Abstract
We have recovered a DNase-protected, chloroform-resistant molecule of DNA from the cell-free supernatant of a Borrelia burgdorferi culture. The DNA is a 32-kb double-stranded linear molecule that is derived from the 32-kb circular plasmids (cp32s) of the B. burgdorferi genome. Electron microscopy of samples from which the 32-kb DNA molecule was purified revealed bacteriophage particles. The bacteriophage has a polyhedral head with a diameter of 55 nm and appears to have a simple 100-nm-long tail. The phage is produced constitutively at low levels from growing cultures of some B. burgdorferi strains and is inducible to higher levels with 10 μg of 1-methyl-3-nitroso-nitroguanidine (MNNG) ml−1. In addition, the prophage can be induced with MNNG from some Borrelia isolates that do not naturally produce phage. We have isolated and partially characterized the phage associated with B. burgdorferi CA-11.2A. To our knowledge, this is the first molecular characterization of a bacteriophage of B. burgdorferi.
Phage-like particles associated with Borrelia burgdorferi were first observed shortly after the bacterium was identified as the causative agent of Lyme disease (10, 19). Identical-looking phages from a culture of Borrelia hermsii, a relapsing fever agent, were later described (5). Two structurally different bacteriophages have been detected in cultures of clinical isolates of B. burgdorferi that were treated with subinhibitory concentrations of the DNA gyrase inhibitor ciprofloxacin (30, 38). Despite the number of bacteriophages observed by electron microscopy in association with this bacterium, to our knowledge no bacteriophage of B. burgdorferi has been isolated and characterized for nucleic acid content or other properties.
B. burgdorferi has a genome consisting of a linear chromosome and both linear and circular plasmids (4, 17). Repeated elements are found throughout the genome (13, 27, 31, 40, 44, 50), and the 32-kb circular plasmid, cp32, has several distinct but homologous forms that coexist in a single bacterium (13, 44). Homologs of cp32 are also found on a large linear plasmid, lp56 (49, 50), the small circular plasmid of B. burgdorferi sensu lato, cp8.3 (15, 50), and a truncated circular plasmid of B. burgdorferi N40, cp18 (43). One or more members of the cp32 family have been found in all isolates of B. burgdorferi and nearly all isolates of closely related Borrelia species (13, 44). Casjens and colleagues have suggested that the ubiquitous nature of the 32-kb circular plasmids and related sequences may be due to a temperate bacteriophage that packages cp32 and exists as an autonomous replicon (12, 13).
Phage particles have been observed in association with many different spirochetes (5, 7, 11, 19–21, 29, 30, 32–34). Only three apparently lytic phages of Leptospira biflexa (34) and a small, inducible transducing phage of Serpulina hyodysenteriae (20, 21) have been isolated and characterized to any degree. We now report the isolation and initial molecular characterization of a bacteriophage of the spirochete B. burgdorferi.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. burgdorferi sensu stricto strain CA-11.2A (28), a clone of CA-11, was kindly provided by P. Rosa (Rocky Mountain Laboratories, Hamilton, Mont.). All other isolates used in this study were kindly provided by R. Marconi (Medical College of Virginia at Virginia Commonwealth University, Richmond) except for B. burgdorferi sensu stricto strains B31, HB19 (41), and CA-11 (39) (the latter were kindly provided by T. Schwan, Rocky Mountain Laboratories). Bacterial isolates were routinely cultivated in Barbour-Stoenner-Kelly (BSK) complete medium (Sigma) at 34°C with a 5% CO2 atmosphere. Culture density was determined by spectrophotometry as described previously (36), except that 1 ml of culture was used and the A600 was multiplied by 1.4 × 109 to calculate the number of cells ml−1.
Bacteriophage recovery.
CA-11.2A cells were cultured in volumes of 10 to 250 ml, as described above, until they reached log phase (>107 cells ml−1; A600 ≥ 0.05), approximately 3 to 5 days after an inoculation with a 1:100 dilution. All subsequent steps in the recovery of phage were performed at 4°C. The cultures were centrifuged at 6,000 × g for 10 min, and the supernatant was collected. A polyethylene glycol (PEG) precipitation of the culture supernatant was done in aliquots of up to 250 ml by a modification of a previously described protocol for phage concentration (35). NaCl was added to a 1 M final concentration and the culture supernatant was rotated for 1 h, followed by centrifugation at 5,000 × g for 10 min. The supernatant was retained, and 10% (wt/vol) PEG 8000 (Sigma) was added. The culture was rotated for 1 h, and the precipitate was recovered by centrifugation at 6,000 × g for 10 min. The supernatant was decanted, and the precipitate was resuspended in suspension medium (100 mM NaCl, 10 mM MgSO4, 50 mM Tris-HCl [pH 7.5]; no gelatin). The resuspension volume was 400 μl of suspension medium per 10 ml of original culture supernatant. The resuspended material was extracted once with an equal volume of chloroform, and the aqueous layer was recovered. The sample was extracted a second time with a 10% volume of chloroform, and the aqueous layer, which contained the phage, was recovered a second time. Samples were stored at 4°C.
DNA extraction.
