Abstract
A cluster of several newly occurring mutations on Omicron is found at the β-core region of the spike protein’s receptor-binding domain (RBD), where mutation rarely happened before. Notably, the binding of SARS-CoV-2 to human receptor ACE2 via RBD happens in a dynamic airway environment, where mechanical force caused by coughing or sneezing occurs. Thus, we used atomic force microscopy-based single-molecule force spectroscopy (AFM-SMFS) to measure the stability of RBDs and found that the mechanical stability of Omicron RBD increased by ∼20% compared with the wild type. Molecular dynamics (MD) simulations revealed that Omicron RBD showed more hydrogen bonds in the β-core region due to the closing of the α-helical motif caused primarily by the S373P mutation. In addition to a higher unfolding force, we showed a higher dissociation force between Omicron RBD and ACE2. This work reveals the mechanically stabilizing effect of the conserved mutation S373P for Omicron and the possible evolution trend of the β-core region of RBD.
Keywords: single-molecule force spectroscopy, SARS-CoV-2, Omicron variant, SMD, biophysics
Introduction
The COVID-19 pandemic, caused by SARS-CoV-2, has been spreading worldwide for more than 2 years, primarily due to the continuous mutation of the virus.1,2 As a single-stranded RNA virus that has infected a very large population, SARS-CoV-2 has undergone numerous mutations and rapidly adapts to the host. As the primary protein of the virus that binds to the angiotensin-converting enzyme 2 (ACE2) receptor of the host cell, the spike protein plays an essential role in the infectivity and transmissibility of the virus, showing an exceptionally high evolutionary rate (Figure 1a).3,4 Mutations in the spike protein’s receptor-binding domain (RBD) are the most dangerous because this domain is the direct contact point with ACE2. In addition, the majority of neutralizing antibodies target the RBD.5−7 Indeed, all SARS-CoV-2 variants of concern (VOCs) that have been announced by the World Health Organization (WHO) possess several mutations in the RBD.8,9 These mutations lead to higher transmission, and these VOCs completely replace the original strain. Thus, knowledge of the effect of each mutation on the RBD is of great importance for understanding and fighting the virus.
Figure 1.
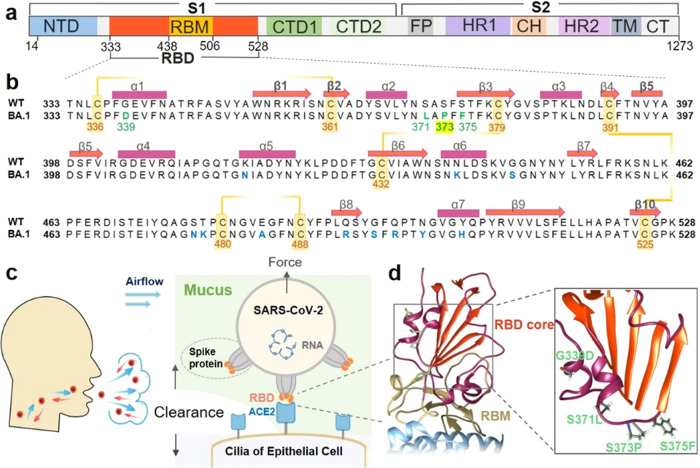
Newly occurring mutations at the β-core region of RBD in the spike protein subjected to mechanical force. (a) Domain architecture of the full-length SARS-CoV-2 spike (S) protein. (b) Sequence alignment of the Omicron variant (BA.1) with the wild-type RBD (333–528). The mutations are highlighted in green or blue. The orange line indicates the four disulfide bonds with the residue number shown. The secondary structure elements of the RBD (PDB code: 6ZGE) are marked with rectangles (helices) and arrows (β-strands) above the sequence. (c) The spike protein trimers (gray) on the SARS-CoV-2 virus particles could bind to ACE2 (light blue) on the human cell surface via the RBDs (orange). (d) Crystal structure of the RBD–ACE2 complex (PDB code: 7wbl). The RBD β-core region and RBM are colored orange and yellow, respectively. Part of ACE2 is represented in light blue. Details of the Omicron variant structure are illustrated in the right panel. Four mutations that first appeared in Omicron are colored green.
The very recent VOC, Omicron (BA.1 or B.1.1.529), has more than 30 mutations in the spike protein, 15 of which are in the RBD, and shows the highest transmission thus far (Figure 1a).10−12 Among these mutations, many with known effects are present on the flexible receptor-binding motif (RBM; residues 438–506), which makes direct interactions with the ACE2 receptor.13 For example, mutations such as E484A and Q493R evade antibody neutralization, and Q498R and N501Y enhance spike/ACE2 binding. However, although containing so many mutations, the binding affinity of Omicron’s spike protein/RBD to ACE2 does not increase significantly, only similar to previous VOC α and β. Thus, understanding the consequence of new mutations in Omicron RBD is still of great interest.14
A cluster of several new mutations, G339D, S371L, S373P, and S375F (Figure 1b,c), is located close to the β-core region in the Omicron RBD (OmicronRBD, refers to BA.1).10 The effects of these mutations on the virus are still largely unknown. RBD has two subdomains with different stabilities and functions. The first is the flexible RBM with a concave surface for direct binding with ACE2. Mutations within RBM have the potential to considerably alter its binding affinity for ACE2. The second is a structured five-stranded antiparallel β-sheet core region that is covered with two connecting α-helices on both sides (Figure 1d, colored in red). This region serves as a stably folded scaffold for the RBM, and these newly occurring mutations (green) in Omicron with undetermined functions are found in this mechanically stable region.
