethanol (g/cm3), Vh is the volume of the CA bioink (cm3), and Ve is the volume of absolute ethanol in the hydrogel pores (cm3).
The morphological characteristics of the CA bioinks were observed using a scanning electron microscope (JEOL) operated at 20 kV. Before this observation, the freeze-dried CA bioinks were mounted on stubs and sputter-coated with gold. The pore structures of the matrices were examined, and the pore sizes were determined using the NIH Image J software (n = 3/group).
2.5. Biological characterization of bioinks
An indirect cell counting method was used to determine the adhesion of HepG2 cells on the CA bioinks. Briefly, the suspended HepG2 cells (1 × 105) were seeded onto the crosslinked CA bioinks. The cell-seeded CA bioinks were placed on non-adhesive culture plates and transferred to a CO2 incubator. The plates were incubated for different durations, and the unadhered cells were collected and counted using a phase-contrast microscope and a Neubauer hemocytometer. The number of adhered cells was determined using the following equation:
| (5) |
The suspended HepG2 cells were mixed with various bioinks prepared using different C:A ratios, crosslinked using CaCl2 (2%), and cultured in complete DMEM (with 10% FBS) maintained at 37 °C in a humidified 5% CO2 atmosphere. The proliferation of HepG2 cells encapsulated in the CA bioinks was determined using a cell-counting kit – 8 (Dojindo, Korea). The viability of the HepG2 cells encapsulated in the CA bioinks was determined by staining them with calcein-AM. The cells were observed on the fifth day of cell encapsulation under a fluorescence microscope (Nikon, Japan).
2.6. Cell labelling and encapsulation
The cells (HepG2 and HUVEC) were labeled to determine their deposition and positions in the fabricated LSOC. To label the cells, each sample was incubated at 37 °C in a 5% CO2 incubator for 15 min with DiI (Cell tracker Red) and CDFDA (Cell tracker Green) fluorescent dyes. The green-stained HepG2 cells were encapsulated in the CA bioink, while the red-stained HUVECs were encapsulated in the 3% EGM-based gelatin bioink. A fluorescence microscope (Nikon, Japan) was used to visualize the cells.
2.7. Fabrication of lumen-based 3D liver sinusoid-on-a-chip
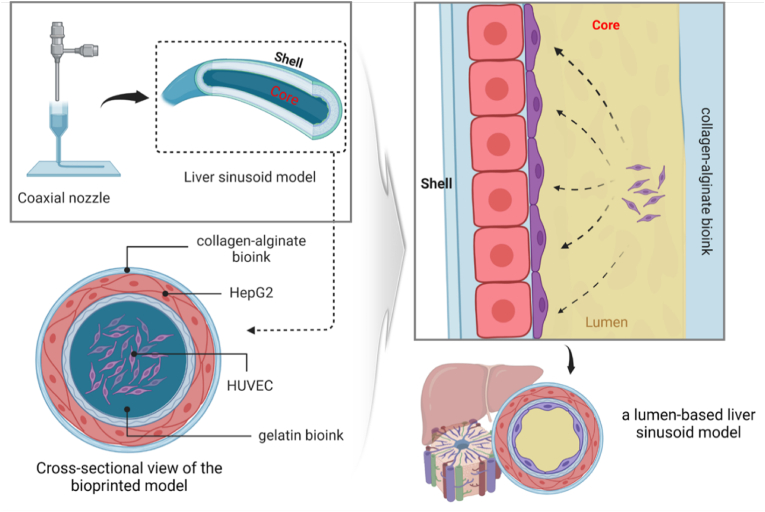
The in-house developed extrusion-based integrated 3D bioprinting system (ITBS) was used to construct a PEVA-based chip platform, followed by cell printing of the liver sinusoid 3D model directly onto the fabricated construct. Fig. 1 shows a schematic diagram of the steps involved in the LSOC printing process. To print the chip platform, the temperature-controlled dispensing module was loaded with PEVA polymer. To print the core/shell liver sinusoidal 3D model, we used a co-axial nozzle linked to two syringes. The syringe linked to the outer nozzle was loaded with HepG2-laden CA bioinks, whereas the syringe linked to the inner nozzle was loaded with a HUVEC-laden gelatin bioink. The cell density in the bioprinting process was 1 × 106 cells/ml for the HUVECs and 2 × 106 cells/ml for the HepG2 cells. The bioprinting process was carried out using the optimized parameters, where the dispensing pressures of the HUVEC- and HepG2-laden bioinks were 70 kPa and 50 kPa, respectively, and the printing speed was 80 mm/min. The printed 3D model was crosslinked using a CaCl2 solution.
Fig. 1.
Schematic illustration of extrusion bioprinting of the core/shell 3D liver sinusoid model using co-axial printing and the creation of a hollow lumen-based structure using bioinks. After the endothelial cell attachment, the bioink present at the center of the 3D model slowly flows out to create a lumen-structure. The figure was created with BioRender.com.
2.8. Culture condition of perfusable liver sinusoid-on-a-chip
The entire LSOC setup was transferred onto a rocker and cultured at 37 °C in a humidified 5% CO2 atmosphere. The free movement of the aforementioned media through the endothelial-lined hollow lumen of the 3D model to simulate a perfusion-based liver sinusoid was driven by gravity. The flow rate (Q) of the media was calculated using the following equations:
| (6) |
| (7) |
where ΔP, ρ, g, and, Δh denote pressure, fluid density, gravity constant, and height difference (L × sin [10°]), respectively. ΔP, R, μ, and L denote pressure drop, the hydraulic radius of the channel, dynamic fluid viscosity (0.78 × 10−3 Pa s), and channel length (20 mm), respectively. The flow rate through the lumen-based LSOC was 25 μl/min. Media for cells were changed after every two days of culture. To evaluate the performance of the developed lumen-based perfused LSOC, the study presents a comparison with (i) a similar 3D bioprinted liver sinusoid core/shell model under static media (LSOC-S) and, (ii) the HepG2-seeded 2D sandwich culture system (SW group) to compare. The fabrication of the HepG2-based sandwich culture system is described in Supplementary section 1.3.
2.9. Viability and proliferation assay of the LSOC
The proliferation of HepG2 cells and HUVECs in the bioprinted structure was quantified using a cell counting kit – 8 (Dojindo, Korea). Briefly, cell counting kit-8 solution was added to the culture medium containing the 3D liver sinusoid model, followed by incubation for 2 h at 37 °C in a humidified 5% CO2 atmosphere. The incubated medium was transferred to a 96-well plate, and absorbance readings were obtained at 405 nm. Furthermore, the viability and morphology of hepatocytes and HUVECs of the LSOC were assessed using a Live/Dead™ viability/cytotoxicity kit (Invitrogen, USA). The LSOC was briefly rinsed with PBS. Then, it was stained for 30 min at 37 °C with a solution composed of calcein-AM and ethidium homodimer. The live (green) and dead (red) stained cells in the printed construct were analyzed using a fluorescent microscope (Nikon, Japan) on the seventh day of the LSOC culture.
2.10. Liver functionality test
To conduct the liver function test, medium from the LSOC culture (seventh day of culture) was collected to determine the secretion of albumin, urea, aspartate aminotransferase (AST), alanine aminotransferase (ALT), and lactate dehydrogenase (LDH). The preparation of standards and quantification of the mentioned secreted proteins were performed following the manufacturer's protocols for the respective calorimetric-based assay kits (Biovision, USA).