Total chromosomal DNA was extracted from B. burgdorferi cells based on a protocol described previously (36). To purify the small linear plasmid of B. burgdorferi, plasmid DNA was extracted from B. burgdorferi B31 cells with the Wizard Plus Midipreps DNA purification system (Promega) as instructed by the manufacturer. Plasmids were resolved by electrophoresis (see below) and sized with the λ monocuts marker (New England Biolabs). The linear 17-kb plasmid was excised from the gel and extracted with the QIAEX II gel extraction kit (QIAGEN) as instructed by the manufacturer.
Prior to the extraction of phage DNA, samples were treated with RQ1-DNase (Promega) as instructed by the manufacturer. After DNase treatment, 100 mM EDTA was added and the sample was treated with a final concentration of 0.3% sodium dodecyl sulfate (SDS) and 100 μg of proteinase K ml−1 at 65°C for 10 min (45). The sample was extracted twice, once with an equal volume of phenol-chloroform and a second time with an equal volume of chloroform. The aqueous layer was recovered, and the DNA was precipitated with NaCl and absolute ethanol as described previously for cellular DNA (36). The DNA pellet was resuspended in 20 μl of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) per 100 μl of original PEG precipitate.
To denature the phage DNA, 10 μl of sample was treated with an equal volume of 0.2 N NaOH and incubated at 25°C for 10 min. Four microliters of 1 M Tris-HCl (pH 8.0) was added, and the sample was incubated at 25°C for 5 min.
Agarose gel electrophoresis.
DNA samples were heated for 3 to 5 min at 65°C in 1% N-laurylsarcosine, 10 mM EDTA, 3% Ficoll 400, 0.05 mg of bromophenol blue ml−1, and 0.05 mg of xylene cyanol ml−1, cooled briefly, and resolved on 0.5% agarose gels (SeaKem LE; FMC Bioproducts) in TAE (40 mM Tris-acetate, 1 mM EDTA) at 30 V (3 V cm−1) for 5 h. Gels were stained with 0.5 μg of ethidium bromide (EtBr) ml−1 for 0.5 to 1 h and destained in water for 1 to 2 h. The DNA was visualized on a UV transilluminator, and images were captured on a Gel Doc 1000 system (Bio-Rad). For field inversion gel electrophoresis, DNA samples were prepared as described above and resolved on 0.8% agarose gels (SeaKem GTG; FMC Bioproducts) in TBE (45 mM Tris-borate, 2 mM EDTA) at 80 V (5 V cm−1) for 16 h with program 2 on the PPI-200 programmable power inverter (MJ Research) per the manufacturer’s instructions. Gels were stained with EtBr and visualized as described above.
Two-dimensional gel electrophoresis was performed as described previously (36). A 20-μl sample of total B. burgdorferi DNA was fractionated on a 0.35% agarose gel in TAE at 20 V (1.25 V cm−1) for 16 h. After 16 h, the gel was rotated 90° and equilibrated with 15 μM chloroquine for 5 h. Electrophoresis was continued in the second dimension in the presence of 15 μM chloroquine at 20 V for another 16 h. The gel was soaked in three changes of water (>1 h each) to remove the chloroquine before staining with EtBr as described above.
Southern hybridization.
Gels were vacuum blotted to Hybond N+ membranes (Amersham Pharmacia) and cross-linked as described previously (25). Probes used were either total phage DNA, prepared as described above, or small B. burgdorferi cp32-specific probes designated probe 2 and probe 4 (13). Probe 4 (∼300 bp) and probe 2 (∼250 bp) were generated from total phage DNA by PCR (25 cycles of 92°C for 1 min, 50°C for 30 s, and 72°C for 1 min; diluted probe at 1:100; and repeat PCR) with primer pairs CP-4–CP-5 and erp177-erp178 (13), respectively. Additionally, a probe (408 bp) encompassing the blyB gene on cp32 was generated by PCR as above, using rev8 (5′-CCAAAGATAATGTTG-3′) and rev06 (5′-GATCTATGTTTGTATC-3′) kindly provided by Don Oliver (Wesleyan University, Middletown, Conn.) (18). One hundred nanograms of DNA to be used as a probe was labeled with [α-32P]dATP with a random primer kit (Prime-it II; Stratagene) as instructed by the manufacturer. Radiolabeled probes were purified from unincorporated label by passage through G50 spin columns as instructed by the manufacturer (Boehringer Mannheim). The blots were hybridized in 15 to 20 ml of QuikHyb (Stratagene) supplemented with 1 mg of salmon sperm DNA for 15 to 20 min at 68°C. After prehybridization, the radioactive probe was added directly to the hybridization buffer and hybridization was conducted for 1 to 2 h at 68°C. The blots were washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 25°C (15 min each) and once at 50°C in 0.1× SSC–0.1% SDS (30 min), wrapped in cellophane, and exposed to Hyperfilm ECL (Amersham Pharmacia) for 16 to 24 h at −80°C with intensifying screens.
Induction of the bacteriophage.
Various isolates were cultured in 10 ml of BSK complete medium as described above, until a density of approximately 5 × 107 cells ml−1 was reached. Cells were pelleted at 6,000 × g, and the supernatant was collected for PEG precipitation. The cell pellet was resuspended in a volume of BSK complete medium equal to that of the original culture, and the culture was split into equal aliquots. One aliquot was treated with 10 μg of 1-methyl-3-nitroso-nitroguanidine (MNNG) ml−1 (stock concentration is 50 mg ml−1 in dimethyl sulfoxide, stored at −20°C). Both the treated and untreated cultures were incubated at 34°C for 2 h. The cultures were centrifuged as described above, and the supernatant was discarded as waste. The cells were resuspended in an equal volume of BSK complete medium and allowed to recover for 60 h at 34°C. After 60 h, the supernatants were collected from both the treated and untreated cultures. Phage was precipitated, and the DNA was extracted and resolved by conventional electrophoresis as described above. The gel was stained with EtBr, followed by the more sensitive GelStar nucleic acid gel stain (FMC Bioproducts) as instructed by the manufacturer. After visualization, the gel was rinsed in water for >1 h to remove excess GelStar and then blotted and probed with a cp32-specific probe as described above.
