Abstract
Transposon mutagenesis in bacteria generally requires efficient delivery of a transposon suicide vector to allow the selection of relatively infrequent transposition events. We have developed an IS903-based transposon mutagenesis system for diverse gram-negative bacteria that is not limited by transfer efficiency. The transposon, IS903φkan, carries a cryptic kan gene, which can be expressed only after successful transposition. This allows the stable introduction of the transposon delivery vector into the host. Generation of insertion mutants is then limited only by the frequency of transposition. IS903φkan was placed on an IncQ plasmid vector with the transposase gene located outside the transposon and expressed from isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoters. After transposase induction, IS903φkan insertion mutants were readily selected in Escherichia coli by their resistance to kanamycin. We used IS903φkan to isolate three catalase-deficient mutants of the periodontal pathogen Actinobacillus actinomycetemcomitans from a library of random insertions. The mutants display increased sensitivity to hydrogen peroxide, and all have IS903φkan insertions within an open reading frame whose predicted product is closely related to other bacterial catalases. Nucleotide sequence analysis of the catalase gene (designated katA) and flanking intergenic regions also revealed several occurrences of an 11-bp sequence that is closely related to the core DNA uptake signal sequence for natural transformation of Haemophilus influenzae. Our results demonstrate the utility of the IS903φkan mutagenesis system for the study of A. actinomycetemcomitans. Because IS903φkan is carried on a mobilizable, broad-host-range IncQ plasmid, this system is potentially useful in a variety of bacterial species.
Transposons are remarkably effective tools for the genetic characterization and manipulation of bacterial genomes. They provide a means to interrupt, mark, identify, characterize, clone, and insert genes of interest (2, 28). Several different transposon mutagenesis systems are available to generate essentially random insertions into host chromosomes (3, 28). In general, these systems require that the transposon be introduced into a target bacterium by a plasmid or phage suicide vector that is unable to replicate or integrate into the bacterial chromosome. Cells that have undergone rare transposition events are then selected by stable expression of a selective marker within the transposon. One disadvantage of such systems is that transfer of the suicide vector into the target host must occur at a high enough efficiency to allow the detection of rare transposition events (1 event per 104 to 106 cells).
IS903 is an insertion sequence of 1,057 bp that contains 18-bp inverted repeats at its ends and a single gene for transposase (tnp). IS903 transposes predominantly by a simple insertion pathway and generates 9-bp target duplications on insertion (18). IS903 transposition has two requirements: (i) the inverted repeat sequences must be present at the ends, and (ii) the transposase gene must be present in cis to the transposon for efficient transposition (9). This simple system is easily manipulated to construct insertion mutagenesis vectors that facilitate stable insertions in host genomes. For example, an IS903 derivative has been used for insertion mutagenesis in Legionella pneumophila, the causative agent of Legionnaires’ disease (7, 49). Here we describe a new derivative, IS903φkan, which allows direct selection of insertion mutations into actively expressed genes. The system does not require efficient delivery of the suicide vector to the host to detect large numbers of transposon insertion mutants, a property particularly important for the mutagenesis of bacterial species that lack efficient systems for the introduction of transposon vectors.
One such bacterium is Actinobacillus actinomycetemcomitans, a gram-negative facultative anaerobe that is believed to cause severe localized juvenile periodontitis, as well as other human infections, including endocarditis, meningitis, pneumonia, septicemia, urinary tract infections, vertebral osteomyelitis, and abscesses (13, 48). Very little is known about the genetics of A. actinomycetemcomitans colonization and virulence factors, in part because it has been difficult to generate transposon insertion mutations in this organism; transposition frequencies are extremely low (10−7), and no efficient delivery system has been developed (29, 40). In this work, we demonstrate the utility of IS903φkan for efficient, random mutagenesis of A. actinomycetemcomitans. Expression of catalase activity is a defining characteristic of A. actinomycetemcomitans, and it may be an important defense mechanism against oxidative killing by phagocytic cells. From a library of 4,000 IS903φkan insertion mutants of A. actinomycetemcomitans, we isolated mutants defective in catalase activity and identified the gene for catalase (katA). Examination of the intergenic regions flanking katA revealed multiple copies of a potential DNA uptake sequence for natural transformation.
MATERIALS AND METHODS
Cell growth and storage.
Escherichia coli strains were grown on Luria-Bertani (LB) plates and broth as previously described (39). Where appropriate, media were supplemented with 20 μg of chloramphenicol per ml, 50 μg of kanamycin per ml, and 0.01, 0.1, or 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The E. coli strains used included DH1 (F− supE44 hsdR17 recA1 gyrA96 relA1 endA1 thi-1 λ) (20) and a spontaneous rifampin-resistant mutant of JA221 (F− lacY leuB6 trpE5 hsdR recA1 λ−) (received from C. Yanofsky). A. actinomycetemcomitans strains were grown in AAGM broth (17) containing 30 g of Trypticase soy broth (BBL), 6 g of yeast extract (BBL), 0.75% glucose, and 0.4% NaHCO3 per liter. The glucose and NaHCO3 were added to the medium after autoclaving. AAGM plates were made similarly, except that 40 g of Trypticase soy agar was substituted for the Trypticase soy broth. Where appropriate, media for A. actinomycetemcomitans were supplemented with 20 μg of kanamycin per ml, 2 μg of chloramphenicol per ml, 20 μg of nalidixic acid per ml, and 1 or 10 mM IPTG. The plates were incubated at 37°C in a CO2-enriched environment in a sealed Gaspak container (BBL) for 48 to 72 h. Broth cultures of A. actinomycetemcomitans were grown in screw-cap plastic tubes at 37°C for 15 to 20 h. A. actinomycetemcomitans strains can be stored by concentrating overnight AAGM broth cultures 10-fold and placing them at −70°C in AAGM broth containing 10% dimethyl sulfoxide. A. actinomycetemcomitans Y4Nal was isolated as a spontaneous Nalr mutant of strain Y4 (ATCC 43718) after plating of dense suspensions of cells on medium containing nalidixic acid. To determine the effect of H2O2 on A. actinomycetemcomitans, strains were grown overnight in AAGM broth. Each culture was diluted to a final absorbance at 600 nm (A600) of 0.030 in 10 ml of fresh medium with or without 0.1 mM H2O2 in screw-cap plastic tubes and grown at 37°C. During growth, 1-ml aliquots were removed and the A600 was monitored.
Plasmid construction.