A permeability test for HUVEC-based lumen formation was conducted by adding FITC-conjugated dextran to the outer compartment of the chip platform. Media flow allows the FITC-dextran into the lumen of the model. FITC-dextran solute permeates from the inner lumen towards the extravascular space into the hepatocyte media present in the middle compartment. At specific time points, the hepatocyte media from the central compartment of the chip was collected and measured by a fluorescence reader at 485 nm. The 3D liver sinusoid model was further imaged by fluorescence microscopy (Nikon, Japan). Furthermore, immunocytochemistry-based quantification was performed to determine the secretion of albumin and expression of VE-cadherin using a standard staining protocol.
2.11. Hepatic gene expression analysis
Samples from the LSOC were taken after seven days of culture, and total RNA was extracted from each sample using TRIzol® reagent (Thermo Scientific). cDNA was synthesized using a random hexamer premix (Takara). A gene expression study based on real-time polymerase chain reaction (PCR) was performed using TB Green Premix Ex Taq (Tli RNaseH Plus, Takara) in a Thermal Cycler Dice Real Time System PCR machine (Applied Biosystems, Foster City, CA, USA). All the forward and reverse primers of the hepatocyte-specific genes are listed in Supplementary section 1.6. Human glyceraldehyde-3-phosphate (GAPDH) was used as the housekeeping gene, and the expression levels of all the genes were normalized relative to that of GAPDH. All the primers were purchased from Pioneer (Korea).
2.12. Drug screening evaluation of the LSOC
For toxicological evaluation, the LSOC was incubated with different concentrations of acetaminophen (0.1, 1, 2, 5, 10, 20, 40, and 80 mM) (Sigma) and diclofenac (1, 2, 5, 10, 20, 50, 100, 200, 400, and 800 μM) (Sigma) for 96 h. After incubation, hepatocyte viability was assessed using a CCK-8 assay. Live/dead imaging of the hepatocytes in the LSOC was performed by staining with a calcein-AM/ethidium homodimer and visualizing the stained specimen under a fluorescence microscope (Nikon, Japan). Furthermore, the functionality of the system was determined after it was treated with hepatotoxins by analyzing the secretions of albumin, urea, AST, ALT, LDH, and bile in a calorimetric-based assay.
2.13. Statistical analysis
All data were expressed as mean ± standard deviations. Statistical analysis of the variables was performed using Student's t-test. The data with p < 0.05 were considered statistically significant.
3. Results and discussion
3.1. Characterization of collagen-alginate composite bioink
For successful fabrication of the in vitro liver sinusoidal model via 3D bioprinting, it is essential to develop suitable bioinks that not only possess the required mechanical and rheological properties but also provide a functionally supportive microenvironment for the encapsulated cells [27]. In this regard, alginate hydrogels have been used as bioinks for 3D bioprinting [28]. However, as indicated in previous studies, native alginate is a bioinert material (lacking cell-adhesive motifs) with limited biological properties [29]. In this study, we used a composite bioink by mixing alginate with another natural polymer, collagen type I. Collagen is a highly abundant structural protein that provides intrinsic cell binding motifs and the nanoarchitecture for efficient cell-matrix interactions [30]. Furthermore, collagen is the most abundant type of protein expressed in the liver, and thus, it provides the encapsulated cells of the in vitro liver model with a more suitable surrounding matrix for growth and proliferation. Initially, we prepared CA bioinks of three different compositions (v/v) (C:A 1:1, C:A 2:1, and C:A 1:2) and determined the optimal formulation of the composite bioink in terms of its bioprintability, rheological properties, and cellular viability and growth.
3.1.1. Physicochemical characterization of collagen-alginate bioink
The rheological properties, namely viscosity and dynamic modulus, of the CA composite bioinks were evaluated (Fig. 2). The viscosity of the CA bioink was determined at 15 °C, which is the estimated temperature of the 3D cell printing process. With increasing shear stress, the graph presented in Fig. 2 indicates a clear decreasing trend of viscosity. Moreover, all the bioink compositions exhibited a similar shear-thinning trend. Shear-thinning is an important factor in bioink development because shear-thinning behavior confers adequate viscosity to the bioinks, which allows them to remain physically unobstructed during nozzle ejection in the cell-printing process and the subsequent deposition, as well as for maintaining the shapes of integral structures with great structural fidelity [31]. Among the blended CA bioinks, the bioink with the C:A composition ratio of 1:2 exhibited the highest viscosity, followed by the CA 1:1 and CA 2:1 bioinks (Fig. 2A). These results indicate that the CA 1:2 bioink required a higher printing force to extrude the same quantity of bioink compared to the other blended bioinks.
Fig. 2.
Rheological study of bioinks. (A) Viscosity of alginate hydrogel and bioinks with different composition ratios of blended collagen and alginate. (B) Storage and loss moduli of the bioinks at various angular frequencies (rad/s).
A dynamic frequency sweep analysis was conducted to measure the frequency-dependent storage (G′) and loss (G″) moduli of the composite hydrogels. Before the evaluation, all the composite bioinks were crosslinked by treating them with a calcium chloride solution. The dynamic moduli were evaluated to check the stability of the bioink-printed structures at 37 °C. The storage moduli of all the crosslinked gels were higher than their loss moduli, which indicates that the crosslinked bioinks possessed gel-like mechanical properties instead of liquid-like properties. Apart from 4% alginate, the CA 1:2 gel exhibited the highest modulus (Fig. 2B). These results show that after the gelation of the hydrogel, the printed structures can maintain their shapes, which is crucial in the fabrication of in vitro model. Notably, because of the presence of collagen as a component, the bioinks exhibited thermal sensitivity during the crosslinking process at 37 °C. Even without the calcium chloride treatment, all the bioinks showed a sol-gel transition, where the bioinks appeared as solutions at room temperature, underwent crosslinking, and entered the gel phase after incubation at 37 °C for 30 min. Additionally, it has been reported that by attaining the biomechanical properties of the native tissue, the tissue specific functionality of the printed structure is enhanced. In this regard, the crosslinked CA 1:1 had a modulus of 1.8 kPa at the angular frequency of 62 rad/s, which is similar to the modulus of native liver tissue (1.5 kPa). Furthermore, we also studied the printability of the various ratios of CA bioink to determine the structural fidelity of the core/shell model (Supplementary section 1.1). We observed mixing of both the CA and gelatin bioinks forming an unclear core/shell interface for CA 2:1. However, CA 1:1 presented an ideal structural property and maintained a stable core/shell form after printing.
Porosity and pore size are important features of any biomaterial because they govern the diffusion of gases, inflow of media and nutrients, and outflow of waste metabolites. Fig. 3 (A–D) shows SEM images revealing the pore sizes of all the CA composite bioinks. From these results, it is evident that the CA-based hydrogels have larger pore sizes compared to those of the alginate hydrogels. Additionally, we determined the pores size distributions of the bioinks, and the graphical data (Fig. 3E) revealed that the CA composite bioinks had a larger average pore size (280–450 μm), which was approximately two to three times higher than the average pore size of the alginate hydrogel (∼140 μm). The porosity of the CA composite bioinks was measured using the absolute ethanol absorption technique. Fig. 3F shows the porosity values of the bioinks composed of various ratios of collagen to alginate after crosslinking with calcium chloride. The results indicate the CA 2:1 bioink had the highest porosity among the CA composite bioinks, followed by the CA 1:1 and CA 1:2 bioinks; the 4% alginate bioink had the lowest porosity.
Fig. 3.
Scanning-electron-microscopy-based morphological study of lyophilized alginate and CA bioink. (A) 4% Alginate, (B) CA 1:1, (C) CA 2:1, and (D) CA 1:2. The scale bar represents 100 μm. (E) Average pore size distribution plot of alginate and CA blended bioinks. (F) Average percentage porosity of alginate and CA bioinks. (∗p < 0.05).