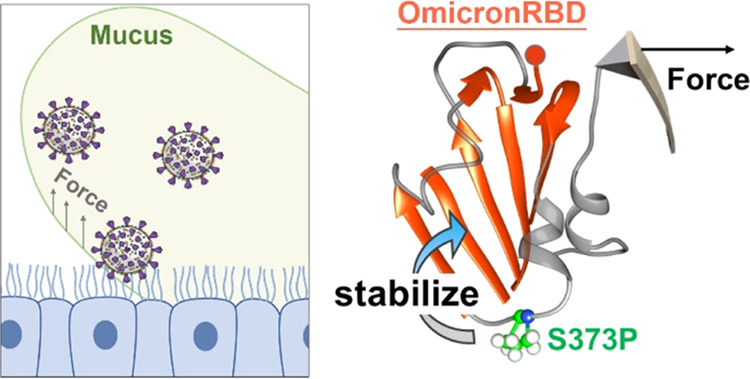
In fact, as the critical component of the virus, the stability of the spike protein plays an important role in its entry efficiency and viral transmission.15 Moreover, the SARS-CoV-2 virus particles invade the human body by binding to ACE2 receptors on the ciliary surface of respiratory epithelial cells.16 Consequently, the act of coughing or sneezing, aimed at clearing mucus within the respiratory tract,17 will apply mechanical force not only to the cell surface but also to the virus particles adhering to the ciliary surface (Figure 1c). Thus, it is possible that RBD will be subject to mechanical force under these harsh conditions. The structural stability of RBD relies on the β-core region, which is maintained by intramolecular interactions. On the other hand, the binding between RBD and ACE2 occurs in the RBM region and relies on intermolecular interactions. Despite their distinct positions and functional roles, both intramolecular and intermolecular interactions could be influenced to varying degrees when the viral particle is subjected to external forces. Thus, viral attachment via RBD will be subject to the perturbations caused by mechanical force. Thus, we hypothesized that these new mutations modified the mechanical stability of Omicron, and a new strategy to study its stability is needed (Figure 1d).
To this end, we used atomic force microscopy-based single-molecule force spectroscopy (AFM-SMFS) to investigate the mechanical stability of the RBDs from the wild-type (wtRBD) and Omicron variants. SMFS has been used to manipulate protein molecule(s) mechanically and studies the interaction between RBD/spike protein and its receptor in particular.18,19 This technique mimics the external force from the dynamic airway on the protein during measurement, which the RBD may experience during binding to ACE2.20 Previously, several groups have used SMFS to study the binding strength between the RBD and ACE2. They found that SARS-CoV-2 shows a higher binding force and binding energy to ACE2 than SARS-CoV-1.21,22 We studied the effects of SARS-CoV-2 mutations and found that the N501Y mutation enhances RBD binding to ACE2, while K417N and E484K do not.23 Similar conclusions have also been validated through molecular dynamics simulations.24 In addition to measuring protein–protein interaction, SMFS has been most widely used to measure the mechanical stability of the protein by (un)folding experiments.25−32 Here, we used it to measure both the mechanical stability of RBDs and the dissociation force between RBD and ACE2 interaction. Notably, we found a significant unfolding force increase of Omicron RBD compared to WT. Moreover, computational simulations were used to gain mechanistic insights into the mechanical unfolding process, and the results revealed the key mutations for the observed stability increment and the underlying mechanism.
