Abstract
ApbE is a lipoprotein in Salmonella typhimurium, and mutants unable to make this protein have a reduced ability to make thiamine (vitamin B1) and require it as a supplement for optimal growth in minimal glucose medium. Polyclonal antibodies specific to ApbE were used to determine that wild-type ApbE is located exclusively in the inner membrane. The periplasmic, monotopic topology of ApbE was determined by using computer-based hydrophobicity plots, LacZ and PhoA gene fusions, and proteinase protection experiments. This extracellular location of ApbE is required for its function, since a cytoplasmic form (ApbEcyto) did not allow an apbE mutant to grow in the absence of thiamine. A periplasmic form of ApbE (ApbEperi) lacking the lipoprotein modification allowed an apbE mutant to grow in the absence of thiamine, indicating that soluble ApbE could function in thiamine synthesis and that lipoation and membrane association were not required. Alteration of the amino acid implicated in membrane sorting for other lipoproteins did not result in a relocalization of ApbE to the outer membrane, suggesting that additional sorting determinants exist for ApbE.
Thiamine is synthesized in Salmonella typhimurium by a low-flux biosynthetic pathway that has recently been shown to be sensitive to a number of subtle metabolic perturbations (7, 8, 10, 11, 13). This sensitivity is manifested as a thiamine auxotrophy and has been exploited to identify new aspects of metabolic integration. Through this approach integration between thiamine synthesis, carbon catabolism, coenzyme A biosynthesis, and the redox state of the cell has been uncovered (7, 8, 10, 11, 13). It was possible to rationalize many of the metabolic interactions, since precursors to vitamin biosynthetic pathways are often metabolites diverted from major anabolic-catabolic pathways (5, 11, 16, 26).
A number of loci that affect thiamine synthesis have been identified based on a thiamine requirement of the respective mutants. Unlike those alluded to above, several of these loci were uncharacterized open reading frames, and thus an explanation for the resulting thiamine requirement was not readily apparent. Of significance to this study was the identification of two such loci, rseC (3) and apbE (4), whose products have been shown to be membrane proteins. The latter locus encodes a 36-kDa lipoprotein. These loci do not appear to encode biosynthetic enzymes involved directly in thiamine synthesis, although membrane association of such enzymes might not be unexpected. Compartmentalization of enzymes to the cytoplasm, cytoplasmic membrane (the inner or outer face), periplasm, or outer membrane has been shown to allow closer interactions with substrates, and such compartmentalization might be warranted when substrate levels are relatively low, as would be expected in a low-flux pathway such as those involved in vitamin synthesis (20).
Membrane proteins are involved in a wide range of cellular functions, including import and export of compounds, sensing environmental cues, and electron transfer for energy generation and redox catalysis. Lipoproteins represent a specific class of membrane proteins that are anchored to the lipid bilayer through their lipid modification. ApbE is the first lipoprotein whose absence results in a nutritional requirement. Although the number of known bacterial lipoproteins has increased to more than 130, the membrane location, topology, and function of most lipoproteins remain unknown (24, 28, 31).
Experiments presented here initiate the structural characterization of the ApbE lipoprotein and reveal it to be completely exposed to the periplasmic space but anchored to the inner membrane. Additionally, we show that the function of ApbE in thiamine synthesis is dependent on its extracellular location. Further, the membrane sorting signals reported for other lipoproteins are not sufficient to alter the location of ApbE, suggesting additional determinants contribute to membrane localization of lipoproteins.