3.1.2. Biological characterization of collagen-alginate bioink
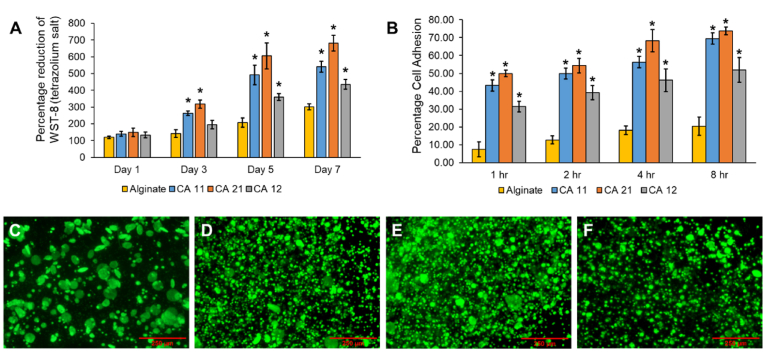
Biological studies were conducted to investigate and determine the most suitable composite bioink environment for maintaining hepatocyte growth and proliferation. Fig. 4A presents the proliferation data obtained over a period of seven days. The proliferation data from Day 1 up to Day 3 indicate a 200% increase in the hepatocytes in all the CA composite bioinks. By contrast, the cells encapsulated in the alginate hydrogel exhibited less than a 75% increase in proliferation. The CA bioinks contained collagen protein as one of their constituents, which is a well-known pro-adhesion polymer that directs early cell adhesion and growth [32]. Furthermore, the presence of collagen protein ensured the formation of large pores and a high level of porosity in the bioinks because collagen materials did not participate in the calcium-based crosslinking of the sodium alginate polymer. This facilitated easy cell migration and proliferation throughout the CA composite bioinks and ensured exponential cell growth within the first three days. Fig. 4B shows the cell adhesion data of the composite bioinks over a period of 8 h. The results indicate the potential of CA 2:1 and CA 1:1 as superior candidate bioinks relative to CA 1:2 in terms of cell adhesion capability. The viability of the encapsulated HepG2 cells was determined by means of fluorescent calcein-AM staining as well. Because the alginate hydrogel had fewer pores and lower porosity, hepatocyte migration was hindered. This hindrance caused the cells to grow and divide in a limited space, leading to the formation of large cell spheroids, as can be seen in the calcein-AM-stained fluorescence images (Fig. 4C–F). Among the CA composite bioinks, the CA 2:1 bioink exhibited the highest proliferation, followed closely by the CA 1:1 bioink. By contrast, HepG2 growth in the CA 1:2 bioink was comparatively slower. The presence of abundant cell interaction motifs in the CA 2:1 and CA 1:1 bioinks, as well as their large pore sizes and increased porosity levels, facilitated relatively rapid hepatocyte adhesion and, subsequently, proliferation. Based on the all the properties, including optimal dynamic moduli, printability, presence of porosity as well as cellular adhesion, growth, and proliferation, the CA 1:1 bioink emerged as the preferred choice for fabrication of the LSOC.
Fig. 4.
(A) Hepatocyte proliferation in different bioinks determined as the percentage of the reduction of formazan crystals. (B) The cellular adhesion capability of the bioinks was measured by means of hemocytometer counting. (C) Fluorescence microscopy images depicting hepatocyte growth (stained in calcein-AM) on Day 5 in alginate, (D) CA 1:1, (E) CA 2:1, and (F) CA 1:2. The scale bar represents 250 μm. (∗p < 0.05).
3.2. Fabrication of lumen-based 3D liver sinusoid-on-a-chip
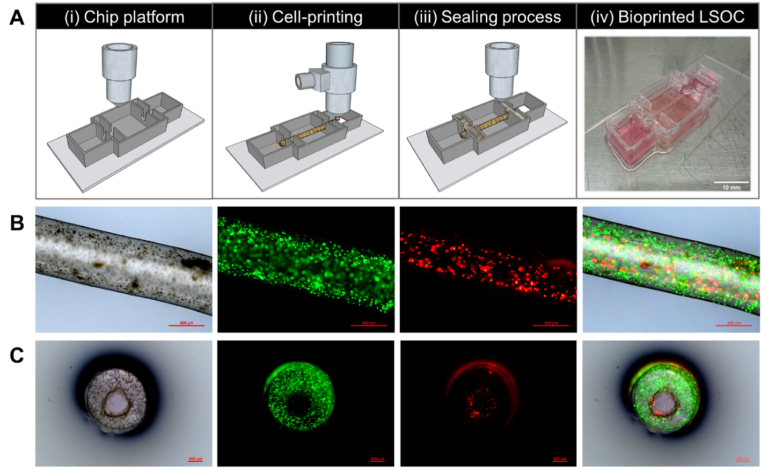
In the development of the lumen-based LSOC, the design of the chip platform is an important aspect because it will contain all the necessary arrangements to enhance the desired functionalities of the in vitro 3D model (Supplementary Fig. S2 shows the platform design of the perfusable LSOC.). The design includes two endothelial blocks on both sides of the chip and a central space for the hepatocyte media. We used PEVA as the housing material to fabricate the LSOC platform. The platform was printed on sterilized and transparent glass slide plates, which could be used directly for further imaging analysis. PEVA is a non-biodegradable thermoplastic polymer that becomes pliable or moldable at higher temperatures and solidifies upon cooling. Before starting a cell-based co-axial printing, we perform an investigation on the variability of lumen dimension of the core/shell 3D model against changing nozzle pressure and speed (Supplementary section 1.2). Based on the study outcome, we determine the optimize printing parameters (inner nozzle pressure – 70 kPa, outer nozzle pressure – 50 kPa, nozzle speed – 80 mm/min) for fabricating the 3D core/shell model. Thereafter, two different bioinks containing HepG2 and HUVECs were simultaneously bioprinted through a co-axial nozzle to form a two-layer cylindrical structure. As mentioned earlier, the outer layer of this structure was composed of HepG2 cells encapsulated in CA 1:1 bioink, and the inner core region was composed of HUVECs encapsulated in EGM-based 3% gelatin bioink. HepG2 cells are widely used in in vitro toxicology studies because of their functional resemblance to primary hepatocytes [33]. Furthermore, HUVECs are a popular source of endothelial cells for the development of various in vitro models, and they have been used to create liver sinusoids and livers-on-a-chip [34]. Thereafter, both ends of the 3D model were fixed to the chip platform by printing a layer of alginate (Fig. 5A, Supplementary video 1). It is well known that gelatin solution solidifies at lower temperatures, but at 37 °C, it is restored to its liquid form. When the bioprinted core/shell structure was transferred into the CO2 incubator at 37 °C for 24 h, the sacrificial 3% gelatin bioink at the center of the 3D model flowed out, creating a hollow lumen-based cylindrical construct. Thus, the fabricated tubular 3D liver sinusoid model consisted of a core compartment with an endothelial lining surrounded by hepatocyte plates to closely resemble the physiological unit of liver sinusoids.
Fig. 5.