Results
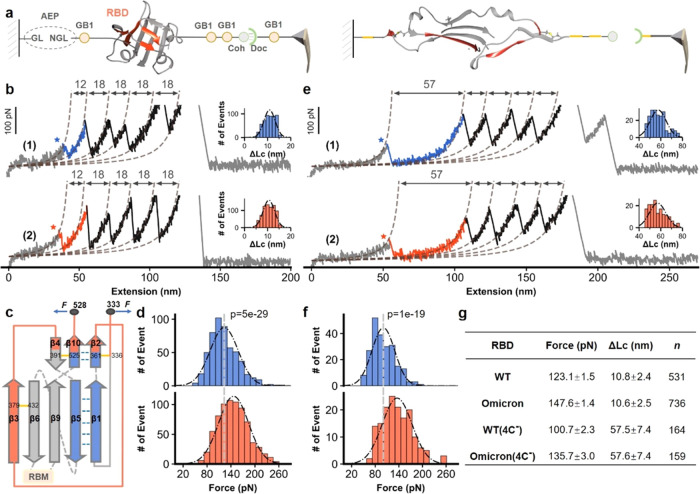
We used the AFM-SMFS system to measure the stability of the RBD.33,34 For an accurate comparison, the target protein RBD is fused with other marker proteins with known stabilities that are site-specifically immobilized in the system. Asparaginyl endopeptidase (AEP)-mediated protein ligation was used for covalent RBD immobilization on a peptide-coated glass coverslip, in which enzymatic ligation occurred between their specific N-terminal NH2-GL (Gly-Leu) dipeptide sequence and C-terminal NGL-COOH (Asp-Gly-Leu) tripeptide sequence (Figure 2a, left panel).35 In addition, an ELP (elastin-like polypeptide) linker with a defined length of ∼50 nm was incorporated into the peptide, serving as a spacer to avoid the short-range nonspecific interaction. Finally, marker protein GB1 with a known unfolding force was used as an internal force caliper, and a dockerin–cohesin (Doc:Coh) noncovalent protein–protein interaction with a rupture force of ∼500 pN was used as a single-molecule pulling handle.33,36 Accordingly, the fused polyprotein Coh-(GB1)2-RBD-GB1-NGL was designed and immobilized for precise AFM measurements (Figure 2a).
Figure 2.
AFM unfolding experiment of wtRBD and OmicronRBD. (a) High-precision AFM-SMFS system to measure the mechanical stability of the RBD using the fused protein Coh-(GB1)2-RBD-GB1-NGL. The protein was site-specifically immobilized to the glass surface using ligase AEP and could be picked up with a Doc-functionalized AFM tip via the Doc:Coh interaction (left panel). Then, the RBD and all other proteins were unfolded by stretching, and their mechanical stability was measured accordingly (right panel). (b) Representative curves showing a series of sawtooth-like peaks from the unfolding of the RBD with a ΔLc of ∼11 nm and marker protein GB1 with a ΔLc of ∼18 nm. The Omicron and WT variants are colored orange and blue, respectively. The histogram of ΔLc is shown in the inset. (c) The unfolded fragments are depicted by their secondary structures. (d) The unfolding force of the OmicronRBD is ∼20% higher than that of the wtRBD. P < 1 × 10–5. P values were determined by a two-sample t-test. (e) Unfolding of the RBD mutants with the deletion of two disulfide bonds (C391–C525, C379–C432) resulted in an elongated ΔLc from 12 to 57 nm, as expected. (f) The force of the disulfide bond-deleted Omicron RBD mutant was still higher than that of the WT. (g) Detailed AFM data including force (ave. ± SEM) and ΔLc (ave. ± SD) are shown.
Then, an AFM cantilever with a GB1-Doc-functionalized tip was used. By moving the tip toward the protein-immobilized surface, the RBD was picked up through the specific Doc:Coh interaction. Then, the tip retracted, and all of the proteins/domains sequentially stretched and unfolded, ending with the rupture of the strongest Doc:Coh interaction. The mechanical stability of the RBD has been measured accordingly (Figure 2a, right). Finally, the tip moved to another spot on the surface and the cycle was repeated on different RBDs thousands of times. This system enables highly reliable and efficient AFM measurements and data analysis from a single molecule.
The fused protein containing wtRBD or OmicronRBD was built for AFM-SMFS measurements. Stretching led to a force–extension curve with an initial ∼50 nm length featureless curve from the extension of the ELP linke, then sawtooth-like peaks from the unfolding of GB1 (18 nm), and a peak from the rupture of the Doc:Coh interaction (Figures 2b and S1). In addition to these auxiliary signals, a force peak with a ΔLc of ∼11 nm was observed for both constructs, which suggests that the signal must result from RBD unfolding. Statistically, the value was 10.8 ± 2.4 nm for the wtRBD and 10.6 ± 2.5 nm for the OmicronRBD.
To confirm these results, we calculated the theoretical number of extendable residues upon RBD unfolding. First, four disulfide bonds are present in the RBD, which cannot be broken, and the residues between them are locked.37 Thus, RBD unfolding will lead to the extension of only 31 residues, from the first structured residue C361 to the last residue C391 (Figure 2c, red). Indeed, the shielding effect of disulfide bonds in RBDs prevents complete unfolding of the protein, which has also been observed in a recent study.20 They discovered that the dissociation of a linked RBD–ACE2 complex is accompanied by partial unfolding of the RBD, resulting in a collective force signal. As a result, the theoretical ΔLc value is 10.3 nm (31 × 0.365 nm/aa–1.0 nm), agreeing well with the experimental value of ∼11 nm and confirming that the peak was attributed to the RBD. Thus, the average unfolding force was 123.1 pN for wtRBD and 147.6 pN for OmicronRBD (Figure 2d). These results support our hypothesis that the OmicronRBD has increased mechanical stability upon mutation.