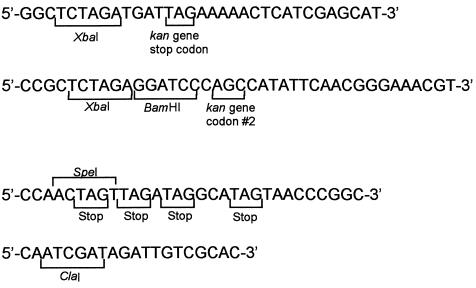
pVJT128 (Fig. 1) is based on a derivative of the mobilizable IncQ expression plasmid pMMB67HE (16), into which a Cmr marker was cloned to generate pJAK17 (30). pVJT128 contains the IS903 transposase gene and inverted repeat sequences from pKD368 (7); located between the inverted repeat sequences is the kanamycin resistance gene (kan) beginning with the second codon. An 838-bp PCR product containing the kan gene flanked by XbaI was generated with primers OKD110 and OKD115 (Table 1) and the template pKD100 (9). OKD115 anneals to the 5′ end of the Tn903 kan gene beginning with the second codon and contains both XbaI and BamHI sites at the 5′ end. OKD110 anneals to the 3′ end of the Tn903 kanamycin gene and contains the stop codon and an XbaI site. The construct was further modified by replacing an SpeI-ClaI fragment, including the 5′ end of the kan gene and the region immediately upstream, with a sequence which introduced three out-of-frame stop codons and eliminated any translational start sites between the stop codons and codon 2 of the kan gene. Primers OKD121 and OKD122 (Table 1) were used to generate the replacement fragment, a 193-bp PCR product flanked by SpeI and ClaI sites, which incorporates the three out-of-frame stop codons and a TAG stop codon instead of an in-frame ATG codon. OKD121 anneals upstream of the kan gene and the inverted repeat sequence. OKD122 anneals within the kan gene.
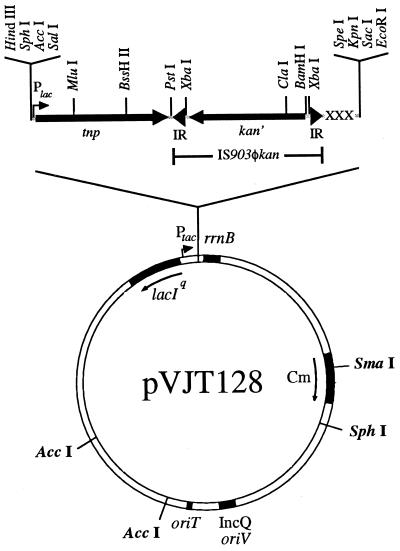
FIG. 1.
Map of the pVJT128 plasmid. tnp is the IS903 transposase gene from pKD368, and kan′ is the kanamycin resistance gene from Tn903 beginning with the second codon. IR indicates the 18-bp inverted-repeat sequence of IS903. The three amber stop codons in different reading frames are indicated by XXX. The boundaries of the transposable element IS903φkan are noted. The MluI and BssHII sites used to create the transposase-minus derivative, pVJT131, are shown.
TABLE 1.
Properties of primers used in the construction of pVJT128 and pVJT131 plasmids
A tnp deletion derivative of pVJT128 was constructed by removing 443 bp between the MluI and BssHII sites within the transposase coding sequence. The resulting construct, pVJT131, is otherwise isostructural to pVJT128 and is used as a transposase-minus control.
Transposition in E. coli.
CaCl2-competent DH1 was transformed with pVJT128 and pVJT131 (39). Transformants were selected on LB containing chloramphenicol and IPTG (1 mM). After overnight growth, two transformants from each plasmid were picked and resuspended in 1 ml of LB and plated on LB containing kanamycin and LB containing chloramphenicol. To determine the frequency of transposition within the colony, the number of kanamycin-resistant colonies was divided by the number of chloramphenicol-resistant colonies.
Transposition in A. actinomycetemcomitans.
The IncP oriT-minus RK2 derivative, pRK21761 (42), was used to mobilize the IncQ plasmids pJAK17, pVJT128, and pVJT131 from E. coli to A. actinomycetemcomitans (17). The E. coli donor strains were constructed by CaCl2 transformation of JA221Rif(pRK21671) with the three IncQ plasmids. Donor strains and the recipient strain, Y4Nal, were grown to stationary phase, mixed in a ratio of 1:10, spotted on AAGM plates, and incubated at 37°C for 16 h. The mating mixtures were scraped from the plates with a cell scraper (Fisher Scientific), resuspended in 1 ml of AAGM, and plated on AAGM plates supplemented with nalidixic acid and chloramphenicol to select for transconjugants. The presence of the transferred plasmids in the transconjugants was confirmed by DNA extraction (Qiagen Midi column), restriction endonuclease digestion, and electrophoresis in a 0.8% agarose gel. Y4Nal containing the plasmid pJAK17, pVJT128, or pVJT131 was grown in AAGM-chloramphenicol broth for 15 h. A 10−6 dilution was plated on AAGM-chloramphenicol plates containing 0, 1, or 10 mM IPTG and incubated at 37°C for 48 h to allow transposition. Four colonies from each of the three strains were picked, resuspended in 1 ml of AAGM broth, and plated on AAGM and AAGM-kanamycin. To determine the frequency of transposition within the colony, the number of kanamycin-resistant cells per colony was divided by the total number of cells per colony.
Curing A. actinomycetemcomitans strains of the IncQ plasmids.
To cure strains of the transposon donor plasmid, kanamycin-resistant potential transposon mutants were picked and streaked onto AAGM-kanamycin. Kanamycin-resistant colonies were picked and grown in 5 ml of AAGM-kanamycin broth for 15 h. Cultures were then streaked on AAGM-kanamycin plates. Individual colonies were then picked and restreaked on both AAGM-kanamycin and AAGM-chloramphenicol. More than half of the colonies were Kmr and Cms, indicating curing of the transposon donor plasmid.
Southern blot analysis.
Genomic DNA was isolated from A. actinomycetemcomitans by following the large-scale protocol of Ausubel et al. (1). A 1.5-kb HaeII fragment containing the Tn903 kan gene of pMK20 (25) was electroeluted (39) from a Tris-acetate-EDTA (TAE)–0.8% agarose gel to use as a probe (kan probe) for the presence of the transposon. Based on a partial sequence from the A. actinomycetemcomitans Genome-Sequencing Project database (35), primers 5′-GTGGATAATGACAACACCATG-3′ (positions 558 to 578 in Fig. 4) and 5′-GCCGTTGTGATCTACGCGCAT-3′ (positions 1640 to 1620 in Fig. 4) were designed to amplify a putative catalase gene to use as a probe (cat probe). Both the kan and cat probes were labeled with digoxigenin by using the digoxigenin labeling kit (Boehringer-Mannheim) as specified by the manufacturer. Southern blotting was done by standard procedures (39).