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All strains used in this study are derivatives of S. typhimurium LT2 (except Escherichia coli CC118) and are listed with their genotypes in Table 1. Tn10d(Tc) refers to the transposition-defective mini-Tn10 described by Way et al. (30). Growth curves were performed by using cells that were grown overnight in rich medium and then resuspended in saline. Cell suspensions (10 μl) were used to inoculate 190 μl of medium in individual wells of a 96-well microtiter dish. Growth was monitored as the increase in absorbance at 650 nm by using a SpectraMAX Plus Microplate Spectrophotometer (Molecular Devices, Sunnyvale, Calif.). The culture plate was maintained at 37°C with periodic shaking for aeration. NCE medium supplemented with MgSO4 (1 mM) (9) and glucose (11 mM) was used as minimal medium. Difco nutrient broth (8 g/liter) with NaCl (5 g/liter) added was used as rich medium. Difco BiTek agar was added to a final concentration of 1.5% for solid medium. Unless otherwise stated, the final concentrations of adenine and thiamine were 0.4 mM and 100 nM, respectively. The final concentrations of antibiotics in rich/minimal medium were as follows: tetracycline (20/10 μg/ml), kanamycin (50/125 μg/ml), ampicillin (30/15 μg/ml), and chloramphenicol (20/4 μg/ml).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| CC118 | araD139 Δ(ara leu)7697 ΔlacZ74 phoAΔ20 galE galK thi rpsE rpoB argE(Am) recA1 | 17 |
| LT2 | S. typhimurium (wild-type) | |
| DM271 | apbE42::Tn10d(Tc) | |
| Plasmids | ||
| pSU19 | Intermediate-copy-number cloning vector; Cmr | 2 |
| pApbE1 | apbE, 1.15-kb insert in pSU19 that encodes ApbE; Cmr | |
| pET20b | Overexpression vector containing T7-dependent promoter and cloning site for C-terminal His6 fusions; Ampr | Novagen |
| pApbE9 | apbE with NdeI start site, no lipoprotein signal peptide sequence, inserted into SmaI site of pSU19; Cmr | |
| pApbE10 | Encodes ApbEcyto; apbE without the lipoprotein signal peptide sequence under the expression of a T7 promoter, NdeI-HindIII fragment from pApbE9 ligated into pET20b; Ampr | |
| pApbE11 | apbE with NdeI start site inserted into SmaI site of pSU19; Cmr | |
| pApbE12 | Encodes ApbE; apbE under the expression of a T7 promoter, NdeI-HindIII fragment from pApbE11 ligated into pET20b; Ampr | |
| pApbE(D21S) | apbE(D21S), 1.15-kb insert in pSU19 that encodes ApbE(D21S); Cmr | |
| pT7-5 | Overexpression vector containing T7-dependent promoter; Ampr | |
| pApbE3 | EcoRI-HindIII-fragment from pApbE1 containing apbE ligated into pT7-5; Ampr | |
| pOmpF | 73-bp fragment encoding the signal peptide of ompF with NdeI and PstI sites; cloned into the PstI site of pSU19; Cmr | |
| pApbE(C20A) | 900-bp fragment cloned into SmaI site of pSU19; Cmr fragment encodes apbE amino acids 21 to 350 with a C20A change | |
| pApbEperi | Encodes ApbEperi; ompF-apbE(C20A) gene fusion expressed by the T7 promoter of pT7-7; Ampr |
Plasmid construction. (i) Cytoplasmic ApbE (ApbEcyto).
The coding sequence of the apbE gene excluding the signal sequence (amino acids 1 to 20) was amplified from the S. typhimurium LT2 chromosome by standard PCR techniques. An NdeI site was designed into the upstream (5′-end) primer and the downstream (3′-end) primer contained the stop codon for apbE. After blunt-end ligation into pSU19 (pApbE9) the 1.1-kb NdeI-HindIII fragment was cloned into pET20b (Novagen, Madison, Wis.), yielding pApbE10. The full-length apbE gene was cloned into pET20b by a similar method with a distinct upstream primer and resulted in pApbE12.
(ii) Periplasmic ApbE (ApbEperi).
The coding sequence of the apbE gene that corresponded to amino acid positions 20 to 350 was amplified from the S. typhimurium chromosome by using standard PCR techniques. The upstream (5′) primer contained a PstI site which replaced the cysteine at position 20 with an alanine; the downstream (3′) primer was as described above. The amplified product was ligated into the SmaI site of pSU19, yielding pApbE(C20A). The ompF sequence for the OmpF-ApbE fusion was generated by annealing complementary sequences of the OmpF signal peptide that contained NdeI and PstI sites. The DNA encoding the OmpF signal peptide was ligated into the SmaI site of pSU19, yielding pOmpF. A 0.9-kb PstI fragment from pApbE(C20A) was ligated into pOmpF that had been digested with PstI. Sequence analysis was used to confirm a clone with the desired ompF::apbE(C20A) fusion. This fusion was then cloned into pT7-7 as a NdeI-HindIII fragment, resulting in pApbE14.
(iii) ApbE(D21S).
The aspartate residue at position 21 of ApbE (+2 in the processed protein) was changed to a serine by the megaprimer method of Barik (1). Chromosomal DNA from S. typhimurium LT2 was used as the DNA template in this two-step amplification method. All plasmid constructs were confirmed by sequence analysis.
Construction of ApbE-His6 and Western analysis.