Microscopy of phage particles.
A culture of B. burgdorferi CA-11.2A was induced, and the phage particles were precipitated as described above. A drop of precipitated phage suspension was applied to a grid (copper 300-mesh, carbon coated; Ted Pella). The sample was stained with 2% phosphotungstic acid and examined on a Hitachi 7100 transmission electron microscope (TEM).
RESULTS
Identification of bacteriophage DNA.
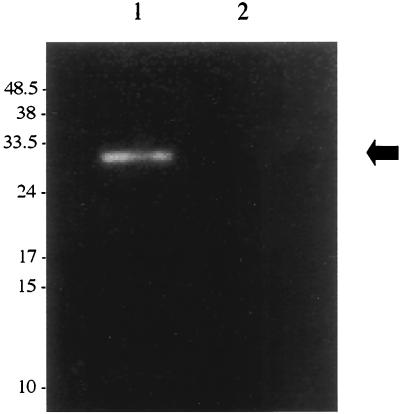
During a biochemical analysis of protein extracts of B. burgdorferi CA-11.2A, we serendipitously discovered a molecule of DNase-resistant nucleic acid in cell samples that had been sonicated to disrupt cell membranes. We found that this nucleic acid could also be recovered from the cell-free supernatants of late-log-phase B. burgdorferi CA-11.2A cultures (Fig. 1). The DNase protection persists throughout the PEG precipitation protocol, which involves two chloroform treatments (Fig. 1, lane 1). The protection is alleviated when a sample of phage is first treated with SDS and proteinase K, extracted with organic solvents, and subsequently treated with DNase (Fig. 1, lane 2). The nucleic acid is resistant to RNase treatment at every step (data not shown). The nucleic acid migrates as a 32-kb molecule with both field inversion (Fig. 1, lane 1) and conventional (Fig. 2, lane 1) gel electrophoresis, indicating the nucleic acid is a double-stranded, linear DNA molecule. When observed by electron microscopy, the phage nucleic acid also appears to be a double-stranded, linear ∼32-kb DNA molecule with no gross secondary structure (24).
FIG. 1.
DNase protection of extracellular bacteriophage DNA. After PEG precipitation, phage samples were extracted twice with chloroform. Samples were subjected to digestion with DNase I prior to DNA extraction. After DNA isolation, the samples either were loaded onto a 0.8% agarose gel (lane 1) or were subjected to another digestion with DNase I and then loaded directly onto the agarose gel (lane 2). The DNA was resolved with field inversion gel electrophoresis. The gel was stained with EtBr. Molecular sizes are in kilobase pairs.
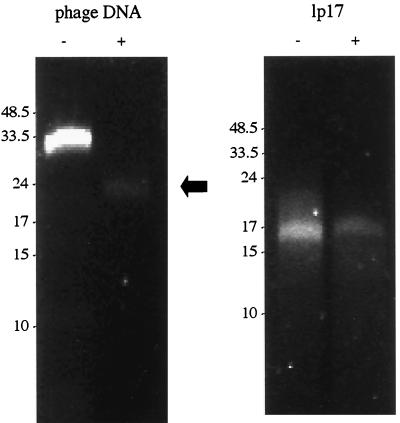
FIG. 2.
Denaturation of phage DNA. DNA denatured with 0.2 M NaOH (+) and an untreated control (−) were resolved on a 0.5% agarose gel. The denatured phage DNA did not “snap back” and regenerate a double-stranded DNA molecule, as did the denatured covalently closed linear plasmid lp17. The arrow indicates single-stranded DNA products generated by the denaturation of non-covalently closed double-stranded DNA. The gel was stained with EtBr. Molecular sizes are in kilobase pairs.
B. burgdorferi cells can be grown in solid medium, but they do not readily form a lawn (16), thus making plaque assays infeasible. The most efficient way of evaluating phage production and presence in a sample is DNA extraction and agarose gel electrophoresis. From an uninduced culture of B. burgdorferi CA-11.2A, phage DNA is usually visible by EtBr staining (approximately 200 ng) when extracted from 100 to 400 μl of phage precipitate (equivalent to 2.5 to 10 ml of original culture supernatant).
The ends of the linear DNA molecules of the B. burgdorferi genome are covalently closed hairpin loops, similar to the ends of the vaccinia virus (4). To characterize the nature of the ends of the linear phage DNA, a sample was denatured with NaOH, producing single-stranded products (Fig. 2). When DNA lacks covalently closed ends, the single-stranded molecules cannot reanneal rapidly (Fig. 2, left). As a control, the small linear plasmid of the B. burgdorferi genome, lp17, was exposed to the same conditions (Fig. 2, right). lp17 has covalently closed ends, and reannealing occurred rapidly during a brief recovery period after denaturation. The phage DNA did not rapidly reanneal, indicating both its double-stranded nature and its lack of covalently closed ends.