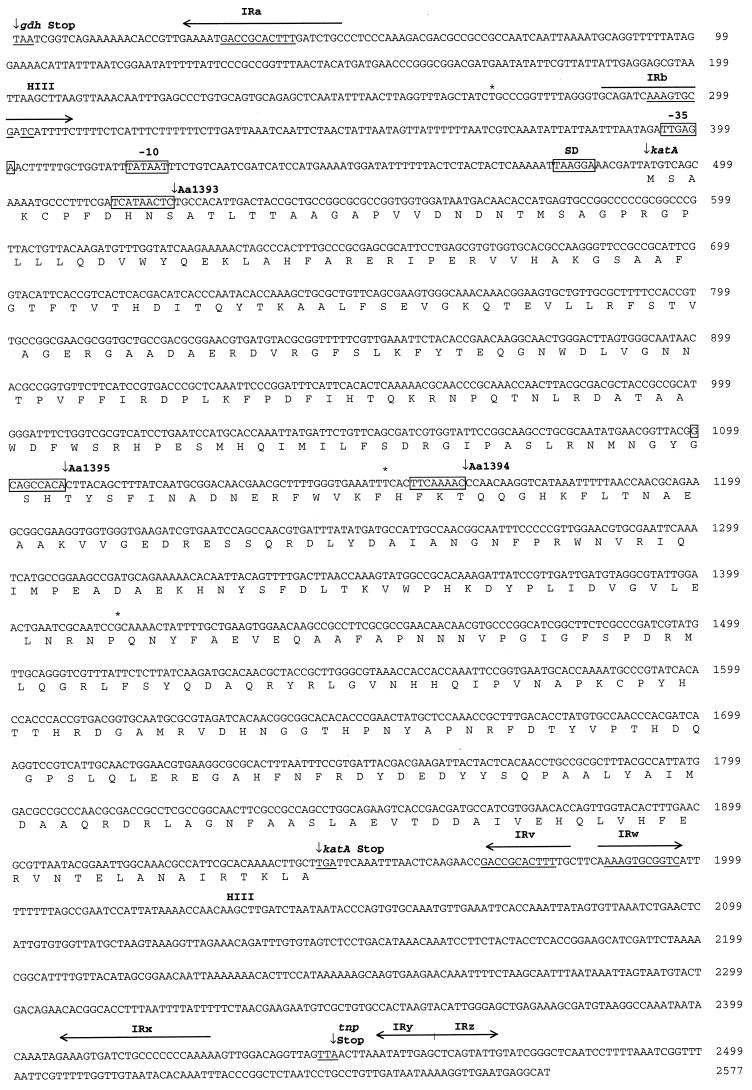
FIG. 4.
Nucleotide sequence of katA and flanking regions. Shown is the 5′→3′ sequence of a 2,577-nucleotide segment of Y4Nal genomic DNA starting from the stop codon of a predicted glutamate dehydrogenase gene (GenBank accession no. AF162654). The predicted amino acid sequence of the katA product is also shown. The −35 and −10 promoter sequences and the Shine-Dalgarno sequence (SD) are boxed. The insertion sites for IS903φkan are shown by vertical arrows above the sequence. The nucleotides duplicated at the sites of IS903φkan insertions are also boxed. The 11-bp putative DNA uptake sequences are underlined. Inverted repeats are indicated by horizontal arrows above the sequence. Asterisks indicate nucleotide differences with respect to the Genome Sequence Project database. None of the differences result in a change of amino acid. The two HindIII sites present in the region are indicated above the sequence.
Catalase activity.
To screen for catalase activity, individual colonies were picked with a toothpick and dipped into a beaker of 3% H2O2 or smeared on a slide and tested with one drop of 3% H2O2. Catalase-positive colonies gave visible oxygen bubbles. Loss of catalase activity was confirmed by a quantitative assay. A 1-ml volume of an overnight broth culture was centrifuged at 2,000 × g in an Eppendorf microcentrifuge for 5 min. The pellet was washed twice with 0.85% phosphate-buffered saline (PBS) and then resuspended in 0.1 ml of PBS. A 0.9-ml volume of 18 mM H2O2 in PBS was added, and the suspension was incubated at 25°C for 5 min. Cells were centrifuged at 16,000 × g for 1 min, and the A240 of the supernatant was measured to determine the concentration of remaining H2O2. The molar concentration of H2O2 was based on an extinction coefficient of 81 cm−1 M−1 (34).
Inverse PCR.
Inverse PCR was done by published methods (47). A 1-μg portion of HindIII-digested genomic DNA was ligated with 10 U of DNA ligase (New England Biolabs) in a 50-μl reaction mixture at 14°C for 16 h. The large reaction volume was used to facilitate intramolecular ligation. The A. actinomycetemcomitans genomic DNA adjacent to the transposon was amplified by PCR with primers which hybridize within the transposon and are oriented outward: kanStart (5′-GTTTCCCGTTGAATATGGCTGGG-3′) and kanStop (5′-GCAGTTTCATTTGATGCTCGA-3′).
Sequencing.
PCR products were cut out of ethidium bromide-stained 1% Tris-acetate gels, and DNA was electroeluted (39). The purified DNA was sequenced at the Columbia University DNA Sequencing Facility by using Perkin-Elmer Applied Biosystems automated DNA sequencer 373A. Three overlapping PCR products were made to sequence the catalase region of Y4Nal. Both strands were sequenced by using the same primers that were used to produce each PCR product. The primer pairs used for PCR and for sequencing were 5′-ATCGCCGGTTTCGTGAAAGTC-3′ (starting 51 bases and ending 30 bases upstream of the catalase region [see Fig. 4]) and 5′-CAGCGCAGCTTTGGTGTATTG-3′ (positions 749 to 729 in Fig. 4), yielding an 800-bp PCR product; 5′-GTGGTGCACGCCAAGGGTTCC-3′ (positions 669 to 689) and 5′-GTGTGTGCCGCCGTTGTGATC-3′ (positions 1649 to 1629), yielding an 981-bp PCR product; and 5′-TGCCCGTATCACACCACCCAC-3′ (positions 1587 to 1607) and 5′-TGACCTGCTGCAAGTGTCTGA-3′ (starting 23 bases and ending 2 bases downstream of the catalase region), yielding a 1,013-bp PCR product. To further confirm some of the sequences in the 1,013-bp PCR product, the internal primer 5′-ACTCAAGAACCGACCGCACT-3′ (positions 1957 to 1976) was also used for sequencing. To look for frameshift mutations in the kanamycin gene in one of the catalase mutants, Aa1394, primers 5′-CAGCGATCGTGGTATTCCGG-3′ (positions 1052 to 1071) and 5′-TAACCAACGCAGAAGCGGCG-3′ (positions 1186 to 1205), hybridizing upstream and downstream of the insertion site, respectively, were used for PCR and were used for sequencing.
RESULTS
Construction of IS903φkan for selection of insertions by expression of kan gene fusions.