A C-terminal His6 fusion to ApbE lacking the leader sequence was constructed by standard PCR amplification with primers specific to the ends of apbE. The appropriate fragment was cloned into the NdeI-HindIII sites of pET20b and protein expression was confirmed utilizing methods described by the manufacturer (Novagen). Purification of ApbE-His6 was achieved by passing extracts expressing the fusion protein over an Ni2+-affinity column (Novagen). ApbE-His6 was obtained in >90% homogeneity by purification under denaturing conditions. Rabbit polyclonal antibodies against ApbE-His6 were generated at the animal care unit of the University of Wisconsin Medical School. Antisera was prepared and titered according to the method of Harlow and Lane (14). Standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used and Western analysis were performed as described previously with alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Promega, Madison, Wis.) used to detect anti-ApbE bound to the membrane support (14).
Isolation of apbE::phoA fusions.
Plasmid pApbE1 was introduced into E. coli CC118. The method of Manoil and Beckwith (17) was used to isolate alkaline phosphatase (phoA) gene fusions to apbE, in addition to unsuccessful attempts to isolate β-galactosidase (lacZ) gene fusions to the same gene. PhoA insertions in the apbE gene were identified on agar plates supplemented with 40 μg of the chromagenic indicator XP (5-bromo-4-chloro-3-indolyl phosphate; Fluka, Milwaukee, Wis.) per ml. The location of the insertions was estimated by PCR amplification with a primer specific to the sequence upstream of the 5′ end of the apbE gene (5′-TATTCCGGCGTACAAATACG-3′) and a primer that annealed within the TnphoA sequence (5′-AATATCGCCCTGAGCAGCCCG-3′). These primers were also used to sequence the junction of the fusions and thus precisely determine the location of the fusion. The alkaline phosphatase activities of the E. coli CC118 cells expressing ApbE::PhoA fusions were determined by the procedure described by Brickman, with a control strain containing no fusion set to zero (6).
Protease accessibility of ApbE in intact spheroplasts.
The accessibility of ApbE from S. typhimurium LT2 whole cells, spheroplasts (generated as described by Osborn and Munson [22]), and lysed spheroplasts to proteinase K was determined by a modification of the method of Randall and Hardy (25). After a 20-min treatment with proteinase K (100 μg/ml) at 4°C, protein was precipitated with 5% trichloroacetic acid. Each sample (equivalent to 0.05 A650) was analyzed by SDS-PAGE and Western hybridization.
Protein analysis and cell fractionation.
Separation of cell protein into cytoplasmic and total membrane fractions and subsequently into inner and outer membrane fractions was carried out as described by Osborn and Munson (22). Briefly, cells were grown to mid-log phase (ca. 55 Klett units [red filter]), spheroplasted, and lysed by osmotic shock. Low-speed centrifugation was used to remove unbroken cells, and then the supernatant was centrifuged at 40,000 rpm for 2 h in a Beckman 70 Ti rotor. The pellet was saved as the total membrane fraction, and the supernatant contained the cytoplasmic-periplasmic fraction. The total membrane fraction was loaded onto a 50 to 35% stepwise sucrose gradient with a 55% sucrose cushion and centrifuged at 36,000 rpm for 16 h in a Beckman SW40 Ti rotor. Fractions were collected dropwise, and the location of the inner and outer membranes within the gradient was visualized by Coomassie staining and confirmed by detecting NADH oxidase activity (29) and 2-keto-deoxyoctonate (data not shown) (15), respectively.
Isolation of periplasmic and cytoplasmic fractions was performed by a modification of the osmotic shock procedures developed by Neu and Heppel (21) and Thorstenson et al. (29). A 5-ml culture was grown in minimal medium to mid-log phase, and 1.0 ml of the cells was resuspended in 100 μl of 0.5 M sucrose–0.2 M Tris–0.5 mM EDTA and kept on ice for 15 min. Then, 400 μl of TE (10 mM Tris [pH 8.0], 1 mM EDTA) was mixed into the suspension, which was kept on ice for 30 min. The cells were pelleted, and the supernatant was saved as the periplasmic fraction, while the pellet was further processed. The cell pellet was washed and resuspended in 0.1 M sucrose–40 mM Tris–0.1 mM EDTA and sonicated for 10 s at a 50% duty cycle by using a model 550 sonic dismembrator (Fisher, Itasca, Ill.), and a low-speed centrifugation was used to remove the unbroken cells. The supernatant was centrifuged at 100,000 rpm for 30 min in a Beckman TLA 100.2 rotor. The supernatant contained the cytoplasmic fraction, and the pellet was washed once with TE buffer to remove any contaminating periplasmic and peripheral proteins and then pelleted again. The final pellet was saved as the total membrane fraction. The relative purity of the periplasmic and membrane fractions was determined by using β-lactamase or NADH oxidase, respectively, as marker enzymes.
