Abstract
Introduction
Despite the benefit of adjuvant systemic therapy for patients with resected non-small cell lung cancer (NSCLC), the risk of postoperative recurrence remains high. Our objective was to characterize temporal genetic heterogeneity between primary resected and recurrent tumors, and its impact on treatment outcomes.
Methods
In this study, next-generation sequencing (NGS) testing was performed on tissue specimens and circulating tumor DNA (ctDNA) collected at postoperative recurrence, and results were compared to the genotypes of initial surgical specimens.
Results
Of forty-five patients with matched primary and post-operative recurrent tumors, EGFR status switched in 17 patients (37.8%) at post-operative recurrence and 28 patients (62.2%) had no genotype change (17 mutant, 11 wild-type). Based on the changes of EGFR status, patients were divided into 4 groups. Following subsequent treatment with EGFR TKI o chemotherapy: In group A, with sustained sensitive mutation, the percentage achieving partial response (PR) was the highest, at 72.2%, the median progression-free survival (PFS) was 17 months, and the median overall survival (OS) was 44.0 months respectively; In group B, with genotype changed from wild-type to mutant, 50% achieved PR, PFS was 10 months, and OS was 35 months; In group C, in which mutant status shifted to wild-type or new co-mutation emerged, the percentage achieving PR was 30%, PFS was 9 months, and OS was 35 months. In group D, with sustained wild type, the percentage achieving PR was 27.3%, PFS was 8 months, and OS was 22 months.
Discussion
Genotypic shift between paired primary and post-operative recurrent tumors was not infrequent, and this temporal genomic heterogeneity substantially impacted subsequent treatment outcomes.
Keywords: dynamic gene sequencing, genotypic switch, temporal heterogeneity, EGFR, NSCLC
Introduction
Adjuvant therapy improves the survival of patients with resected non-small cell lung cancer (NSCLC), although 25–80% of patients experience postoperative recurrence (1, 2). Subsequent treatment strategy for patients with postoperative recurrences often depends on the genomic data determined from the initial surgical specimens. However, studies have shown that there is genomic temporal heterogeneity in recurrent and metastatic lung cancer after surgery. The coexistence of intratumoral driver gene heterogeneity and subcloning leads to clonal evolution, potentially resulting in genotypic changes and temporal heterogeneity (3). Lung cancer recurrence usually manifests as intrathoracic lesions or as extrapulmonary metastasis, such as in the brain or bone. Repeat genomic sequencing at recurrence or second primary has been used at some cases to guide treatment. At the same time, repeat tissue biopsy may not be feasible for some patients, so an important alternative is next-generation sequencing (NGS) of blood-derived circulating tumor DNA (ctDNA) (4).
In this study, NGS testing was performed on tissue specimens and ctDNA collected at postoperative recurrence, and results were compared to the genotypes of initial surgical specimens. The purpose of this study was to evaluate postoperative genotype changes and to identify the prevalence of clonal evolution in order to more accurately guide individualized treatment at recurrence.
Materials and methods
Patients
The study was conducted at the Department of Oncology at Shanghai Pulmonary Hospital from September 1, 2015 to April 1, 2018, including 45 NSCLC patients who experienced recurrence after surgery. The study was reviewed and approved by the institutional review boards, and in accordance with the Declaration of Helsinki and Good Clinical Practice. Informed consent was obtained for all subjects. The collected clinicopathological parameters included age, sex, smoking history, histology, TNM classification [based on the 8th edition of the American Joint Committee on Cancer (AJCC)/Union for International Cancer Control (UICC)], EGFR mutational status and types of subsequent treatment (including EGFR-TKIs). A person who had smoked <100 cigarettes in his or her lifetime was deemed as a never-smoker.
Sample collection and processing
Initial surgical specimens and tissue and blood samples, and those at recurrence, were collected. Tumor content on hematoxylin and eosin-stained slides were assessed by board-certified pathologists for all formalin-fixed, paraffin-embedded tissue specimens. Tissue specimens were deemed evaluable if tumor cells were identified. Eight sections of FFPE samples 5-10 μm were extracted using the QIamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Ten millimeters of fasting peripheral blood was collected into Streck blood collection tubes (BCT; Streck, Omaha, NE, US). Samples were transported at room temperature and processed at Shanghai Smartquerier Biomedicine Co. Ltd within 48 hours according to Streck BCT protocol. Samples were centrifuged at 2500g for 10 minutes. The supernatant serum was then collected and centrifuged at 16,000g for an additional 10 minutes. The resultant 3-5mL of supernatant was collected and circulating tumor DNA (ctDNA) was extracted using a QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany). Germline DNA was extracted from the supernatant after the first centrifugation using a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). NGS was used to detect gene sequencing in surgical specimens and in plasma at recurrence. Sequencing of ctDNA by NGS was performed according to a previously published study to identify genetic mutations (5). A plasma and germline DNA library was used for NGS. The list of 156 genes is provided in Supplementary Table 1 .
Data collection
The median follow up period was thirty-seven months. No second primary cancers were found at the end of follow-up. All patients had physical examinations monthly, as well as chest CT scans, abdominal color Doppler ultrasound, bone scan, and brain MRI at 3-month intervals. Tumor response were determined by Response Evaluation Criteria in Solid Tumors (RECIST)1.1 criteria as complete response (CR), partial response (PR), stable disease (SD), or progressive disease (PD). Disease control rate (DCR) was defined as the proportion of cases with complete, partial response, or stable disease. The objective response rate (ORR) was defined as the proportion of patients with complete or partial response. The endpoint of the study was progression-free survival (PFS). Recurrent progression-fee survival (rPFS) was calculated from the date of the treatment after recurrence (1st EGFR-TKI treatment or chemotherapy) until progressive disease (PD) or death due to any cause. Recurrent overall survival OS (rOS) was defined as the time of recurrence to death from any cause. Patients with EGFR-sensitive mutations were treated with EGFR-TKI as first-line treatment. The EGFR-TKIs used included gefitinib (250 mg, once a day), erlotinib (150 mg, once a day) and icotinib (125 mg, three times a day). Platinum-based doublet chemotherapy was given to patients without driver mutations at recurrence.
