Abstract
Spontaneous intracerebral hemorrhage (ICH) is a devastating disease with high morbidity and mortality worldwide. We have previously shown that ferroptosis contributes to neuronal loss in ICH mice. The overload of iron and dysfunction of glutathione peroxidase 4 (GPx4) promote neuronal ferroptosis post-ICH. However, how epigenetic regulatory mechanisms affect the ferroptotic neurons in ICH remains unclear. In the current study, hemin was used to induce ferroptosis in N2A and SK-N-SH neuronal cells to mimic ICH. The results showed that hemin-induced ferroptosis was accompanied by an increment of global level of trimethylation in histone 3 lysine 9 (H3K9me3) and its methyltransferase Suv39h1. Transcriptional target analyses indicated that H3K9me3 was enriched at the promoter region and gene body of transferrin receptor gene 1 (Tfr1) and repressed its expression upon hemin stimulation. Inhibition of H3K9me3 with inhibitor or siRNA against Suv39h1 aggravated hemin- and RSL3-induced ferroptosis by upregulating Tfr1 expression. Furthermore, Suv39h1-H3K9me3 mediated repression of Tfr1 contributes to the progression of ICH in mice. These data suggest a protective role of H3K9me3 in ferroptosis post ICH. The knowledge gained from this study will improve the understanding of epigenetic regulation in neuronal ferroptosis and shed light on future clinical research after ICH.
Keywords: Intracerebral hemorrhage, neurons, ferroptosis, epigenetics, transferrin receptor
Introduction
Spontaneous intracerebral hemorrhage (ICH) accounts for only 10–15% in all stroke subtypes, but holds the highest mortality. 1 ICH affects more than 1 million people each year worldwide, and most survivors suffer from severe sequelae due to a lack of effective therapeutic options.1,2 Hematoma formation and expansion lead to intracranial pressure elevation that causes brain damage and neuronal death. Subsequently, toxins, such as hemoglobin (Hb), heme, and iron, released from the red blood cells further contribute to neuronal death. Extracellular iron is transported into neurons mainly through the transferrin (TF)-TF receptor 1 (TFR1, the protein encoded by Tfr1 gene) system. 3 Overloaded free iron in the neuron can react with hydrogen peroxide (H2O2) to form more toxic hydroxyl radicals (·OH) via the Fenton reaction, which causes damages to DNA, proteins, or lipids, and may induce several forms of cell death, including apoptosis, necroptosis, autophagy, and newly identified ferroptosis.4–6
Ferroptosis, a non-apoptotic and regulated type of cell death, is caused by severe lipid peroxidation-mediated membrane damage in an iron-dependent manner.7,8 Factors that affect the metabolism of iron and/or lipids, as well as those that affect the redox systems are potential regulators of ferroptotic cell death. Erastin (the cysteine/glutamate antiporter inhibitor), RSL3 (inhibitor of glutathione peroxidase 4, GPx4), iron, and hemin (to increase intracellular iron and decrease GPx4 activity) are widely used to induce ferroptosis,7,9–11 whereas iron chelator deferoxamine (DFO) and lipid reactive oxygen species (ROS) scavenger ferrostatin-1 (Fer-1) are ferroptosis inhibitors. 7 We and others have demonstrated that ferroptosis occurs during ICH and contributes to neuronal death in vitro and in vivo.5,11 More importantly, rescuing neuronal death, including ferroptosis, reduces brain injury, and improves functional outcomes of ICH animals effectively. Inhibiting ferroptosis thus represents a new target for the treatment of ICH. 12 However, administration of DFO didn’t obtain optimistic results from clinical trials, and for Fer-1, as a small molecule compound, there is still a long way to go before clinical applications. Further investigation into the molecular mechanism of ferroptosis may give rise to new therapeutic options for treating ICH.
Epigenetic modification of DNA and histones at specific loci on chromatin plays fundamental roles in the regulation of gene expression and related biological processes. The post-translational modification of histones is extremely complicated that involves methylation, acetylation, ubiquitination, etc. The combination of different types of modification at specific histone residues constitutes the “histone code” to dictate gene expression. 13 Among those, histone 3 lysine 9 trimethylation (H3K9me3) is a well-studied and conserved epigenetic modification. Its enrichment promotes constitutive heterochromatin formation and gene repression.14,15 The suppressor of variegation 3–9 homolog 1 and 2 (Suv39h1/2) and SET domain bifurcated histone lysine methyltransferase 1 (SETDB1/ESET) are methyltransferases of H3K9me3, while jumonji D2/lysine-specific demethylase 4A (JMJD2/KDM4) family members are the corresponding demethylases.16–20 H3K9me3 has been shown to participate in a broad range of biological processes, including cell differentiation, embryonic development, cellular senescence, and malignancies.21–24 However, whether epigenetic regulation, especially H3K9me3, participates in neuronal ferroptosis during ICH is unknown.
In this study, we describe a novel role of H3K9me3 in neuronal ferroptosis. The level of H3K9me3 was increased in neuronal N2A cells after hemin treatment. H3K9me3 enriched at the promoter of the Tfr1 gene and repressed its expression. Decreasing H3K9me3 levels by repressing the activity or depleting the expression of its methyltransferase Suv39h1, or knocking down Tfr1 all effectively rescued neuronal ferroptosis induced by hemin and RSL3. Suppression of Suv39h1 and up-regulation of Tfr1 aggravates ICH in mice. Together, these findings will advance the understanding of neuronal ferroptosis following ICH and help to improve functional recovery using epigenetic strategies after stroke.
Material and methods
Animals
All male C57BL/6J (6–8 weeks) mice were obtained from Charles River Laboratories. Mice were housed at 20–26°C, 40–70% humidity and kept on a 12 h light/dark cycle. Food and water were available ad libitum. Animal experiments were performed according to National Institutes of Health Guide and reported in compliance with ARRIVE guidelines (Animals in Research: Reporting In vivo Experiments). All animal experimental protocols were approved by the Institutional Animal Care and Use Committee of Capital Medical University.
Intracerebroventricular (i.c.v.) injection of siRNA
A mixture of three different siRNAs (200 μM, Science artis, China) against Suv39h1 or the scramble control siRNA were delivered 48 h before ICH modeling. The mice were anesthetized using 1–2% isoflurane (R510-22, RWD, China) inhalation and fixed on a stereotactic frame (E04545, RWD). Then, a burr hole was drilled in the left skull and 0.5 μL siRNA was injected at a rate of 0.1 μL/min at the following coordinates relative to bregma: 0.5 mm anterior, 0.9 mm left lateral and 2.5 mm in the depth. The needle was held for 10 min after the injection. The craniotomy was sealed with Super Glue (1469SB, 3 M, USA). The sequences of the siRNAs are listed in Supplementary table 3.
Mouse ICH model
The mouse ICH model was established as previously described. 25 In brief, mice were anesthetized using 1–2% isoflurane inhalation and fixed on a stereotactic frame. A 0.6 mm burr hole was drilled in the left skull and 0.5 μL collagenase (C2399, Sigma-Aldrich, USA) was injected at a rate of 0.1 μL/min at the following coordinates relative to bregma: 0.8 mm anterior and 2.0 mm left lateral and 3.15 mm in the depth. The needle was held for 10 min after the injection and then the craniotomy was sealed with Super Glue (1469SB, 3M). Sham control mice received only needle insertion. Animal body temperature was maintained at 37°C throughout with a heating blanket.
Neurologic function assessment
Behaviors of the mice were tested at 12 h post ICH as previously reported. 25 Neurologic deficit scores included six parts, body symmetry, gait, climbing, circling behavior, front limb symmetry, and compulsory circling. Each test was graded from 0 to 4, and 4 indicates the most severe impairment. Forelimb placing test was used to assess the forelimb movements evoked by vibrissae-touching. The mice were placed on the edge of the table and brushed the vibrissae. Normal animals quickly placed their forelimbs on the table. The result was the probability of success for ten rounds of the trial. For hindlimb placing test, the mice were placed on the edge of the table and contralateral hind limb were pulled down. Behaviors were divided into three categories: immediate pullback (0 point), delayed pullback (1 point), inability to pull back (2 points). The results were the sum of the scores from 10 rounds of trials.
MDA measurement
Amount of MDA was measured by using MDA Assay kit (ab118970, Abcam) according to the manufacturer’s instructions. Briefly, 10 mg ipsilateral striatum was collected and reacted with thiobarbituric acid (TBA) to generate an MDA-TBA adduct. The reaction mix was transferred to wells of microplate. The absorbance was measured immediately on a microplate reader (SpectraMax iD5, Molecular devices, USA) at OD 532 nm for colorimetric assay.
Cell cultures
The N2A and SK-N-SH cells were cultured in Dulbecco's modified eagle medium (DMEM) of high glucose (C11995500BT, Gibco, USA) containing 10% fetal bovine serum (FBS, 10099141, Gibco) and 1% penicillin-streptomycin mixture (KGY0023, Keygen, China) at 37°C, 5% CO2, 95% saturated humidity in an incubator. Cells were passaged every 3 days.
Drug administration
Unless specifically indicated, N2A and SK-N-SH cells were exposed to 15 μM hemin (16009-13-5, Frontier Scientific, USA), 5 μM RSL3 (S8155, Selleck, USA), Erastin (S7242, Selleck) 2 μM Fer-1 (S7243, Selleck), 15 μM DFO (D9533, Sigma-Aldrich), or 0.5 μM Chaetocin (S8068, Selleck) for 24 hours.