Identifying the prophage DNA.
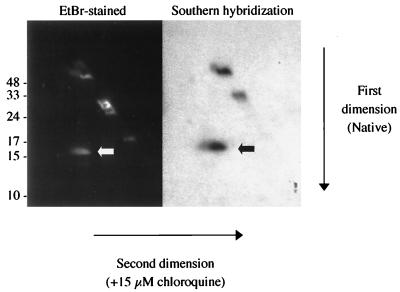
To locate the prophage in the B. burgdorferi genome, total cellular B. burgdorferi CA-11.2A DNA was resolved with two-dimensional electrophoresis. With this method, the second dimension is electrophoresed in the presence of chloroquine, a DNA intercalater. Chloroquine introduces positive writhe, relaxing the negatively supercoiled circular DNA molecules (36). The migration of these relaxed circular DNA molecules in the second dimension was retarded (Fig. 3, left). On a Southern blot of the gel, total phage DNA hybridized with cp32 (Fig. 3, right) and its linearized and nicked forms. A small probe specific to the 26-kb circular plasmid, cp26, was used to localize the circular form of this plasmid (data not shown). A comparison of the hybridization patterns of these two probes demonstrated that the phage DNA hybridized to the larger cp32 and not to the smaller cp26 (data not shown). Additional evidence for the prophage being at least one cp32 is the hybridization of three distinct cp32-specific probes, probe 4 (Fig. 4), probe 2, and the blyB probe (data not shown), to phage DNA. Also, several fragments generated by HindIII digestion of phage DNA from B. burgdorferi CA-11.2A have been partially sequenced (16). A comparison to known B. burgdorferi B31 cp32 sequences indicated that most of the fragments had ≥95% sequence identity to at least one cp32, and all clones had more than 85% sequence identity to one or more cp32 plasmids.
FIG. 3.
Genomic location of prophage DNA. Total cellular DNA from B. burgdorferi CA-11.2A was resolved by two-dimensional gel electrophoresis (left). The large circular plasmid (white arrow) was retarded in its migration in the second dimension, and the linear plasmids migrated on the diagonal. A Southern blot of the gel was probed with total phage DNA that was extracted and radiolabeled (right). The phage DNA hybridized to the circular 32-kb plasmid (black arrow). Additionally, the phage DNA hybridized to the nicked (upper band) and linearized (middle band) forms of cp32 that were generated during DNA extraction. Molecular sizes are in kilobase pairs.
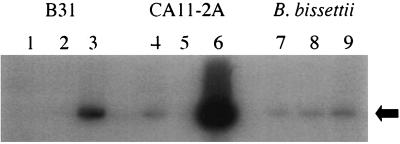
FIG. 4.
Induction of prophage from different Borrelia strains. Phage DNA was extracted from 10 ml of cell-free supernatants from B. burgdorferi B31 (lanes 1 to 3), B. burgdorferi CA-11.2A (lanes 4 to 6), and B. bissettii DN127 (lanes 7 to 9) cultures. The DNA was collected from log-phase starter cultures (lanes 1, 4, and 7), untreated controls (lanes 2, 5, and 8), and cultures treated with 10 μg of MNNG ml−1 (lanes 3, 6, and 9) and was electrophoresed on a 0.5% agarose gel. The gel was blotted and probed with cp32-specific probe 4 to enhance detection of phage DNA.
Induction of the prophage.
The reversion of a temperate prophage from a quiescent state to an active state is often achieved by stressing the host bacterial cell by chemical or physical means (8). Previously, mitomycin C has been used to induce a prophage of the spirochete S. hyodysenteriae (20, 21), and bacteriophages have been observed in Borrelia cultures treated with ciprofloxacin (30, 38). Neither of these chemicals was successful in inducing the prophage from B. burgdorferi CA-11.2A. Previously, several phages have been induced with MNNG, a potent DNA alkylating agent (8), though none from spirochetes.
The concentration of MNNG required for induction was evaluated over a range from 0.1 to 500 μg ml−1 (data not shown). Using 10 μg of MNNG ml−1, we were able to induce the prophage from B. burgdorferi B31 (Fig. 4, lanes 1 to 3) and Borrelia bissettii DN127 (Fig. 4, lanes 7 to 9), as well as B. burgdorferi strain CA-11.2A (Fig. 4, lanes 4 to 6). The induction of prophage from B. burgdorferi B31 is notable because this strain rarely produces phage spontaneously. DN127, another California isolate (39), releases low levels of phage when uninduced and can be treated with MNNG to consistently produce slightly higher levels. B. burgdorferi CA-11.2A can be induced to produce much larger quantities of phage than are naturally released (Fig. 4, lanes 4 to 6). We have assayed several Borrelia isolates for the induction of phage, including B. burgdorferi sensu stricto strains CA-11, CA-2, CA-9, HB19, and N40, as well as strains from the closely related genospecies Borrelia afzelii, Borrelia garinii, Borrelia andersonii, Borrelia japonica, and Borrelia valaisiana. We have also examined culture supernatants from the relapsing fever spirochetes B. hermsii, Borrelia turicatae, and Borrelia parkeri. We have seen no evidence of either constitutive production of phage or MNNG induction of the prophage from any of these strains.
