Abstract
Ovarian cancer (OC), particularly high-grade serous cancer (HGSC), is the leading cause of mortality among gynecological cancers owing to the treatment difficulty and high recurrence probability. As therapeutic drugs approved for OC, poly ADP-ribose polymerase inhibitors (PARPi) lead to synthetic lethality by inhibiting single-strand DNA repair, particularly in homologous recombination-deficient cancers. However, even PARPi have distinct efficacies and are prone to have drug resistance, the molecular mechanisms underlying the PARPi resistance in OC remain unclear. A patient-derived organoid platform was generated and treated with a PARPi to understand the factors associated with PARPi resistance. PARPi significantly inhibits organoid growth. After 72 h of treatment, both the size of organoids and the numbers of adherent cells decreased. Moreover, immunofluorescence results showed that the proportion of Ki67 positive cells significantly reduced. When the PARPi concentration reached 200 nM, the percentage of Ki67+/4′,6-diamidino-2-phenylindole (DAPI) cells decreased approximately 50%. PARPi treatment also affected the expression of genes involved in base excision repair and cell cycle. Functional assays revealed that PARPi inhibits cell growth by upregulating early apoptosis. The expression levels of several key genes were validated. In addition to previously reported genes, some promising genes FEN1 and POLA2, were also be founded. The results demonstrate the complex effects of PARPi treatment on changes in potential genes relevant to PARPi resistance, and provide perspectives for further research on the PARPi resistance mechanisms.
Keywords: apoptosis, base excision repair, cell cycle, ovarian cancer organoid, PARPi
Introduction
Ovarian cancer (OC) is a common cancer in females, accounting for 2.5% of all females and has an extremely high mortality rate 1 . Owing to the difficulty of early diagnosis, OC is often diagnosed at an advanced stage and considered as intractable. Poly ADP-ribose polymerase (PARP) plays an important role in single-stranded DNA repair and base excision repair pathways 2 . PARP inhibitors (PARPi) prevent single-strand DNA breaks (SSB) repair and allows SSB accumulation to develop into double-strand breaks (DSB) that require homologous recombination (HR)3,4. This results in synthetic lethality in HR-deficient cancer cells, and has been widely studied 3 . Several PARPis, including olaparib, niraparib, and rucaparib 3 , have been approved as a novel therapeutic option for treating OC in the clinical setting and presented significant excitement.
However, clinical treatment with PARPi remains intractable. PARPis are effective as maintain therapy for OC patients with BRCA mutations, but more than 40% of these patients still do not benefit from PARPi 5 . Platinum resistance has been reported in a patient with gBRCAmt recurrent OC after PARPi resistance 6 . Thus, the increasing use of PARPi in clinical practice has increased PARPi resistance. Therefore, the mechanisms of PARPi resistance should be identified to address this problem and provide guidance for early prevention.
HR restoration, DNA replication fork stabilization, BRCA reversion mutations, increased drug efflux, PARP1 and PAR glycohydrolase (PARG) dissociation, and epigenetic molecular modifications are the most common mechanisms of PARPi resistance. Although multiple mechanisms are involved, these are only some mechanisms contributing to PARPi resistance 7 . As an increasing number of patients receive initial therapy and potential re-treatment with PARPi, a drug-susceptibility testing platform and a clear understanding of the mechanisms by which tumors acquire PARPi resistance are required.
By exploiting our patient-derived organoid platform, we developed organoid models with high similarity to clinical OC tissues. PARPi successfully inhibited organoid growth and reduced the proportion of Ki67-positive cancer cells; however, some Ki67-positive cells survived at high PARPi concentrations, indicating that PARPi resistance was reproduced on organoid platforms. In this study, we investigated the impact of PARPi treatment on OC organoids and attempted to identify the factors associated with PARPi resistance. Several genes involved in base excision repair and cell cycle pathways were also altered.
Materials and Methods
Patient Characteristics
We derived organoids from several representative patients and enrolled one patient with stage IV high-grade serous cancer (HGSC), who underwent surgery a bilateral salpingectomy in October 2022 and did not undergo any radiotherapy or chemotherapy before surgery. In this study, surgical samples were used in the experiments after collecting informed consent and approval from the Ethics Committee of the Obstetrics and Gynecology Hospital of Fudan University.
Organoid Culture
Fresh obtained clinical tissues were washed three times with an organoid washing buffer (LSTO00100201; Shanghai LiSheng Biotech, China) to rinse the mucus and debris. Then, the tissues were mechanically sheared using and Ovarian Cancer Tissue Sampling Kit (LSTO00100101, Shanghai LiSheng Biotech, China). Next, 3- to 6-mm tissue debris was seeded in 6-well culture plates (#3516, Costar) and cultured with 6-mL OC organoid medium (LSTO001004, Shanghai LiSheng Biotech, China) in a Heracell™ Vios 160i CR CO2 incubator (51033770, ThermoFisher) at 37℃ under a humidified atmosphere with 5% CO2. Approximately 50% medium was changed every 5 days, to provide sufficient nutrients for the organoids. After the tumor pieces developed into organoids, they were passaged every 15 days according to the manufacturer’s instructions. If the pieces were circular and presented the characteristics of the parental tumor within 7 days, the culture was regarded as successful.