(A) Schematics of the perfusion-based liver sinusoid-on-a-chip (LSOC) fabrication procedure. ((i) 3D printing of a PEVA-based chip platform, (ii) extrusion bioprinting of the core/shell 3D liver sinusoid model with encapsulated HepG2/HUVECs using a co-axial nozzle, (iii) sealing of the core/shell bioprinted liver sinusoid model, and (iv) the single-step bioprinted LSOC. (B) Longitudinal view of the 3D liver sinusoid model containing HepG2 cells (green) and HUVECs (red), as visualized under a fluorescence microscope. (C) Cross-sectional view of the 3D liver sinusoid model. HepG2 cells and HUVECs were stained with CDFDA (Cell tracker Green) and DiI (Cell tracker Red), respectively.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.mtbio.2023.100723
The following is the supplementary data related to this article:
2
To determine the cell adhesion and position in the 3D model after bioprinting, we stained the LSOC using cell tracker dyes. HepG2 cells and HUVECs were stained with CMFDA cell tracker dye (Green) and CM-DiI cell tracker dye (Red), respectively. The liver sinusoid model containing stained hepatocytes and endothelial cells is presented in the longitudinal and cross-sectional formats in Fig. 5B and C, respectively. The cross-sectional images confirmed the formation of a hollow core with HUVEC attachment on the inner side of the lumen-based construct. Thus, the present method of an integrated single-step co-axial bioprinting technique is a favorable approach for fabricating a structurally relevant lumen-based liver sinusoid model with a perfusable vascularized core. The developed LSOC was then transferred onto a rocker to facilitate the gravity-induced flow of culture media between the two endothelial blocks of the chip platform through the 3D model to mimic the fluidic condition of the liver.
3.3. Viability and proliferation assay of the developed LSOC
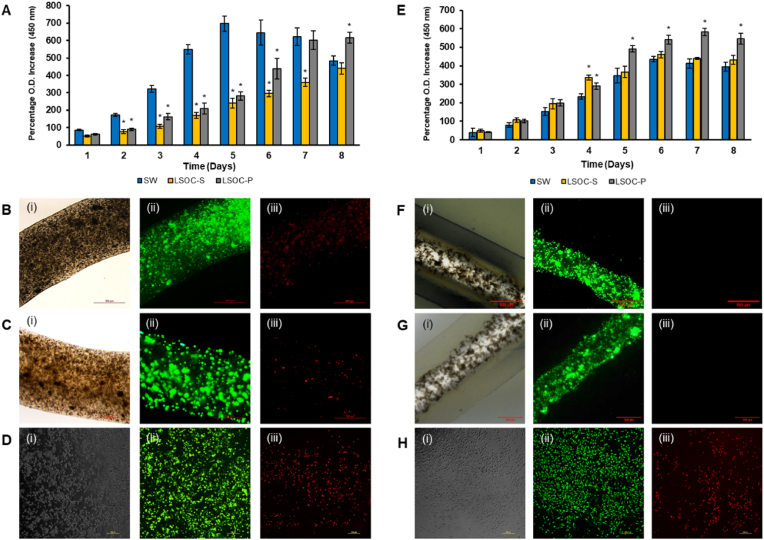
The hepatocyte and HUVEC activity and performance of the perfused LSOC (LSOC-P) were compared to a static media-based LSOC (LSOC-S) and a 2D SW culture model (the HepG2-based sandwich culture system is described in Supplementary section 1.3). The sandwich culture of hepatocytes between two layers of a hydrogel disc is a well-established in vitro model. The hepatocyte SW culture has been used in liver physiology studies, as well as for drug metabolism/toxicity testing [35]. Hepatocyte growth on all the three culture types were compared using a proliferation assay for a period of eight days (Fig. 6A–D). Although the O.D. values on the starting day were similar across all three platforms, a significant difference was observed between both LSOC models and the SW platform. The O.D. values of the SW model increased until Day 5 and were greater compared to both the LSOC-S and LSOC-P models. The reason for this significant increase in proliferation values is the monolayer culture of hepatocytes in the SW model. In a 2D/monolayer culture, cells are in direct contact with the medium supplements, which allows the predominance of proliferative cell populations. In the next few days (Days 6–8), hepatocyte proliferation in the SW culture started to decline, indicating increased cell death, probably because of the overcrowding of cells leading to the limited availability of medium and growth space. By contrast, hepatocytes in the LSOC-S and LSOC-P cultures remained proliferative until Day 8. Among the 3D LSOC models, the proliferation of cells in the LSOC-P culture showed enhanced growth in contrast to the LSOC-S culture. The slower growth rate of hepatocytes in the LSOC-S model might be due to the stationary phase of culture media, which limits the diffusion of essential growth supplements to the hydrogel-embedded cells.
Fig. 6.
(A) WST-8 assay to investigate hepatocyte proliferation in LSOC-P, LSOC-S, and SW models. A live/dead assay was performed on Day 7 on hepatocytes on the (B) LSOC-P, (C) LSOC-S, and (D) SW platforms. (E) HUVEC proliferation on the three platforms. A live/dead assay was performed on Day 7 on endothelial cells on the (F) LSOC-P, (G) LSOC-S, and (H) SW platforms. (i) Microscopic phase-contrast images. (ii) Green color represents live cells, (iii) red color represents dead cells. Scale bar = 500 μm for B, C, F, and G; 200 μm for D & H; ∗p < 0.05.
Interestingly, in the case of HUVEC growth, the cells were continually proliferative in all three cultures until Day 7, and this growth was significantly greater in the LSOC-P culture compared to the SW and LSOC-S cultures. This significant difference in cell growth is due to the continuous movement of growth media through the lumen-based core region of the liver model in the LSOC-P culture. On the contrary, HUVECs in the SW and LSOC-S cultures were present under static media, wherein the growth started to decline after Day 7, indicating cell death. The microscopy results showed the presence of a higher number of live-stained HepG2 cells and HUVECs in the LSOC-P model, whereas more HepG2 and HUVEC cell death was observed in the LSOC-S and SW models.
3.4. Liver functionality studies
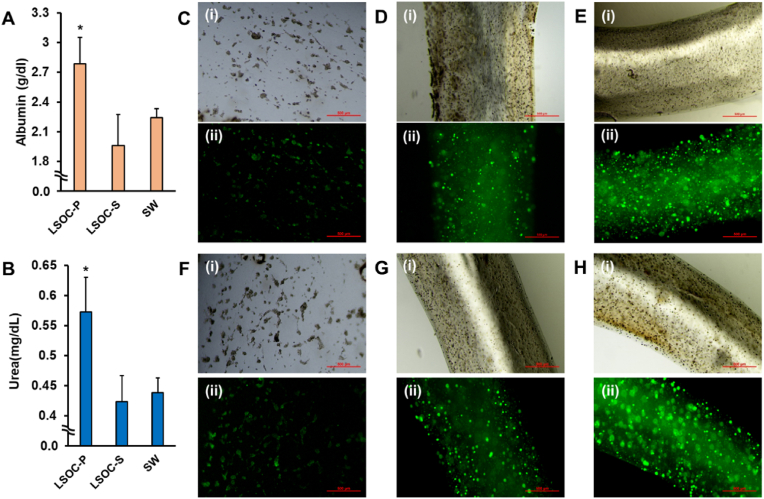
For any in vitro model, it is highly essential to persistently mimic cellular functionality under physiological conditions. Herein, hepatocyte functionality was assessed by determining the production of albumin and urea among the LSOC-P, LSOC-S, and SW platforms for a period of seven days. The graphical data indicate increased albumin and urea production in the LSOC-P culture relative to the LSOC-S and SW cultures (Fig. 7A and B). The increased hepatocyte production in the LSOC-P indicates the healthy functional state of the cells, which depends on the immediate microenvironment available to the cells (cell-cell interactions, cell-matrix interactions, matrix mechanical properties, matrix biochemical composition, and spatiotemporal gradient of biochemical cues). Also, it has been widely reported that a suitable microenvironment, such as 3D culture, is important for liver cell culture [36,37]. The LSOC-P platform not only provides the encapsulated hepatocytes with a suitable mechanical and biochemical environment but also provides the much-required 3D microarchitecture that allows them to achieve superior functional states compared to those in a 2D monolayer culture. Furthermore, the core/shell structure of the LSOC-P allows for easy perfusion of media and gas exchange, thereby enhancing the physiological functional state of the cells. However, oxygen and nutrient diffusion to the encapsulated cells was limited and may have led to the suboptimal production of albumin and urea in the LSOC-S model. The functional expression of albumin was visualized by immunostaining (Fig. 7C–E) and indicated clear albumin expression in the LSOC-P model compared to the LSOC-S and SW models.