Statistics
The categorical variables were compared with the Chi-square test or Fisher exact test when expected count in each category were less than 5. Survival analysis was determined by Kaplan-Meier curves with two-sided log rank tests. The Cox proportional hazards model with calculated hazard ratios (HR) and 95% confidence interval (CI) was applied to adjust for potential confounders. Statistical significance was defined as Two-sided P < 0.05. All statistical analyses were performed and displayed using SPSS statistical software, version 22.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 9.0.0.
Results
Clinical characteristics
Of the 45 patients, 17 (37.8%) were female and 28 (62.2%) were male. The median age at post-operative recurrence was 58 years (range 43-79 years. There were 43 cases of adenocarcinoma nd 2 case of squamous cell carcinoma. Patients had stages ranging from IB to IIIA. Thirteen of the total patients with more advanced disease received 4-cycles of adjuvant platinum-based doublet chemotherapy, and the majority of patients with N2 disease received mediastinal radiotherapy after operation. Twenty-two patients developed postoperative recurrence in the lungs, while 12 patients had disease after recurrence in the bone, and 11 patients had local recurrence in mediastinal lymph nodes. None of the patients had adjuvant targeted therapy ( Table 1 ).
Table 1.
Baseline clinical characteristics of Chinese patients with non-small cell lung cancer (n=45).
| Chacteristic | Number (%) | |
|---|---|---|
| Gender | Male | 28 (62.2) |
| Female | 17 (37.8) | |
| Smoking history | Never smoker | 27 (60.0) |
| Smoker | 18 (40.) | |
| Family history | 2 (4.4) | |
| TNM Stage | IA | 13 (28.9) |
| IB | 11 (24.5) | |
| IIA | 5 (11.1) | |
| IIB | 2 (4.4) | |
| IIIA | 14 (31.1) | |
| Histopathology | Adenocarcinoma | 43 (95.6) |
| Squamous cell carcinoma | 2 (4.4) | |
| Site of post-operative recurrence | Pleura | 9 (20.) |
| Lung | 22 (48.9) | |
| Bone | 12 (26.7) | |
| Brain | 8 (17.8) | |
| Liver/spleen | 2 (4.4) | |
| Mediastinal lymph node | 11 (24.4) |
Shifts of EGFR mutation status at recurrence
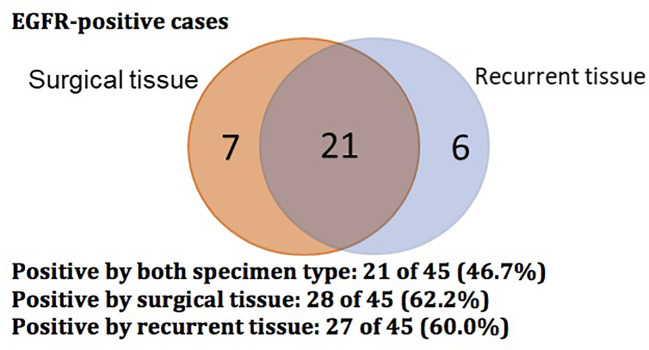
The EGFR mutational status changed in 17 patients (37.8%) at post-operative recurrence. The rest of 28 patients (62.2%) had no change (17 mutations, 11 wild-type; Tables 2 , 3 ; Figure 1 ). We divided these cases into 4 groups by comparing results of sequencing matched primary and post-operative recurrent NSCLC tumors ( Table 3 ):
Table 2.
Alteration of EGFR Mutation Status.
| Surgical Specimen | ||
|---|---|---|
| Recurrence | Mutant (N, %) | Wild type (N, %) |
| Mutant | 21 (46.7) | 6 (13.3) |
| Wildtype | 7 (15.5) | 11 (24.5) |
Table 3.
Transformation of genome.
| No. | Gender | Age | pTNM | Surgical tissue | Postoperative adjuvant therapy | Site(s) of recurrence | Recurrence specimen | Current treatment | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | Blood | |||||||||
| Wild type→Mutant | 1 | Female | 61 | IIB | Wild type | Chemotherapy | Bilateral lung Mediastinal lymph node Pleura |
EGFR exon21 p.L858R | EGFR exon21 p.L858R | Gefitinib |
| 2 | Male | 53 | IA | Wild type | None | Bone | EGFR exon21 L858R | Gefitinib | ||
| 3 | Female | 53 | IIA | Wild type | Chemotherapy | Bilateral lung Mediastinal lymph node Brain |
EGFR exon 21 p.L858R | EGFR exon 21 p.T854A | Gefitinib | |
| 4 | Male | 52 | IIIA | Wild type | Chemoradiotherapy | Brain | EGFR exon19 del | Gefitinib | ||
| 5 | Female | 70 | IIIA | Wild type | Chemoradiotherapy | Bone Bilateral lung |
EGFR exon 21 p.L858R | EGFR exon 21 p.L858R | Gefitinib | |
| 6 | Male | 43 | IB | Wild type | Chemotherapy | Pleural Lung |
EGFR exon19 del | EGFR exon19 del | Icotinib | |
| Mutant→Wild type | 7 | Male | 70 | IA | EGFR exon19 del | None | Pleura | EGFR p.A767delinsAQRG | EGFR p.A767delinsAQRG | Gefitinb |
| 8 | Female | 48 | IA | EGFR exon19 del | None | Bilateral lung | Wild type | Wild type | Icotinib | |
| 9 | Male | 58 | IIB | EGFR exon19 del | Chemotherapy | Bilateral lung | Wild type | Wild type | Gefitinib | |
| 10 | Female | 54 | IIA | EGFR exon19 del | Chemoradiotherapy | Mediastinal lymph node | Wild type | Wild type | Gefitinib | |
| 11 | Male | 56 | IA | EGFR exon19 del | None | Bone | Wild type | Gefitinbi | ||