Drug Treatment Assay
The mechanically disrupted organoids were cultured in 6- and 24-well plates (#3516 and #3337, respectively; Costar) for 8 h and treated with the indicated concentrations of olaparib (AZD2281, Selleck). For gene expression analysis, RNA was extracted from a portion of the cells in 6-well plates cultured for 48 h and sequenced. The cells cultured for 24 and 72 h were analyzed by flow cytometry. To measure the growth and morphological changes in organoids, adherent cells in 24-well plates and organoids in 6-well plates were observed daily using a Leica DMi1 inverted microscope and imaged at 24, 48, and 72 h. Organoids in 6-well plates were sliced into frozen sections for immunofluorescence and histological analyses. The results were normalized to those of the dimethyl sulfoxide (DMSO) controls.
Histology and Immunofluorescence
Tissues and organoids were fixed in 4% paraformaldehyde (PFA) (BL539A, Biosharp) for 30 min, dehydrated with sucrose, and embedded in 7.5% gelatin for standard histology and immunofluorescence (IF). Frozen sections (10 mm) of the embedded samples were retrieved using citric acid (PH6.0). The adherent cells were fixed using 4% PFA for 20 min. Both adherent cells and frozen sections were permeabilized using 0.25% Triton X-100 in PBST, and blocked with Primary Antibody Dilution Buffer (E674004, Sangon Biotech, China). After overnight incubation with 1:1000 primary antibody Rabbit anti-PAX8 (10336-1-AP, Proteintech) and anti-Ki67 (MA5-14520, ThermoFisher) at 4°C, sections were washed two times in 0.125% PBST and incubated at room temperature with 1:1000 secondary antibody anti-rabbit (Cy3) (711-165-152, Jackson). Then, they were incubated with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) (D1306, ThermoFisher) stain solution at room temperature. After washing three times with 0.125% PBST, they were scanned using a Keyence BZ-X810 Fluorescence Microscope and the proportions of immunoreactive cells were counted. For H&E staining, the sections were washed twice and stained using a hematoxylin-eosin (HE) Stain Kit (G1120, Solarbio, China) according to the manufacturer’s instructions.
Flow Cytometry
The adherent cells were collected and resuspended in fresh PBS. Apoptosis was analyzed by flow cytometry using an Agilent NovoCyte Penteon instrument with an Annexin V/PI kit (BD Biosciences) according to the manufacturer’s instructions.
RNA Sequencing
RNA was extracted from adherent cells and several floating organoids culture for 48 h using the RNA Easy Fast Tissue/Cell Kit (4992732, Tiangen, China) and the purity was assessed using Qubit 3.0 Fluorometer (ThermoFisher, Waltham, England). After constructing the RNA library followed by NEBNext Ultra RNA library for Illumina (Biolab, England), RNA was sequenced on Illumina Hiseq 4000 using 150 bp paired-end reads. The sequencing results of fragments per kilobase of transcript per million mapped reads (FPKM) were analyzed. Differential gene expression analysis was performed using online software (Morpheus, https://software.broadinstitute.org/morpheus). An online database (g: Profiler) was used for analyzing the molecular functions, biological processes, and cell cycles.
Gene Expression Analysis