To further verify that the observed data were from RBD unfolding, we deleted two disulfide bonds (C379–C432 and C391–C525) by mutating the four cysteines to alanines. As a result, 190 residues that the two disulfide bonds had locked in the RBD and previously inextensible would be unfolded in this construct, which led to a larger ΔLc of 58.1 nm (162 × 0.365 nm/aa–1.0 nm; Supporting Figure S2). Then, we performed the same AFM experiments on these two constructs. As expected, the 11 nm peak mostly disappeared, while a new peak with a ΔLc of ∼58 nm was observed (Figure 2e), confirming that the previous 11 nm peak was from the RBD. These values were 57.5 nm for the WT and 57.6 nm for the Omicron. Moreover, the unfolding force of OmicronRBD (135.7 pN) was higher than that of wtRBD (100.7 pN) for this mutant (Figure 2f,g), again demonstrating the stronger stability of OmicronRBD than that of wtRBD.
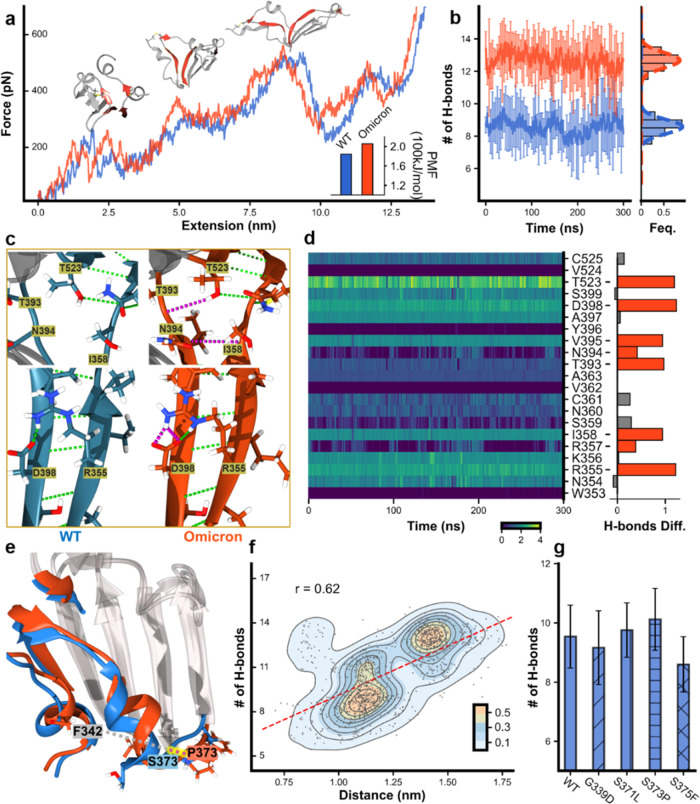
To gain mechanistic insights into the considerable stability enhancement and pin down the exact mutation(s) responsible for this effect, we performed two different all-atom molecule dynamics simulations, focusing on the hydrogen bond in the β-core region of the RBD.38 First, we performed steered molecular dynamics (SMD) simulations to visualize the mechanical unfolding process of the RBD and reveal the unfolding barrier/key force-bearing motif. Similar to the above experiment, the RBD was stretched with a constant pulling speed under a simulated force field and showed the force–extension trajectory (Figure 3a, Video S1 for WT, and Video S2 for Omicron). Indeed, the unfolding force of the OmicronRBD was generally higher than that of the wtRBD over the trajectory. This difference was quantitatively reflected by the potential of mean force analysis from irreversible pulling through Jarzynski’s equality (Figure S3), which gave a value of 205 versus 184 kJ/mol (Figure 3a, inset). Moreover, the SMD trajectory demonstrated that the major contributing factors to the unfolding barrier are from the peeling between β-strands 2 and 10 and β-strands 1 and 5, as shown by snapshots of the three key unfolding events (Figure 3a, red). Therefore, the interaction between these β-strands determined the mechanical stability of the RBD.
Figure 3.
MD simulation results for wtRBD and OmicronRBD focusing on the four force-bearing β strands. (a) SMD trajectories of stretching wtRBD (blue) and OmicronRBD (orange). The three key snapshots show the renderings of the RBDs at different times. The four force-bearing β-strands, β2/β10 and β1/β5, in the β-sheet are colored red. Inset: the potential of mean force (PMF) calculated. (b) MD simulations show the number of hydrogen bonds formed in the four β-strands. (c) The enlarged structure shows the hydrogen bonds (dashed line) in the four β-strands. Additional H-bonds in Omicron are colored pink. (d) The evolution of the number of hydrogen bonds for each residue involved in β2/β10 and β1/β5 during the MD simulations. The difference in hydrogen bonds between Omicron and WT was shown in the right panel, and the red bars indicate a noticeable increment in Omicron compared to WT. (e) Overlapped structure of Omicron and WT focusing on the β-core region of the RBD shows that the conformation of α2 in Omicron has changed. The distance between P373 and F342 in Omicron is longer (red dash line) than that between S373 and F342 in the WT. (f) The relationship between the distances of residues 373 near α2 and F342 near α1 (as a reference point) and the number of H-bonds was extracted from multiple MD simulations. (g) By introducing a single mutation into the wtRBD, MD simulations show that S371L and S373P increase the number of hydrogen bonds in the region.