RESULTS AND DISCUSSION
ApbE is located in the inner membrane.
Our characterization of ApbE was driven by two assumptions: (i) by understanding its location and structural features, we could better address the role of this protein in thiamine synthesis, and (ii) we could further the general understanding of bacterial lipoproteins by topological and mutational characterization of another lipoprotein. Analysis of ApbE sequence in the context of characterized lipoproteins suggested that the mature protein would be lipoated and localized to the inner membrane. Immunoblot analyses were performed on wild-type and mutant strains to validate and extend this prediction. Comparison of whole-cell extracts of strain DM271(apbE) and wild-type strain LT2 blotted with antibodies specific to ApbE identified an appropriately sized band that was detected in the LT2 extract but not in extracts from the apbE mutant strain (data not shown). From these comparisons, we concluded this band corresponded to ApbE.
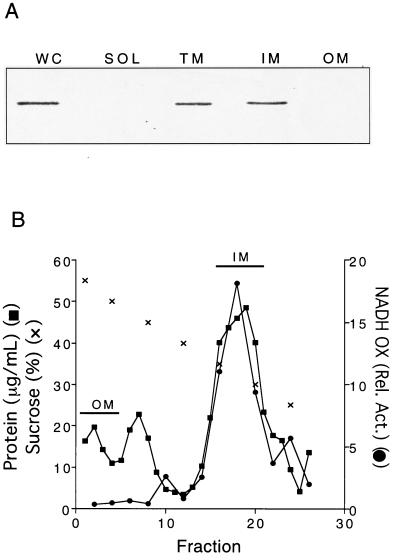
Western blot analyses with LT2 with ApbE-specific antisera showed that ApbE clearly separated with the membrane fraction, and none was detected in the soluble fraction (Fig. 1A). When the membrane was separated into inner and outer membrane fractions, ApbE was found exclusively in the inner membrane. Biochemical characterization of the membrane separation is documented in Fig. 1B and is representative of the separations in subsequent experiments.
FIG. 1.
Chromosomally encoded ApbE from S. typhimurium LT2 is located in the inner membrane. (A) Immunoblot of whole-cell (WC), soluble cytoplasmic and periplasmic (SOL), total membrane (TM), inner membrane (IM), and outer membrane (OM) fractions. Cellular fractions were prepared as described in Materials and Methods. Western analysis with polyclonal antibodies against ApbE (1:5,000 dilution) was performed (14). (B) Representative membrane separation profile from S. typhimurium LT2. Inner and outer membranes were fractionated by sucrose density equilibration. The protein concentration, NADH oxidase activity, and sucrose concentration values are shown. The fractions pooled and used in Fig. 1A are indicated.
ApbE is exposed on the periplasmic face of the inner membrane.
Although the lipoated N terminus of ApbE would be expected to be located in the inner membrane (31), the presence of a cytoplasmic domain was tested. Based on analyses with a number of computer programs (i.e., TMPRED, TMAP, and Kyte-Doolittle), ApbE does not have an apparent membrane-spanning region (data not shown), suggesting that the entire ApbE protein was located in the periplasm with the fatty acids tethering it to the cell membrane.
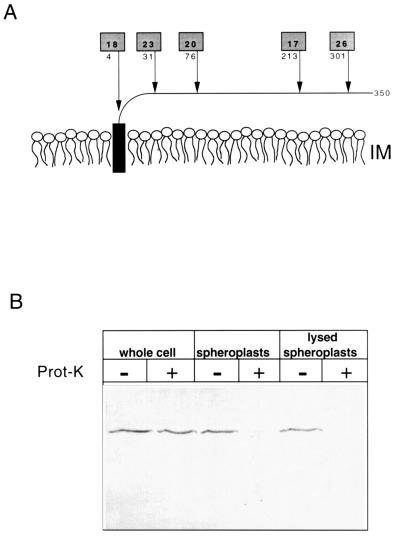
Two lines of genetic evidence confirmed that ApbE did not contain a cytoplasmic domain. Attempts to isolate translational fusions between ApbE and LacZ, producing functional β-galactosidase that would indicate a cytoplasmic domain, were unsuccessful. Conversely, five active PhoA fusions to ApbE were isolated, and the fusion junctions determined by DNA sequencing (Fig. 2A). Since the activity of alkaline phosphatase requires the protein to be extracellular, these results were consistent with the topology predicted by computer analysis.