An assay of supernatants from Borrelia anserina, the causative agent of avian spirochetosis, and Borrelia coriaceae, the causative agent of epizootic bovine abortions (5), did indicate the presence of both a naturally released phage (of B. anserina) and an induced phage (of both B. anserina and B. coriaceae). B. anserina has a genome that apparently lacks circular DNA (16, 26), suggesting that the putative bacteriophage released from this species and the bacteriophage that packages the 32-kb circular plasmid of B. burgdorferi are different. Furthermore, the DNA isolated from the supernatants of B. anserina cultures has been sized at approximately 42 kb (data not shown). Neither B. burgdorferi phage DNA nor probe 4, highly conserved among Lyme disease spirochetes (13), hybridizes to the DNA released from either B. anserina or B. coriaceae. The DNA isolated from the supernatants of these two species is detected by EtBr or GelStar staining. The presumptive bacteriophages produced from these two isolates appear to be unrelated to the B. burgdorferi phage that packages cp32.
Electron microscopy of bacteriophages.
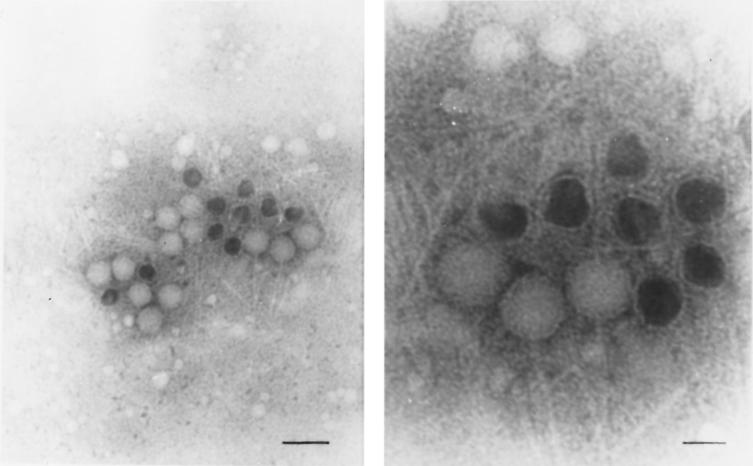
Phage ultrastructure was examined by TEM. Samples were prepared from uninduced (data not shown) or MNNG-induced (Fig. 5) cultures. The phage heads are apparently polyhedral with a diameter of 50 to 60 nm. The tails are approximately 100 nm without neck or baseplate. The tails do not appear to be contractile, although our analysis does not rule out that possibility. In the PEG-precipitated preparation, both empty (no DNA packaged) and full, electron-dense (DNA packaged) heads were observed (Fig. 5). The bacteriophage we report here has not yet been seen in association with B. burgdorferi cells. Previously, other researchers reported a cubic Borrelia phage (5, 19), and still others have described two isometric phages with contractile and noncontractile tails (30, 38).
FIG. 5.
Microscopy of B. burgdorferi phage particles. Samples were collected from PEG-precipitated cell-free supernatants of an induced culture of B. burgdorferi CA-11.2A and viewed by TEM. (Left) Tailless heads, headless tails, and intact phage particles are visible, including both full and empty heads. Phosphotungstic acid stain; magnification, ×75,000 (bar = 150 nm). (Right) Close-up of the intact phage particles. Phosphotungstic acid stain; magnification, ×250,000 (bar = 40 nm).
DISCUSSION
Although bacteriophages have been occasionally observed in cultures of B. burgdorferi (19, 30, 38) and one of the circular plasmids has features of a possible prophage (13), until this study no phage of B. burgdorferi had been isolated and characterized. We now report the isolation of a bacteriophage of B. burgdorferi that has a polyhedral head of 50 to 60 nm and a simple, noncontractile tail of 100 nm. This phage is structurally different from the cubic phages (19) and one of the ciprofloxacin-inducible phages (30, 38) described previously. The phage that we isolated may be structurally similar to the phage with B-1 morphology reported by Neubert and colleagues, but the capsid size of the phage we have described is larger than that of the phage from previous work (30 nm) (30, 38). Additionally, the phage described by these investigators was inducible with ciprofloxacin (30, 38), while we have seen no evidence of phage induction with this antibiotic. These differences lead us to conclude that we have isolated a new bacteriophage of B. burgdorferi.
Based on the highly conserved size of the chromosome of widely distributed members of the Lyme disease complex, Casjens et al. have suggested that a prophage of B. burgdorferi would likely replicate as an extrachromosomal element (12, 13). Because of the ubiquitous nature of the cp32s and the highly conserved size of these different, but related, molecules, cp32 seemed to be a likely candidate for a prophage (13, 44). The phage we describe here packages a linear 32-kb molecule that lacks covalently closed ends and hybridizes under high-stringency conditions to the cp32 family of the B. burgdorferi genome. Additionally, several cp32-specific probes hybridize under high-stringency conditions with phage DNA and partial sequences from several cloned phage DNA fragments are nearly identical to known B. burgdorferi B31 cp32 sequences.
No phage-specific proteins have been purified to date, despite intensive efforts. B. burgdorferi is grown in a protein-rich, serum-based medium, and extensive purification attempts have failed to remove protein contaminants from phage preparations. The structural gene products of tailed phages usually lack similarity at the amino acid sequence level (1), hindering a sequence comparison of the predicted cp32 open reading frames to those of known structural proteins of other tailed phages. Two proteins encoded by cp32, BlyA and BlyB, were initially identified as hemolytic proteins with a possible role in B. burgdorferi pathogenesis (18). However, more recently, BlyA and BlyB have been proposed to constitute a holin-like system (14). Bacteriophage-encoded holins, which promote cell lysis for the release of bacteriophages, have been identified in almost all known tailed phages (47).