Thus, we performed MD simulations to determine the number of hydrogen bonds in these four β-strands of the wtRBD (Video S3) and OmicronRBD (Video S4). After 300 ns of 10 times simulations, ∼9 hydrogen bonds were present in the wtRBD, while ∼13 bonds were found in the OmicronRBD, on average (Figure 3b). Moreover, a detailed analysis of the hydrogen bond number for each residue along the simulation for both wtRBD and OmicronRBD was also performed, showing that several residues participated in more hydrogen bonds in Omicron (Figures 3c,d, S4, and S5). For the region of the force-bearing β-strands, we also analyzed the energy terms (Figure S6). The short-range LJ (Lennard-Jones) potential has no obvious change between wtRBD and OmicronRBD. However, the Coulomb interaction of OmicronRBD is stronger than that of wtRBD. Thus, the Coulomb interaction might play an important role in the stability of the β-strands and can be affected by the sodium chloride in the solvent.
To rationalize the origin of this difference in the number of hydrogen bonds, we overlapped the structures of the WT and Omicron RBDs, focusing on the β-core region. Notably, α-helix 2 in Omicron moved toward the β-sheet core, as reflected in the cryo-electron microscopy (cryo-EM) structure (Supporting Figure S7). Although this movement is not apparent in the cryo-EM structure with a fixed conformation, it can be readily inspected through the structure from MD simulations with multiple dynamic conformations (Figure 3e, highlighted in red). To validate the relationship between this structural change and the number of hydrogen bonds, we performed 10 times of MD simulations (300 ns) and averaged the results. Then, we chose every six ns to calculate the number of hydrogen bonds in each simulation. Since the position of α-helix 1 is relatively consistent, we chose it as a reference point and measured the distance between residue 373 in α-helix 2 and residue F342 in α-helix 1 at each section. These distances were plotted along with the number of hydrogen bonds. As shown in Figure 3f, the longer the distance between them, the more hydrogen bonds are generally observed. The relationship between the distance of residue 371/375 to residue F432 and the number of hydrogen bonds is not very significant (Supporting Figure S8).
The new mutations G339D, S371L, S373P, and S375P in the core region of the RBD are close to α-helix 2, suggesting their roles in this structural change. Thus, we built a single mutation into the wtRBD for MD simulations (n = 10) and analyzed the corresponding number of hydrogen bonds. On average, wtRBD(S373P) showed the highest increase of two hydrogen bonds, and wtRBD(S371L) showed the modest addition of one hydrogen bond, and the effects for the other two mutations were trivial (Figure 3g). Thus, it appears that the S373P mutation rigidifies the local polypeptide chain, leading to the inward movement of helix 2 from the structure. As shown in Supporting Figures S9 and S10, rigid proline 373 in OmicronRBD might open the loop between the nearby helix and the β-strand, so that it can maintain a relatively large distance from the α-helix. Since the helix is directly connected to β1/β2, this conformational change may allow these β-strands to form more hydrogen bonds in a better orientation. Consequently, MD simulations revealed that S371L and S373P most likely account for the increased stability of the OmicronRBD.
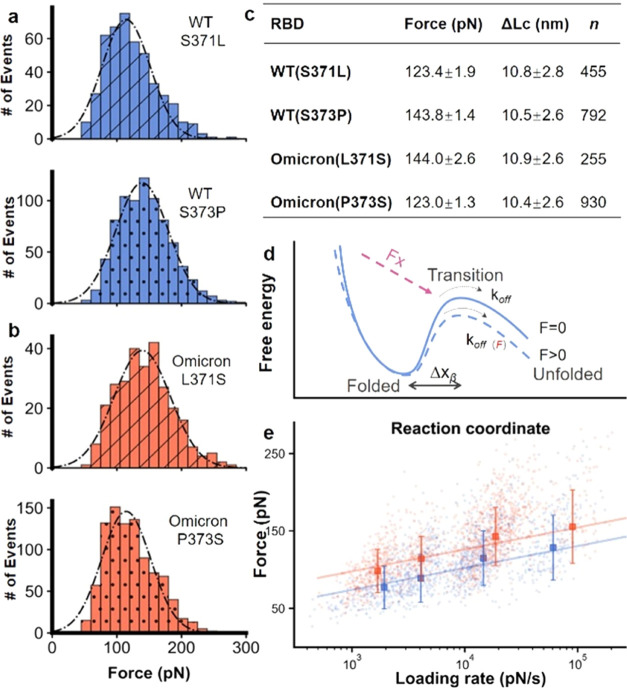
Inspired by the simulation results, we built four wtRBD mutants in the polyprotein constructs, focusing on residues 371 and 373 for experimental confirmation. First, wtRBD(S371L) and wtRBD(S373P) were constructed, in which these two mutations found in Omicron were added to the wtRBD. As expected, the unfolding signal of the RBD was the same except for the force (Figures 4a and S11). The AFM results for wtRBD(S371L) showed no force increment (123.4 pN), while wtRBD(S373P) showed a considerable increment of 20 pN (143.8 pN).