We constructed an IS903-based transposon with a cryptic kan gene (Fig. 1). The 5′-end of the kan open reading frame in this element, named IS903φkan, extends through one of the IS903 inverted-repeat sequences flanking the transposon. kan is not expressed, because it lacks a start codon, ribosome binding site, and promoter. In addition, the IncQ plasmid carrying IS903φkan (pVJT128) has three tandem stop codons in different reading frames as well as the rrnB transcriptional terminator, all located immediately upstream of kan and outside IS903φkan to further reduce any residual expression from the vector. Activation of the cryptic kan gene would occur by transposition of IS903φkan into an expressed gene in the appropriate reading frame to generate a gene fusion with kan.
Transposition of IS903φkan from pVJT128 was designed to be inducible by IPTG. Transcription of IS903 tnp is initiated from the Lac repressor-regulated promoters lacUV5 and tac (Fig. 1). To further increase the expression of transposase, we replaced the GTG start codon of the tnp gene with ATG and altered a strong transcriptional terminator that occurs within the gene, as described previously (7). The tnp gene is located outside the transposon and immediately adjacent to IS903φkan; thus, it becomes physically separated from IS903φkan following transposition. Subsequent transposition events should be less likely because IS903 transposase is predominantly cis acting (10).
We expected that expression of Kmr by pVJT128-containing cells would be dependent upon transposition of IS903φkan. To test this, the E. coli recA strain DH1 was transformed with pVJT128 and Cmr transformants were selected in the presence of 1 mM IPTG to induce transposase expression. Individual colonies were picked and resuspended in broth, and cells were plated on kanamycin-containing medium to select for cells that may have undergone a transposition event. The colonies each contained an average of 5.4 × 103 Kmr cells in a total of 2.3 × 107 cells, yielding a frequency of Kmr mutants of 2.3 × 10−4. As a control, the experiment was repeated with cells carrying plasmid pVJT131, which is identical to pVJT128 except for a 443-bp MluI-BssHII deletion in tnp, which prevents the expression of functional transposase. With pVJT131, Kmr cells occurred at a frequency of 10−7 or lower. Thus, the 103-fold-higher-frequency appearance of Kmr mutants in the pVJT128-containing colonies was dependent on expression of the IS903 transposase, as expected if transposition of IS903φkan had occurred. The apparent transposition frequency of 2.3 × 10−4, as reflected by the appearance of Kmr mutants, is consistent with transposition frequencies of other IS903 derivatives assayed by conjugation (8).
Transposition of IS903φkan in A. actinomycetemcomitans.
The IncQ plasmid replicon of the IS903φkan-containing plasmid pVJT128 is functional in a wide variety of gram-negative bacteria, including the periodontal pathogen A. actinomycetemcomitans (17). To determine if IS903φkan could be used for the isolation of insertion mutants of this bacterial species, plasmids pVJT128 (IS903φkan tnp+), pVJT131 (IS903φkan Δtnp), and pJAK17 (vector) were introduced into A. actinomycetemcomitans Y4Nal by conjugation. We used E. coli donor strains containing an oriT mutant of RK2, which is able to mobilize IncQ plasmids but is defective in self-transfer (42). Transconjugants were selected on medium containing chloramphenicol and nalidixic acid. Individual transconjugant colonies for each plasmid were purified and tested for sensitivity to kanamycin to confirm that RK2 was not present and that the kan gene of IS903φkan was not expressed. Southern blot analysis of DNA from the transconjugants with a kan-specific probe confirmed that transposition of IS903φkan had not occurred (Fig. 2).
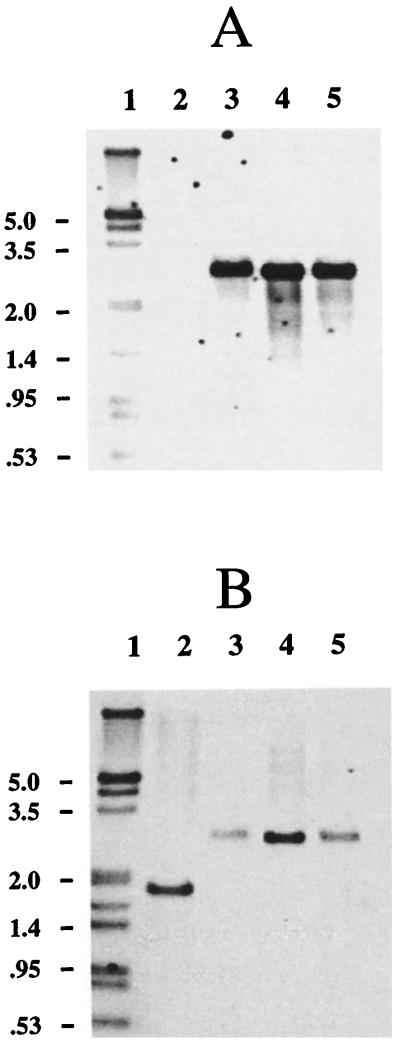
FIG. 2.
Southern blot of IS903φkan insertions. Shown is the Southern blot of EcoRI-digested genomic DNA. Digoxigenin-labeled DNA containing the kan gene was used as a probe. The sizes of DNA markers are shown on the left in kilobases. The control lanes contain DNA from Y4Nal (lane 1), Y4Nal(pJAK17) (lane 2), Y4Nal(pVJT131) (lane 4), and Y4Nal(pVJT128) (lane 6). The bands observed in lanes 4 and 6 are due to hybridization with the pVJT parent plasmids. Kanamycin-resistant mutants were derived from Y4Nal(pJAK17) (lane 3), Y4Nal(pVJT131) (lane 5), and Y4Nal(pVJT128) (lanes 7 to 22). All mutants were cured of the parent plasmid.
To test for transposition of IS903φkan in A. actinomycetemcomitans, the strains were plated for individual colonies on AAGM containing chloramphenicol and 0, 1, or 10 mM IPTG. After 48 h, individual colonies were picked, resuspended in broth, and then titrated on nonselective medium for total cells and on selective medium for Kmr cells. Colonies from the pVJT128-containing strain showed a significant increase in the fraction of Kmr cells with increasing concentration of IPTG (Table 2). In contrast, the low frequency (<10−6) of Kmr cells arising from the plasmidless and pJAK17-containing control strains reflected the spontaneous rate of chromosomal mutation to resistance. For the control strain carrying pVJT131 (Δtnp), Kmr cells occurred at a frequency approximately equivalent to that of the uninduced pVJT128 cells. This frequency is higher than the background due to spontaneous chromosomal mutation, owing to plasmid DNA rearrangements that activate the cryptic kan gene on the plasmid copy of IS903φkan. This conclusion is based on the following evidence: (i) nearly all of the Kmr pVJT131-containing cells became Kms upon loss of the plasmid and (ii) plasmids from these isolates were able to confer Kmr in E. coli (data not shown). In contrast, curing of plasmids from the Kmr mutants isolated after IPTG induction of pVJT128-containing cells rarely resulted in the loss of Kmr, as expected if IS903φkan transposed to the chromosome in these strains (see below).