FIG. 2.
ApbE is periplasmically located. (A) The predicted monotopic structure of ApbE anchored in the outer leaflet of the inner membrane lipid bilayer is schematically shown. Arrows indicate the location of active alkaline phosphatase (PhoA) fusions to ApbE encoded by pApbE1, and the number below the shaded box indicates the amino acid of ApbE that immediately proceeds the fusion junction. The numbers in the shaded boxes represent the relative PhoA activity (in units per minute per optical density at 650 nm) of each fusion compared to a control containing an out-of-frame PhoA fusion. (B) Immunoblot showing the proteinase K accessibility of chromosomally encoded ApbE in whole cells, spheroplasts, and lysed spheroplasts. Samples were treated with (+) or without (−) proteinase K (100 μg/ml). The proteins in each sample (equivalent to 0.05 A650) were separated by SDS-PAGE and immunoblotted with a 1:5,000 dilution of ApbE antisera.
Independent verification of periplasmic exposure was sought by measuring the accessibility of ApbE to proteinase K. The ability of proteinase K to degrade ApbE from S. typhimurium LT2 whole cells, spheroplasts, and lysed spheroplasts was determined and visualized by SDS-PAGE, followed by Western hybridization (Fig. 2B). As anticipated, ApbE in whole cells was protected from proteinase K digestion, presumably by the intact outer membrane. ApbE in spheroplasts was completely sensitive to proteinase K digestion. Taken together, the results in this section clearly demonstrated a periplasmic location for ApbE.
The periplasmic location of ApbE is required for its function.
Having identified the subcellular location of ApbE, we took advantage of the nutritional defect of an apbE mutant to explore the correlation of function with location. The extent of the nutritional defect caused by an apbE mutation is dependent on both the genetic background and the carbon source in the growth medium (4). Specifically, when there is low carbon flux through the purine biosynthetic pathway (e.g., purF), ApbE is required for thiamine synthesis regardless of the carbon source, hence its apb designation (23). In the nutritional studies described here, we took advantage of the fact that apbE mutations also result in a thiamine auxotrophy in an otherwise wild-type genetic background when glucose is the sole carbon and energy source.
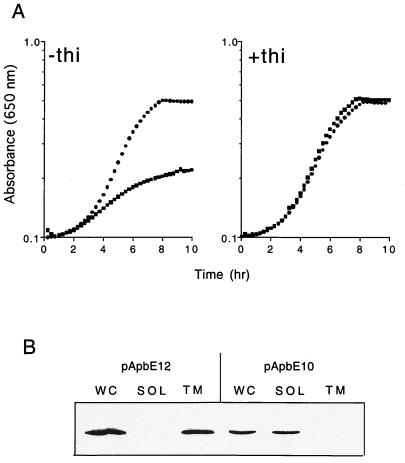
The first experiment tested whether the role of ApbE in thiamine synthesis required export to the membrane or whether the protein might have a cytoplasmic function prior to its export. Two strains, carrying either pApbE10 (expressing ApbE without the signal peptide sequence and lipoation site) or pApbE12 (encoding the full-length ApbE protein) as the only source of ApbE, were assessed for their growth in the absence of thiamine and the cellular location of the respective plasmid-encoded ApbE. Data from Western blot analysis (Fig. 3B) confirmed that when the signal peptide sequence was removed (pApbE10) ApbE was located completely in the cytoplasm, whereas wild-type ApbE (pApbE12) was located solely in the membrane fraction. Significantly, the growth data in Fig. 3A showed that the leaderless construct was unable to restore thiamine-independent growth to an apbE mutant, while the wild-type protein completely complemented the growth defect (Fig. 3A, left panel). While we cannot eliminate the possibility that the cytoplasmic ApbE was partially functional, it did not result in significantly more thiamine-independent growth than strain DM271 carrying the vector alone (data not shown).
FIG. 3.