MTT assay
N2A or SK-N-SH cells (1 × 104) were seeded in 96-well plates and maintained for 24 h (cell density reaches 70–80%). After treatment with different reagents, the medium was removed, and the cells were washed twice with PBS. Cells were incubated with 20 μL of 5 mg/mL MTT solution (0793, Amresco, USA) for 4 hours. One hundred microliter DMSO (D8418, Sigma-Aldrich) was added to cells after removing MTT and incubated at 37°C for 10 min. Finally, absorbance at the wavelength of 570 nm was recorded with a microplate reader (SpectraMax iD5, Molecular DEVICES, USA).
Propidium iodide (PI) staining
N2A cells (2 × 104) were seeded in 48-well plates and cultured for 24 h until the cell density reaches 70–80%. After treating with the drugs, the cells were incubated with 1 μL of 20 μg/mL PI dye (P4170, Sigma-Aldrich) followed with observation under a fluorescence microscope (ECLIPSE CI, NIKON, Japan) and the percentage of PI+ cells were counted blindly.
Chromatin immunoprecipitation (ChIP)
N2A cells (1 × 107) were plated in 15 cm cell culture dishes and cultured for 24 h. Cells were treated with 15 µM hemin or vehicle (0.075% DMSO) for 12 h. After removing the medium, the cells were incubated with 1% formaldehyde solution (F8775, Sigma-Aldrich) at 37°C for 10 min, followed by ultrasonic lysis and incubation with H3K9me3 (ab8898, Abcam, USA) or IgG (C1755, Applygen, China) antibodies at 4°C overnight. The antibody-protein-DNA complex was immunoprecipitated with protein A/G (sc-2003, Santa Cruz), and sequentially washed and eluted. After de-crosslinking at 65°C, the DNA was extracted and purified using a PCR Purification Kit (28106, QIAquick, Germany).
The primer sequences for the mouse Tfr1 promoter and gene body used in the ChIP experiment are listed in Supplementary table 6.
ChIP-sequencing (ChIP-seq)
The chromatin DNA was immunoprecipitated with anti-H3K9me3 or IgG from N2A cells, followed with purification as mentioned above and the DNA sequencing was performed by Beijing igeneCode Biotec (Beijing, China) on the MGISEQ T7 platform. ChIP-seq reads were aligned to the mouse reference genome (GRCm38/mm10) using Soap2.21 (BGI, China) with parameters as: −p 4 −v 2 −s 35 and only the alignments within 2 mismatches were considered in peak calling, for which MACS2 (version 1.4.2) was employed with parameters as: –bw 200 −p 0.005 −w –single-profile –space 50. The genomic distribution of H3K9me3 was analyzed by ChIPseeker, an R package for peak annotation and comparison, while Clusterprofiler was employed for gene ontology (GO) analyses. Integrative Genomics Viewer (IGV) was used for peak visualization at the specific genomic locus. The ChIP-seq data have been deposited in GEO repository under accession number of GSE217972.
Lipid Reactive Oxygen Species (ROS) determination
N2A cells (2 × 104) were seeded in 48-well plates and treated with different drugs for 24 h. The cells were incubated with a lipid peroxidation sensor BODIPY C11 (D3861, Invitrogen, USA) for 30 min according to the manufacturer’s instructions. After the medium was removed, the cells were washed three times with PBS. Fluorescence of different wavelengths was observed under a fluorescence microscope (ECLIPSE CI, NIKON, Japan), and the ratio of the fluorescence intensity emitted at 590 nm and 510 nm was calculated as the level of lipid ROS of the cells.
siRNA transfection in vitro
N2A cells cultured for 12–24 h were transfected with siRNAs when the cell density reached 30–50%. In brief, 10 μL Lipofectamine™ RNAiMAX (13778150, Invitrogen) was added into 1000 μL Opti-MEM™ I reduced serum culture (31985070, Gibco), which were then mixed and incubated at room temperature for 5 min. Meanwhile, another mixture of 10 μL siRNA (storage concentration was 20 μM) with 1000 μL Opti-MEM™ I reduced serum medium was prepared, which was then further mixed with the Opti-MEM™ containing the Lipofectamine™ RNAiMAX and incubated at room temperature for 15–20 min. The medium in the culture plate was discarded, and the cells were washed twice with PBS solution. Two milliliters of the mixed solution mentioned above was added to each well, maintained in the cell incubator. Six hours later, the transfection reagents were replaced with normal high glucose DMEM medium containing 10% FBS, 1% penicillin-streptomycin, and the cells were cultured for 48 h. The sequences of siRNAs are listed in Supplementary table 4.
Iron measurement
Ferrous and total iron were measured with Iron Assay kit (K390-100, Biovision, USA). N2A cells were dissociated by trypsinization. The cell suspension was collected and centrifuged. 100 μL Iron Assay Buffer was added into the cell pellet. The samples were divided into two parts, 5 μL Assay Buffer was added to 50 μL samples for ferrous iron assay, and 5 μL Iron Reducer was added to 50 μL samples for total iron assay. The Iron Probe was then added, mixed and incubated for 60 min at 37 °C.The output was measured immediately on a colorimetric microplate reader (SpectraMax iD5, Molecular devices) at OD 593 nm.
Statistical analysis
Data of experiments in vitro were presented in triplicate, and all statistical analyses were performed with GraphPad Prism 7.0. Before analyses, Shapiro-Wilk normality test was used to assess data distribution. When the data exhibit a normal distribution, two tailed Student’s t test was used for comparison of two groups while one or two-way ANOVA followed by Tukey's multiple comparisons tests were performed for comparisons of multiple groups. Otherwise, nonparametric Kruskal-Wallis test followed by Dunn's multiple comparisons was employed for comparisons of multiple groups. Specific methods were indicated in figure legend. A P value less than 0.05 was considered to be statistically significant.
Results
Hemin induces ferroptosis in neuronal cells
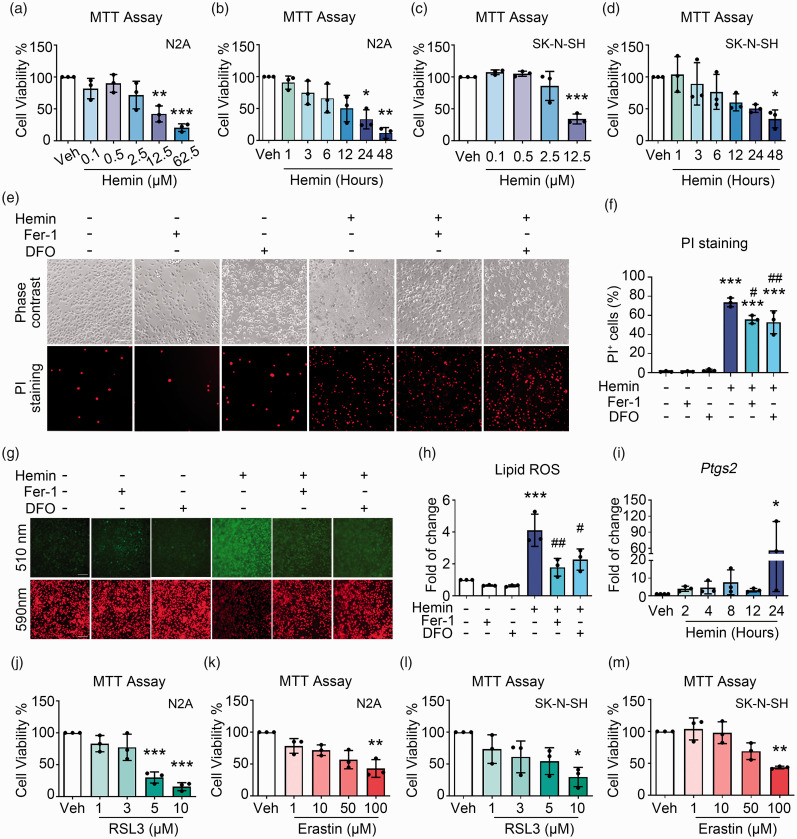
In order to explore the epigenetic regulation of neuronal death after ICH, we used hemin to induce cell death in N2A neuronal cells. 25 Hemin markedly decreased the cell viability in both dose- and time-dependent manners (Figure 1(a) and (b)). The LD50 of hemin was estimated as 15 μM and this dosage was used for the rest of the experiments. Meanwhile, a human neuronal cell line SK-N-SH was also examined, and hemin also decreased its viability in dose- and time-dependent manners (Figure 1(c) and (d)).
Figure 1.
Hemin induces ferroptosis in neuronal cells. (a) N2A cells were incubated with hemin or vehicle, and cell viability was analyzed using MTT assays. (b) N2A cells were treated with 15 μM hemin or vehicle, and MTT assays were performed. (c) SK-N-SH cells were treated with different concentrations of hemin, and MTT assays were performed. (d) SK-N-SH cells were treated with 8 μM hemin for different time points, and MTT assays were performed. (e, f) N2A cells were treated as indicated, and PI staining was performed to detect cell death. Representative images (e) and quantifications (f) are shown. (g, h) N2A cells were treated as indicated, and BODIPY 581/591 C11 reagent was used for lipid ROS detection. Representative images (g) and quantifications (h) are shown. (i) N2A cells were treated with hemin or vehicle as indicated, and the mRNA expression of Ptgs2 was detected using RT-qPCR. GAPDH serves as an internal control. (j, k) N2A cells were treated with RSL3 (j), Erastin (k), or vehicle as indicated, and MTT assays were performed. (l, m) SK-N-SH cells were treated with RSL3 (l), Erastin (m), or vehicle as indicated, and MTT assays were performed. Results are shown as scatter plots (Mean ± SD). n = 3 independent experiments. One-way ANOVA (a, c, d, f, h, i, j, m) or Kruskal-Wallis test (b, k, l) followed by Tukey's or Dunn’s multiple comparisons tests, respectively, were used. *p < 0.05, **p < 0.01, ***p < 0.001 vs vehicle; #p < 0.05, ##p < 0.01 vs Hemin. Scale bar: (e, g) 100 μm.