| 12 | Male | 50 | IA | EGFR exon 21 p.L858R | None | Pleura | Wild type | Wild type | Gefitinbi | |
| 13 | Male | 67 | IIIA | EGFR exon19 del | Chemoradiotherapy | Brain | Wild type | Icotinib | ||
| EGFR Co-Mutation | 14 | Male | 56 | IIA | EGFR exon19 del | Chemotherapy | Mediastinal lymph node Bilateral Lung Liver Spleen |
EGFR exon19 del TP53 p.R273C exon8 |
EGFR exon19 del TP53 p.R273C exon8 |
Gefitinib |
| 15 | Female | 53 | IA | EGFR exon19 del | None | Bilateral lung Bone |
EGFR exon19 del TP53 exon10 p.R342 |
EGFR exon19 del TP53 exon10 p.R342 |
Icotinib | |
| 16 | Male | 54 | IB | EGFR exon 21 p.L858R | Chemotherapy | Pleura |
EGFR p.L858R exon21 PIK3CA p.E542K exon10 |
EGFR p.L858R exon21 PIK3CA p.E542K exon10 |
Gefitinib | |
| Mutation consistent | 17 | Male | 79 | IB | EGFR exon19 del | Chemotherapy | Bilateral lung | EGFR exon21 p.L858R | EGFR exon21 p.L858R | Icotinib |
| 18 | Male | 55 | IA | EGFR exon19 del | None | Bilateral lung | EGFR exon19 del | EGFR exon19 del | Gefitinib | |
| 19 | Male | 59 | IA | EGFR exon19 del | None | Mediastinal lymph node | EGFR exon19 del | EGFR exon19 del | Gefitinib | |
| 20 | Male | 63 | IB | EGFR exon19 del | Chemotherapy | Bone Bilateral lung |
EGFR exon19 del | EGFR exon19 del | Ictonitb | |
| 21 | Male | 60 | IB | EGFR exon19 del | Chemotherapy | Bilateral lung | EGFR exon19 del | EGFR exon19 del | Icotinib | |
| 22 | Male | 72 | IB | EGFR exon 21 p.L858R | Chemotherapy | Mediastinal lymph node Bilateral lung |
EGFR exon 21 p.L858R | EGFR exon 21 p.L858R | Gefitinib | |
| 23 | Female | 50 | IIIA | EGFR exon19 del | Chemoradiothearpy | Bilateral lung | EGFR exon19 del | EGFR exon19 del | Gefitinib | |
| 24 | Female | 58 | IIIA | EGFR exon19 del | Chemoradiotherapy | Mediastinal lymph node | EGFR exon19 del | EGFR exon19 del | Erlotinib | |
| 25 | Male | 46 | IIIA | EGFR exon19 del | Chemoradiotherapy | Bilateral lung | EGFR exon19 del | EGFR exon19 del | Gefitinib | |
| 26 | Male | 49 | IIIA | EGFR exon19 del | Chemoradiothearpy | Bone | EGFR exon19 del | Gefinitib | ||
| 27 | Female | 62 | IIA | EGFR exon 21 p.L858R | Chemotherapy | Bone | APOB exon29 p.I4381L,ARAF exon7 p.R188H, SPTA1 exon37 p.E1761G | Gefitinib | ||
| 28 | Female | 59 | IB | EGFR exon19 del | Chemotherapy | Pleural | EGFR exon19 del | EGFR exon19 del | ||
| 29 | Female | 60 | IA | EGFR exon19 del | None | Bone | EGFR exon19 del | Icotinib | ||
| 30 | Female | 55 | IA | EGFR exon 21 p.L858R | None | Bone Bilateral lung |
EGFR exon 21 p.L858R | EGFR exon 21 p.L858R | Gefitinib | |
| 31 | Female | 61 | IA | EGFR exon19 del | None | Bilateral lung | EGFR exon19 del | EGFR exon19 del | Icotinib | |
| 32 | Male | 62 | IIIA | EGFR exon 21 p.L858R | Chemoradiotherapy | Pleural | EGFR exon 21 p.L858R | EGFR exon 21 p.L858R | Erlotinbi | |
| 33 | Male | 56 | IIIA | EGFR exon19 del | Chemoradiotherapy | Mediastinal lymph node Bilateral lung |
EGFR exon19 del | EGFR exon19 del | Icotinib | |
| 34 | Female | 60 | IIIA | EGFR exon 21 p.L858R | Pleural Bone |
EGFR exon 21 p.L858R | EGFR exon 21 p.L858R | Gefitinib | ||
| Wild type consistent | 35 | Male | 55 | IIA | Wild type | Chemoradiotherapy | Bone Liver |
Wild type | Wild type | Chemotherapy |
| 36 | Male | 55 | IIIA | Wild type | Chemoradiotherapy | Bilateral lung | Wild type | Wild type | Chemotherapy | |
| 37 | Male | 44 | IIIA | Wild type | Chemoradiotherapy | Brain | Wild type | Chemotherapy | ||
| 38 | Male | 57 | IB | Wild type | Chemotherapy | Brain | Wild type | Chemotherapy | ||
| 39 | Male | 64 | IB | Wild type | Chemotherapy | Bilateral lung Mediastinal lymph node |
Wild type | Wild type | Chemotherapy | |
| 40 | Female | 60 | IB | Wild type | Chemothearpy | Bilateral lung Mediastinal lymph node |
Wild type | Wild type | Chemotherapy | |
| 41 | Male | 66 | IIA | Wild type | Chemotherapy | Bilateral lung Mediastinal lymph node |
Wild type | Wild type | Chemotherapy | |
| 42 | Male | 66 | IIA | Wild type | Chemotherapy | Bone | Wild type | Chemotherapy | ||
| 43 | Male | 74 | IA | Wild type | None | Pleural | Wild type, NRAS p.A146T | Wild type, NRAS p.A146T | Chemotherapy | |
| 44 | Male | 60 | IB | Wild type | Chemothearpy | Brain | Wild type | Chemotherapy | ||
| 45 | Female | 54 | IA | Wild type | None | Brain | Wild type | Chemotherapy | ||
Figure 1.