ApbE requires a signal peptide for membrane association and function in thiamine synthesis. (A) The growth of strain DM271 (apbE) carrying pApbE12, encoding wild-type ApbE (●), or pApbE10, encoding ApbE without the lipoprotein signal peptide (■), is shown. Cells were grown in minimal medium with glucose as the sole carbon and energy source and supplemented with 100 nM thiamine where indicated. (B) Immunoblot showing the cellular location of ApbE produced by pApbE10 and pApbE12. Cellular fractions were isolated as described in Materials and Methods. Abbreviations: WC, whole-cell fraction; SOL, soluble cytoplasmic and periplasmic proteins; TM, total membrane proteins.
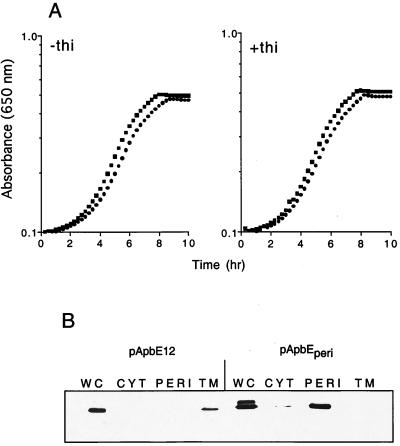
Although the results from the above experiment did not distinguish between a requirement for location and a role for the signal peptide or lipoation site in function, the simplest conclusions from this experiment were that (i) membrane association of ApbE required the presence of the signal peptide and that (ii) membrane association and/or extracellular localization was necessary for function. To determine whether ApbE function required association with the membrane, a periplasmic form of ApbE was constructed by replacing the ApbE lipoprotein signal peptide with the signal peptide of OmpF (ApbEperi). Cleavage of the OmpF signal peptide should result in the release of ApbE into the periplasm, where its function could be assessed in the absence of membrane association as had been done with AcrA, a component of the multi-drug efflux complex AcrAB-TolC of E. coli (33). As shown in Fig. 4, plasmid pApbEperi restored wild-type thiamine-independent growth to strain DM271 (apbE) (Fig. 4A, left panel). Western blot analysis of fractions from cells carrying pApbEperi confirmed that ApbEperi was localized to the periplasm and was completely absent from the membrane fraction (Fig. 4B). We noted two additional features of the Western blot analyses with pApbEperi. First, unlike cells expressing wild-type ApbE, extracts from cells containing pApbEperi contained a significant amount of unprocessed ApbE. Second, a small but detectable amount of ApbEperi was located in the soluble, cytoplasmic fraction. The presence of ApbE in this fraction is most likely ApbEperi that was not released by the osmotic shock but was released upon sonic disruption of the cells. We discounted the role of ApbE in this fraction in the functional studies since the experiments described above showed that ApbE located in the cytoplasm was unable to restore thiamine synthesis in an apbE mutant. From this experiment we concluded that wild-type ApbE does not require membrane anchorage to fulfill its role in thiamine synthesis.
FIG. 4.
A periplasmic form of ApbE complements the thiamine requirement of an apbE mutant. (A) Growth of strain DM271 (apbE) carrying pApbEperi (●) or pApbE12 that produces wild-type ApbE (■) in minimal glucose medium supplemented with 10 nM thiamine where indicated. (B) Immunoblot showing the cellular location of ApbE and ApbEperi expressed from pApbE12 and pApbEperi, respectively. Cellular fractions were isolated as described in Materials and Methods. Abbreviations: WC, whole-cell proteins; CYT, cytoplasmic proteins; PERI, periplasmic proteins; TM, total membrane proteins.
The above conclusion raises questions about the role of lipoation of ApbE. The functional purpose of a lipid modification is most obvious with gram-positive organisms that lack an outer membrane (28). In these organisms, the fatty acylations serve as an anchor for extracellular proteins. However, in gram-negative organisms the presence of an outer membrane may minimize the need for a membrane anchor. Perhaps the lipid anchor serves to optimally localize a protein, thus increasing its potential for interaction with the respective extracellular or membrane components. Such a model suggests that the lipoated form of the protein would be more efficient than the periplasmic form, a prediction that cannot be tested currently because of the lack of assay for ApbE function.
The +2 amino acid is not sufficient to determine membrane target.