Fig. 7.
Functional study of 3D liver sinusoid-on-a-chip with perfusion (LSOC-P), 3D liver sinusoid with static media (LSOC-S) and 2D sandwich culture (SW). (A) Albumin and (B) urea expression in the LSOC-P, LSOC-S and SW platforms (∗p < 0.05). An immunocytochemistry study was conducted to determine the expression of albumin in (C) SW (D) LSOC-S (E) LSOC-P. Immunostained MRP2 expression shown in (F) SW, (G) LSOC-S and (H) LSOC-P. (Scale bar: 500 μm).
Excreted bile acids play a crucial role in the digestion and absorption of nutrients in the intestine. Over accumulation of bile acid has harmful effects on hepatocytes and induces liver toxicity. Therefore, biliary system creation and bile acid removal are important aspects pertaining to the functioning of a liver sinusoid platform. The continuous movement of media supplements through the perfusion system increases the intracellular metabolic activity of hepatocytes, which eventually results in the generation of higher amounts of bile acid compared to the LSOC-S and SW platforms. Multidrug resistance-associated protein 2 (MRP2) is a liver-specific bile transporter. The images in Fig. 7F–H shows that MRP2 was highly expressed in the LSOC-P culture, indicating the effective creation of a biliary system and the removal of bile acids compared to the LSOC-S and SW systems. The higher expression level of biliary-specific protein in the LSOC-P may be attributed to its favorable hepatocyte microenvironment (in terms of both composition and architecture). Furthermore, the functional status of HUVEC in the lumen of 3D LSOC model were established by determining the expression of VE-cadherin (immunostaining assay), as well as through permeability assay (Supplementary section 1.5).
3.5. Gene expression study
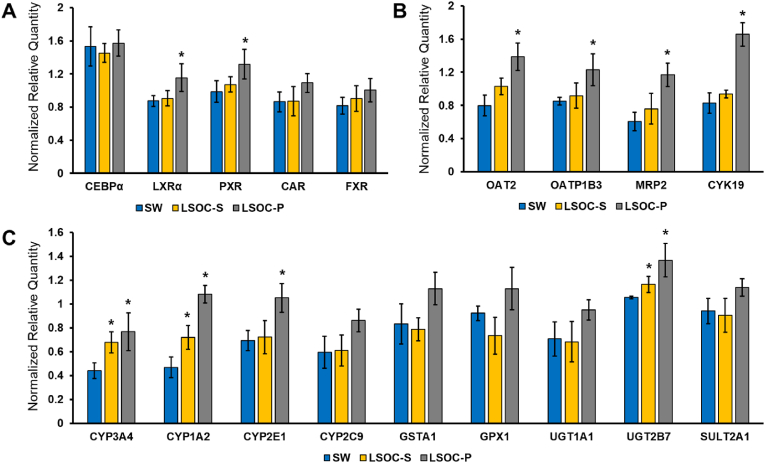
A real-time polymerase chain reaction (PCR) assay was used to evaluate the hepatocyte-specific gene expression in all three culture platforms. The list of liver-specific genes was categorized under the following labels: transcriptional factors (TFs) and nuclear receptors, metabolic enzymes, and transporter proteins. TFs are proteins that bind to specific DNA sequences and regulate gene expression. They have diverse roles such as cell growth and proliferation, maintaining normal cellular functions, and metabolism. The hepatocyte-specific TFs were well-expressed in both culture systems. Among the transcriptional factors, liver X receptor alpha (LXR-α), pregame X receptor (PXR), and constitutive androstane receptor (CAR) exhibited enhanced expression in the LSOC-P culture relative to those in the LSOC-S and SW cultures. LXR-α is an important factor that regulates hepatocytes for maintaining intracellular cholesterol and lipid homeostasis [38], whereas the nuclear receptor PXR is involved in the induction of phase I and II drug-metabolizing enzymes [39]. The expression of both TFs indicated that a perfusion-based chip has an enhanced effect on the regulation of metabolic enzymes and fatty acid homeostasis.
The PCR analysis also indicated higher expression of various phase I and II metabolic enzymes and drug transporters in the LSOC-P culture relative to the LSOC-S and SW cultures. The mRNA transcript levels of phase I xenobiotic-metabolizing enzymes CYP3A4, CYP1A2, CYP2E1, CYP2C9, GSTA1, and GPX1 were systematically analyzed in both culture platforms over a period of seven days. The mRNA levels of several phase II xenobiotic-metabolizing enzymes UGT1A1, UGT2B7, and SULT2A1 increased in the LSOC-P. In addition, phase III drug-transporting enzymes played major roles in the formation of metabolites and drug detoxification. The expression levels of OAT2, OATP1B3, and MRP2, which are major transporters of various clinically relevant drug classes, lipophilic anions, and bile acid, increased substantially in the LSOC-P culture relative to the LSOC-S and SW cultures. The induction of metabolic and transporter enzymes is a complex phenomenon that is mediated by the activation of nuclear receptors and gene transcription. The low levels of metabolic enzymes observed in the SW and LSOC-S cultures were ascribed to lower expressions of the nuclear receptors that regulate their induction. Moreover, the gene expression results agree with the results of the liver functionality study, which showed an enhanced expression of the biliary-specific gene, MRP2, in the LSOC-P culture. CAR and PXR are considered master regulators in drug metabolism [40]. The increased expression of xenobiotic receptors in the LSOC-P makes it a suitable platform for studying drug-drug interactions mediated by the CYP450 enzyme (see Fig. 8).
Fig. 8.
Gene expression study of 3D liver sinusoid-on-a-chip with perfusion (LSOC-P), 3D liver sinusoid with static media (LSOC-S) and 2D sandwich culture (SW). The expression of hepatocyte markers is categorized under (A) transcriptional factors and nuclear receptors, (B) transporter proteins, and (C) liver-specific metabolic enzymes. (∗p < 0.05).