To further identify if the cell death caused by hemin was ferroptosis, 11 cell death was assessed with PI staining. Hemin significantly increased the percentage of PI+ cells, and co-treatment with ferroptosis inhibitors, Fer-1 and DFO, effectively attenuated cell death induced by hemin (Figure 1(e) and (f)), indicating that ferroptosis accounted for part of hemin-induced cell death in N2A cells. The accumulation of lipid reactive oxygen species (ROS) is a hallmark of ferroptosis. By staining with the BODIPY 581/591 C11 fluorescent probes, hemin increased the level of lipid ROS, which was significantly reduced by Fer-1 and DFO (Figure 1(g) and (h)). Hemin also greatly augmented the mRNA level of Ptgs2 time-dependently (Figure 1(i)), another hallmark of ferroptosis. Furthermore, other ferroptosis inducers, RSL3 and Erastin, both induced ferroptosis in N2A cells (Figure 1(j) and (k)) and SK-N-SH cells (Figure 1(l) and (m)) in a dose-dependent manner as hemin did, suggesting that ferroptosis contributed to N2A cell death under hemin treatment.
Suv39h1-mediated H3K9me3 is increased and protects against hemin-induced ferroptosis
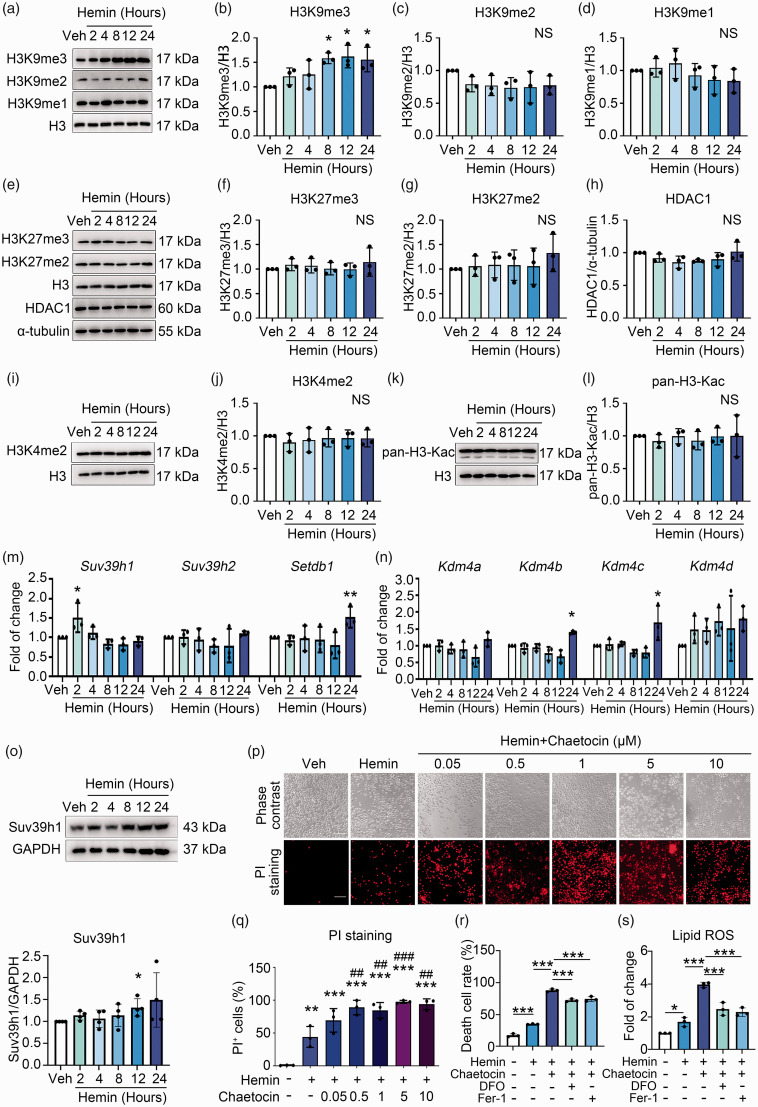
To investigate whether epigenetic regulation was involved in hemin-induced neuronal ferroptosis, N2A cells were treated with hemin and the levels of various well-characterized epigenetic modifications on histone residues were assessed. Compared with vehicle groups, the global level of transcriptional repression marker H3K9me3 was increased in a time-dependent manner, which reached a peak at 8 h and persisted till 24 h post hemin treatment (Figure 2(a) and (b)). The levels of other transcriptional repression markers, including H3K9me2, H3K9me1, H3K27me3, and H3K27me2 were not notably changed upon hemin treatment (Figure 2(a), (c) to (g)). The protein level of histone deacetylase 1 (HDAC1), the enzyme that deacetylates histone acetylation and mediates gene repression, also did not changed markedly (Figure 2(e) and (h)). As gene repression and activation are dynamically and synergistically regulated, we additionally examined the level of markers for gene transcriptional activation, such as H3K4me2 and H3Kac, which did not show significant alterations either (Figure 2(i) to (l)).
Figure 2.
Suv39h1-mediated H3K9me3 elevation protects against hemin-induced neuronal ferroptosis. (a–l) N2A cells were treated with hemin or vehicle as indicated, and the levels of different histone modifications were assessed using Western blot. α-tubulin serves as an internal control. Representative images (a, e, i, k) and quantifications (b–d, f–h, j, l) are shown. (m, n) N2A cells were treated as Continued.indicated. Total mRNA was extracted and Real-time RT-PCR was performed. GAPDH serves as an internal control. (o) N2A cells were treated as indicated. The protein level of Suv39h1 was detected using Western blot (up) and quantified (down). (p, q) N2A cells were treated as indicated, and PI staining was performed. Representative images (p) and quantifications (q) are shown. (r, s) Cell death and lipid ROS were detected. Results are shown as scatter plots (Mean ± SD). n = 3–4 independent experiments. One-way ANOVA followed by Tukey's multiple comparisons tests (b–d, f–h, j, l, m, n, q, r, s) or two-tailed t test (o) was used. *p < 0.05, **p < 0.01, ***p < 0.001 vs vehicle; ##p < 0.01, ###p < 0.001 vs. Hemin; NS, not significant. Scale bar: 100 μm.
To figure out the mechanism(s) underlying the increment of H3K9me3 upon hemin treatment, the mRNA levels of its methyltransferases (Suv39h1, Suv39h2, and Setdb1) and demethylases (Kdm4a, Kdm4b, Kdm4c, and Kdm4d) were detected in hemin-treated N2A cells. Compared with the vehicle group, the mRNA level of Suv39h1 increased significantly as early as 2 h of hemin treatment (Figure 2(m)), while the levels of other enzymes did not change significantly at early stage of treatment (Figure 2(m) and (n)). The protein level of Suv39h1 showed mild increase since 2 h (though without significance), and reached peak at 12 h after hemin treatment (Figure 2(o)). Therefore, upregulation of Suv39h1 responding to hemin treatment might contribute to the increase of H3K9me3 levels during the later time points of hemin-induced ferroptosis.
To confirm the involvement of Suv39h1 in hemin-induced ferroptosis, chaetocin, an inhibitor of Suv39h1/2, was used to inhibit the level of H3K9me3 pharmacologically. PI staining showed that hemin treatment increased the percentage of PI+ cells, while co-treatment with chaetocin further increased the percentage of PI+ cells (Figure 2(p) and (q)). Meanwhile, the cells were rescued when co-treated with DFO or Fer-1 (Figure 2(r) and Figure S1A). The level of lipid ROS also increased after hemin treatment, while more lipid ROS accumulated after co-treatment with chaetocin. The ferroptosis inhibitor DFO and Fer-1 suppressed lipid ROS accumulation induced by hemin and chaetocin stimulation (Figure 2(s) and Figure S1B). These results indicated that H3K9me3 plays a neuronal protective role in hemin induced ferroptosis in N2A cells.