An extension of the above experiment was to determine whether ApbE functioned when transported to the outer membrane. Such a result would impact the kind of models we could consider for the function of ApbE in thiamine synthesis. As noted previously, ApbE contains an aspartate residue at the +2 position in the processed protein (position 21 in the full-length protein). The lipoprotein studies of Yamaguchi et al. (32), Gennity and Inouye (12), and Matsuyama et al. (18, 19) predicted that substitution of a serine residue for this aspartate would target ApbE to the outer membrane. Construction of such a mutant was done to (i) test the significance of this consensus amino acid in another lipoprotein and (ii) if possible address the function of ApbE located in the outer membrane. Oligonucleotide-directed site-specific mutagenesis was used to generate a serine-to-aspartate substitution at position 21 in the full-length protein (+2 in the mature protein), and the resulting mutant ApbE protein was designated ApbE(D21S). The gene encoding the ApbE(D21S) mutant protein was cloned into pSU19 (2).
Growth analyses determined that ApbE(D21S) fully restored thiamine-independent growth to strain DM271(apbE). This result demonstrated that the D21S substitution did not significantly impair the function of the protein compared to wild-type ApbE (μ = 0.45 versus 0.53 h−1, respectively, in the absence of thiamine). Unexpectedly, Western blot analyses showed that ApbE(D21S) was located only in the inner membrane, a location indistinguishable from the location of the wild-type protein (data not shown). The fact that no ApbE(D21S) was detected in the outer membrane demonstrated that, in contrast to some other lipoproteins, in ApbE the +2 position is not the determinant for outer membrane targeting. Efforts by Gennity and Inouye (12) indicate that the amino acids adjacent to position +2 (positions +3 and +4) may influence its role as the sorting determinant. Specifically, a glutamate at position +3 was observed to cause minor but significant inner membrane retention of Lipo-β-lactamase(E3), a hybrid lipoprotein containing a serine at position +2. Since ApbE has a glutamate at position +3, the effect of the serine at position +2 could be minimized, but the complete lack of movement to the outer membrane strongly suggests that there are additional structural determinants involved in the inner membrane localization of ApbE.
Conclusions.
At present our thinking is that ApbE participates in thiamine synthesis indirectly, possibly as part of a complex that mediates a redox-sensitive step in thiamine synthesis. This thinking has been influenced by several recent results. First, RnfF, a membrane protein from Rhodobacter capsulatus, contains sequences orthologous to both ApbE and RseC, an additional inner membrane protein required for optimal thiamine synthesis in S. typhimurium (3). The function of RnfF in R. capsulatus physiology remains unclear but has been suggested to anchor a membrane-associated protein complex that passes electrons to nitrogenase for nitrogen fixation (27). Our finding that RnfF can complement both apbE and rseC mutants (data not shown) makes it tempting to suggest that ApbE and/or RseC may associate and perform a function similar to that of RnfF. Second, recent work in the lab has identified additional loci affecting thiamine synthesis that are involved in maintaining the redox environment of the cell. Lastly, growth under anaerobic conditions eliminates the need for ApbE in thiamine synthesis.
ApbE belongs to a growing class of proteins that are required for thiamine synthesis under conditions that strain this biosynthetic pathway (e.g., low flux through the purine biosynthetic pathway). Thus, these proteins may have a role in thiamine synthesis that can be satisfied by partially redundant cellular functions when substrates are plentiful. We suggest that many of the unknown open reading frames in various genome sequences may encode similar redundancies and that their metabolic role may be uncovered only when the relevant pathways are required to function efficiently. Current work in the lab seeks to address the specific thiamine biosynthetic enzyme(s) affected in apbE mutants and use this knowledge to identify the biochemical role of ApbE.
ACKNOWLEDGMENTS
This work was supported by Hatch grant WIS3734 from the U.S. Department of Agriculture and by National Institutes of Health grant GM47296 to D.M.D.
REFERENCES
- 1.Barik S. Site-directed mutagenesis by double polymerase chain reaction. Mol Biotechnol. 1995;3:1–7. doi: 10.1007/BF02821329. [DOI] [PubMed] [Google Scholar]
- 2.Bartolomé B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 3.Beck B J, Connolly L E, de las Penas A, Downs D M. Evidence that rseC, a gene in the rpoE cluster, has a role in thiamine synthesis in Salmonella typhimurium. J Bacteriol. 1997;179:6504–6508. doi: 10.1128/jb.179.20.6504-6508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck B J, Downs D M. The apbE gene encodes a lipoprotein involved in thiamine synthesis in Salmonella typhimurium. J Bacteriol. 1998;180:885–891. doi: 10.1128/jb.180.4.885-891.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begley T P, Downs D M, Ealick S, McLafferty F, van Loon D, Taylor S, Chiu H, Kinsland C, Reddick J, Xi J, Campobasso N. Thiamin synthesis in prokaryotes. Arch Microbiol. 1999;171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 6.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and φ80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 7.Christian T, Downs D M. Defects in pyruvate kinase cause a conditional increase of thiamine synthesis in Salmonella typhimurium. Can J Microbiol. 1999;45:1–7. [PubMed] [Google Scholar]
- 8.Claas, K., S. Weber, and D. M. Downs. Mutants defective in the energy conserving NADH dehydrogenase are unable to utilize the oxidative pentose phosphate pathway. Submitted for publication.