RNA was reverse-transcribed using FastKing gDNA Dispelling RT SuperMix (KR118, Tiangen, China). For quantitative real-time PCR (qPCR), gene-specific forward and reverse primers were used to amplify the samples with LightCycler 96 Instrument (Roche). The primer sequences are listed in Supplementary Table 1. The target gene expression levels were normalized to those of the housekeeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ribosomal protein 18 (18S). Relative gene expression levels were calculated as 2−(∆∆Ct) values.
Statistical Analysis
To determine the cutoff points for qPCR and IHC, receiver operating characteristic curves were analyzed. The Student’s t-test was used to determine significant differences between multiple statistical comparisons. Data analysis and graph drawing were performed using GraphPad Prism version 8. *P < 0.05 was considered significant.
Results
OC-Derived Organoids Were Highly Similar to Clinical Tissues
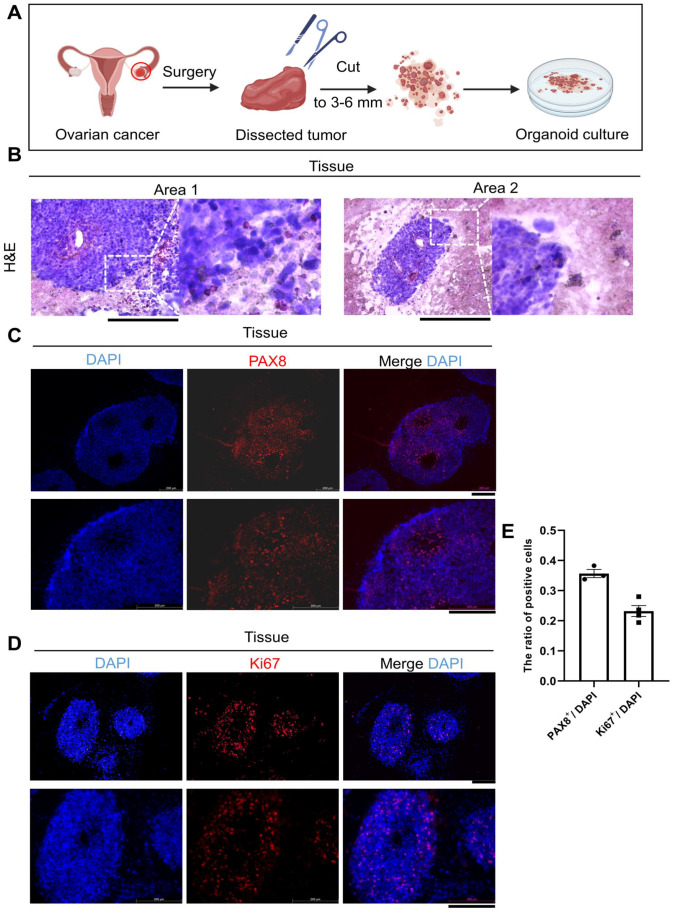
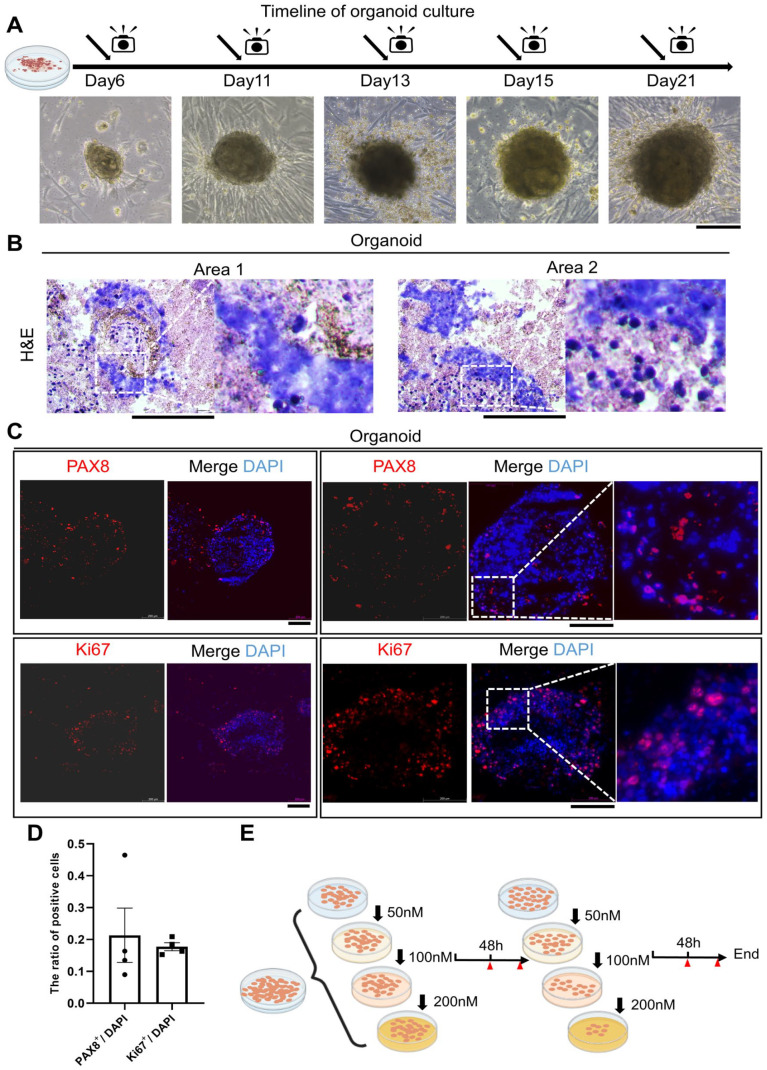
OC tissues obtained from consenting patient were dissociated and seeded in a commercial OC organoid culture medium (Fig. 1A). Histological image and immunohistological analyses were performed for one clinical tissue and one organoid to assess the similarity between the organoid and parental tumors. The morphology of organoids directly cultured from clinical tissues were highly coherent (Figs. 1B, 2A, B). PAX8 and Ki67 expression is frequently detected in ovarian carcinomas, and immunofluorescence is often used to confirm PAX8 and Ki67 positive tumors8,9. Immunofluorescence staining showed that the PAX8 and Ki67 expression levels in organoids remained consistent with those in clinical tissues (Figs. 1C–E, 2C, D). Our results were also consistent with the reported findings about Ki67 and PAX8 positive proportion in OC9,10. This result suggested that the organoids showed similar morphological and proliferative activity to patient tumors, indicating that our organoids have a high degree of consistency in patient tumor tissues. Hence, this organoid model may be useful for drug screening and other clinical tests.
Figure 1.
Detection of patient-derived tumor tissues. (A) Schematic diagram of OC organoid culture processing. (B) Representative H&E staining of clinical tissues. Scale bars: 200 μm. (C, D) Immunofluorescence results of PAX8 and Ki67 were shown. The expression level was showed by merged images. Scale bars: 200 μm. (E) Bar graphs describe the ratio of ovarian cancer marker positive cells to DAPI-stained cells in OC tissues. Dots represent individual data points. Quantitative analysis was presented as the mean ± SEM.
Figure 2.
Detection of patient-derived OC organoids. (A) Bright view pictures of patient-derived ovarian organoids at different time points; (B) Representative H&E staining of OC organoids. (C) immunofluorescence results of PAX8 and Ki67 were shown. The expression level was showed by merged images. Scale bars: 200 μm. (D) Bar graphs describe the ratio of ovarian cancer marker positive cells to DAPI-stained cells in OC organoids. Dots represent individual data points. Quantitative analysis was presented as the mean ± SEM. (E) Schematic diagram of drug sensitivity assay processing.