H3K9me3 represses the expression of Tfr1 transcriptionally
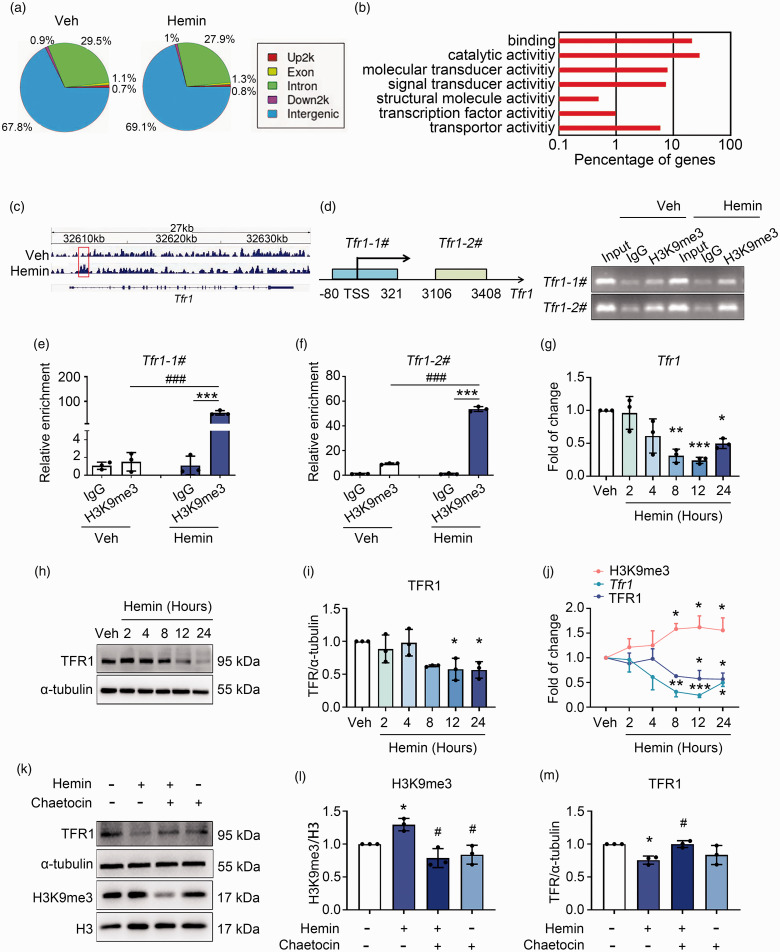
To further investigate how the increased H3K9me3 level regulates ferroptosis, we performed ChIP-Seq to screen potential target genes of H3K9me3. Figure 3(a) showed the genomic distribution of H3K9me3, which preferred to mark intergenic and intron regions on the genome in both vehicle- and hemin-treated N2A cells (Figure 3(a)). GO analyses indicated that H3K9me3 antibody-immunoprecipitated genes were enriched in several different terms, including catalytic activity, signal transducer activity, transcription factor activity, etc. (Figure 3(b)). Among the lists of vehicle- and H3K9me3-enriched genes (supplementary tables 1 and 2), we noticed a prominent enrichment of H3K9me3 at up- and downstream of the transcription start site (TSS) of Tfr1 (also named as Tfrc), the gene coding transferrin receptor 1 (TFR1) which imports transferrin-bound iron into cells (Figure 3(c)). ChIP assays were carried out to confirm the alteration of H3K9me3 level on the promoter region (primer Tfr1-1#) and gene body (primer Tfr1-2#) of Tfr1 in N2A cells with or without hemin treatment. Results of the agarose electrophoresis (Figure 3(d)) and Real-time PCR following ChIP (Figure 3(e) and (f)) both showed that treatment with hemin significantly increased the level of H3K9me3 at both the promoter and the gene body of Tfr1. Consistent with these data, compared with the vehicle group, hemin significantly reduced the mRNA level (Figure 3(g)) and protein level (Figure 3(h) and (i)) of Tfr1 time-dependently. In addition, we compared the levels of H3K9me3, Tfr1 mRNA, and TFR1 protein in N2A cells at each time point after hemin treatment. We found that while hemin treatment increased the level of H3K9me3 in N2A cells, it decreased the Tfr1 mRNA and protein levels correspondingly (Figure 3(j)).
Figure 3.
H3K9me3 represses Tfr1 expression during neuronal ferroptosis. (a) Genomic distribution of the H3K9me3 in vehicle and hemin treated N2A cells that detected with ChIP-Seq. (b) GO analyses of annotated targets of H3K9me3 histone modification in hemin treated group based on ChIP-Seq data. (c) Integrated genome viewer of H3K9me3 ChIP-seq tracings for the Tfr1 locus in vehicle and Hemin groups. Increased enrichment of H3K9me3 signal on the Tfr1 gene is shown in red rectangle. (d) Left: the schematic diagram shows the primers designed to perform ChIP assays at the Tfr1 locus. TSS, transcription start site. Right: DNA Continued.electrophoresis using agarose gels followed by ChIP assays. (e, f) ChIP assays were analyzed with Real-time qPCR. Results are expressed as fold enrichment to IgG. (g) N2A cells were treated as indicated, and the mRNA level of Tfr1 was assessed with Real-time RT-PCR. GAPDH serves as an internal control. (h, i, k–m) N2A cells were treated as indicated, and proteins were extracted for Western blot. α-tubulin serves as an internal control. Representative images (h, k) and quantifications (i, l, m) are shown. (j) The levels of H3K9me3, Tfr1 mRNA, and TFR1 protein in N2A cells at different time points upon hemin or vehicle treatment were compared. Results are shown as scatter plots or line charts (Mean ± SD). n = 3 independent experiments. Two-way ANOVA followed by Tukey's multiple comparisons tests (e, f) or One-way ANOVA followed by Tukey's multiple comparisons tests (g, i, j, l, m) was used. *p < 0.05, **p < 0.01, ***p < 0.001 vs vehicle or corresponding IgG. #p < 0.05, vs Hemin.
These data strongly suggested that hemin induced decrement of Tfr1 expression was through up-regulation of H3K9me3. Therefore, we incubated cells with hemin with or without Suv39h1/2 inhibitor chaetocin. Compared with the vehicle group, hemin-induced increment of H3K9me3 level and decrement of TFR1 protein level, which were totally reversed by the addition of chaetocin (Figure 3(k) to (m)).
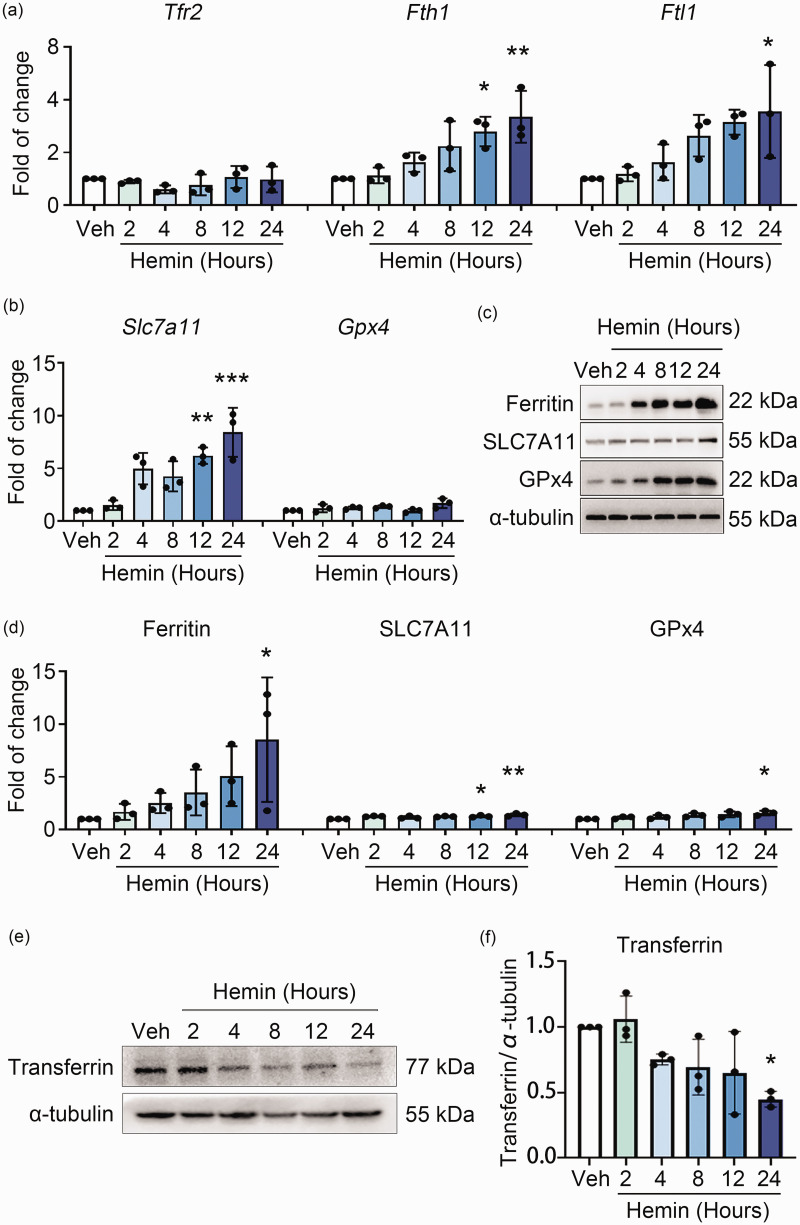
As TFR1 and TFR2 (encoded by the Tfr2 gene) have similar functions, and TFR1 is widely expressed in mammalian cells, 26 while TFR2 is mainly expressed in liver and red blood cells. 27 We next assessed the mRNA expression of Tfr2 and other related genes in iron metabolism and the xCT/GPx4 pathway. As shown in Figure 4(a), hemin treatment neither changed the mRNA expression of Tfr2, nor the Ferritin encoding genes Fth1 and Ftl1 at earlier time points. Additionally, the mRNA level of Slc7a11 (the gene that encodes xCT) increased significantly at later time points of hemin treatment, whereas the mRNA level of Gpx4 did not change markedly (Figure 4(b)). Furthermore, Western blot analyses showed that the protein levels of Ferritin, xCT, and GPx4 increased at 24 h after treating with hemin (Figure 4(c) and (d)). Consistent with the inhibition of TFR1, the intracellular level of transferrin gradually decreased post hemin treatment, which was most significant at 24 h (Figure 4(e) and (f)). These results suggested that Tfr1 was the most probable important target of H3K9me3 among those ferroptosis-related genes tested.