TABLE 2.
Transposition of IS903φkan in A. actinomycetemcomitans
| Plasmid | Description | No. of Kmr CFU/total CFU (106) in presence of IPTG at:
|
||
|---|---|---|---|---|
| 0 mM | 1 mM | 10 mM | ||
| None | <1 | 3.0 ± 2.1a | <1 | |
| pJAK17 | Vector | <1 | <1 | <1 |
| pVJT128 | IS903φkan tnpA+ | 4.4 ± 2.0 | 74 ± 49 | 351 ± 146 |
| pVJT131 | IS903φkan tnpA | 9.6 ± 1.3 | 11.6 ± 1.9 | 1.4 ± 0.1 |
Represents one colony.
To confirm that transposition of IS903φkan had occurred in A. actinomycetemcomitans, Southern blot hybridization analysis was done with genomic DNA of various Kmr mutants after they were cured of pVJT128, pVJT131, or pJAK17 as described in Materials and Methods (Fig. 2). No hybridization was detectable with DNA of Kmr mutants derived from pVJT131-containing and pJAK17-containing control strains (Fig. 2 and data not shown). This confirms that these mutants arose by spontaneous chromosomal mutation rather than by transposition of IS903φkan to the bacterial chromosome.
In contrast, genomic DNA from 15 of 16 Kmr mutants isolated from the IPTG-induced pVJT128-containing strain hybridized to the kan probe (Fig. 2). Each of these isolates showed a single hybridizing band, indicating (i) that multiple transposition events within a single cell are infrequent under the conditions used here and (ii) that none of the mutants contains an insertion of the complete pVJT128 plasmid, as expected from their Cms phenotype. The variable sizes of the hybridizing fragments are consistent with the target specificity expected for IS903 (22). One of the 16 Kmr mutants arising from the pVJT128-containing strain did not hybridize to the kan gene probe (Fig. 2, lane 15). This mutant is likely to have occurred by chromosomal mutation.
Isolation of catalase-deficient mutants of A. actinomycetemcomitans.
A distinguishing feature of A. actinomycetemcomitans is the expression of catalase (43). To identify the catalase gene, we used IS903φkan to obtain insertion mutants defective in catalase activity. Strain Y4Nal(pVJT128) was grown on medium containing chloramphenicol and 10 mM IPTG to induce the transposition of IS903φkan. Seventy Cmr colonies were pooled and plated on kanamycin-containing medium to select a library of transposon insertion mutants. A total of 4 × 103 Kmr colonies were screened individually by a rapid toothpick assay for the ability to hydrolyze H2O2, as described in Materials and Methods. We identified and confirmed three mutants (Aa1393, Aa1394, and Aa1395) with no apparent catalase activity. The frequency of catalase-defective mutants (3/4,000) is consistent with an estimated frequency (1/1,000) based on the assumptions that (i) the size of the genome is 2 × 106 bp, of which half codes for essential genes; (ii) the size of the catalase gene is 1,000 bp; and (iii) IS903φkan inserts randomly. All three mutants grew more slowly than the wild type in broth containing 0.1 mM H2O2, and the MICs of H2O2 are 1 mM for wild-type Y4Nal and 0.3 mM for each of the mutants (data not shown).
Southern blot hybridization analysis with the kan probe showed that all three mutants displayed a hybridizing HindIII fragment of about 2.8 kb whereas the wild-type genomic DNA did not hybridize (Fig. 3A). By searching the raw data from the A. actinomycetemcomitans Genome Sequencing Project database (35) for similarities to catalase sequences from other bacteria, we identified two contigs with sequences predicted to encode catalase-related proteins. Using one primer specific for each contig, we showed by PCR that the two contigs contained nonoverlapping segments of the same putative catalase-encoding gene. The PCR product was then used as a hybridization probe for this gene (kat probe). For all three catalase-deficient mutants, the kat probe hybridized to a HindIII fragment of about 2.8 kb (Fig. 3B), the same size as the kan-hybridizing fragment (Fig. 3A). In wild-type Y4Nal, the fragment that hybridized to the catalase probe was smaller by approximately 1 kb (Fig. 3B), which is the size of IS903φkan. The insertion sites were identified precisely by sequencing the products of inverse PCR with primers pointing outward from the ends of IS903φkan (see Materials and Methods).
FIG. 3.
Southern blots of catalase-defective mutants. Shown are the Southern blots of HindIII-digested genomic DNA from the wild type, Y4Nal (lane 2), and catalase-negative mutants, Aa1393 (lane 3), Aa1394 (lane 4), and Aa1395 (lane 5). Lane 1 contains digoxigenin-labeled DNA molecular weight markers (shown in kilobases) (Boehringer Mannheim). Probes were digoxigenin-labeled DNA containing the kan gene (A) or the katA gene (B).
The A. actinomycetemcomitans Genome Sequencing Project used a different strain from that described here. We have therefore confirmed the sequence of a 2,577-bp region containing the IS903φkan insetion mutations and the putative catalase gene of strain Y4Nal used in our studies (Fig. 4). Apart from a few individual nucleotide differences, our sequence is identical to that of the Genome Sequencing Project (Fig. 4). The region contains only one open reading frame able to encode a polypeptide larger than 60 amino acids. This large open reading frame was interrupted by IS903φkan in all three catalase-deficient mutants. The predicted polypeptide product of the open reading frame has 484 amino acids and shows more than 50% identity to several bacterial catalases (Fig. 5). Upstream of the open reading frame are a reasonable Shine-Dalgarno sequence for ribosome binding and a predicted strong ς70 promoter. We conclude that this open reading frame is the structural gene for catalase in A. actinomycetemcomitans, and we have designated it katA.
FIG. 5.
Alignment of related bacterial catalases. The predicted amino acid sequence of catalase from A. actinomycetemcomitans is aligned with known bacterial catalases that have the highest identity to it: A. actinomycetemcomitans (Aa), Bordetella pertussis (Bp) (accession no. 1345688), Pseudomonas aeruginosa (Pa) (accession no. 2896139), Vibrio fischeri (Vf) (accession no. 3064126), Proteus mirabilis (Pm) (accession no. 1345690 [6]), H. influenzae (Hi) (accession no. 1168784 [4]), and Neisseria gonorrhoeae (Ng) (Accession no. 1016016 [23]).
Multiple copies of a potential DNA uptake sequence in the katA region.
Several significant inverted- and direct-repeat sequences of potential regulatory significance were found in the upstream and downstream noncoding regions (Fig. 4). Two sets of related inverted repeats are particularly interesting. The upstream region contains two nearly perfect 24-bp repeats in inverted orientation that are separated by 237 bp (IRa and IRb in Fig. 4). Downstream of katA is an inverted repeat (IRv and IRw) with 13-bp arms separated by a 3-bp spacer. The inverted repeat arms IRa, IRv, and IRw share an 11-bp sequence (Fig. 6), and IRb differs from this sequence by only a single base. This 11-bp sequence contains the 9-bp core DNA uptake signal sequence (USS) for natural transformation in Haemophilus influenzae, which occurs 1,465 times in the H. influenzae genome (44).