PARPi Treatment Affects OC Organoids Morphology and Growth
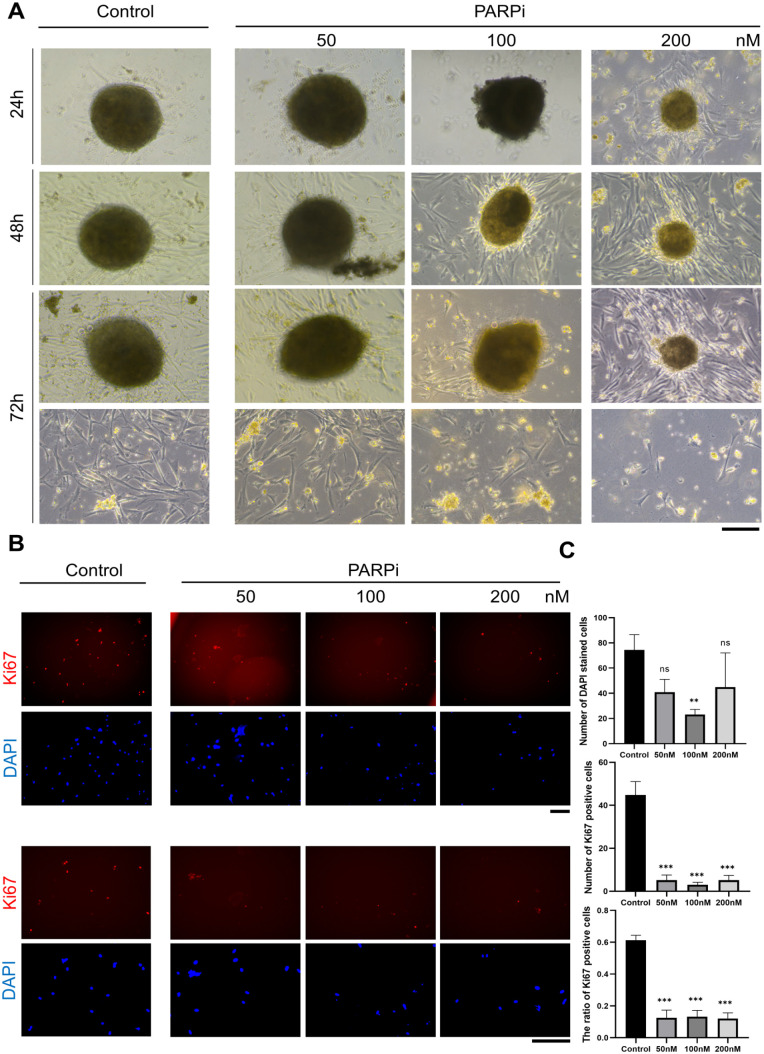
Several organoids from one patient were tested for sensitivity to PARPi, as it is a useful U.S. Food and Drug Administration (FDA)-approved drug for OC treatment (Fig. 2E). This experiment was divided into three groups according to the drug concentration. Morphological changes in organoids or adherent cells were assessed at different time points. As expected, our organoids showed an increased sensitivity to PARPi (Fig. 3A). The organoids were treated with PARPi and cultured for 72 h, which resulted in progressively smaller organoids and less dense structures. Fibroblast-like cells that migrated from the organoids, and adhered to the culture plate were regarded as adherent cells. The number of adherent cells significantly reduced after PARPi treatment (Fig. 3A). Ki67 staining indicated a decreasing Ki67 positive cell proportion among adherent cells (Fig. 3B). Moreover, PARPi treatment decreased total cell number and Ki67 expression (Fig. 3C). Postoperatively, the patient received cisplatin and paclitaxel in accordance with the guidelines of the National Comprehensive Cancer Network and PARPi as candidates. Given the positive results of our studies and the completed chemotherapy of the patient, we suggest that PARPi might be a clinically viable choice for maintain therapy.
Figure 3.
Morphological changes of organoids after PARPi treatment. (A) Representative bright field images of organoids after PARPi treatment were shown. The patient-derived organoids were inhibited by high-concentration PARPi, and treating for a long time did not represent a significant difference. Scale bars, 200 μm. (B) Representative images of Ki67 and DAPI immunofluorescence staining of adherent cells. Three indicated groups were divided to investigate dose sensitivity. (C) The expression level of ovarian cancer marker Ki67 and proportion of Ki67/DAPI in adherent cells after PARPi treatment was measured. Data are presented as the mean ± SEM (n = 3; *P < 0.05; **P < 0.01; ***P < 0.001; student’s test).
PARPi Promotes Early Apoptosis in OC Oragnoids
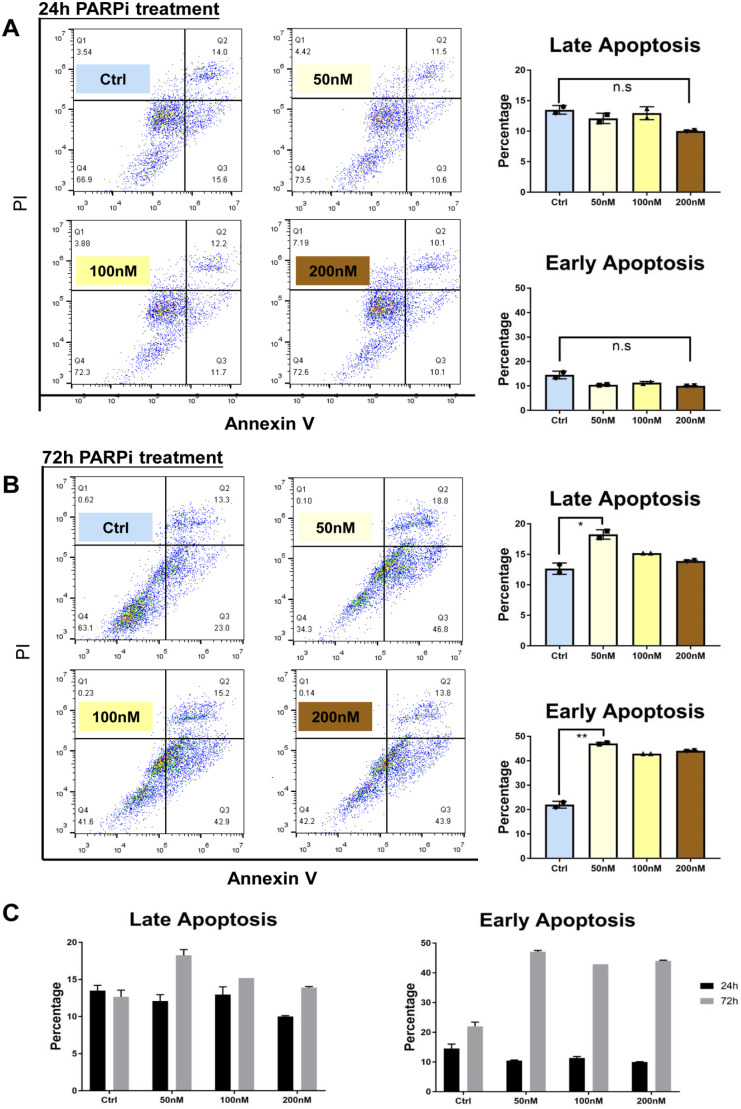
We assessed whether PARPi affected the apoptosis of organoids. After PARPi treatment for 24 h, the percentage of early and late apoptotic organoids was not significantly different among all groups (Fig. 4A). However, after 72 h, the drug treatment substantially induced early apoptosis and had a negligible effect on late apoptosis (Fig. 4B). Regardless of the concentration used, early apoptosis after 72 h treatment was four to five folds higher than that after 24 h treatment (Fig. 4C). These results suggest that early apoptosis significantly increased 72 h after PARPi administration, which may guide clinical practice.