Figure 4.
Genes in iron-metabolism or the xCT/GPx4 pathway other than Tfr1 were not repressed by H3K9me3 during hemin-induced ferroptosis. (a, b) N2A cells were treated as indicated. Total mRNA was extracted and Real-time RT-PCR was performed. The mRNA levels of genes in iron metabolism (a) and xCT/GPx4 pathway (b) are shown. GAPDH serves as an internal control. (c, d) N2A cells were treated as indicated. Proteins were extracted and Western blot was performed. α-tubulin serves as an internal control. Representative images (c) and quantifications (d) are shown. (e, f) N2A cells were treated as indicated. The level of transferrin was detected by using Western blot (e) and quantified (f). Results are shown as scatter plots (Mean ± SD). n = 3 independent experiments. One-way ANOVA (a, b, f) or Kruskal-Wallis test (d) followed by Tukey's or Dunn’s multiple comparisons tests was used. *p < 0.05, **p < 0.01, ***p < 0.001 vs vehicle.
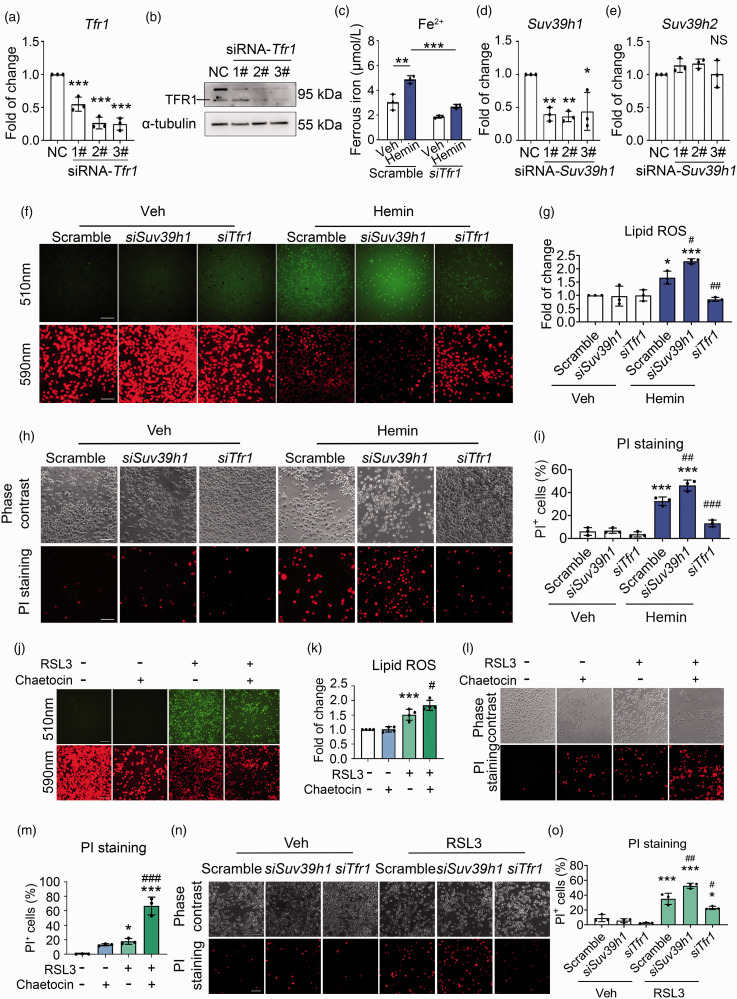
H3K9me3 represses Tfr1 to protect against ferroptosis in N2A cells
To further confirm that H3K9me3 dictates ferroptosis by transcriptionally regulating Tfr1, we knocked down the expression of Tfr1 (Figure 5(a) and (b)). The intracellular Fe2+ increased after hemin stimulation while silencing Tfr1 reduced Fe2+ content at both the basal and hemin treated conditions (Figure 5(c)). We also knocked down the expression of Suv39h1 (Figure 5(d)), whereas the mRNA level of Suv39h2 was not affected (Figure 5(e)). We found that cells transfected with Suv39h1 siRNA significantly increased the level of hemin-induced lipid ROS, while silencing of Tfr1 notably reduced the lipid ROS level induced by hemin treatment (Figure 5(f) and (g)). Consistent with results of lipid ROS, PI staining revealed that knocking down of Suv39h1 aggravated hemin-induced cell death, while Tfr1 siRNAs decreased the percentage of PI+ cells (Figure 5(h) and (i)).
Figure 5.
Suv39h1-mediated H3K9me3 protects against hemin- and RSL3-induced neuronal ferroptosis by repressing Tfr1. (a, b) N2A cells were transfected with Scramble or Tfr1 siRNA for 48 h. (a) Total mRNA was extracted, and Real-time RT-PCR was performed. GAPDH serves as an internal control. (b) Proteins were extracted and Western blot was performed. α-tubulin serves as an Continued.internal control. Representative images are shown. (c) The intracellular ferrous iron was detected. (d, e) N2A cells were transfected with Scramble or Suv39h1 siRNA for 48 h. Total mRNA was extracted, and Real-time RT-PCR was performed. GAPDH serves as an internal control. (f–o) N2A cells were treated as indicated. Lipid ROS that were assessed using BODIPY 581/591 C11 dye (f, g, j, k), and cell death that were assessed using PI staining (h, i, l–o). Representative images (f, h, j, l, n) and quantifications (g, i, k, m, o) are shown. Results are shown as scatter plots (Mean ± SD). n = 3 independent experiments. One-way ANOVA followed by Tukey's multiple comparisons tests (a, d, e, k, m) or Two-way ANOVA followed by Tukey's multiple comparisons tests (c, g, i, o) was used. *p < 0.05, **p < 0.01, ***p < 0.001 vs vehicle. #p < 0.05, ##p < 0.01, ###p < 0.001 vs. Hemin, RSL3, or Scramble; NS: not significant. Scale bar: (f, h, j, l, n) 100 μm.
Inhibition of GPx4 activity is one of the core mechanisms underpinning hemin induced neuronal ferroptosis. 28 We next incubated RSL3-treated cells with or without chaetocin. The results showed that compared with the vehicle group, RSL3 treatment alone increased the intracellular lipid ROS levels (Figure 5(j) and (k)) and the percentage of PI+ cells (Figure 5(l) and (m)), whereas chaetocin aggravated the intracellular lipid ROS accumulation (Figure 5(j) and (k)) and cell death (Figure 5(l) and (m)) caused by RSL3. Furthermore, compared with the scramble siRNA-transfection treatment, Suv39h1 siRNA treatment caused severer cell death, while Tfr1 siRNA treatment attenuated cell death induced by RSL3 (Figure 5(n) and (o)). Altogether, these results indicated that Suv39h1 mediated H3K9me3 represses Tfr1 at the transcription level to protect against neuronal ferroptosis.
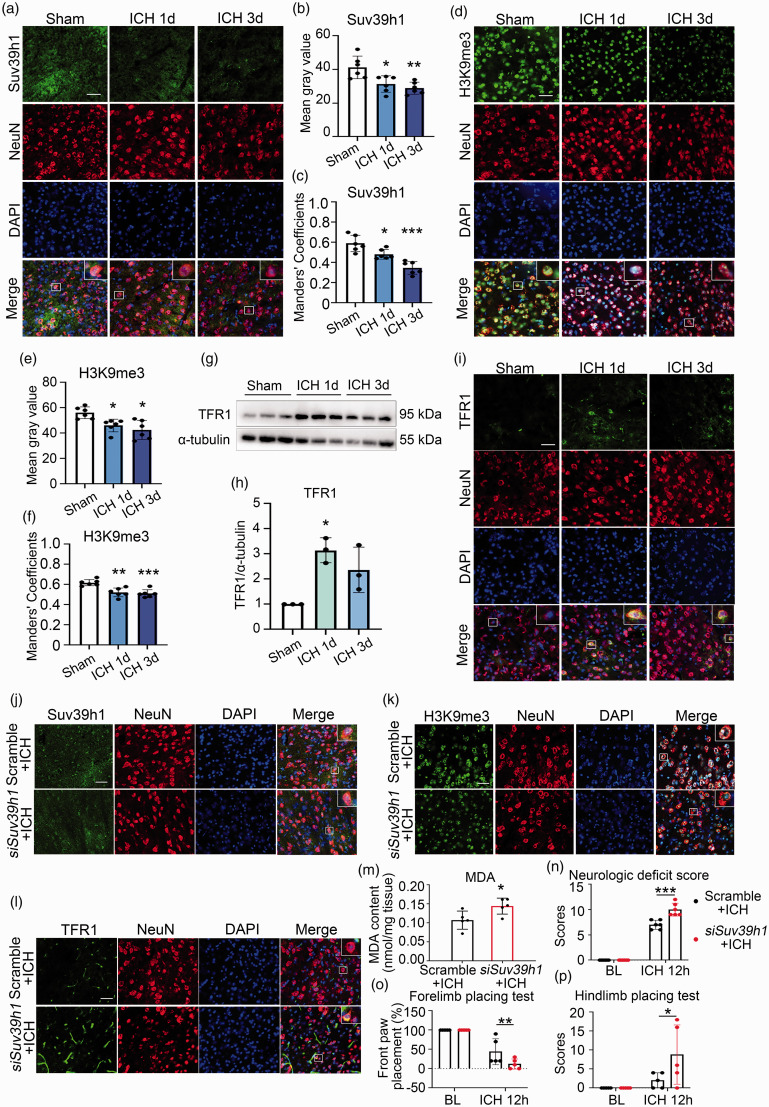
Suv39h1-H3K9me3 mediated repression of TFR1 plays a protective role after ICH
To further test whether Suv39h1-H3K9me3 mediated repression of TFR1 also works in vivo, we used a murine model of ICH. The alteration dynamics for the level of Suv39h1 and H3K9me3 were examined with ICH progression. We first examined the level of Suv39h1 and H3K9me3 at day 1 and 3 post ICH with immunofluorescence staining. Different from in vitro results, the global average level of Suv39h1 and H3K9me3 were both decreased (Figure 6(a), (b), (d) and (e)), together with the reduction of neuronal localization for Suv39h1 or H3K9me3 as demonstrated with Manders’ colocalization coefficient between Suv39h1 or H3K9me3 and the neuron marker NeuN (Figure 6(c) and (f)). On the contrary, the level of TFR1 was increased as determined with western blot and immunofluorescence, which was most prominent at day 1 after ICH (Figure 6(g) to (i)). The number of degenerating neurons around the lesion has been reported to peak at 24 hours after ICH, 5 we postulated that if Suv39h1 mediated H3K9me3 dictates neuronal ferroptosis, their level change should precede massive ferroptosis at day 1. To test this hypothesis, we further detected the level of Suv39h1 and H3K9me3 at earlier time points before 24 h post ICH with immunofluorescence. This time, consistent with the in vitro data, the average level of Suv39h1 and H3K9me3 both gradually increased at 6 h and 12 h post ICH, with the change at 12 h reaching peak and showing statistical significance (Figure S2A-D). These results indicate that early changes of Suv39h1 and H3K9me3 may contribute to neuronal ferroptosis in the ICH model. We also compared the relative level of Suv39h1 in neurons (NeuN+), astrocytes (GFAP+), microglia (Iba-1+) and oligodendrocytes (Olig2+) with immunostaining assays, where the neurons showed the highest expression in both Sham and ICH groups (Figures S3A-D).