3.6. Evaluation of drug response
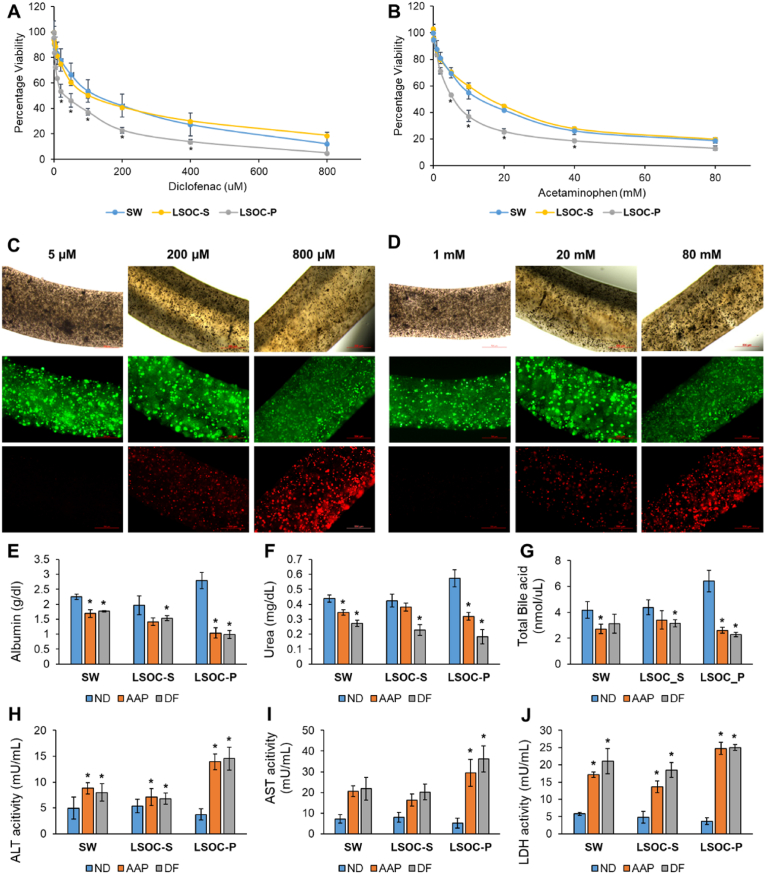
For toxicological evaluations, the LSOC-P, LSOC-S, and SW systems were treated with different concentrations of acetaminophen (AAP; 0–80 mM) and diclofenac (DF; 0–800 μM) on Day 7. DF causes rare but significant cases of severe hepatotoxicity, including liver necrosis, jaundice, fulminant hepatitis with and without jaundice, and liver failure [41,42]. Previous studies have demonstrated high in vitro concentrations of DF alone (>200 μM) induce acute toxicity in primary cultures of human hepatocytes [43]. For AAP, which is the most frequently used pain killer worldwide, there is a large body of literature characterizing its safety in vitro and in vivo, and an acute AAP overdose can cause liver toxicity and liver failure [44,45]. The viability results indicated that both AAP and DF caused dose-dependent toxicity in all three systems (Fig. 9A and B). With 800 μM DF and 80 mM AAP, the hepatocyte viability was 20%, irrespective of the culture system, which clearly indicates acutely toxic concentrations. DF treatment at the concentration of 200 μM resulted in the death of more than 50% of the hepatocytes in the LSOC-S and SW cultures, whereas in the LSOC-P system, this concentration led to the death of more than 75% of the hepatocyte population. Similarly, 20 mM AAP resulted in the death of more than 50% of the hepatocyte populations in the LSOC-S and SW cultures and approximately 75% of the hepatocyte population in the LSOC-P system. These results indicate that compared to the LSOC-S and SW cultures, the perfusion-based LSOC-P system exhibited considerably higher drug sensitivity when subjected to repeated drug treatment. This result was also confirmed when the inhibitory concentration at 50% viability (IC50) was calculated for all the culture systems treated with both DF and AAP. The IC50 values of the LSOC-P system for DF and AAP were 28.58 μM and 6.03 mM, respectively, which was quite similar to the safe levels of therapeutic serum concentration for DF and AAP (6.4 μM and 1 mM, respectively) [46,47]. Furthermore, the live/dead staining confirmed the hepatotoxic effect of both drugs, as indicated by a considerable increase in the dead cell population with increasing drug concentration (Fig. 9C and D). Overall, the results demonstrated that the LSOC-P platform could lead to remarkably improved hepatoxic predictions with close estimates of the therapeutic ranges of safe drug concentrations.
Fig. 9.
Drug toxicity analysis of 3D liver sinusoid-on-a-chip with perfusion (LSOC-P), 3D liver sinusoid model in static media (LSOC-S) and 2D sandwich culture (SW). The graphical plot represents drug-concentration-based cellular viability. Both culture platforms were treated with (A) diclofenac (from 1 μM to 800 μM) and (B) acetaminophen (from 0.1 mM to 80 mM). (∗p < 0.05). Cell viability analysis at different drug concentrations of (C) diclofenac and (D) acetaminophen after calcein-AM/ethidium homodimer-based live (green)/dead (red) staining. The expression levels of liver-specific proteins (E) albumin, (F) urea, and (G) bile acid were calorimetrically determined in all three platforms. The expression levels of liver-specific enzymes (H) aspartate aminotransferase (AST), (I) alanine aminotransferase (ALT), and (J) lactate dehydrogenase (LDH) after treating the three platforms with diclofenac (200 μM) and acetaminophen (20 mM) are shown. (ND: No Drugs; DF: Diclofenac; AAP: Acetaminophen, ∗p < 0.05).
Medicines are recognized as cost-effective interventions because they combat infections and treat disorders that were formerly thought to be incurable, thus saving millions of lives. However, during clinical trials and after product approval, medications/drugs can cause harmful and even fatal reactions. Globally, more than half of acute liver failure cases are caused by drug-induced liver injury (DILI) [48,49]. Therefore, it is also important to investigate the functional effects of liver 3D models when they are subjected to drug treatments. In this regard, we analyzed the expression of liver-specific proteins such as albumin, urea, and total bile acid, in addition to determining the levels of liver enzymes such as AST, ALT, and LDH to further compare the specificity of the 3D liver sinusoid system. The results of this study showed that after treatment with 200 μM DF and 20 mM AAP, the expressions of liver proteins (albumin and urea) and total bile acid production decreased in all the platforms (Fig. 9E–G). However, the reductions in protein expression were more significant in the LSOC-P system compared to the LSOC-S and SW systems. For instance, albumin expression decreased by 63.91 ± 8.77% in the LSOC-P system when treated with DF and by 61.9 ± 9.71% when treated with AAP. By contrast, albumin expression in the LSOC-S system was reduced by 21.7 ± 9.36% and 27.82 ± 4.8%, whereas SW demonstrated a reduction of 20.93 ± 7.17% and 24.11 ± 7.6% with DF and AAP treatment, respectively. These data indicate that liver cells are more susceptible to drug toxicity in the LSOC-P platform than in the LSOC-S and SW platforms. Furthermore, the expression levels of liver enzymes (ALT, AST, and LDH) were higher when they were treated with DF and AAP than in the untreated platforms (ND: No Drugs, Control) (Fig. 9H–J). AST and ALT are abundant liver enzymes usually present at low levels in the serum. Increased levels of AST, ALT, and LDH are indicative of liver cell or tissue damage, where they are released from compromised hepatocyte cell membranes following a necrotic event [50]. The LSOC-P platform showed the highest increase in the expression of liver enzymes compared to the LSOC-S and SW platforms. For instance, when treated with DF, the expression level of AST in the LSOC-P platform increased to more than 5X that of the untreated control. However, AST expression increased up to 1.5X in the LSOC-S platform and 2X in the SW platform. The data obtained from the liver functional study corroborated the fact that drug treatment caused hepatocyte injury. Nevertheless, this study also substantiates the improved relevance of the LSOC-P device over the LSOC-S and SW platforms in terms of drug toxicity and liver-specific function.
The perfusion environment helped with the passive diffusion of medium to provide higher coverage of drugs throughout the 3D liver sinusoid model. Furthermore, the higher metabolic rate of the encapsulated cells in the LSOC-P allowed for easy interaction of drugs. The metabolism of these drugs leads to the production of reactive metabolites, which form complex covalent links with cellular proteins. Excessive accumulation of these complexes causes oxidative stress in the endoplasmic reticulum and mitochondria, resulting in apoptosis and necrosis of the hepatocytes [51]. Overall, these results indicate that the compact LSOC-P platform fabricated through a single-step co-axial bioprinting approach is an efficient system in terms of screening for hepatotoxic drugs.