One of the most well-characterized, extrachromosomally replicating temperate phages is Escherichia coli phage P1, a bacteriophage that replicates autonomously during the lysogenic cycle (8, 22, 42). P1 is known to package via a processive “headful” packaging mechanism, in which a circular concatemer of viral units is generated by rolling circle replication late in the lytic cycle. The large circular intermediate is then cleaved at a specific site (the pac site), and four to five phage heads are filled processively (42). This process generates a cyclically permuted linear phage genome with terminal redundancy (22, 48). Preliminary efforts to label and identify the ends of the phage genome suggest that the B. burgdorferi CA-11.2A phage is also cyclically permuted (16), but efforts to identify concatemers in B. burgdorferi CA-11.2A have not been fruitful.
Previously, both mitomycin C and ciprofloxacin have been used to induce bacteriophages from spirochetes (11, 20, 30). The induction of prophage from B. burgdorferi with MNNG is presumably through the same well-characterized mechanisms of E. coli prophage induction, which involves the damage of DNA, the subsequent activation of the RecA protein, and the reversal of the repressed state of the prophage (8). Previously, MNNG has been used to generate mutant cyanophage (2, 37), induce prophage λ from recA mutants of E. coli (46), and induce prophages through mutation of Haemophilus influenzae (3, 9).
The natural or induced release of a temperate prophage is often associated with a decrease in cell density during the “lytic burst” (8). Barbour and Hayes suggested that this phenomenon might account for the periodicity seen during early attempts at cultivating borreliae (5). We have never witnessed a dramatic decrease in cell density associated with inducing bacteriophages from B. burgdorferi CA-11.2A. Preliminary time course studies have shown that during the recovery period following MNNG treatment, a treated culture grows at the same rate as an untreated culture for 24 h before growth levels at a cell density about 2.5 times lower than that of the untreated culture. There is no dramatic decrease in cell density at 60 h, which is the time of highest phage production (data not shown). This is presumably due to the small population of cells that is releasing phage, even after induction. The decrease in culture density due to phage release may be masked by the decrease in viability and density of the MNNG-treated culture as a whole. Alternatively, the phage may be exiting the cell by means other than lysis.
Considerable work remains to be done on the similarities and differences of the bacteriophages shed from the different Borrelia isolates. Only a limited number of Borrelia species and strains produce phage constitutively or can be induced to produce phage by our methods. The degree of variability of phage production between even CA-11.2A and its parent strain, CA-11, is remarkable. We have performed multiple assays but have observed CA-11 to produce 32-kb extracellular DNA only once, even when treated with MNNG, whereas CA-11.2A, a clone selected based on its outer surface protein profile (28), releases phage continuously with and without induction.
Little is known about the way that B. burgdorferi replicates and partitions its plasmids. Different E. coli plasmids that have the same replication mechanisms and partition machinery are known to be incompatible (6). Some strains of B. burgdorferi, though, are able to maintain five or more homologous cp32 plasmids within a single cell (13). A detailed analysis of the replication and partitioning mechanisms of any of the plasmids of B. burgdorferi has yet to be done. Because of their small size and the inherent similarities in the metabolisms of phage DNA and host DNA, phages have long been considered important tools for studying cellular replication mechanisms (23). Additionally, phages have been developed into manipulatable genetic systems for many bacteria. Bacteriophages have been used successfully to transduce genetic markers between bacteria, including the spirochete S. hyodysenteriae (21). With the identification of cp32 as a prophage genome, we may now begin to dissect the prophage requirements for replication, partitioning, induction, and packaging. We believe that with further characterization, the bacteriophage described here may be useful for analyzing the molecular mechanisms of DNA metabolism in B. burgdorferi.
ACKNOWLEDGMENTS
We thank K. Tilly and S. Casjens for thoughtful and critical review of the manuscript; S. Casjens, K. Tilly, S. F. Hayes, T. Stanton, P. Rosa, D. Mount, C. Garon, L. Lubke, and C. Damman for useful discussions; G. Card for MNNG and advice on its use; B. Stevenson, D. Oliver, and C. Damman for cp32-specific probes; R. Marconi, T. Schwan, and P. Rosa for strains; J. Driver and W. Granath for assistance with microscopy; and D. Emlen and K. Barbian for assistance with figure preparation.
Work in our laboratory is supported by grants from the National Institutes of Health (AI41559 and AI39695), Arthritis Foundation, MONTS (Montana’s NSF EPSCoR program), National Science Foundation (MCB-9722408), and the UM University Grant Program. C.H.E. is a recipient of a Predoctoral Honors Fellowship from The University of Montana.