Figure 4.
AFM-SMFS results of RBDs with a point mutation. (a, b) Unfolding force histograms of wtRBD(S371L), wtRBD(S373P), OmicronRBD(L371S), and OmicronRBD(P373S), showing that residue 373 is the primary site for the fine-tuning of RBD stability with the most significant force change upon mutation. (c) The unfolding force of RBDs is shown. (d) The free-energy potential between the folded state and the transition state of the protein in the absence (solid, blue) and presence of an externally applied pulling force (dashed, blue). (e) The unfolding forces of the WT and Omicron RBDs show a linear relationship with the logarithm of the loading rate.
Moreover, we built OmicronRBD(L371S) and OmicronRBD(P373S), in which the mutation found in Omicron is reverted back to the residues present in the wtRBD while keeping all other mutations. The unfolding force decreased in these RBDs. The unfolding force of OmicronRBD(L371S) decreased slightly to 144.0 pN, while that of OmicronRBD(P373S) decreased to 123.0 pN (Figure 4b,c). Also, we built a similar point mutation on the remaining two residues, 339 and 375, in the cluster and performed the same AFM experiments. Agreeing with the computational simulation, the mutation on these two sites did not change the mechanical stability of the RBD (Supporting Figure S12). Consequently, this experimental measurement showed that the enhanced stability of OmicronRBD can be mainly attributed to the S373P mutation.
Next, the unfolding kinetics of the wtRBD and OmicronRBD were determined by stretching the proteins at different velocities during a force loading rate experiment.39−41 In force spectroscopy measurements, the application of external mechanical force lowers the activation energy of protein unfolding (Figure 4d). Thus, the unfolding force is proportional to the logarithm of the loading rate, which describes the effect of the force applied on the protein over time. As expected, the plots showed a linear relationship (Figures 4e and S13). From the fit, we estimated the unfolding rate (koff) value and the length scale of the energy barrier (Δx) value. koff was 0.59 ± 0.43 s–1 (value and fitting error) for the wtRBD and 0.12 ± 0.12 s–1 for the OmicronRBD. The koff of WT(S373P) decreased to a similar value as that of OmicronRBD, while WT(S371L) remained mostly unchanged. The kinetic results also showed a stabilized OmicronRBD with a lower off rate than the wtRBD, and mutation S373P plays an important role in the increase of mechanical stability.
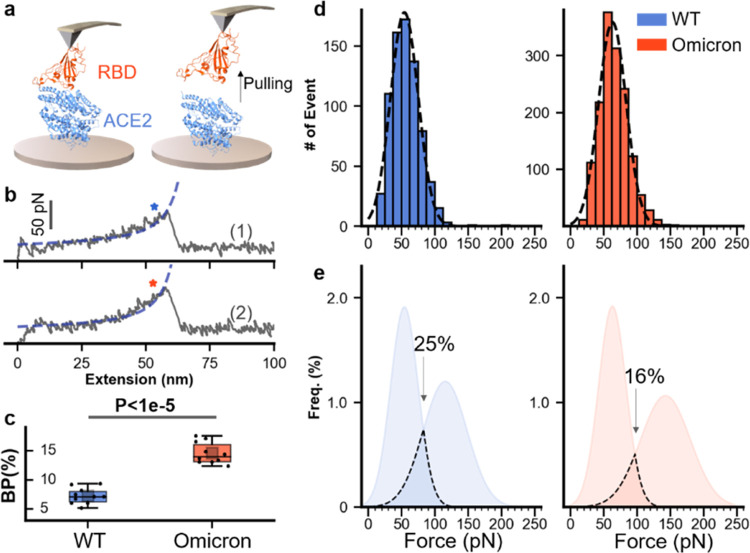
Finally, we performed an RBD–ACE2 unbinding experiment to study the binding ability of RBD to ACE2, which is the primary biological function of RBD. RBDs and ACE2 were ligated on the AFM tip and the glass surface using the previous AEP-based enzymatic method, respectively (Figure 5a). The AFM tip approached the surface, leading to a complex formation. Then, the tip retracted, and the complex unbound upon stretching. The corresponding force–extension curve showed a single peak (Figure 5b) from the unbinding event between ACE2 and RBD. Then, the tip moves to another position and repeats this cycle thousands of times. We then counted their unbinding events from 10 independent experiments. It is found that the binding probability of OmicronRBD is higher than that of wtRBD (14.6 vs 7.3%, Figure 5c).
Figure 5.