Venn diagram showing EGFR-positive status of 45 cases with surgical and recurrent specimens.
Group A (18 cases, 40%): sustained sensitive EGFR mutations in both primary and recurrent tumors. Group B (6 cases, 13.3%): EGFR wild type in the primary tumors but EGFR sensitive mutation in the recurrent tumors.
Group C (10 cases, 22.2%): 7 cases had EGFR mutations in the primary tumors but wild type at recurrence, of which, 1 case had EGFR p.A767delinsAQRG, and 3 patients developed an EGFR-co-mutation in the recurrent specimens: 1) EGFR exon19 del and TP53 p.R273C exon8; 2) EGFRexon19 del and TP53 exon10 p.R342, and 3) EGFR p.L858R exon21 and PIK3CA p.E542K exon10.
Group D (11 cases, 24.4%): Sustained EGFR wild-type in both primary and recurrent specimens.
Clinical responses in groups with different EGFR mutation shift
At recurrence, patients with a sensitive EGFR mutation were treated with first-line EGFR-TKI, and those in the wild-type group received chemotherapy. Twenty-two patients had partial response, 21 patients had stable disease, and 2 cases showed disease progression.
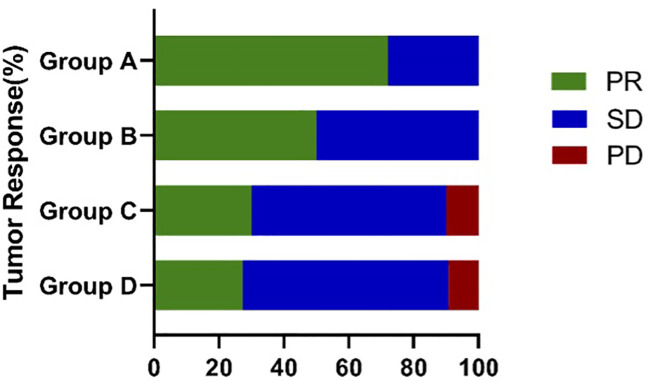
After receiving the first-line treatment, the disease control rates were high: 100% (Group A), 100% (Group B), 90% (Group C), and 90.0% (Group D). The highest PR rate of 72.2% was observed in Group A with sustained sensitive EGFR mutations and received EGFR TKIs. Patients in group B had a PR rate of 50%. In group C, the PR rate was 30%. In group D with sustained EGFR wild-type, the PR rate with chemotherapy was 27.3% ( Table 4 ; Figure 2 ). Likely due to small sample size, there was no significant difference in PR rate among the four groups (χ2 = 7.273, P=0.061; Table 5 ), but patients in group A had higher PR rates than those in group D 185 (χ2 = 5.730, P=0.027).
Table 4.
Tumor response of first-line EGFR-TKI or chemotherapy in patients.
| Tumor Response | Total No. | |||||||
|---|---|---|---|---|---|---|---|---|
| Partial Response | Stable Disease | Progressive Disease | ||||||
| Group | Treatment | No. | % | No. | % | No. | % | |
| A | EGFR-TKI | 13 | 72.2 | 5 | 27.8 | 0 | 0.0 | 18 |
| B | EGFR-TKI | 3 | 50.0 | 3 | 50.0 | 0 | 0.0 | 6 |
| C | EGFR-TKI | 3 | 30.0 | 6 | 60.0 | 1 | 10.0 | 10 |
| D | Chemotherapy | 3 | 27.3 | 7 | 63.6 | 1 | 9.1 | 11 |
| Total | 22 | 48.9 | 21 | 46.7 | 2 | 4.4 | 45 | |
Figure 2.
Response outcome between different groups.
Table 5.
PR rate among groups.
| Groups | χ2 | P |
|---|---|---|
| A group vs B group | 0.965 | 0.362 |
| A group vs C group | 4.755 | 0.050 |
| A group vs D group | 5.730 | 0.027 |
| B group vs C group | 0.635 | 0.607 |
| B group vs D group | 0.866 | 0.600 |
| C group vs D group | 0.019 | 1.000 |
Management of brain metastases at time of recurrence
Most patients with brain metastases at recurrence had mild symptoms only. Patients with EGFR mutations and brain metastases detected at time of recurrence were started on EGFR targeted therapy and had repeat MRI brain at 1 month following treatment initiation. Patients did not receive brain radiotherapy if the brain metastases had significantly improved following one month of EGFR TKI therapy. Wild type patients with brain metastases were treated with radiation therapy in addition to chemotherapy if symptomatic.
Survival outcomes in groups with different EGFR mutation shift
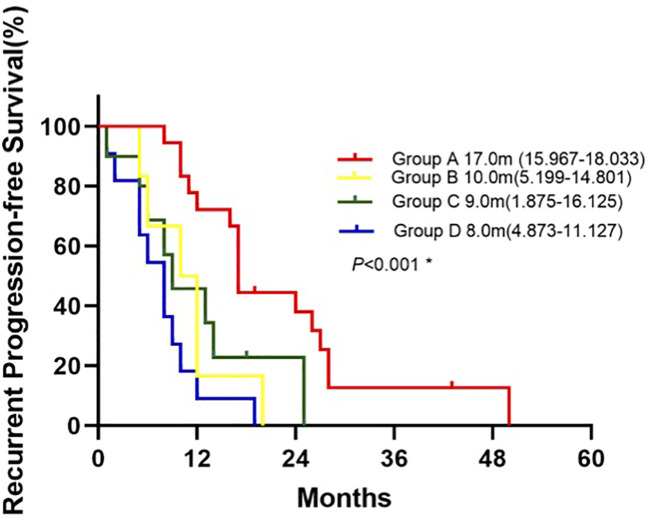
Patients in Group A had the longest median rPFS of the four groups at 17.0 months [95% CI 15.967-18.033], compared to 10.0 months with group B [95% CI 5.199-14.801], 9.0 months with group C [95% CI 1.875-16.125], and 8.0 months with group D [95% CI 4.873-11.127]. The difference in rPFS was statistically significant between patients in Group A and Group B(P=0.008), Group A and Group C (P=0.016), Group A and Group D (P = 0.001) ( Figure 3 ). Univariate analysis identified mutations in both surgical and recurrent specimens as being significantly associated with better PFS (HR 0.172, 95% CI 0.071-0.421; P <0.001) ( Table 6 ). After adjusting for confounding factors, the multivariate analysis showed that sustained EGFR mutations remained as predictors of better PFS (HR 0.163, CI 0.064-0.415; P <0.001).