FIG. 6.
Alignment of putative DNA uptake sequences. The four arms of inverted repeats that contain the 11-bp consensus sequence (see Fig. 4) were aligned. Shown below are the 11-bp sequence that is repeated 848 times in the incomplete A. actinomycetemcomitans genome sequence and the 9-bp DNA uptake sequence (USS) that is repeated 1,465 times in the H. influenzae genome.
DISCUSSION
We constructed a transposon, IS903φkan, that allows direct selection of insertions without the need for either a suicide vector or an efficient DNA transfer system. Because the kan gene on IS903φkan is cryptic, the transposon can be maintained in a bacterial population without expressing Kmr. Following induction of IS903 transposase expression by IPTG, Kmr insertion mutants are readily detected within the population whenever IS903φkan transposition results in the fusion of kan to an expressed open reading frame in the target genome. To demonstrate the utility of IS903φkan, we generated a library of random insertion mutants of the periodontal pathogen A. actinomycetemcomitans and screened for mutants defective in catalase activity. All three mutants isolated contained an IS903φkan insertion in an open reading frame whose predicted product displays greater than 50% identity to several known catalases from other bacterial species. We conclude that this open reading frame is the structural gene for catalase (katA) in A. actinomycetemcomitans.
The genetic analysis of the human pathogen A. actinomycetemcomitans has been hindered by the absence of a suitable transposon mutagenesis system. Tn5 (29) and Tn916 (40) transpose in A. actinomycetemcomitans, but their utility has been limited by the efficiency of DNA transfer in this bacterial species. The IncQ plasmid-based system used here to carry IS903φkan can replicate in a broad range of gram-negative bacteria and can be mobilized to an even wider range of bacterial species by the conjugative transfer systems of promiscuous IncP plasmids (19). Furthermore, the simple genetic requirements of IS903 transposition (9) (Fig. 1), combined with its ability to transpose randomly in a variety of bacteria (L. pneumophila, E. coli, and A. actinomycetemcomitans), suggest that IS903φkan may prove a potentially useful tool for mutagenesis of diverse bacterial species. If delivery of the IS903φkan donor plasmid to the target host is efficient by either conjugation or transformation, Kmr insertion mutants can be selected immediately, as was done in the E. coli transposition assay described in above. However, the greatest advantage of the IS903φkan system will be conferred on hosts that do not transform well and conjugate poorly with the known plasmid transfer systems. For these hosts, it will be necessary only to establish the IS903φkan-containing plasmid in a single transformant or transconjugant. After growth of the clone, induction of transposase should yield large numbers of IS903φkan transposon insertion mutants. We have been particularly interested in “rough,” adherent, clinical isolates of A. actinomycetemcomitans (12, 37), which are exceedingly poor recipients in conjugation and are even more difficult to transform than “smooth,” nonadherent, laboratory variants such as the Y4Nal strain used here. As a consequence, rough A. actinomycetemcomitans have been refractory to genetic analysis. Recently, we successfully used the IS903φkan system to isolate mutants of rough A. actinomycetemcomitans without difficulty (24).
A limitation of the IS903φkan mutagenesis system is that only expressed, nonessential genes can be targeted. In addition, the expression of some kan fusions may not be sufficient to confer kanamycin resistance. Transcription or translation of the gene fusion may be too low, or the fusion protein may be unstable. We also expected that only one in six insertions should be in the proper reading frame to make a functional kan fusion. In practice, the frequency appears to be higher. The fusions in Aa1393, Aa1394, and Aa1395 are out of frame with katA. No secondary frameshifts were found in katA or the 5′ end of kan in these mutants suggesting that kan translation is initiated at an alternative start site. In another study, six of nine Kmr IS903φkan insertion mutants of A. actinomycetemcomitans were out of frame (24). Thus, the selection for Kmr may select for compensatory mutations to allow kan to be expressed from initially out-of-frame insertions and/or the kan fusions may be expressed from alternative translational start sites.
A. actinomycetemcomitans is a facultative anaerobe that resides in the subgingival plaque (43), where it is exposed to oxygen and by-products of oxygen metabolism generated by host cells such as phagocytic leukocytes (33, 36, 43). Catalase is likely to be part of an important defense mechanism for A. actinomycetemcomitans against oxidative killing by phagocytic cells. H2O2 is the major bactericidal agent that affects A. actinomycetemcomitans in periodontal pockets (11, 34). The toxicity of H2O2 is indirect and results from the intracellular generation of hydroxy radicals, which can cause DNA strand scission (15). Therefore, protective enzymes such as catalases and peroxidases should be important for the resistance of A. actinomycetemcomitans to H2O2. Indeed, the catalase-defective mutants isolated in this study are more sensitive to hydrogen peroxide than the wild type. However, they remain able to grow in the presence of 0.3 mM hydrogen peroxide, suggesting that there are other mechanisms of resistance, consistent with previous observations (11). Perhaps this residual resistance to H2O2 is encoded by catalase genes that are normally not expressed. Their activity would therefore not be detected in the initial screening assay, but their expression would be induced by the presence of H2O2 in the growth media.
The A. actinomycetemcomitans katA gene is predicted to code for a protein of 484 amino acids with a molecular weight of 54,961. The predicted polypeptide product is closely related to several other heme-binding bacterial catalases in molecular weight, pI, and amino acid sequence (Fig. 5 and data not shown). The katA structural gene is 52% G+C, which is close to the value of 48% G+C calculated from a sample of A. actinomycetemcomitans genes involved in basic cellular functions (26). The upstream and downstream intergenic regions have a significantly lower G+C content (34 and 36%, respectively). Overall, the katA region is 44% G+C, which is consistent with the reported overall value of 43% G+C for the genome (27). katA is likely to be expressed from a monocistronic mRNA. No other open reading frames of any significance occur immediately upstream or downstream of katA (Fig. 4). On the basis of sequences available in the Genome Sequencing Project database, the closest putative upstream gene is predicted to encode a glutamate dehydrogenase. The closest downstream gene, which is predicted to encode an ATP-binding cassette transporter, is oriented opposite to katA and terminates 1,021 bp from the katA stop codon (results not shown). A predicted strong ς70-type promoter in the upstream region of katA is consistent with expression of catalase during logarithmic growth of A. actinomycetemcomitans. Catalase expression in some bacteria is inducible (4, 5, 31, 45) or growth-phase regulated (21, 32, 38). We have not yet investigated the possible regulation of catalase expression in A. actinomycetemcomitans, although the upstream region contains inverted and direct repeats that could act as targets for regulatory factors.