AFM-SMFS unbinding experiment of the RBD–ACE2 complex. (a) Schematic of the measurement of the unbinding between RBDs and ACE2 using SMFS. RBD and ACE2 are immobilized to the AFM tip and the substrate, respectively. (b) Representative force–extension curves show the unbinding peak from RBD–ACE2 interaction. WT is colored blue (top), and Omicron is orange (bottom). (c) Box plot of the binding probabilities shows a higher value for Omicron than that for WT (14.6 vs 7.3%). One data point belongs to the BP from one experiment (10 in total). The box indicates the 25th and 75th percentiles. P values were determined by a two-sample t-test. (d) The unbinding force distribution of the RBD–ACE2 complex. Dashed lines represent the Gaussian fit of the result. (e) The merged results of unfolding and unbinding for WT and Omicron RBDs. Overlap of the force distribution is observed for both WT (25%) and Omicron (16%) under experimental conditions.
In addition, the average rupture force was 54 ± 0.77 pN (ave. ± SEM) for wtRBD–ACE2 interaction and 63 ± 0.54 pN for OmicronRBD–ACE2 interaction, determined from the force distribution (Figure 5d). The larger dissociation force of OmicronRBD–ACE2 than that of wtRBD–ACE2 has been further validated by SMD simulations (Figure S14). Finally, we fit the distribution by the Gaussian function and merged the unfolding force of RBD and unbinding force (Figure 5e). The overlapped area between two distributions is 25% for wtRBD and 16% for OmicronRBD, implying that the enhanced mechanical stability of RBD may contribute to the protection of the protein structure from unbinding events under our experimental conditions.
Discussion
Coronaviruses are large, single-stranded RNA viruses evolving with a remarkable mutation rate, as evidenced by the transmission of several VOCs of SARS-CoV-2 in only the past 2 years. In addition to neutral mutations, the effect of many accumulated mutations has been revealed. In this study, we combined experimental and computational approaches to study the mechanical stability of the OmicronRBD and determine the effect of mutation S373P in the β-core region. First, the AFM measurements showed an unfolding force for that OmicronRBD that was 20% higher than its characteristic unfolding. To the best of our knowledge, this is the first experimental work to show higher mechanical stability of the OmicronRBD quantitatively.
In addition to the experimental results, the computational data provide essential mechanistic insights into the OmicronRBD, and these simulations were essential parts of our work.42,43 MD simulations uncovered its detailed unfolding pathway and revealed the underlying mechanism of the stability increment, which is due to the increased number of hydrogen bonds caused by mutations S373P and S371L.
From a structural perspective, two subdomains with different mechanical stabilities are present in the RBD. The RBM dominates ACE2 binding and receptor specificity. Before Omicron, most accumulating mutations in the RBD occurred in the RBM. In addition, mutations in the other subdomain, the β-core region consisting of a central β-sheet flanked by α-helices, were rare. In fact, there are four disulfide bonds in the RBD, strongly supporting the overall structure of the RBD and its function. To date, no naturally occurring variant with mutations to these cysteine residues has been observed. Intended cysteine mutation results in the dysfunction of the RBD binding to ACE2, highlighting the importance of a stable RBD structure supported by the disulfide bonds. In this work, we found that a mutation in the β-core domain can fine-tune the stability of the RBD. There is indeed an overlap between the unfolding force of RBD and the RBD–ACE2 unbinding force. Thus, it is possible that the enhanced mechanical stability of RBD contributes to its function and virus transmission.15,44 Nevertheless, this should be investigated using viruses at the cellular level and model animals.
In addition to discovering the effect of mutation S373P for SARS-CoV-2, this work revealed a new way that nature used to increase protein’s mechanical stability by using proline mutation. Proline mutation is often used to decrease protein’s stability by disrupting the hydrogen bond networks in the β-sheet region. Here, by introducing proline in the loop close to the β-sheet region, more hydrogen bonds are formed and stabilize the region. Thus, a new strategy to design proteins with enhanced stability is demonstrated. In fact, the importance of mechanical force in immunity has been demonstrated, such as tuning T-cell receptor sensitivity to mechanical force for better CAR T-cell therapy.45 So many single-molecule studies of protein, DNA, and RNA show the relationship between mechanical force and their folding and function.46−51 Thus, mechanical stability can be a factor in SARS-CoV-2 mutation selection.
Finally, mutation S373P in the RBD of the spike protein is a conserved mutation in Omicron. It is found in most Omicron’s sister strains, including BA.2 (B.1.1.529.2), BA.3 (B.1.1.529.3), BA.4 (B.1.1.529.4), and BA.5 (B.1.1.529.5).52−55 Moreover, it is found in the very recent strains BQ1 and XBB. Interestingly, the mutations found in the β-core region of BA.1 remain in the other Omicron strains. Mutation S373P is identical and relatively conservative, while S371 is mutated to the more hydrophobic residue leucine in BA.1 and phenylalanine in BA.2–5, further demonstrating the importance of this mutation and suggesting its effect on higher transmission. Since we cannot predict future mutations and emerging variants of SARS-CoV-2, it is critical to maintain the surveillance of existing variants and mutations and study their effects.