Figure 3.
Kaplan-Meier curve for recurrent progression-free survival of patients between groups.*Log-rank test: Group A vs. Group B(P=0.008), Group A vs. Group C (P=0.016), Group A vs. Group D(P<0.001), Group B vs. Group C(P=0.620), Group B vs. Group D(P=0.163), Group C vs. Group D(P=0.156).
Table 6.
Predictors on prognostic factors in Cox Regression Analysis for 45 NSCLC Patients.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| rPFS | ||||||
| Gender | ||||||
| Female | 1.000 | 1.000 | ||||
| Male | 1.320 | 0.690-2.526 | 0.402 | 0.981 | 0.402-2.393 | 0.967 |
| Smoking history | ||||||
| NO | 1.000 | 1.000 | ||||
| YES | 1.570 | 0.833-2.958 | 0.163 | 1.404 | 0.577-3.416 | 0.454 |
| Age Group | ||||||
| Group D | 1.000 | |||||
| Group A | 0.172 | 0.071-0.421 | 0.000 | 0.163 | 0.064-0.415 | 0.000 |
| Group B | 0.583 | 0.213-1.595 | 0.293 | 0.535 | 0.182-1.567 | 0.254 |
| Group C | 0.491 | 0.194-1.243 | 0.133 | 0.515 | 0.201-1.321 | 0.167 |
| rOS | ||||||
| Gender | ||||||
| Female | 1.000 | 1.000 | ||||
| Male | 1.497 | 0.665-3.367 | 0.329 | 1.396 | 0.451-4.321 | 0.563 |
| Smoking history | ||||||
| NO | 1.000 | 1.000 | ||||
| YES | 1.499 | 0.691-3.249 | 0.305 | 0.895 | 0.289-2.773 | 0.848 |
| Age Group | ||||||
| Group D | 1.000 | 1.000 | ||||
| Group A | 0.368 | 0.141-0.961 | 0.041 | 0.361 | 0.131-1.000 | 0.050 |
| Group B | 0.693 | 0.212-2.267 | 0.544 | 0.817 | 0.236-2.833 | 0.750 |
| Group C | 0.463 | 0.154-1.388 | 0.169 | 0.556 | 0.170-1.821 | 0.332 |
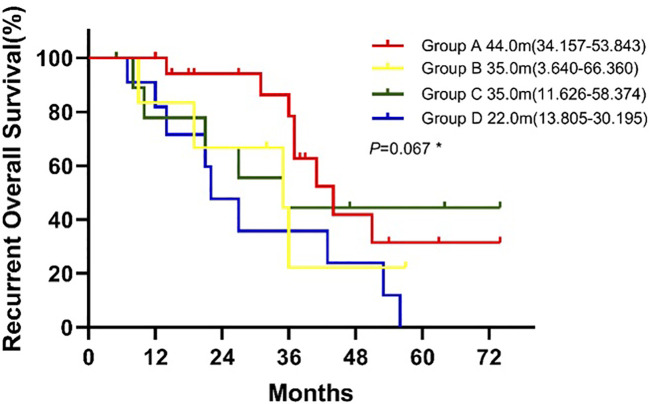
The median rOS in four groups were 44.0 months (95% CI 34.157-53.843 for group A, 35.0 months (95% CI, 3.640-66.360) for group B, 35.0 months (95% CI 11.626-58.374) for group C, and 22.0 months (95% 13.805-30.195) for group D. Patients in group A had a significantlyy longer OS than patients in group D (P = 0.039; Figure 4 ). Compared with group D, univariate analysis showed mutations in both surgical and recurrent specimens as being significantly associated with favorable OS (HR 0.368, 95% CI 0.141-0.961; P=0.041).
Figure 4.
Kaplan-Meier curve for recurrent overall survival of patients between groups. * Log-rank test: Group A vs. Group B(P=0.171), Group A vs. Group C (P=0.641), Group A vs. Group D(P=0.039), Group B vs. Group C(P=0.715), Group B vs. Group D(P=0.482), Group C vs. Group D(P=0.182).
Discussion
Tumor heterogeneity, with genetic and non-genetic diversity, is a key feature of cancer (3). Spatial heterogeneity may be observed in different parts of a given tumor, and temporal heterogeneity may occur with recurrent or metastatic disease. Tumor heterogeneity may result from distinct clonal evolution, mutational processes, and selection pressures. NSCLC, especially lung adenocarcinoma, is one of the most heterogeneous cancers. Drugs, environment, and other factors promote dynamic, competitive evolution of tumor subclones, and dominant clones arise by survival of the fittest (6). Failure to recognize and detect tumor heterogeneity may lead to treatment failure in lung cancer (7). Therefore, it is important to monitor the dynamics of oncogenic driver mutations to guide the treatment of lung cancer in an individualized and precise manner.