Figure 6.
Suv39h1-mediated H3K9me3 regulated ferroptosis after ICH. (a–f) Brain slices obtained at day 1 and 3 post ICH were stained with Suv39h1, H3K9me3, NeuN antibody and DAPI (a, d). The mean fluorescence intensity of Suv39h1 and H3K9me3 were quantified respectively (b, e). The Manders' Colocalization Coefficients (M1) were calculated (c, f). (g, h) The protein of TFR1 was detected from ipsilateral striatum using Western blot (g) and quantified (h). (i) Immunostaining was performed using TFR1 antibody at 1 day and 3 days post ICH. (j–l) Immunostaining was performed using Suv39h1 (j), H3K9me3 (k) or TFR1 antibody (l) at day 3 post ICH. (m) MDA content was measured at day 3 after ICH. (n–p) Neurological function was assessed at 12 h post ICH. Results are shown as scatter plots (Mean ± SD). n = 6 images from 3 mice (b, c, e, f). n = 3 mice (h). n = 5–6 mice (m–p). One-way ANOVA (b, c, f, h, n–p) or Kruskal-Wallis test (e) followed by Tukey's or Dunn’s multiple comparisons tests, respectively or two-tailed t test (m) was used. *p < 0.05, **p < 0.01, ***p < 0.001 vs Sham or Scramble or baseline (BL) group. Scale bar: (a, d, i, j, k, l) 75 μm.
To further explore the function of neuronal Suv39h1-mediated H3K9me3 in ICH, we knocked down the expression of Suv39h1 in ICH mice by intracerebroventricular injection of Suv39h1-specific and scramble siRNAs. The expression of Suv39h1 was decreased the most in neurons as demonstrated with decreased colocalization between NeuN and Suv39h1 around the hematoma, while its colocalization coefficients in glial cells were less affected (Figure 6(j) and S3E-H). Compared with scramble group, the level of H3K9me3 was decreased (Figure 6(k)) while TFR1 increased (Figure 6(l)) with Suv39h1 knockdown. The level of malondialdehyde (MDA), one main product of lipid peroxidation,29,30 was measured to represent ferroptosis in ICH tissue, which showed overt increment with knocking down Suv39h1 (Figure 6(m)). To be of pathophysiological importance, neurologic functions were assessed with neurologic deficit score, forelimb placing test and hindlimb placing test, and the results showed that the neurological deficit was even worse, and forelimb and hindlimb movement were more severely impaired after knocking down Suv39h1 (Figure 6(n) to (p)).
Discussion
Labile iron accumulation promotes ferroptosis, and genes involved in iron metabolism are vital regulators of ferroptosis. 29 Transferrin receptors (TFRs) are cell surface receptor proteins encoded by Tfr1 or Tfr2, whose main function is incorporating Fe3+-transferrin (TF) complex into cells or regulating systematic iron homeostasis and erythropoiesis, respectively. 31 The level of TFR1 was much more sensitive to alterations of intracellular iron levels. When the cellular iron increases, the expression TFR1 decreases in a negative feedback manner, and vice versa. 31 It thus no wonder that TFR1 was down regulated upon hemin treatment, which significantly increases the level of intracellular iron. In line with its iron-uptake function, TFR1 has been reported to play an important role in ferroptosis promotion in several disease models, including myocardial I/R injury, 32 central presbycusis, 33 and cerebral ischemic injury. 34 In those models, the level of TFR1 can be directly regulated in a p53-dependent manner at the transcription level and by the upregulation of iron regulatory protein 2 (IRP-2) at the translation level, or be regulated by GPx4 and FTH1 indirectly.33,34 Interestingly, in some types of cancer like high-grade ovarian cancer and MYCN-amplified neuroblastoma, TFR1 is overexpressed, and underlies the predilection of these cells to a high level of intracellular iron and renders them more susceptible to inhibition of the system χc−/glutathione (GSH) system.35,36 Thus, the role and regulation of TFR1 in ferroptosis is complicated in various cells/diseases, but how epigenetics participate in the regulation of Tfr1 in ferroptosis was unknown.
In our study, Tfr1 was down-regulated during hemin-induced neuronal ferroptosis, and further down-regulating its expression level attenuated ferroptosis notably. Therefore, down-regulation of Tfr1 acted as an endogenous self-protective mechanism to buffer intracellular iron overload induced by hemin degradation. Consistent with this hypothesis, treating cells with chaetocin, an inhibitor of methyltransferases Suv39h1/2 of H3K9me3, upregulated Tfr1 expression level, and aggravated ferroptosis. In vivo model showed similar results that down-regulation of Tfr1 through Suv39h1 mediated H3K9me3 protects neurons from ferroptosis post ICH.
Chromatin remodeling,37–39 DNA methylations,40–42 histone modifications,40,43,44 and non-coding RNAs45–48 regulate ferroptosis in cancer cells. However, other than a study where inhibitors of HDAC induce neuronal ferroptosis by deacetylation H4ac, 49 how epigenetic regulates ferroptosis in neurons remains to be explored. In this study, through a screening of histone modifications in hemin-treated neuronal cells, we focused on the role of H3K9me3 in mediating ferroptosis. H3K9me3 is the best-known histone modification for its role in facilitating constitutive heterochromatin formation and in the repression of repetitive DNA. 15 Three methyltransferases and 4 demethylases are responsible for the dynamic changes of H3K9me3 level. 50 In our study, we found that the level of global H3K9me3 showed a concomitant upregulation during hemin treatment, and the upregulation of Suv39h1 mRNA was observed before H3K9me3 accumulation, suggesting its contribution to H3K9me3. How hemin treatment increases the level of Suv39h1 is currently unknown. To be of functional importance, decreasing global H3K9me3 through pharmacologic and genetic approaches towards Suv39h1 effectively aggravated hemin- and RSL3-induced ferroptosis. Consistent with our observation, a recent report indicates that Suv39h1 deficiency induces ferroptosis in clear cell renal cell carcinoma through targeting DPP4 (dipeptidyl-peptidase-4), 51 suggesting protection against ferroptosis in different cell types. Suv39h1-mediated H3K9me3 was supposed to regulate Tfr1 and lots of other genes, however, repressing Tfr1 gene expression may be the one of the most important functions of H3K9me3 increment in restricting neuronal ferroptosis.
Interestingly, all methyltransferases of H3K9me3, including Suv39h1, cannot recognize specific DNA sequences of target genes to bind directly. Therefore, additional scaffold proteins or transcription factors are needed, such as Krüppel-associated box (KRAB)-containing zinc-finger proteins (KRAB-ZFPs) 52 or retinoblastoma (Rb) protein. 53 In the future study, how Suv39h1 recognizes and binds to Tfr1, and mediates H3K9me3 increment at Tfr1 promoter and gene body are worthy to be investigated. Meanwhile, whether inhibition of Tfr1 by H3K9me3 functions as common mechanism that underlies the feedback down-regulation of Tfr1 upon increased iron level in other pathophysiological models also needs further investigation. Furthermore, besides H3K9me3, other epigenetic modifications must also participate in the neuronal ferroptosis in orchestration with H3K9me3. Although other histone modifications levels did not show significant global alterations in our screening, the re-distribution of these modifications on the chromosome may also interfere with hemin-induced ferroptosis.