Figure 4.
Analysis of apoptosis after PARPi treatment. (A) The apoptosis after 24 h PARPi treatment was evaluated and quantified. (B) The apoptosis after 72 h PARPi treatment was evaluated and quantified. (C) The percentages of apoptotic cells with different treatment concentration in early apoptosis and later apoptosis.
Gene Expression of Base Excision Repair and Cell Cycle Were Affected by PARPi
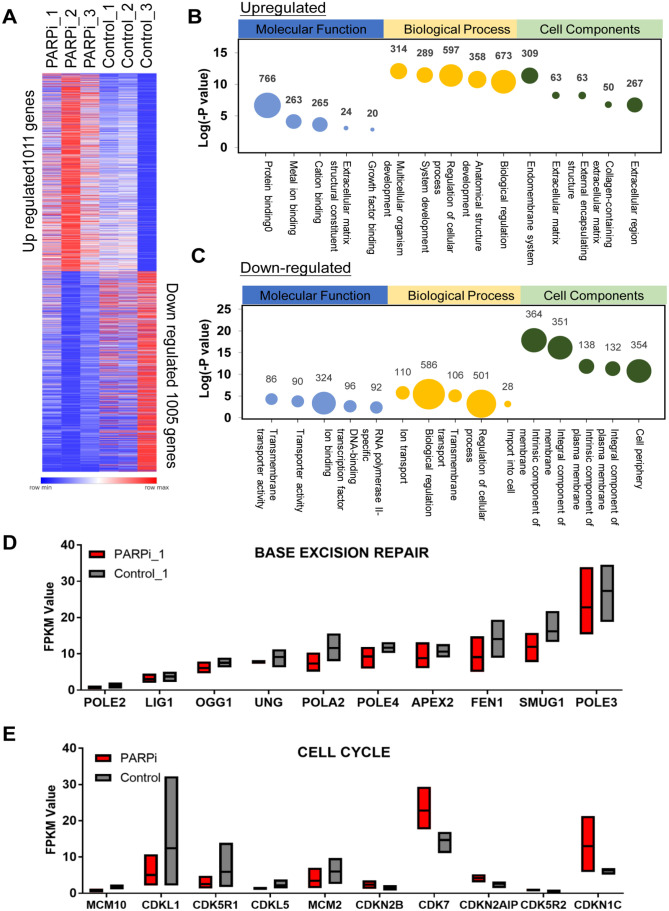
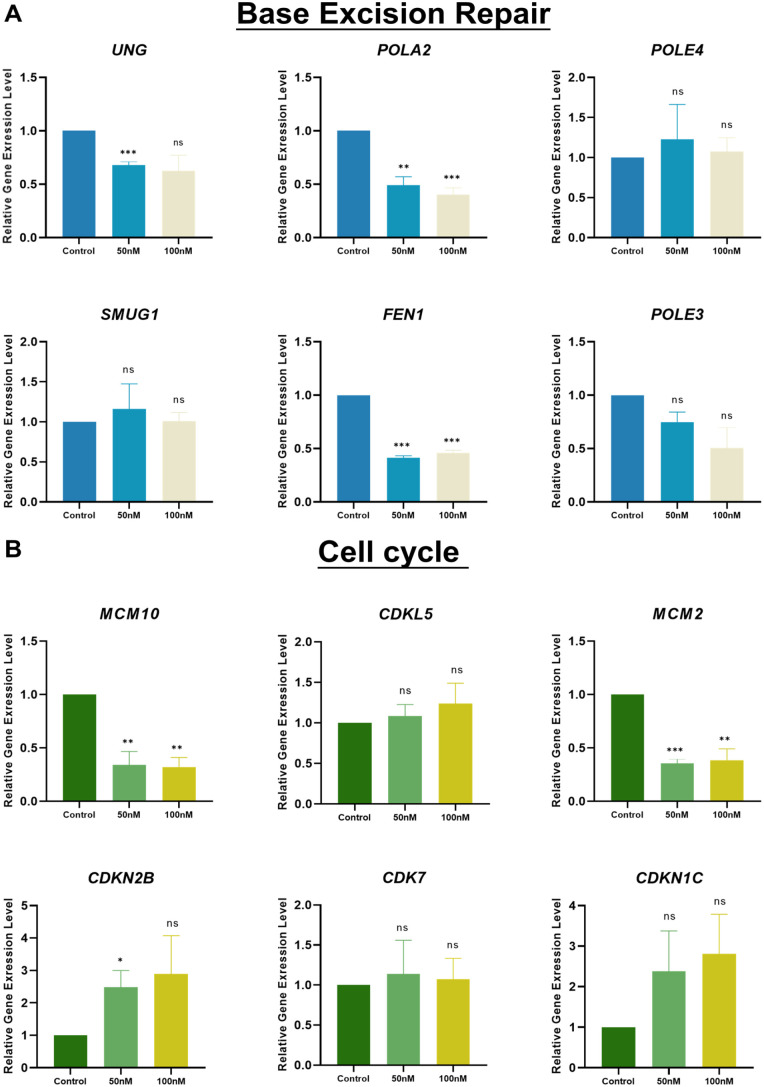
This study aimed to enhance the understanding of the effects of PARPi treatment on gene modulation. RNA sequencing (RNA-seq) was performed to determine changes in the organoid gene expression profile in one parental tumor sample and one derived organoid sample after treatment with 200 nM PARPi. We compared the transcriptomes of the PARPi-treated and control samples and found that at least 1,011 and 1,005 genes were upregulated and downregulated, respectively (Fig. 5A). Gene ontology (GO) was used to classify these differentially expressed genes (DEGs) into biological processes, molecular functions, and cellular components. The molecular function associated with upregulated genes was protein binding (Fig. 5B). The regulation of cellular progress in biological processes was also dramatically affected (Fig. 5C). Several pathways, such as the cell periphery, integral component of the membrane, and intrinsic component of the membrane, were downregulated. Genes belonging to these groups may be associated with PARPi treatment. Some PARPi-related upregulated and downregulated genes have never been reported. Considering the mechanism of PARP inhibition, we selected several genes related to base excision repair and the cell cycle for FPKM analysis11,12 (Fig. 5D, E). Among the identified upregulated genes, the expression of the cell proliferation gene MCM2 decreased after treatment, which was consistent with gene expression level analysis because MCM2 and PARP synergistically regulate cell proliferation 13 (Figs. 5E, 6B). To further verify the gene expression signatures before and after PARPi treatment, several gene expression levels were analyzed using qPCR with specific primers. FEN1 and SMUG1 are highly expressed and regarded as promising biomarkers in OC14,15. Therefore, changes in the expression of FEN1 and SMUG1, as well as uracil-DNA glycosylases encoding gene UNG, after PARPi treatment were also evaluated to identify potential drug targets. FEN1 and UNG were decreased after PARPi treatment. This suggests that these genes may be associated with the pathways targeted by PARPi. Interestingly, MCM10 has no widely identified as a potential biomarker in OC, the decreasing trend of MCM10 may provide evidence that MCM10 is correlated with OC. POLA2 is a therapeutic target in lung cancer, and POLA2 mutation even causes gemcitabine resistance in lung cancer cells. Therefore, POLA2 downregulation after PARPi treatment may have biological significance16,17. Representative DEGs of base excision repair and cell cycle were also validated by qRT-PCR analysis (Fig. 6A, B).