Activating somatic mutations in the EGFR receptor tyrosine kinase are the most common targetable driver alterations identified in NSCLC, occurring in up to 16% patients with adenocarcinoma (8). EGFR belongs to a family of receptor tyrosine kinases known as ERBB. Under physiologic conditions, EGFR activates multiple downstream signaling pathways including mitogen-activated protein kinases (MAPK), PI3K/AKT/mTOR, and JAK/STAT leading to cellular proliferation and oncogenesis (9). There are three generations of EGFR TKIs currently in clinical practice with varying mechanisms of action and efficacy for particular mutation subtypes (10). First generation EGFR TKIs, including gefitinib and erlotinib, reversibly compete with ATP for binding to the EGFR tyrosine kinase receptor preventing downstream signaling. Second generation EGFR TKIs including neratinib, dacomitinib and afatinib irreversibly bind to the EGFR receptor tyrosine kinase domain and target other members of the ERBB family. While NSCLC with EGFR exon 19 deletions of exon 21 L858R mutations, those harboring the T90M mutation are associated with resistance. The third generation EGFR TKIs such as osimertinib form an irreversible covalent bond with the cysteine-797 residue in the EGFR ATP binding site, with potent activity against the EGFR-T790M mutation. Identification of EGFR mutations and development of selective EGFR inhibitors has revolutionized the management of NSCLC for a molecularly-defined group of patients leading to improved clinical outcomes (11). In addition, previous studies implicated that EGFR mutation status may differ between paired primary and metastatic NSCLC tumors in a portion of patients. A meta-analysis found that the discordance rate was 14.5% (12), and another study reported a discordance rate of 5%–17% (13). These results indicate that primary or metastatic samples alone are insufficient to reflect genomic features due to temporal heterogeneity.
In our study, we also found discordance between the primary tumor at initial resection and at the time of recurrence. Genotypic changes were observed in 17 patients (37.8%) at post-operative recurrence. Six patients had a mutation from wild type, 7 patients changed to wild type from mutant, 3 patients had developed new EGFR-co-mutation in recurrent specimens, and 1 patient had a sensitive mutation with changed from 19del to L858R. Twenty-eight patients (62.2%) had no change in genotype (17 mutations, 11 wild-type). One possible explanation for the observed genotypic shift from EGFR-mutant to wild type might be due to spatial heterogeneity of the tumor, which could result in an uneven distribution of genetic subpopulations within a single tumor or across disease sites. Another possibility is the potential for a false negative result, which could exist due to low tumor load or inadequate tissue sampling. Collection of additional tissue specimens or blood samples for repeated NGS testing would be one means to more effectively rule this out. As no patients received adjuvant EGFR TKIs after surgical resection, we would not expect the change in mutation status to be related to targeted therapy. Of note, our discordance rate was higher than the ones in previous reports, which may be due to the use of ctDNA analysis at time of recurrence in our study. At disease progression, insufficient material obtained by tissue biopsy may preclude complete sequencing, and this may occur in up to 20–25% of needle biopsies (14, 15). In this case, ctDNA analysis can provide otherwise complimentary sequencing data that could significantly impact treatment decisions.
Importantly, given the potential for tumor heterogeneity, diverse genetic alterations leading to drug resistance may be detected by ctDNA analysis across different metastatic sites that may not be detected by biopsy of a single site (16). Using ctDNA analysis, Paweletz et al. found two mutations (exon19 of EGFR, high levels of MET amplification), which was not previously detected in tumor tissue (17). Previous work also compared mutations found in plasma and in tumor samples from NSCLC patients, and additional mutations were found in ctDNA analysis in several genes: EGFR, KRAS, PIK3CA, and TP53 (18). Another study showed that only 1 of 9 patients had a plasma ctDNA mutational profile that was completely consistent with the mutational profile of the biopsied tumor (19). Nevertheless, there is limitation to characterize tumor heterogeneity using tumor biopsy tissues, whereas plasma ctDNA analysis appears to be a powerful tool to monitor the dynamic changes or heterogeneity in lung cancer. One advantage of ctDNA compared to tissue based NGS is its potential for overcoming the spatial heterogeneity that could exist within a particular tumor, or across various metastatic sites. As a result, ctDNA may characterize the genomic profile of a tumor more comprehensively than a single tissue biopsy alone.
In addition to detecting genetic changes, the dynamic monitoring of mutational status and tumor heterogeneity with ctDNA analysis also predicted response to therapy and helped to guide personalized treatment. Group A, in which there were sensitizing EGFR mutations at initial resection and at recurrence, had the highest PR rate of 72.2%, with a median rPFS of 17.0 months and rOS of 41.0 months. Patients from group B were newly found to have sensitizing EGFR mutations and gained access to EGFR-TKI therapy at recurrence; These patients had a PR rate of 50%, rPFS of 10 months, and rOS of 35.0 months, which appeared to be numerically inferior to group A and tumor heterogeneity may play a role. Group C, which shifted from having a sensitizing EGFR mutation to wild-type, achieved PR in 30%, rPFS of 9 months, and rOS of 35.0 months. In group D, patients received chemotherapy, the PR rate was only 27.3%, with rPFS of 8 months, and rOS of 22.0 months. Overall, patients in group A had significantly better rPFS, rOS and a higher PR rate than those in group D. For patients treated with EGFR-TKI, patients of group A owned longer rPFS than group B and group C, that indicated the low heterogeneity of tumors may associated with better survival although there was no significant difference in survival among other groups, possibly due to small sample size.
NGS analysis can help to identify resistance mechanisms to EGFR-TKI ( 20, 21), such as certain EGFR co-mutations. In such patients harboring EGFR co-mutations, the response to EGFR-targeted therapy was reported to be significantly lower, and survival appeared shorter (22–24). In group C, we found a TP53 co-mutation in 2 patients. Mutated TP53 not only loses its tumor suppressor function but also are often associated with poor prognosis (25), regardless of EGFR status. In one study of 43 patients with TP53 mutations, PFS with first-line EGFR-TKI was significantly shorter compared to those with wild type TP53, but OS was not significantly different (26). The survival of patients taking first-line EGFR-TKI with EGFR and TP53 co-mutations, were shorter than those with only EGFR mutations (PFS 4.2 months vs. 12.5 months and OS 16.2 months vs. 32.3 months) (27). In our study, one patient whose TP53 p.R273C exon 8 allele frequency was 32.6% had taken first-line EGFR-TKI for only 5 months before disease progression and had an OS of 10 months. Another patient had TP53 exon10 p.R342 with an allele frequency of 5.7% and achieved PFS of 8 months.