4. Conclusion
In this work, we successfully developed a lumen-based 3D LSOC with a perfusable vascularized channel. The entire device, which includes the chip platform and the core/shell structure of the liver sinusoid model, was fabricated using a single-step co-axial bioprinting process. A bioink composed of equal fractions of collagen and alginate was used to fabricate the shell component for encapsulating the hepatocytes, whereas an endothelial-media-based gelatin was used as a sacrificial bioink for perfusable lumen formation. The novel and improved single-step fabrication approach results in an in vitro model with a close structural similarity to a liver sinusoid, where hepatocyte growth and proliferation in the fabricated platform enabled the stable construction of the system. Additionally, the expression levels of liver-specific proteins and genes in the LSOC-P platform were superior to those of the static 3D liver model (LSOC-S) and standard liver sandwich (SW) culture. Furthermore, the LSOC-P platform showed improved liver-specific functional activity and exhibited a high level of sensitivity to drugs; thus, it outperformed the control platforms. In conclusion, the development of the LSOC-P herein, fabricated by an integrated single-step bioprinting system, is a promising in vitro liver test platform that is a valuable tool for drug development applications.
CRediT authorship contribution statement
Somnath Maji: Methodology, Validation, Data curation, Writing – original draft. Minkyoung Lee: Validation, Investigation. Jooyoung Lee: Resources, Validation. Jaehee Lee: Resources, Visualization. Hyungseok Lee: Conceptualization, Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2020R1C1C1011147).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2023.100723.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- 1.Cho S., Yoon J.-Y. Organ-on-a-chip for assessing environmental toxicants. Curr. Opin. Biotechnol. 2017;45:34–42. doi: 10.1016/j.copbio.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hutmacher D.W., Holzapfel B.M., De-Juan-Pardo E.M., Pereira B.A., Ellem S.J., Loessner D., et al. Convergence of regenerative medicine and synthetic biology to develop standardized and validated models of human diseases with clinical relevance. Curr. Opin. Biotechnol. 2015;35:127–132. doi: 10.1016/j.copbio.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Zhang B., Radisic M. Organ-on-a-chip devices advance to market. Lab Chip. 2017;17:2395–2420. doi: 10.1039/c6lc01554a. [DOI] [PubMed] [Google Scholar]
- 4.Tao T., Peng J. Liver development in zebrafish (Danio rerio) J. Genet. Genom. 2009;36:325–334. doi: 10.1016/S1673-8527(08)60121-6. [DOI] [PubMed] [Google Scholar]
- 5.Holt M.P., Ju C. Mechanisms of drug-induced liver injury. AAPS J. 2006;8:E48–E54. doi: 10.1208/aapsj080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du Y., Li N., Yang H., Luo C., Gong Y., Tong C., et al. Mimicking liver sinusoidal structures and functions using a 3D-configured microfluidic chip. Lab Chip. 2017;17:782–794. doi: 10.1039/c6lc01374k. [DOI] [PubMed] [Google Scholar]
- 7.Lee P.J., Hung P.J., Lee L.P. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol. Bioeng. 2007;97:1340–1346. doi: 10.1002/bit.21360. [DOI] [PubMed] [Google Scholar]
- 8.Prodanov L., Jindal R., Bale S.S., Hegde M., McCarty W.J., Golberg I., et al. Long-term maintenance of a microfluidic 3D human liver sinusoid. Biotechnol. Bioeng. 2016;113:241–246. doi: 10.1002/bit.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhise N.S., Manoharan V., Massa S., Tamayol A., Ghaderi M., Miscuglio M., et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/1/014101. [DOI] [PubMed] [Google Scholar]
- 10.Lee S.-A., Kang E., Ju J., Kim D.-S., Lee S.-H. Spheroid-based three-dimensional liver-on-a-chip to investigate hepatocyte–hepatic stellate cell interactions and flow effects. Lab Chip. 2013;13:3529–3537. doi: 10.1039/c3lc50197c. [DOI] [PubMed] [Google Scholar]
- 11.Yeon J.H., Park J.-K. Microfluidic cell culture systems for cellular analysis. Biochip J. 2007;1:17–27. [Google Scholar]
- 12.GhavamiNejad A., Ashammakhi N., Wu X.Y., Khademhosseini A. Crosslinking strategies for 3D bioprinting of polymeric hydrogels. Small. 2020;16 doi: 10.1002/smll.202002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kilian D., Ahlfeld T., Akkineni A.R., Lode A., Gelinsky M. Three-dimensional bioprinting of volumetric tissues and organs. MRS Bull. 2017;42:585–592. [Google Scholar]
- 14.Wang X., Yan Y., Pan Y., Xiong Z., Liu H., Cheng J., et al. Generation of three-dimensional hepatocyte/gelatin structures with rapid prototyping system. Tissue Eng. 2006;12:83–90. doi: 10.1089/ten.2006.12.83. [DOI] [PubMed] [Google Scholar]
- 15.Lee K.H., Shin S.J., Kim C.-B., Kim J.K., Cho Y.W., Chung B.G., et al. Microfluidic synthesis of pure chitosan microfibers for bio-artificial liver chip. Lab Chip. 2010;10:1328–1334. doi: 10.1039/b924987g. [DOI] [PubMed] [Google Scholar]
- 16.Mi S., Yi X., Du Z., Xu Y., Sun W. Construction of a liver sinusoid based on the laminar flow on chip and self-assembly of endothelial cells. Biofabrication. 2018;10 doi: 10.1088/1758-5090/aaa97e. [DOI] [PubMed] [Google Scholar]
- 17.Lan S.-F., Safiejko-Mroczka B., Starly B. Long-term cultivation of HepG2 liver cells encapsulated in alginate hydrogels: a study of cell viability, morphology and drug metabolism. Toxicol. Vitro. 2010;24:1314–1323. doi: 10.1016/j.tiv.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 18.Miller J.S., Stevens K.R., Yang M.T., Baker B.M., Nguyen D.-H.T., Cohen D.M., et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 2012;11:768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H., Han W., Kim H., Ha D.-H., Jang J., Kim B.S., et al. Development of liver decellularized extracellular matrix bioink for three-dimensional cell printing-based liver tissue engineering. Biomacromolecules. 2017;18:1229–1237. doi: 10.1021/acs.biomac.6b01908. [DOI] [PubMed] [Google Scholar]
- 20.Lee H., Chae S., Kim J.Y., Han W., Kim J., Choi Y., et al. Cell-printed 3D liver-on-a-chip possessing a liver microenvironment and biliary system. Biofabrication. 2019;11 doi: 10.1088/1758-5090/aaf9fa. [DOI] [PubMed] [Google Scholar]
- 21.van Genderen A.M., Valverde M.G., Capendale P.E., Kersten M.V., Garví E.S., Schuurmans C.C., et al. Co-axial printing of convoluted proximal tubule for kidney disease modeling. Biofabrication. 2022;14(4) doi: 10.1088/1758-5090/ac7895. [DOI] [PubMed] [Google Scholar]
- 22.Singh N.K., Kim J.Y., Lee J.Y., Lee H., Gao G., Jang J., et al. Coaxial cell printing of a human glomerular model: an in vitro glomerular filtration barrier and its pathophysiology. Biofabrication. 2023 Jan 13;15(2) doi: 10.1088/1758-5090/acad2c. [DOI] [PubMed] [Google Scholar]