Figure 5.
OC organoid have distinct transcriptomes after PARPi treatment. (A) Significantly upregulated and downregulated genes from control and PARPi treatment samples for the RNA sequencing results. (B, C) Gene ontology classification of organoid. Representatively upregulated or downregulated genes closely related molecular function, biological process, and cell components. The size of the bubbles indicates the number of associated genes. (D, E) Significantly changed genes linked base excision repair and cell cycle.
Figure 6.
PARPi reduce cell proliferation and cell cycle in patient-derived organoid. (A, B) Expression level of 6 candidate genes linked to base excision repair pathways or cell cycle pathways after 48 h with different dosages of PARPi (0/50/100 nM) treatment. Data are presented as the mean ± SEM (n = 3; ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001; student’s test).
Discussion
We cultured organoids from pathological HGSC specimens and verified that the clinical and biochemical profiles of the organoids were similar to those of clinical tissues. PARPi treatment significantly inhibited the growth of HGSC organoids with increasing drug concentration and extended culture time. This is consistent with previous reports18,19. Immunohistochemical staining and immunofluorescence confirmed that PARPi treatment caused necrosis in the HGSC organoids. Further analysis revealed a significant increase in early and late apoptosis after 72 h of treatment. RNA-seq analysis revealed that PARPi treatment either upregulated or downregulated the expression of multiple genes. The expression of UNG, POLA2, FEN1, and POLE3, which are involved in base excision repair 20 , were downregulated. The expression of MCM10, CDKL5, MCM2, CDKN2B, CDK7, CDKN1C related to the cell cycle21,22, were also altered after PARPi treatment. Some of these results were confirmed by qPCR.
PARP plays a central role in single-stranded DNA base excision and repair23,24. Loss-of-function BRCA1/2 and HR result in DSBs are not normally repaired through HR repair (HRR). PARPi generally cause synthetic lethality by blocking SSB repair in tumor cells 23 . Therefore, given the mechanism of PARPi, BRCA 1/2 mutation or HR deficiency (HRD) indicate the application of PARPi25,26. However, some patients who meet these criteria experience rapid recurrence after PARPi treatment 27 . Therefore, a potentially efficient platform for assessing PARPi-sensitive in patients is required.