PIK3CA alterations are also associated with poor prognosis in NSCLC, with significantly shorter PFS and OS compared to wild type (28). Alterations in PIK3CA are associated with both acquired and primary resistance to EGFR-TKIs in 1–3% of patients (28). One patient in our study had a PIK3CA p.E542K exon 10 alteration (allele frequency 0.155%) and was treated with first-line EGFR TKI, but had disease progression after 6 months, with an OS of 21 months. In another case, EGFR exon 19 del was no longer detectable but EGFR P. A767 delins AQRG (allele frequency 5.0%) was found by ctDNA analysis at the time of post-operative recurrence. The patient had PD just one month after initiating EGFR TKI, with OS of 8 months.
Genetic sequencing of the initial tumor specimen is often used to determine therapy at recurrence although tumor spatial heterogeneity may lead to poorer efficacy to targeted therapy (29, 30). In our study, through the analysis of matched primary and post-operative recurrence in 45 patients, we observed two key findings: first, temporal genetic heterogeneity occurred relatively commonly between the primary and post-operative recurrent NSCLC tumors; Second, temporal heterogeneity might influence the therapeutic efficacy of first-line EGFR TKI treatment at relapse. Therefore, the genetic sequencing of surgical specimens should not be used as the sole guide for targeted therapy either at initial diagnosis or recurrence. The addition of ctDNA analysis, which is noninvasive and sensitive, can be used to assess tumor heterogeneity and to guide personalized treatment. In our study, ctDNA results were not available for patients at time of initial resection as ctDNA was not widely used (years 2013-2016); however, it would be interesting to study how ctDNA results at recurrence would compare to those obtained at time of initial resection.
Conclusion
In this study, through dynamic analysis of matched primary and post-operative recurrence in 45 patients, we found that genotypic shift was not infrequent in the relapsed tumors, and this temporal genomic heterogeneity substantially impacted subsequent treatment outcomes. Our study suggests that dynamic evaluation of genomic profile, especially oncogenic drivers such as EGFR mutational status, at cancer recurrence or relapse (or second primary) is warranted to tailor subsequent individualized therapy.
Data availability statement
The data analyzed in this study is subject to the following licenses/restrictions: Data not available due to ethical restrictions. Requests to access these datasets should be directed to songwenzhou2017@vip.126.com.
Ethics statement
The studies involving human participants were reviewed and approved by Shanghai Pulmonary Hospital. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
Study conception and design: SZ, CZ, BX, YL. Acquisition of data: QF, XW, QD and HS. Analysis and interpretation of data: QF, XW, QD, HS,WG, AD, EL, and HC. Drafting and revising: QF, XW, QD, SZ, CZ, BX, AD, EL, HC and YL. Approving final version of manuscript: All authors. SZ, CZ, BX, YL take responsibility for the integrity of the data analysis. All authors contributed to the article and approved the submitted version.
Funding Statement
This study was funded by National Natural Science Foundation of China (81301994); Fundamental Research Funds for the Central Universities (22120180369).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1116809/full#supplementary-material
References
- 1. van den Berg LL, Klinkenberg TJ, Groen HJM, Widder J. Patterns of recurrence and survival after surgery or stereotactic radiotherapy for early stage NSCLC. J Thorac Oncol (2015) 10(5):826–31. doi: 10.1097/JTO.0000000000000483 [DOI] [PubMed] [Google Scholar]
- 2. Yamazaki K, Sugio K, Yamanaka T, Hirai F, Osoegawa A, Tagawa T, et al. Prognostic factors in non-small cell lung cancer patients with postoperative recurrence following third-generation chemotherapy. Anticancer Res (2010) 30(4):1311–5. [PubMed] [Google Scholar]
- 3. Ramón Y Cajal S, Sesé M, Capdevila C, Aasen T, De Mattos-Arruda L, Diaz-Cano SJ, et al. Clinical implications of intratumor heterogeneity: challenges and opportunities. J Mol Med (Berl) (2020) 98(2):161–77. doi: 10.1007/s00109-020-01874-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rolfo C, Mack P, Scagliotti GV, Aggarwal C, Arcila ME, Barlesi F, et al. Liquid biopsy for advanced NSCLC: a consensus statement from the international association for the study of lung cancer. J Thorac Oncol (2021) 16(10):1647–62. doi: 10.1016/j.jtho.2021.06.017 [DOI] [PubMed] [Google Scholar]
- 5. Deng Q, Fang Q, Sun H, Singh AP, Alexander M, Li S, et al. Detection of plasma EGFR mutations for personalized treatment of lung cancer patients without pathologic diagnosis. Cancer Med (2020) 9(6):2085–95. doi: 10.1002/cam4.2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Amirouchene-Angelozzi N, Swanton C, Bardelli A. Tumor evolution as a therapeutic target. Cancer Discov (2017) 7:805–17. doi: 10.1158/2159-8290.CD-17-0343 [DOI] [PubMed] [Google Scholar]
- 7. Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med (2012) 4(127):127ps10. doi: 10.1126/scitranslmed.3003854 [DOI] [PubMed] [Google Scholar]
- 8. Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med (2009) 361(10):958–67. doi: 10.1056/NEJMoa0904554 [DOI] [PubMed] [Google Scholar]