- 9.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 10.Enos-Berlage J L, Downs D M. Mutations in sdh (succinate dehydrogenase genes) alter the thiamine requirement of Salmonella typhimurium. J Bacteriol. 1997;179:3989–3996. doi: 10.1128/jb.179.12.3989-3996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frodyma M, Downs D M. The panE gene, encoding ketopantoate reductase, maps at 10 minutes and is allelic to apbA in Salmonella typhimurium. J Bacteriol. 1998;180:4757–4759. doi: 10.1128/jb.180.17.4757-4759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gennity J M, Inouye M. The protein sequence responsible for the lipoprotein membrane localization in Escherichia coli. J Biol Chem. 1991;266:16458–16464. [PubMed] [Google Scholar]
- 13.Gralnick, J., and D. M. Downs. Unpublished data.
- 14.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 15.Keleti G, Lederer W H. Handbook of micromethods for the biological sciences. New York, N.Y: Van Nostrand Reinhold Co.; 1974. [Google Scholar]
- 16.Lam H M, Winkler M E. Metabolic relationships between pyridoxine (vitamin B6) and serine biosynthesis in Escherichia coli K-12. J Bacteriol. 1990;172:6518–6528. doi: 10.1128/jb.172.11.6518-6528.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manoil C, Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 18.Matsuyama S, Tajima T, Tokuda H. A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO J. 1995;14:3365–3372. doi: 10.1002/j.1460-2075.1995.tb07342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuyama S, Yokota N, Tokuda H. A novel outer membrane lipoprotein, LolB (HemM) involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 1997;16:6947–6955. doi: 10.1093/emboj/16.23.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer F. “Compartments” in the bacterial cell and their enzymes. ASM News. 1993;59:346–350. [Google Scholar]
- 21.Neu H C, Heppel L A. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]
- 22.Osborn M J, Munson R. Separation of the inner (cytoplasmic) and outer membranes of gram-negative bacteria. Methods Enzymol. 1974;31:642–652. doi: 10.1016/0076-6879(74)31070-1. [DOI] [PubMed] [Google Scholar]
- 23.Petersen L A, Enos-Berlage J E, Downs D M. Genetic analysis of metabolic crosstalk and its impact on thiamine synthesis in Salmonella typhimurium. Genetics. 1996;143:37–44. doi: 10.1093/genetics/143.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Randall L L, Hardy S J S. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell. 1986;46:921–928. doi: 10.1016/0092-8674(86)90074-7. [DOI] [PubMed] [Google Scholar]
- 26.Rondon M R, Trzebiatowski J R, Escalante-Semerena J C. Biochemistry and molecular genetics of cobalamin biosynthesis. Vol. 56. New York, N.Y: Academic Press, Inc.; 1997. [DOI] [PubMed] [Google Scholar]
- 27.Schmehl M, Jahn A, Meyer zu Vilsendorf A, Hennecke S, Masepohl B, Schuppler M, Marxer M, Oelze J, Klipp W. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol Gen Genet. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 28.Sutcliffe I C, Russell R R B. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorstenson Y R, Zhang Y, Olson P S, Mascarenhas D. Leaderless polypeptides efficiently extracted from whole cells of osmotic shock. J Bacteriol. 1997;179:5333–5339. doi: 10.1128/jb.179.17.5333-5339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Way J C, Davis M A, Morisato D, Roberts D E, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 31.Wu H C. Biosynthesis of lipoproteins. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. [Google Scholar]
- 32.Yamaguchi K, Yu F, Inouye M. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell. 1988;53:423–432. doi: 10.1016/0092-8674(88)90162-6. [DOI] [PubMed] [Google Scholar]
- 33.Zgurskaya H I, Nikaido H. ArcA is a highly asymmetric protein capable of spanning the periplasm. J Mol Biol. 1999;285:409–420. doi: 10.1006/jmbi.1998.2313. [DOI] [PubMed] [Google Scholar]