The remnant of a possible IS element is found 460 bp after the termination codon of katA (Fig. 4). The region contains a small open reading frame whose predicted product is related to a C-terminal portion of the IS150 transposase of E. coli. A complete copy of this putative transposase gene is present on another contig in the A. actinomycetemcomitans genome database. The predicted complete transposase has 38% identity to the IS150 transposase of E. coli (41) and 47% identity to a putative transposase from H. influenzae (14). The complete transposase gene is contained within a putative IS element of 1,262 bp with 22-bp inverted repeats at the termini. A copy of the terminal sequence with 21 matching nucleotides (IRx) is present near the transposase gene remnant in the region downstream of katA (Fig. 4). For the next 100 nucleotides further downstream of katA, including a portion of the putative transposase gene, this remnant is 90% identical to the analogous portion of the predicted IS element.
The intergenic upstream and downstream regions of katA also revealed three copies of an 11-bp sequence (5′-AAAGTGCGGTC-3′) and a fourth copy with one mismatch (Fig. 6). We note that this 11-bp sequence contains the 9-bp core USS of H. influenzae used to facilitate specific uptake of DNA in natural transformation (Fig. 6) (44). There are 1,465 copies of the USS in the H. influenzae genome. The H. influenzae 9-bp core USS is part of a larger 29-bp consensus sequence, which includes an AT-rich region adjacent to one side of the core. Nearly all closely spaced USS in H. influenzae are in inverted orientation relative to each other, and they occur predominantly in intergenic regions. Downstream inverted repeats are thought to function additionally in termination of transcription or stability of mRNA. Like H. influenzae, A. actinomycetemcomitans is capable of natural transformation (40, 46). The 11-bp repeats found in the A. actinomycetemcomitans katA region share the properties of the H. influenzae USS, including an adjacent AT-rich region (Fig. 4). These findings strongly suggest that the 11-bp sequence may function as a DNA uptake signal for transformation in A. actinomycetemcomitans. The A. actinomycetemcomitans Genome Sequencing Project database contains 848 exact occurrences of the 11-bp sequence in 1.9 × 106 bases of DNA sequence. This 11-bp sequence is expected to occur randomly only once in every 4.1 × 106 bp. We predict that the 11-bp sequence constitutes the core USS for DNA uptake in A. actinomycetemcomitans. The close relationship of this sequence to the DNA uptake sequence of H. influenzae and the close relatedness of A. actinomycetemcomitans to the genus Haemophilus raise the possibility that these organisms readily exchange DNA by natural transformation.
ACKNOWLEDGMENTS
We thank Angelina Kouroubali for her help in initiating the use of IS903φkan in A. actinomycetemcomitans and Wen-Yuan Hu for his help with computer graphics. We are grateful to Tom Rosche and Scott Kachlany for helpful discussions and their comments on the manuscript. We also thank Bruce Roe, Fares Z. Najar, Sandy Clifton, Tom Ducey, Lisa Lewis, and Dave Dyer for the use of unpublished nucleotide sequence data from the A. actinomycetemcomitans Genome Sequencing Project at the University of Oklahoma. We acknowledge the use of the Wadsworth Center molecular genetics core facilities for oligonucleotide synthesis and DNA sequencing.
This work was supported in part by NIH grants R03 DE10562 to D.H.F. and GM50699 to K.M.D. and NIH NRSA fellowship 1 F32 GM17762-01A1 to V.J.T.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 2.Berg C M, Berg D E, Groisman E A. Transposable elements and the genetic engineering of bacteria. In: Berg C M, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 879–925. [Google Scholar]
- 3.Berg C M, Berg D E. Transposable element tools for microbial genetics. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2588–2612. [Google Scholar]
- 4.Bishai W R, Smith H O, Barcak G J. A peroxide/ascorbate-inducible catalase from Haemophilus influenzae is homologous to the Escherichia coli katE gene product. J Bacteriol. 1994;176:2914–2921. doi: 10.1128/jb.176.10.2914-2921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown S M, Howell M L, Vasil M L, Anderson A J, Hassett D J. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buzy A, Bracchi V, Sterjiades R, Chroboczek J, Thibault P, Gagnon J, Jouve H, Hudry-Clergeon G. Complete amino acid sequence of Proteus mirabilis PR catalase. Occurrence of a methionine sulfone in the close proximity of the active site. J Protein Chem. 1995;14:59–72. doi: 10.1007/BF01888363. [DOI] [PubMed] [Google Scholar]
- 7.Derbyshire K M. An IS903-based vector for transposon mutagenesis and the isolation of gene fusions. Gene. 1995;165:143–144. doi: 10.1016/0378-1119(95)00512-5. [DOI] [PubMed] [Google Scholar]
- 8.Derbyshire K M, Grindley N D. cis preference of the IS903 transposase is mediated by a combination of transposase instability and inefficient translation. Mol Microbiol. 1996;21:1261–1272. doi: 10.1111/j.1365-2958.1996.tb02587.x. [DOI] [PubMed] [Google Scholar]
- 9.Derbyshire K M, Hwang L, Grindley N D. Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. Proc Natl Acad Sci USA. 1987;84:8049–8053. doi: 10.1073/pnas.84.22.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derbyshire K M, Kramer M, Grindley N D. Role of instability in the cis action of the insertion sequence IS903 transposase. Proc Natl Acad Sci USA. 1990;85:4048–4052. doi: 10.1073/pnas.87.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dongari A I, Miyasaki K T. Sensitivity of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus to oxidative killing. Oral Microbiol Immunol. 1991;6:363–372. doi: 10.1111/j.1399-302x.1991.tb00508.x. [DOI] [PubMed] [Google Scholar]
- 12.Fine D, Furgang D, Schreiner H, Goncharoff P, Charlesworth G, Ghazwan G, Fitzgerald-Bocarsly P, Figurski D H. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology. 1999;145:1335–1347. doi: 10.1099/13500872-145-6-1335. [DOI] [PubMed] [Google Scholar]
- 13.Fives-Taylor P, Meyer D, Mintz K, Brissette C. Virulence factors of Actinobacillus actinomycetemcomitans. Periodontol 2000. 1999;20:136–167. doi: 10.1111/j.1600-0757.1999.tb00161.x. [DOI] [PubMed] [Google Scholar]
- 14.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Lu L, Glodek A, Kelley J M, Weideman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 15.Friedberg E, Walker G, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 16.Fürste J, Pansegrau W, Frank R, Blöcker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 17.Goncharoff P, Yip J K K, Wang H, Schreiner H C, Pai J A, Furgang D, Stevens R H, Figurski D H, Fine D H. Conjugal transfer of broad-host-range incompatibility group P and group Q plasmids from Escherichia coli to Actinobacillus actinomycetemcomitans. Infect Immun. 1993;61:3544–3547. doi: 10.1128/iai.61.8.3544-3547.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grindley N, Joyce C M. Analysis of the structure and function of the kanamycin-resistance transposon Tn903. Cold Spring Harbor Symp Quant Biol. 1981;45:125–133. doi: 10.1101/sqb.1981.045.01.021. [DOI] [PubMed] [Google Scholar]
- 19.Guiney D. Broad host-range conjugation and mobilizable plasmids in gram-negative bacteria. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Publishing Corp.; 1993. pp. 75–103. [Google Scholar]
- 20.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 21.Hassan H M, Fridovich I. Regulation of the synthesis of catalase and peroxidase in Escherichia coli. J Biol Chem. 1978;253:6445–6420. [PubMed] [Google Scholar]
- 22.Hu W-Y, Derbyshire K M. Target choice and orientation preference of the insertion sequence IS903. J Bacteriol. 1998;180:3039–3048. doi: 10.1128/jb.180.12.3039-3048.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson S R, Steiner B M, Perkins G H. Cloning and characterization of the catalase gene of Neisseria gonorrhoeae: use of the gonococcus as a host organism for recombinant DNA. Infect Immun. 1999;64:2627–2634. doi: 10.1128/iai.64.7.2627-2634.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kachlany, S. C., P. J. Planet, D. Fine, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 1999. Unpublished data.