A three-dimensional clinical tumor-derived organoid model was used to investigate the sensitivity of the patient to PARPi. After treatment with a PARPi for 72 h, we determined that the patient was highly sensitive to olaparib. The results showed that our organoid culture method is applicable for determining drug sensitivity and can provide guidance for clinical practice. This organoid platform can be combined with next generation sequencing (NGS) to detect drug therapy 28 . We recommend patients with BRCA1/2 mutations or HRD and still resistant to PARPi doing drug sensitivity test by organoid model. Therefore, improper PARPi treatment-mediated exacerbation and delay in optimal treatment could be avoided.
In this study, a small fraction of the cells and organoids remained viable after PARPi treatment, indicating PARPi resistance. Various mechanisms are involved in PARPi resistance23,25. The role of the tumor microenvironment in PARPi-resistant cancers remains unclear. Organoid models are one of the best methods for investigating the tumor microenvironment29,30. Studies on PARPi-resistant organoids can lay the foundation for further detailed research and provide effective strategies to overcome PARPi resistance.
Conclusion
Patient-derived organoids with certain OC marker expression levels and morphology similar to the original cancer tissues were used to investigate the effects of PARPi treatment on organoids and adherent cells from organoid cultures. The proportion of Ki67 positive cells significantly reduced after PARPi treatment. Functional assays revealed that PARPi inhibits cell growth by upregulating apoptosis. Several pathways, such as cell cycle and base excision repair, have been highlighted as downstream targets of PARPi. Although further studies are needed, these results indicate the utility of our organoid platform and some potential genes related to PARPi resistance. Furthermore, these data provided clues for clarifying the PARPi resistance mechanisms and solving clinical problems.
Supplemental Material
Supplemental material, sj-docx-1-cll-10.1177_09636897231187996 for PARPi Decreased Primary Ovarian Cancer Organoid Growth Through Early Apoptosis and Base Excision Repair Pathway by Qi Cao, Lanyang Li, Yuqing Zhao, Chen Wang, Yanghua Shi, Xiang Tao, Chunhui Cai and Xin-Xin Han in Cell Transplantation
Acknowledgments
The authors thank all the colleges in their laboratory. Figure 1A and 2E was generated using BioRender.
Footnotes
Author Contributions: H.X.X. and C.C.H. conceived and designed the experiments; C.Q., L.L.Y., W.C., S.Y.H., and T.X. performed the experiments; C.Q., L.L.Y., and C.C.H. analyzed the data; and C.Q., L.L.Y., C.C.H. and H.X.X. wrote the manuscript. All the authors approved the final manuscript.
Availability of Data and Material: All relevant data and materials used in this article and the Supplementary Materials are available from the corresponding author, CCH, on reasonable request.
Ethical Approval: This study was approved by the Ethics Committee of the Obstetrics and Gynecology Hospital of Fudan University (Obstetrics and Gynecology ethical approval number: 2022-04).
Statement of Human and Animal Rights: All human specimens were performed according to the Declaration of Helsinki and the Chinese guidelines of GB/T4352.1-2021 and GB/T38736-2020. Approved by the Ethics Committee of the Obstetrics and Gynecology Hospital of Fudan University, patients/subjects signed a written informed consent. All human tissues were collected with patient consent according to clinical SOPs.
Statement of Informed Consent: Patient consent was obtained from each patient.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Chunhui Cai  https://orcid.org/0000-0002-8700-4383
https://orcid.org/0000-0002-8700-4383
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5): E359–86. [DOI] [PubMed] [Google Scholar]
- 2. Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10(4): 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035): 913–7. [DOI] [PubMed] [Google Scholar]
- 4. Franzese E, Centonze S, Diana A, Carlino F, Guerrera LP, Di Napoli M, De Vita F, Pignata S, Ciardiello F, Orditura M. PARP inhibitors in ovarian cancer. Cancer Treat Rev. 2019;73:1–9. [DOI] [PubMed] [Google Scholar]
- 5. Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, Scott C, Weitzel JN, Oaknin A, Loman N, Lu K, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376(9737): 245–51. [DOI] [PubMed] [Google Scholar]
- 6. Zhao Q, Ni J, Dong J, Cheng X, Xiao L, Xue Q, Xu X, Guo W, Chen X. Platinum resistance after PARPi resistance in a gBRCAmt recurrent ovarian cancer patient: a case report. Reprod Sci. 2022;30:615–21. [DOI] [PubMed] [Google Scholar]
- 7. Giudice E, Gentile M, Salutari V, Ricci C, Musacchio L, Carbone MV, Ghizzoni V, Camarda F, Tronconi F, Nero C, et al. PARP inhibitors resistance: mechanisms and perspectives. Cancers. 2022;14(6):1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laury AR, Perets R, Piao H, Krane JF, Barletta JA, French C, Chirieac LR, Lis R, Loda M, Hornick JL, Drapkin R, et al. A comprehensive analysis of PAX8 expression in human epithelial tumors. Am J Surg Pathol. 2011;35(6): 816–26. [DOI] [PubMed] [Google Scholar]