- 23.Gao G., Park J.Y., Kim B.S., Jang J., Cho D.W. Coaxial cell printing of freestanding, perfusable, and functional in vitro vascular models for recapitulation of native vascular endothelium pathophysiology. Adv. Healthc. Mater. 2018;7(23) doi: 10.1002/adhm.201801102. [DOI] [PubMed] [Google Scholar]
- 24.Gao G., Lee J.H., Jang J., Lee D.H., Kong J.S., Kim B.S., et al. Tissue engineered bio-blood-vessels constructed using a tissue-specific bioink and 3D coaxial cell printing technique: a novel therapy for ischemic disease. Adv. Funct. Mater. 2017;27(33) [Google Scholar]
- 25.Taymour R., Kilian D., Ahlfeld T., Gelinsky M., Lode A. 3D bioprinting of hepatocytes: core–shell structured co-cultures with fibroblasts for enhanced functionality. Sci. Rep. 2021;11(1):1–8. doi: 10.1038/s41598-021-84384-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zu Y., Zhang Y., Zhao X., Shan C., Zu S., Wang K., et al. Preparation and characterization of chitosan–polyvinyl alcohol blend hydrogels for the controlled release of nano-insulin. Int. J. Biol. Macromol. 2012;50:82–87. doi: 10.1016/j.ijbiomac.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Montero F.E., Rezende R.A., Da Silva J.V., Sabino M.A. Development of a smart bioink for bioprinting applications. Front. Mech. Eng. 2019;5:56. [Google Scholar]
- 28.Jia J., Richards D.J., Pollard S., Tan Y., Rodriguez J., Visconti R.P., et al. Engineering alginate as bioink for bioprinting. Acta Biomater. 2014;10:4323–4331. doi: 10.1016/j.actbio.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Billiet T., Vandenhaute M., Schelfhout J., Van Vlierberghe S., Dubruel P. A review of trends and limitations in hydrogel-rapid prototyping for tissue engineering. Biomaterials. 2012;33:6020–6041. doi: 10.1016/j.biomaterials.2012.04.050. [DOI] [PubMed] [Google Scholar]
- 30.Cheema U. Standardisation in Cell and Tissue Engineering. Elsevier; 2013. Three-dimensional collagen biomatrix development and control; pp. 18–33. [Google Scholar]
- 31.Ferris C.J., Gilmore K.J., Beirne S., McCallum D., Wallace G.G. Bio-ink for on-demand printing of living cells. Biomater. Sci. 2013;1:224–230. doi: 10.1039/c2bm00114d. [DOI] [PubMed] [Google Scholar]
- 32.Heino J. The collagen family members as cell adhesion proteins. Bioessays. 2007;29:1001–1010. doi: 10.1002/bies.20636. [DOI] [PubMed] [Google Scholar]
- 33.Kammerer S., Küpper J.-H. Human hepatocyte systems for in vitro toxicology analysis. J. Cell. Biotechnol. 2018;3:85–93. [Google Scholar]
- 34.Jiménez N., Krouwer V.J., Post J.A. A new, rapid and reproducible method to obtain high quality endothelium in vitro. Cytotechnology. 2013;65:1–14. doi: 10.1007/s10616-012-9459-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang K., Guo C., Woodhead J.L., Claire R.L.S., III, Watkins P.B., Siler S.Q., et al. Sandwich-cultured hepatocytes as a tool to study drug disposition and drug-induced liver injury. J. Pharmaceut. Sci. 2016;105:443–459. doi: 10.1016/j.xphs.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kostrzewski T., Cornforth T., Snow S.A., Ouro-Gnao L., Rowe C., Large E.M., et al. Three-dimensional perfused human in vitro model of non-alcoholic fatty liver disease. World J. Gastroenterol. 2017;23:204. doi: 10.3748/wjg.v23.i2.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamada K.M., Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Musso G., Gambino R., Cassader M. Cholesterol metabolism and the pathogenesis of non-alcoholic steatohepatitis. Prog. Lipid Res. 2013;52:175–191. doi: 10.1016/j.plipres.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Urquhart B.L., Tirona R.G., Kim R.B. Nuclear receptors and the regulation of drug-metabolizing enzymes and drug transporters: implications for interindividual variability in response to drugs. J. Clin. Pharmacol. 2007;47:566–578. doi: 10.1177/0091270007299930. [DOI] [PubMed] [Google Scholar]
- 40.Modica S., Bellafante E., Moschetta A. Master regulation of bile acid and xenobiotic metabolism via the FXR, PXR and CAR trio. Front. Biosci.-Landmark. 2009;14:4719–4745. doi: 10.2741/3563. [DOI] [PubMed] [Google Scholar]
- 41.Boelsterli U.A. Diclofenac-induced liver injury: a paradigm of idiosyncratic drug toxicity. Toxicol. Appl. Pharmacol. 2003;192:307–322. doi: 10.1016/s0041-008x(03)00368-5. [DOI] [PubMed] [Google Scholar]
- 42.Bort R., Ponsoda X., Jover R., Gómez-Lechón M.J., Castell J.V. Diclofenac toxicity to hepatocytes: a role for drug metabolism in cell toxicity. J. Pharmacol. Exp. Therapeut. 1999;288:65–72. [PubMed] [Google Scholar]
- 43.Lauer B., Tuschl G., Kling M., Mueller S.O. Species-specific toxicity of diclofenac and troglitazone in primary human and rat hepatocytes. Chem. Biol. Interact. 2009;179:17–24. doi: 10.1016/j.cbi.2008.10.031. [DOI] [PubMed] [Google Scholar]
- 44.Lane J.E., Belson M.G., Brown D.K., Scheetz A. Chronic acetaminophen toxicity: a case report and review of the literature. J. Emerg. Med. 2002;23:253–256. doi: 10.1016/s0736-4679(02)00526-7. [DOI] [PubMed] [Google Scholar]
- 45.Shear N.H., Malkiewicz I.M., Klein D., Koren G., Randor S., Neuman M.G. Acetaminophen-lnduced toxicity to human epidermoid cell line A431 and hepatoblastoma cell line hep G2, in vitro. Is diminished by silymarin. Skin Pharmacol. Physiol. 1995;8:279–291. doi: 10.1159/000211359. [DOI] [PubMed] [Google Scholar]
- 46.Davern T.J., II, James L.P., Hinson J.A., Polson J., Larson A.M., Fontana R.J., et al. Measurement of serum acetaminophen–protein adducts in patients with acute liver failure. Gastroenterology. 2006;130:687–694. doi: 10.1053/j.gastro.2006.01.033. [DOI] [PubMed] [Google Scholar]
- 47.Hinz B., Chevts J., Renner B., Wuttke H., Rau T., Schmidt A., et al. Bioavailability of diclofenac potassium at low doses. Br. J. Clin. Pharmacol. 2005;59:80–84. doi: 10.1111/j.1365-2125.2005.02226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fontana R.J. Acute liver failure due to drugs. Semin. Liver Dis. 2008:175–187. doi: 10.1055/s-2008-1073117. ©Thieme Medical Publishers. [DOI] [PubMed] [Google Scholar]
- 49.Verma N., Kumar P., Mitra S., Taneja S., Dhooria S., Das A., et al. Drug idiosyncrasy due to pirfenidone presenting as acute liver failure: case report and mini-review of the literature. Hepatol. Commun. 2018;2:142–147. doi: 10.1002/hep4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nyblom H., Berggren U., Balldin J., Olsson R. High AST/ALT ratio may indicate advanced alcoholic liver disease rather than heavy drinking. Alcohol Alcohol. 2004;39:336–339. doi: 10.1093/alcalc/agh074. [DOI] [PubMed] [Google Scholar]
- 51.Gómez-Lechón M.J., Ponsoda X., O'Connor E., Donato T., Castell J.V., Jover R. Diclofenac induces apoptosis in hepatocytes by alteration of mitochondrial function and generation of ROS. Biochem. Pharmacol. 2003;66:2155–2167. doi: 10.1016/j.bcp.2003.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2
Data Availability Statement
No data was used for the research described in the article.