REFERENCES
- 1.Ackerman H-W. Tailed bacteriophages: the order caudovirales. Adv Virus Res. 1998;51:135–201. doi: 10.1016/S0065-3527(08)60785-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amla D V. Mutagenesis of free and intracellular cyanophage AS-1 by ultraviolet, N-methanyl-N′-nitro-N-nitrosoguanidine and acriflavin. Mutat Res. 1979;59:147–155. doi: 10.1016/0027-5107(79)90152-0. [DOI] [PubMed] [Google Scholar]
- 3.Balganesh M, Setlow J K. Prophage induction in Haemophilus influenzae and its relationship to mutation by chemical and physical agents. Mutat Res. 1984;125:15–22. doi: 10.1016/0027-5107(84)90027-7. [DOI] [PubMed] [Google Scholar]
- 4.Barbour A G, Garon C F. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987;237:409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- 5.Barbour A G, Hayes S F. Biology of Borrelia species. Microbiol Rev. 1986;50:381–400. doi: 10.1128/mr.50.4.381-400.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergquist P L. Incompatibility. In: Hardy K G, editor. Plasmids: a practical approach. Oxford, England: IRL Press; 1987. pp. 37–78. [Google Scholar]
- 7.Berthiaume L, Elazhary Y, Alain R, Ackerman H-W. Bacteriophage-like particles associated with a spirochete. Can J Microbiol. 1979;25:114–116. doi: 10.1139/m79-017. [DOI] [PubMed] [Google Scholar]
- 8.Birge E A. Bacterial and bacteriophage genetics. 3rd ed. New York, N.Y: Springer-Verlag; 1994. [Google Scholar]
- 9.Boling M E, Kimball R F. Mutation induction by MNNG in a bacteriophage of Haemophilus influenzae. Mutat Res. 1976;37:1–10. doi: 10.1016/0027-5107(76)90050-6. [DOI] [PubMed] [Google Scholar]
- 10.Burgdorfer W, Barbour A G, Hayes S F, Benach J L, Grunwaldt E, Davis J P. Lyme disease—a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 11.Calderaro A, Dettori G, Collini L, Ragni P, Grillo R, Cattani P, Fadda G, Chezzi C. Bacteriophages induced from weakly beta-haemolytic human intestinal spirochaetes by mitomycin C. J Basic Microbiol. 1998;38:323–335. [PubMed] [Google Scholar]
- 12.Casjens S, Huang W M. Linear chromosomal physical and genetic map of Borrelia burgdorferi, the Lyme disease agent. Mol Microbiol. 1993;8:967–980. doi: 10.1111/j.1365-2958.1993.tb01641.x. [DOI] [PubMed] [Google Scholar]
- 13.Casjens S, van Vugt R, Tilly K, Rosa P A, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J Bacteriol. 1997;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damman, C. J., and D. B. Oliver. Characterization of Borrelia burgdorferi BlyA and BlyB: a plasmid-encoded system with holin-like properties. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 15.Dunn J J, Buchstein S R, Butler L-L, Fisenne S, Polin D S, Lade B N, Luft B J. Complete nucleotide sequence of a circular plasmid from the Lyme disease spirochete, Borrelia burgdorferi. J Bacteriol. 1994;176:2706–2717. doi: 10.1128/jb.176.9.2706-2717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eggers, C. H., and D. S. Samuels. Unpublished data.
- 17.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 18.Guina T, Oliver D B. Cloning and analysis of a Borrelia burgdorferi membrane-interactive protein exhibiting haemolytic activity. Mol Microbiol. 1997;24:1201–1213. doi: 10.1046/j.1365-2958.1997.4291786.x. [DOI] [PubMed] [Google Scholar]
- 19.Hayes S F, Burgdorfer W, Barbour A G. Bacteriophage in the Ixodes dammini spirochete, etiological agent of Lyme disease. J Bacteriol. 1983;154:1436–1439. doi: 10.1128/jb.154.3.1436-1439.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphrey S B, Stanton T B, Jensen N S. Mitomycin C induction of bacteriophages from Serpulina hyodysenteriae and Serpulina innocens. FEMS Microbiol Lett. 1995;134:189–194. doi: 10.1111/j.1574-6968.1995.tb07921.x. [DOI] [PubMed] [Google Scholar]
- 21.Humphrey S B, Stanton T B, Jenson N S, Zuerner R L. Purification and characterization of VSH-1, a generalized transducing bacteriophage of Serpulina hyodysenteriae. J Bacteriol. 1997;179:323–329. doi: 10.1128/jb.179.2.323-329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda H, Tomizawa J I. Prophage P1, an extrachromosomal replication unit. Cold Spring Harbor Symp Quant Biol. 1969;30:791–798. doi: 10.1101/sqb.1968.033.01.091. [DOI] [PubMed] [Google Scholar]
- 23.Kornberg A, Baker T A. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman and Company; 1992. [Google Scholar]
- 24.Lubke, L. L., and D. S. Samuels. Unpublished data.