- 9. Roskoski R. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res (2014) 79:34–74. doi: 10.1016/j.phrs.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 10. Chhouri H, Alexandre D, Grumolato L. Mechanisms of acquired resistance and tolerance to EGFR targeted therapy in non-small cell lung cancer. Cancers (Basel) (2023) 15(2):504. doi: 10.3390/cancers15020504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kitadai R, Okuma Y. Treatment strategies for non-small cell lung cancer harboring common and uncommon EGFR mutations: drug sensitivity based on exon classification, and structure-function analysis. Cancers (Basel) (2022) 14(10):219. doi: 10.3390/cancers14102519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang S, Wang Z. Meta-analysis of epidermal growth factor receptor and KRAS gene status between primary and corresponding metastatic tumours of non-small cell lung cancer. Clin Oncol (R Coll Radiol) (2015) 27(1):30–9. doi: 10.1016/j.clon.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 13. Sherwood J, Dearden S, Ratcliffe M, Walker J. Mutation status concordance between primary lesions and metastatic sites of advanced non-small-cell lung cancer and the impact of mutation testing methodologies: a literature review. J Exp Clin Cancer Res (2015) 34:92. doi: 10.1186/s13046-015-0207-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zill OA, Greene C, Sebisanovic D, Siew LM, Leng J, Vu M, et al. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov (2015) 5(10):1040–8. doi: 10.1158/2159-8290.CD-15-0274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meric-Bernstam F, Brusco L, Shaw K, Horombe C, Kopetz S, Davies MA, et al. Feasibility of Large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J Clin Oncol (2015) 33(25):2753–62. doi: 10.1200/JCO.2014.60.4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med (2018) 379(18):1754–65. doi: 10.1056/NEJMra1706174 [DOI] [PubMed] [Google Scholar]
- 17. Paweletz CP, Sacher AG, Raymond CK, Alden RS, O'Connell A, Mach SL, et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res (2016) 22(4):915–22. doi: 10.1158/1078-0432.CCR-15-1627-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu S, Lou F, Wu Y, Sun DQ, Zhang JB, Chen W, et al. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett (2016) 370(2):324–31. doi: 10.1016/j.canlet.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vanni I, Coco S, Truini A, Rusmini M, Dal Bello MG, Alama A, et al. Next-generation sequencing workflow for NSCLC critical samples using a targeted sequencing approach by ion torrent PGM™ platform. Int J Mol Sci (2015) 16(12):28765–82. doi: 10.3390/ijms161226129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee CK, Kim S, Lee JS, Lee JE, Kim SM, Yang IS, et al. Next-generation sequencing reveals novel resistance mechanisms and molecular heterogeneity in EGFR-mutant non-small cell lung cancer with acquired resistance to EGFR-TKIs. Lung Cancer (2017) 113:106–14. doi: 10.1016/j.lungcan.2017.09.005 [DOI] [PubMed] [Google Scholar]
- 21. Hinrichs JW, van Blokland WT, Moons MJ, Radersma RD, Radersma-van Loon JH, de Voijs CM, et al. Comparison of next-generation sequencing and mutation-specific platforms in clinical practice. Am J Clin Pathol (2015) 143(4):573–8. doi: 10.1309/AJCP40XETVYAMJPY [DOI] [PubMed] [Google Scholar]
- 22. Barnet MB, O'Toole S, Horvath LG, Selinger C, Yu B, Ng CC, et al. EGFR-Co-Mutated advanced NSCLC and response to EGFR tyrosine kinase inhibitors. J Thorac Oncol (2017) 12(3):585–90. doi: 10.1016/j.jtho.2016.09.001 [DOI] [PubMed] [Google Scholar]
- 23. Kim EY, Cho EN, Park HS, Hong JY, Lim S, Youn JP, et al. Compound EGFR mutation is frequently detected with co-mutations of actionable genes and associated with poor clinical outcome in lung adenocarcinoma. Cancer Biol Ther (2016) 17(3):237–45. doi: 10.1080/15384047.2016.1139235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen JY, Cheng YN, Han L, Wei F, Yu WW, Zhang XW, et al. Predictive value of K-ras and PIK3CA in non-small cell lung cancer patients treated with EGFR-TKIs: a systemic review and meta-analysis. Cancer Biol Med (2015) 12(2):126–39. doi: 10.7497/j.issn.2095-3941.2015.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Canale M, Petracci E, Delmonte A, Chiadini E, Dazzi C, Papi M, et al. Impact of. Clin Cancer Res (2017) 23(9):2195–202. doi: 10.1158/1078-0432.CCR-16-0966 [DOI] [PubMed] [Google Scholar]
- 26. Labbé C, Cabanero M, Korpanty GJ, Tomasini P, Doherty MK, Mascaux C, et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer (2017) 111:23–9. doi: 10.1016/j.lungcan.2017.06.014 [DOI] [PubMed] [Google Scholar]
- 27. Jakobsen JN, Santoni-Rugiu E, Grauslund M, Melchior L, Sørensen JB. Concomitant driver mutations in advanced. Oncotarget (2018) 9(40):26195–208. doi: 10.18632/oncotarget.25490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eng J, Woo KM, Sima CS, Plodkowski A, Hellmann MD, Chaft JE, et al. Impact of concurrent PIK3CA mutations on response to EGFR tyrosine kinase inhibition in EGFR-mutant lung cancers and on prognosis in oncogene-driven lung adenocarcinomas. J Thorac Oncol (2015) 10(12):1713–9. doi: 10.1097/JTO.0000000000000671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yao S, Zhi X, Wang R, Qian K, Hu M, Zhang Y. Retrospective study of adjuvant icotinib in postoperative lung cancer patients harboring epidermal growth factor receptor mutations. Thorac Cancer (2016) 7(5):543–8. doi: 10.1111/1759-7714.12365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takenaka T, Inamasu E, Yoshida T, Toyokawa G, Nosaki K, Hirai F, et al. Post-recurrence survival of elderly patients 75 years of age or older with surgically resected non-small cell lung cancer. Surg Today (2016) 46(4):430–6. doi: 10.1007/s00595-015-1200-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed in this study is subject to the following licenses/restrictions: Data not available due to ethical restrictions. Requests to access these datasets should be directed to songwenzhou2017@vip.126.com.