- 25.Kahn M, Kolter R, Thomas C, Figurski D H, Meyer R, Remaut E, Helinski D R. Plasmid cloning vehicles derived from plasmids ColE1, F, R6K, and RK2. Methods Enzymol. 1979;68:268–280. doi: 10.1016/0076-6879(79)68019-9. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan J, Fine D. Codon usage in Actinobacillus actinomycetemcomitans. FEMS Microbiol Lett. 1998;163:31–36. doi: 10.1111/j.1574-6968.1998.tb13022.x. [DOI] [PubMed] [Google Scholar]
- 27.Kilian M. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J Gen Microbiol. 1976;93:9–62. doi: 10.1099/00221287-93-1-9. [DOI] [PubMed] [Google Scholar]
- 28.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 29.Kolodrubetz D, Kraig E. Transposon Tn5 mutagenesis of Actinobacillus actinomycetemcomitans via conjugation. Oral Microbiol Immunol. 1994;9:290–296. doi: 10.1111/j.1399-302x.1994.tb00073.x. [DOI] [PubMed] [Google Scholar]
- 30.Kornacki, J. 1999. Unpublished data.
- 31.Loewen P C, Switala J, Triggs-Raine B L. Catalases HPI and HPII in Escherichia coli are induced independently. Arch Biochem Biophys. 1985;243:144–149. doi: 10.1016/0003-9861(85)90782-9. [DOI] [PubMed] [Google Scholar]
- 32.Loewen P C, Triggs B L. Genetic mapping of katF, a locus that with katE affects the synthesis of a second catalase species in Escherichia coli. J Bacteriol. 1984;160:668–675. doi: 10.1128/jb.160.2.668-675.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marquis R E. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J Ind Microbiol. 1995;15:198–207. doi: 10.1007/BF01569826. [DOI] [PubMed] [Google Scholar]
- 34.Miyasaki K T, Wilson M E, Reynolds H S, Genco R J. Resistance of Actinobacillus actinomycetemcomitans and differential susceptibility of oral Haemophilus species to the bactericidal effects of hydrogen peroxide. Infect Immun. 1984;46:644–648. doi: 10.1128/iai.46.3.644-648.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roe, B., F. Najar, S. Clifton, T. Ducey, L. Lewis, and D. Dyer. October 1999, revision date. Actinobacillus Genome Sequencing Project. [Online.] http://www.genome.ou.edu/act.html. [25 October 1999, last date accessed.]
- 36.Roos M, Roos D. Differences in oxygen metabolism of phagocytosing monocytes and neutrophils. J Clin Investig. 1978;61:480–488. doi: 10.1172/JCI108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosan B, Slots J, Lamont R J, Listgarten M A, Nelson G M. Actinobacillus actinomycetemcomitans fimbriae. Oral Microbiol Immunol. 1988;3:58–63. doi: 10.1111/j.1399-302x.1988.tb00082.x. [DOI] [PubMed] [Google Scholar]
- 38.Sak B D, Eisenstark A, Touati D. Exonuclease III and the catalase hydroperoxidase II in Escherichia coli are both regulated by the katF gene product. Proc Natl Acad Sci USA. 1989;86:3271–3275. doi: 10.1073/pnas.86.9.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sato S, Takamatsu N, Okahashi N, Matsunoshita N, Inoue I, Takehara T, Koga T. Construction of mutants of Actinobacillus actinomycetemcomitans defective in serotype b-specific polysaccharide antigen by insertion of transposon Tn916. J Gen Microbiol. 1992;138:1203–1209. doi: 10.1099/00221287-138-6-1203. . (Erratum, 138:1992.) [DOI] [PubMed] [Google Scholar]
- 41.Schwartz E, Kroger M, Rak B. IS150: distribution, nucleotide sequence and phylogenetic relationships of a new E. coli insertion element. Nucleic Acids Res. 1988;16:6789–6802. doi: 10.1093/nar/16.14.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sia E, Kuehner D M, Figurski D H. The mechanism of retrotransfer in conjugation: prior transfer of the conjugative plasmid is required. J Bacteriol. 1996;178:1457–1464. doi: 10.1128/jb.178.5.1457-1464.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slots J. The predominant cultivable organism in juvenile periodontitis. Scand J Dent. 1976;84:1–10. doi: 10.1111/j.1600-0722.1976.tb00454.x. [DOI] [PubMed] [Google Scholar]
- 44.Smith H O, Tomb J F, Dougherty B A, Fleischmann R D, Venter J C. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269:538–540. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 45.Storz G, Tartaglia L A, Ames B N. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science. 1990;248:189–194. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- 46.Tonjum T, Bukholm G, Bovre K. Identification of Haemophilus aphrophilus and Actinobacillus actinomycetemcomitans by DNA-DNA hybridization and genetic transformation. J Clin Microbiol. 1990;28:1994–1998. doi: 10.1128/jcm.28.9.1994-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Triglia T, Peterson M, Kemp D. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Winkelhoff A, Slots J. Actinobacillus actinomycetemcomitans and Prophorymonas gingivalis in non-oral infections. Periodontol 2000. 1999;20:122–135. doi: 10.1111/j.1600-0757.1999.tb00160.x. [DOI] [PubMed] [Google Scholar]
- 49.Wiater L, Sadosky A B, Shuman H A. Mutagenesis of Legionella pneumophila using Tn903dII/lacZ: identification of a growth-phase regulated pigmentation gene. Mol Microbiol. 1994;11:641–653. doi: 10.1111/j.1365-2958.1994.tb00343.x. [DOI] [PubMed] [Google Scholar]