Although multiple differences exist between experimental conditions, like difference in cellular context (neuron only in vitro versus neuron and glia cells, etc. in vivo), and difference in stimuli types (hemin only in vitro versus intracranial pressure, and many types of toxins including hemin in vivo), both in vitro and in vivo results showed that Suv39h1 and H3K9me3 repressed the expression of Tfr1, and more importantly, early up-regulation of Suv39h1-mediated H3K9me3 before massive ferroptosis played a protective role for the neuron. These results also indicated that early interventions targeting Suv39h1-H3K9me3-TFR1 axis may represent a novel therapeutic option for ICH.
Epigenetic regulation of neuronal ferroptosis might cross talk with other protective or destructive mechanisms during cerebral hemorrhage injury. For example, inhibition of HDAC could reduce white matter damage after ICH by modulating the polarization of microglia and macrophages and improve ICH-mediated neuroinflammation. 54 However, inhibition of HDAC induces neuronal ferroptosis, 49 how to properly manipulating those mechanisms to most benefit the whole brain or organism will need more intensive investigation. The mechanism of ferroptosis in tumors has been extensively studied, some of which also works in the neurons. Acyl-CoA synthetase long chain family member 4 (ACSL4), a promoter of ferroptosis in tumor cells, also exacerbate early brain damage caused by subarachnoid hemorrhage by mediating ferroptosis. 55 The interrelationship between these metabolic and epigenetic specific enzymes in neuronal ferroptosis during ICH also needs further investigation, which may hold powerful therapeutic potentials for ICH treatment.
In our in vivo experimental design, we delivered siRNAs intracerebroventricularly to reduce the Suv39h1 expression. We knocked down Suv39h1 in neurons with less perturbation in the glial cells, possibly due to their different expression levels. From a point view of basic research, the use of neuronal conditional knockout mice or the expression of shRNAs/siRNAs in viral vectors under the control of a promoter like Syn1 are more specific strategies to address the in vivo involvement of Suv39h1 in ICH, as will be demonstrated in the future. Consistent with our method, several recent papers also used siRNA to examine the function of target genes, and successfully decreased the expression of genes in the cell types of interest in vivo.56,57 With respect to the translation from basic research to clinical application, direct siRNA delivery in non-viral vectors such as nanoparticles may be a better option, which should provide a more prompt and less toxic treatment for ICH cases. 58 Further studies in the two directions above would be worthwhile to better elucidate the function of the neuronal Suv39h1-H3K9me3-TFR1 axis during ICH and their potential clinical applications.
In summary, Suv39h1 mediated H3K9me3 repressed Tfr1 gene expression in neurons. Attenuation of Suv39h1-mediated H3K9me3 aggravated hemin-induced neuronal ferroptosis and ICH damage. Our findings shed new light on ferroptosis’ molecular mechanisms and may help improve patient recovery after ICH.
Supplemental Material
Supplemental material, sj-xls-1-jcb-10.1177_0271678X231165653 for H3K9 trimethylation dictates neuronal ferroptosis through repressing Tfr1 by Ting Lan, Liye Hu, Tingting Sun, Xuechun Wang, Zhongnan Xiao, Danmin Shen, Weihua Wu, Zhaoli Luo, Chao Wei, Xiaotong Wang, Meng Liu, Yi Guo, Liyong Wang, Yamei Wang, Yabin Lu, Yan Yu, Fei Yang, Chenguang Zhang and Qian Li in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xls-2-jcb-10.1177_0271678X231165653 for H3K9 trimethylation dictates neuronal ferroptosis through repressing Tfr1 by Ting Lan, Liye Hu, Tingting Sun, Xuechun Wang, Zhongnan Xiao, Danmin Shen, Weihua Wu, Zhaoli Luo, Chao Wei, Xiaotong Wang, Meng Liu, Yi Guo, Liyong Wang, Yamei Wang, Yabin Lu, Yan Yu, Fei Yang, Chenguang Zhang and Qian Li in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-3-jcb-10.1177_0271678X231165653 for H3K9 trimethylation dictates neuronal ferroptosis through repressing Tfr1 by Ting Lan, Liye Hu, Tingting Sun, Xuechun Wang, Zhongnan Xiao, Danmin Shen, Weihua Wu, Zhaoli Luo, Chao Wei, Xiaotong Wang, Meng Liu, Yi Guo, Liyong Wang, Yamei Wang, Yabin Lu, Yan Yu, Fei Yang, Chenguang Zhang and Qian Li in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-4-jcb-10.1177_0271678X231165653 for H3K9 trimethylation dictates neuronal ferroptosis through repressing Tfr1 by Ting Lan, Liye Hu, Tingting Sun, Xuechun Wang, Zhongnan Xiao, Danmin Shen, Weihua Wu, Zhaoli Luo, Chao Wei, Xiaotong Wang, Meng Liu, Yi Guo, Liyong Wang, Yamei Wang, Yabin Lu, Yan Yu, Fei Yang, Chenguang Zhang and Qian Li in Journal of Cerebral Blood Flow & Metabolism
Acknowledgements
We thank Dr. Zhiqing Xu from Capital Medical University for kindly providing the N2A cell line. We thank Dr. Xi Wang from Capital Medical University for providing Suv39h1/2 antibodies. We thank Dr. Lin Shan from Capital Medical University for providing histone modification related antibodies. We thank Wenzhu Wang, Ting Li and Fan Bai from Chinese Institute of Rehabilitation Science, China Rehabilitation Science Institute, Beijing, China, and Beijing Key Laboratory of Neural Injury and Rehabilitation, China Rehabilitation Research Center, Beijing, China for providing technical support.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: this work was supported by the National Natural Science Foundation of China (32070735 to Q. Li, 81971037 to F. Yang).
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions: Lan T., Hu L., Zhang C., and Li Q. designed the experiments, wrote, and revised the manuscript. Lan T. and Hu L. performed experiments; Lan T., Hu L., Sun T., Wang X., Xiao Z., Shen D., Wu W., Luo Z., Wei C., Wang X., Liu M., Guo Y., Wang L., Wang Y., Lu Y., Yu Y., Yang F., Zhang C., and Li Q. collected and analyzed data. All authors have agreed on the final version to be published.
ORCID iD: Yamei Wang https://orcid.org/0000-0002-8240-3991
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Supplemental material
Supplemental material for this article is available online.
References
- 1.Keep RF, Hua Y, Xi G. Intracerebral haemorrhage: mechanisms of injury and therapeutic targets. Lancet Neurol 2012; 11: 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Asch CJ, Luitse MJ, Rinkel GJ, et al. Incidence, case fatality, and functional outcome of intracerebral haemorrhage over time, according to age, sex, and ethnic origin: a systematic review and meta-analysis. Lancet Neurol 2010; 9: 167–176. [DOI] [PubMed] [Google Scholar]
- 3.Graeber MB, Raivich G, Kreutzberg GW. Increase of transferrin receptors and iron uptake in regenerating motor neurons. J Neurosci Res 1989; 23: 342–345. [DOI] [PubMed] [Google Scholar]
- 4.Zhu X, Tao L, Tejima-Mandeville E, et al. Plasmalemma permeability and necrotic cell death phenotypes after intracerebral hemorrhage in mice. Stroke 2012; 43: 524–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q, Han X, Lan X, et al. Inhibition of neuronal ferroptosis protects hemorrhagic brain. JCI Insight 2017; 2: e90777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Y, Ma L, Luo CL, et al. IL-33 Exerts neuroprotective effect in mice intracerebral hemorrhage model through suppressing inflammation/apoptotic/autophagic pathway. Mol Neurobiol 2017; 54: 3879–3892. [DOI] [PubMed] [Google Scholar]
- 7.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012; 149: 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang WS, Stockwell BR. Ferroptosis: death by lipid peroxidation. Trends Cell Biol 2016; 26: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang WS, SriRamaratnam R, Welsch ME, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014; 156: 317–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon MY, Park E, Lee SJ, et al. Heme oxygenase-1 accelerates erastin-induced ferroptotic cell death. Oncotarget 2015; 6: 24393–24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zille M, Karuppagounder SS, Chen Y, et al. Neuronal death after hemorrhagic stroke in vitro and in vivo shares features of ferroptosis and necroptosis. Stroke 2017; 48: 1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren H, Han R, Chen X, et al. Potential therapeutic targets for intracerebral hemorrhage-associated inflammation: an update. J Cereb Blood Flow Metab 2020; 40: 1752–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- 14.Ninova M, Fejes Toth K, Aravin AA. The control of gene expression and cell identity by H3K9 trimethylation. Development 2019; 146: dev181180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker JS, Nicetto D, Zaret KS. H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet 2016; 32: 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz DC, Ayyanathan K, Negorev D, et al. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 2002; 16: 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicetto D, Donahue G, Jain T, et al. H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 2019; 363: 294–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rea S, Eisenhaber F, O'Carroll D, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000; 406: 593–599. [DOI] [PubMed] [Google Scholar]
- 19.Loh YH, Zhang W, Chen X, et al. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev 2007; 21: 2545–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couture JF, Collazo E, Ortiz-Tello PA, et al. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol 2007; 14: 689–695. [DOI] [PubMed] [Google Scholar]