- 9. Giurgea LN, Ungureanu C, Mihailovici MS. The immunohistochemical expression of p53 and Ki67 in ovarian epithelial borderline tumors: correlation with clinicopathological factors. Rom J Morphol Embryol. 2012;53(4): 967–73. [PubMed] [Google Scholar]
- 10. Zhang S, Iyer S, Ran H, Dolgalev I, Gu S, Wei W, Foster CJR, Loomis CA, Olvera N, Dao F, Levine DA, et al. Genetically defined, syngeneic organoid platform for developing combination therapies for ovarian cancer. Cancer Discov. 2021;11(2): 362–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mittica G, Ghisoni E, Giannone G, Genta S, Aglietta M, Sapino A, Valabrega G. PARP inhibitors in ovarian cancer. Recent Pat Anticancer Drug Discov. 2018;13(4): 392–410. [DOI] [PubMed] [Google Scholar]
- 12. Vanacker H, Harter P, Labidi-Galy SI, Banerjee S, Oaknin A, Lorusso D, Ray-Coquard I. PARP-inhibitors in epithelial ovarian cancer: actual positioning and future expectations. Cancer Treat Rev. 2021;99:102255. [DOI] [PubMed] [Google Scholar]
- 13. Xiao G, Lundine D, Annor GK, Canar J, Ellison V, Polotskaia A, Donabedian PL, Reiner T, Khramtsova GF, Olopade OI, Mazo A, et al. Gain-of-function mutant p53 R273H interacts with replicating DNA and PARP1 in breast cancer. Cancer Res. 2020;80(3): 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abdel-Fatah TM, Russell R, Albarakati N, Maloney DJ, Dorjsuren D, Rueda OM, Moseley P, Mohan V, Sun H, Abbotts R, Mukherjee A, et al. Genomic and protein expression analysis reveals flap endonuclease 1 (FEN1) as a key biomarker in breast and ovarian cancer. Mol Oncol. 2014;8(7): 1326–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raja S, Van Houten B. The multiple cellular roles of SMUG1 in genome maintenance and cancer. Int J Mol Sci. 2021;22(4): 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koh V, Kwan HY, Tan WL, Mah TL, Yong WP. Knockdown of POLA2 increases gemcitabine resistance in lung cancer cells. BMC Genomics. 2016;17(Suppl 13): 1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fan Z, Bai Y, Zhang Q, Qian P. CircRNA circ_POLA2 promotes lung cancer cell stemness via regulating the miR-326/GNB1 axis. Environ Toxicol. 2020;35(10): 1146–56. [DOI] [PubMed] [Google Scholar]
- 18. Tao M, Wu X. The role of patient-derived ovarian cancer organoids in the study of PARP inhibitors sensitivity and resistance: from genomic analysis to functional testing. J Exp Clin Cancer Res. 2021;40(1): 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tao M, Sun F, Wang J, Wang Y, Zhu H, Chen M, Liu L, Liu L, Lin H, Wu X. Developing patient-derived organoids to predict PARP inhibitor response and explore resistance overcoming strategies in ovarian cancer. Pharmacol Res. 2022;179:106232. [DOI] [PubMed] [Google Scholar]
- 20. Yagüe-Capilla M, García-Caballero D, Aguilar-Pereyra F, Castillo-Acosta VM, Ruiz-Pérez LM, Vidal AE, González-Pacanowska D. Base excision repair plays an important role in the protection against nitric oxide- and in vivo-induced DNA damage in Trypanosoma brucei. Free Radic Biol Med. 2019;131:59–71. [DOI] [PubMed] [Google Scholar]
- 21. Cui F, Hu J, Ning S, Tan J, Tang H. Overexpression of MCM10 promotes cell proliferation and predicts poor prognosis in prostate cancer. Prostate. 2018;78(16): 1299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Z, Li J, Chen J, Shan Q, Dai H, Xie H, Zhou L, Xu X, Zheng S. MCM family in HCC: MCM6 indicates adverse tumor features and poor outcomes and promotes S/G2 cell cycle progression. BMC Cancer. 2018;18(1): 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Liu ZY, Wu N, Chen YC, Cheng Q, Wang J. PARP inhibitor resistance: the underlying mechanisms and clinical implications. Mol Cancer. 2020;19(1): 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355(6330): 1152–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Noordermeer SM, van Attikum H. PARP inhibitor resistance: a tug-of-war in BRCA-mutated cells. Trends Cell Biol. 2019;29(10): 820–34. [DOI] [PubMed] [Google Scholar]
- 26. Stover EH, Fuh K, Konstantinopoulos PA, Matulonis UA, Liu JF. Clinical assays for assessment of homologous recombination DNA repair deficiency. Gynecol Oncol. 2020;159(3): 887–98. [DOI] [PubMed] [Google Scholar]
- 27. Biegała Ł, Gajek A, Marczak A, Rogalska A. PARP inhibitor resistance in ovarian cancer: underlying mechanisms and therapeutic approaches targeting the ATR/CHK1 pathway. Biochim Biophys Acta Rev Cancer. 2021;1876(2): 188633. [DOI] [PubMed] [Google Scholar]
- 28. Morice PM, Coquan E, Weiswald LB, Lambert B, Vaur D, Poulain L. Identifying patients eligible for PARP inhibitor treatment: from NGS-based tests to 3D functional assays. Br J Cancer. 2021;125(1): 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, Liu IH, Chiou SH, Salahudeen AA, Smith AR, Deutsch BC, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175(7): 1972–88.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xia T, Du WL, Chen XY, Zhang YN. Organoid models of the tumor microenvironment and their applications. J Cell Mol Med. 2021;25(13): 5829–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-cll-10.1177_09636897231187996 for PARPi Decreased Primary Ovarian Cancer Organoid Growth Through Early Apoptosis and Base Excision Repair Pathway by Qi Cao, Lanyang Li, Yuqing Zhao, Chen Wang, Yanghua Shi, Xiang Tao, Chunhui Cai and Xin-Xin Han in Cell Transplantation