- 25.Marconi R T, Samuels D S, Garon C F. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J Bacteriol. 1993;175:926–932. doi: 10.1128/jb.175.4.926-932.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marconi R T, Samuels D S, Schwan T G, Garon C F. Identification of a protein in several Borrelia species which is related to OspC of the Lyme disease spirochetes. J Clin Microbiol. 1993;31:2577–2583. doi: 10.1128/jcm.31.10.2577-2583.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marconi R T, Sung S Y, Hughes C A N, Carlyon J A. Molecular and evolutionary analyses of a variable series of genes in Borrelia burgdorferi that are related to ospE and ospF, constitute a gene family, and share a common upstream homology box. J Bacteriol. 1996;178:5615–5626. doi: 10.1128/jb.178.19.5615-5626.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margolis N, Rosa P. Regulation of expression of major outer surface proteins in Borrelia burgdorferi. Infect Immun. 1993;61:2207–2210. doi: 10.1128/iai.61.5.2207-2210.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masuda K, Kawata T. Bacteriophage-like particles induced from the Reiter treponeme by mitomycin C. FEMS Microbiol Lett. 1979;6:29–31. [Google Scholar]
- 30.Neubert U, Schaller M, Januschke E, Stolz W, Schmieger H. Bacteriophages induced by ciprofloxacin in a Borrelia burgdorferi skin isolate. Zentbl Bakteriol. 1993;279:307–315. doi: 10.1016/s0934-8840(11)80363-4. [DOI] [PubMed] [Google Scholar]
- 31.Porcella S F, Popova T G, Akins T G, Li M, Radolf J D, Norgard M V. Borrelia burgdorferi supercoiled plasmids encode multicopy tandem open reading frames and a lipoprotein gene family. J Bacteriol. 1996;178:3293–3307. doi: 10.1128/jb.178.11.3293-3307.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie A E, Robinson I M, Joens L A, Kinyon J M. A bacteriophage for Treponema hyodysenteriae. Vet Rec. 1978;103:34–35. doi: 10.1136/vr.103.2.34. [DOI] [PubMed] [Google Scholar]
- 33.Saheb S A. Spirochetal organisms from pigs. III. Preliminary observations on bacteriophage particles associated with spirochetes of the genus Treponema. Rev Can Biol. 1974;33:67–70. [PubMed] [Google Scholar]
- 34.Saint Girons I, Margarita D, Amouriaux P, Baranton G. First isolation of bacteriophages for a spirochete: potential genetic tools for Leptospira. Res Microbiol. 1990;143:615–621. doi: 10.1016/0923-2508(90)90086-6. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Samuels D S, Garon C F. Coumermycin A1 inhibits growth and induces relaxation of supercoiled plasmids in Borrelia burgdorferi, the Lyme disease agent. Antimicrob Agents Chemother. 1993;37:46–50. doi: 10.1128/aac.37.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarma T A, Singh R. Isolation and characterization of temperature-sensitive mutants of cyanophage N-1. Acta Virol. 1994;38:11–16. [PubMed] [Google Scholar]
- 38.Schaller M, Neubert U. Bacteriophages and ultrastructural alteration of Borrelia burgdorferi induced by ciprofloxacin. J Spirochet Tickborne Dis. 1994;1:37–40. [Google Scholar]
- 39.Schwan T G, Schrumpf M E, Karstens R H, Clover J R, Wong J, Daugherty M, Struthers M, Rosa P A. Distribution and molecular analysis of Lyme disease spirochetes, Borrelia burgdorferi, isolated from ticks throughout California. J Clin Microbiol. 1990;31:3096–3108. doi: 10.1128/jcm.31.12.3096-3108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson W J, Garon C F, Schwan T G. Borrelia burgdorferi contains repeated DNA sequences that are species specific and plasmid associated. Infect Immun. 1990;58:847–853. doi: 10.1128/iai.58.4.847-853.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steere A C, Grodzicki R L, Kornblatt A N, Craft J E, Barbour A G, Burgdorfer W, Schmid G P, Johnson E, Malawista S E. The spirochetal etiology of Lyme disease. N Engl J Med. 1983;308:733–740. doi: 10.1056/NEJM198303313081301. [DOI] [PubMed] [Google Scholar]
- 42.Sternberg N. Recognition and cleavage of the bacteriophage P1 packaging site (pac). I. Differential processing of the cleaved ends in vivo. J Mol Biol. 1987;194:453–468. doi: 10.1016/0022-2836(87)90674-7. [DOI] [PubMed] [Google Scholar]
- 43.Stevenson B, Casjens S, van Vugt R, Porcella S F, Tilly K, Bono J L, Rosa P. Characterization of cp18, a naturally truncated member of the cp32 family of Borrelia burgdorferi plasmids. J Bacteriol. 1997;179:4285–4291. doi: 10.1128/jb.179.13.4285-4291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevenson B, Tilly K, Rosa P A. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J Bacteriol. 1996;178:3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su M-T, Venkatesh T V, Bodmer R. Large- and small-scale preparation of bacteriophage λ lysate and DNA. BioTechniques. 1998;25:44–46. doi: 10.2144/98251bm08. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto K, Shinagawa H, Kondo S. Induction of prophage λ in Escherichia coli recA-strain by N-methyl-N′-nitro-N-nitrosoguanidine. Mutat Res. 1983;107:33–40. doi: 10.1016/0027-5107(83)90076-3. [DOI] [PubMed] [Google Scholar]
- 47.Young R, Blasi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]
- 48.Yun T, Vapnek D. Electron microscopic analysis of bacteriophage P1, P1Cm, and P7. Determination of genome sizes, sequence homology, and location of antibiotic resistance determinants. Virology. 1977;77:376–385. doi: 10.1016/0042-6822(77)90434-2. [DOI] [PubMed] [Google Scholar]
- 49.Zückert W R, Filipuzzi-Jenny E, Meister-Turner J, Ståhlhammar M, Carlemalm, Meyer J. Repeated DNA sequences on circular and linear plasmids of Borrelia burgdorferi sensu lato. In: Axford J S, Rees D H E, editors. Lyme borreliosis. New York, N.Y: Plenum Press; 1994. pp. 253–260. [Google Scholar]
- 50.Zückert W R, Meyer J. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J Bacteriol. 1996;178:2287–2298. doi: 10.1128/jb.178.8.2287-2298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]