- 21.Sabbattini P, Canzonetta C, Sjoberg M, et al. A novel role for the Aurora B kinase in epigenetic marking of silent chromatin in differentiated postmitotic cells. EMBO J 2007; 26: 4657–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puschendorf M, Terranova R, Boutsma E, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet 2008; 40: 411–420. [DOI] [PubMed] [Google Scholar]
- 23.Wood JG, Hillenmeyer S, Lawrence C, et al. Chromatin remodeling in the aging genome of Drosophila. Aging Cell 2010; 9: 971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiencke JK, Zheng S, Morrison Z, et al. Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells. Oncogene 2008; 27: 2412–2421. [DOI] [PubMed] [Google Scholar]
- 25.Xiao Z, Shen D, Lan T, et al. Reduction of lactoferrin aggravates neuronal ferroptosis after intracerebral hemorrhagic stroke in hyperglycemic mice. Redox Biol 2022; 50: 102256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gammella E, Buratti P, Cairo G, et al. The transferrin receptor: the cellular iron gate. Metallomics 2017; 9: 1367–1375. [DOI] [PubMed] [Google Scholar]
- 27.Kawabata H, Yang R, Hirama T, et al. Molecular cloning of transferrin receptor 2. A new member of the transferrin receptor-like family. J Biol Chem 1999; 274: 20826–20832. [DOI] [PubMed] [Google Scholar]
- 28.Karuppagounder SS, Alin L, Chen Y, et al. N-acetylcysteine targets 5 lipoxygenase-derived, toxic lipids and can synergize with prostaglandin E2 to inhibit ferroptosis and improve outcomes following hemorrhagic stroke in mice. Ann Neurol 2018; 84: 854–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol 2021; 22: 266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su LJ, Zhang JH, Gomez H, et al. Reactive oxygen species-induced lipid peroxidation in apoptosis, autophagy, and ferroptosis. Oxid Med Cell Longev 2019; 2019: 5080843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawabata H. Transferrin and transferrin receptors update. Free Radic Biol Med 2019; 133: 46–54. [DOI] [PubMed] [Google Scholar]
- 32.Tang LJ, Zhou YJ, Xiong XM, et al. Ubiquitin-specific protease 7 promotes ferroptosis via activation of the p53/TfR1 pathway in the rat hearts after ischemia/reperfusion. Free Radic Biol Med 2021; 162: 339–352. [DOI] [PubMed] [Google Scholar]
- 33.Chen X, Li D, Sun HY, et al. Relieving ferroptosis may partially reverse neurodegeneration of the auditory cortex. FEBS J 2020; 287: 4747–4766. [DOI] [PubMed] [Google Scholar]
- 34.Li G, Li X, Dong J, et al. Electroacupuncture ameliorates cerebral ischemic injury by inhibiting ferroptosis. Front Neurol 2021; 12: 619043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Floros KV, Cai J, Jacob S, et al. MYCN-amplified neuroblastoma is addicted to iron and vulnerable to inhibition of the system Xc-/glutathione axis. Cancer Res 2021; 81: 1896–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basuli D, Tesfay L, Deng Z, et al. Iron addiction: a novel therapeutic target in ovarian cancer. Oncogene 2017; 36: 4089–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang Y, He Y, Liu S, et al. Chromatin remodeling factor lymphoid-specific helicase inhibits ferroptosis through lipid metabolic genes in lung cancer progression. Chin J Cancer 2017; 36: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao C, Wang X, Liu Y, et al. A G3BP1-interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res 2018; 78: 3484–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang D, Li Q, Sun X, et al. CRL4(DCAF8) dependent opposing stability control over the chromatin remodeler LSH orchestrates epigenetic dynamics in ferroptosis. Cell Death Differ 2021; 28: 1593–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang Y, Mao C, Yang R, et al. EGLN1/c-Myc induced lymphoid-specific helicase inhibits ferroptosis through lipid metabolic gene expression changes. Theranostics 2017; 7: 3293–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Sui S, Wang L, et al. Inhibition of tumor propellant glutathione peroxidase 4 induces ferroptosis in cancer cells and enhances anticancer effect of cisplatin. J Cell Physiol 2020; 235: 3425–3437. [DOI] [PubMed] [Google Scholar]
- 42.Lee JY, Nam M, Son HY, et al. Polyunsaturated fatty acid biosynthesis pathway determines ferroptosis sensitivity in gastric cancer. Proc Natl Acad Sci U S A 2020; 117: 32433–32442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Shi J, Liu X, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol 2018; 20: 1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Zhuang L, Gan B. BAP1 suppresses tumor development by inducing ferroptosis upon SLC7A11 repression. Mol Cell Oncol 2019; 6: 1536845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qi W, Li Z, Xia L, et al. LncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress during erastin-induced ferroptosis in HepG2 hepatocellular carcinoma cells. Sci Rep 2019; 9: 16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomita K, Fukumoto M, Itoh K, et al. MiR-7-5p is a key factor that controls radioresistance via intracellular Fe(2+) content in clinically relevant radioresistant cells. Biochem Biophys Res Commun 2019; 518: 712–718. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z, Chen X, Liu N, et al. A nuclear long non-coding RNA LINC00618 accelerates ferroptosis in a manner dependent upon apoptosis. Mol Ther 2021; 29: 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma Q, Dai X, Lu W, et al. Silencing long non-coding RNA MEG8 inhibits the proliferation and induces the ferroptosis of hemangioma endothelial cells by regulating miR-497-5p/NOTCH2 axis. Biochem Biophys Res Commun 2021; 556: 72–78. [DOI] [PubMed] [Google Scholar]
- 49.Zille M, Kumar A, Kundu N, et al. Ferroptosis in neurons and cancer cells is similar but differentially regulated by histone deacetylase inhibitors. eNeuro 2019; 6: 0263-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell 2007; 25: 1–14. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Yin X, He W, et al. SUV39H1 deficiency suppresses clear cell renal cell carcinoma growth by inducing ferroptosis. Acta Pharm Sin B 2021; 11: 406–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsui T, Leung D, Miyashita H, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 2010; 464: 927–931. [DOI] [PubMed] [Google Scholar]
- 53.Sdek P, Zhao P, Wang Y, et al. Rb and p130 control cell cycle gene silencing to maintain the postmitotic phenotype in cardiac myocytes. J Cell Biol 2011; 194: 407–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang H, Ni W, Wei P, et al. HDAC inhibition reduces white matter injury after intracerebral hemorrhage. J Cereb Blood Flow Metab 2021; 41: 958–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qu XF, Liang TY, Wu DG, et al. Acyl-CoA synthetase long chain family member 4 plays detrimental role in early brain injury after subarachnoid hemorrhage in rats by inducing ferroptosis. CNS Neurosci Ther 2021; 27: 449–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen F, Xu X, Yu Z, et al. Rbfox-1 contributes to CaMKIIalpha expression and intracerebral hemorrhage-induced secondary brain injury via blocking micro-RNA-124. J Cereb Blood Flow Metab 2021; 41: 530–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.You M, Long C, Wan Y, et al. Neuron derived fractalkine promotes microglia to absorb hematoma via CD163/HO-1 after intracerebral hemorrhage. Cell Mol Life Sci 2022; 79: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Almarghalani DA, Boddu SHS, Ali M, et al. Small interfering RNAs based therapies for intracerebral hemorrhage: challenges and progress in drug delivery systems. Neural Regen Res 2022; 17: 1717–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xls-1-jcb-10.1177_0271678X231165653 for H3K9 trimethylation dictates neuronal ferroptosis through repressing Tfr1 by Ting Lan, Liye Hu, Tingting Sun, Xuechun Wang, Zhongnan Xiao, Danmin Shen, Weihua Wu, Zhaoli Luo, Chao Wei, Xiaotong Wang, Meng Liu, Yi Guo, Liyong Wang, Yamei Wang, Yabin Lu, Yan Yu, Fei Yang, Chenguang Zhang and Qian Li in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xls-2-jcb-10.1177_0271678X231165653 for H3K9 trimethylation dictates neuronal ferroptosis through repressing Tfr1 by Ting Lan, Liye Hu, Tingting Sun, Xuechun Wang, Zhongnan Xiao, Danmin Shen, Weihua Wu, Zhaoli Luo, Chao Wei, Xiaotong Wang, Meng Liu, Yi Guo, Liyong Wang, Yamei Wang, Yabin Lu, Yan Yu, Fei Yang, Chenguang Zhang and Qian Li in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-3-jcb-10.1177_0271678X231165653 for H3K9 trimethylation dictates neuronal ferroptosis through repressing Tfr1 by Ting Lan, Liye Hu, Tingting Sun, Xuechun Wang, Zhongnan Xiao, Danmin Shen, Weihua Wu, Zhaoli Luo, Chao Wei, Xiaotong Wang, Meng Liu, Yi Guo, Liyong Wang, Yamei Wang, Yabin Lu, Yan Yu, Fei Yang, Chenguang Zhang and Qian Li in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-4-jcb-10.1177_0271678X231165653 for H3K9 trimethylation dictates neuronal ferroptosis through repressing Tfr1 by Ting Lan, Liye Hu, Tingting Sun, Xuechun Wang, Zhongnan Xiao, Danmin Shen, Weihua Wu, Zhaoli Luo, Chao Wei, Xiaotong Wang, Meng Liu, Yi Guo, Liyong Wang, Yamei Wang, Yabin Lu, Yan Yu, Fei Yang, Chenguang Zhang and Qian Li in Journal of Cerebral Blood Flow & Metabolism
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.