Materials and Methods
Engineering of the Recombinant Protein, Experimental Procedure, and Data Analysis
All of the different RBD constructs are built by standard molecular biology methods. Their expression, purification, and immobilization in the AFM-SMFS system can be found in SI Appendix for details. AFM-SMFS experiments were performed by a Nanowizard 4 atomic force microscope, and the details can be found in SI Appendix. (S)MD simulations were used to reveal the underlying molecular mechanism, and the details are also provided in SI Appendix.
Acknowledgments
The numerical calculations in this paper have been done on the computing facilities in the High-Performance Computing Center (HPCC) of Nanjing University. The authors thank Prof. Thomas T. Perkins and Prof. Michael M. Nash for inspiring discussion.
Data Availability Statement
All data are available in the main text or the Supporting Information.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.3c00142.
Author Contributions
CRediT: Bin Zheng data curation, formal analysis, investigation, methodology, software, writing-original draft; Yuelong Xiao data curation, investigation; Bei Tong formal analysis, resources; Yutong Mao investigation; Rui Ge investigation; Fang Tian investigation, methodology; Xianchi Dong formal analysis, resources; Peng Zheng conceptualization, funding acquisition, project administration, resources, supervision, writing-original draft.
The National Natural Science Foundation of China (Grant Nos. 21977047 and 22222703 (P.Z.)), the Natural Science Foundation of Jiangsu Province (Nos. BK20200058 and BK20202004 (P.Z.)), and Fundamental Research Funds for the Central Universities (No. 020514380301).
The authors declare no competing financial interest.
Supplementary Material
References
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.; Zhao S.; Yu B.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J.; Ge J.; Yu J.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Ksiazek T. G.; Goldsmith C. S.; Zaki S. R.; et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Starr T. N.; Greaney A. J.; Hilton S. K.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310. 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C. B.; Farzan M.; Chen B.; Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.; Yang Z.; Hu H.; et al. Covalently Engineered Protein Minibinders with Enhanced Neutralization Efficacy against Escaping SARS-CoV-2 Variants. J. Am. Chem. Soc. 2022, 144, 5702–5707. 10.1021/jacs.1c11554. [DOI] [PubMed] [Google Scholar]
- Harvey W. T.; Carabelli A. M.; Jackson B.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y.; Schmidt F.; Zhang F.; et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 2020, 9, e61312 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W.; Xu Y.; Xu P.; et al. Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody. Science 2022, 375, 1048–1053. 10.1126/science.abn8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott P.; Bodinier B.; Eales O.; et al. Rapid increase in Omicron infections in England during December 2021: REACT-1 study. Science 2022, 375, 1406–1411. 10.1126/science.abn8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z.; Liu P.; Wang N.; et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell 2022, 185, 860–871. 10.1016/j.cell.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Cai Y.; Lavine C. L.; et al. Structural and functional impact by SARS-CoV-2 Omicron spike mutations. Cell Rep. 2022, 39, 110729 10.1016/j.celrep.2022.110729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.; Zhou J.; Tian M.; et al. Omicron SARS-CoV-2 mutations stabilizes spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape. Nat. Commun. 2022, 13, 4958 10.1038/s41467-022-32665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.; Zhang Y.; Fei P.; et al. Mechanical activation of spike fosters SARS-CoV-2 viral infection. Cell Res. 2021, 31, 1047–1060. 10.1038/s41422-021-00558-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-T.; Lidsky P. V.; Xiao Y.; et al. SARS-CoV-2 replication in airway epithelia requires motile cilia and microvillar reprogramming. Cell 2023, 186, 112–130. 10.1016/j.cell.2022.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button B.; Goodell H. P.; Atieh E.; et al. Roles of mucus adhesion and cohesion in cough clearance. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 12501–12506. 10.1073/pnas.1811787115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar-Camacho J. L.; Bhatia S.; Reiter-Scherer V.; et al. Quantification of Multivalent Interactions between Sialic Acid and Influenza A Virus Spike Proteins by Single-Molecule Force Spectroscopy. J. Am. Chem. Soc. 2020, 142, 12181–12192. 10.1021/jacs.0c02852. [DOI] [PubMed] [Google Scholar]
- Dufrêne Y. F.; Evans E.; Engel A.; et al. Five challenges to bringing single-molecule force spectroscopy into living cells. Nat. Methods 2011, 8, 123–127. 10.1038/nmeth0211-123. [DOI] [PubMed] [Google Scholar]
- Bauer M. S.; Gruber S.; Hausch A.; et al. A tethered ligand assay to probe SARS-CoV-2:ACE2 interactions. Proc. Natl. Acad. Sci. U.S.A. 2022, 119, e2114397119 10.1073/pnas.2114397119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W.; Dong C.; Kim S.; et al. Biomechanical characterization of SARS-CoV-2 spike RBD and human ACE2 protein-protein interaction. Biophys. J. 2021, 120, 1011–1019. 10.1016/j.bpj.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Petitjean S. J. L.; Koehler M.; et al. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020, 11, 4541 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F.; Tong B.; Sun L.; et al. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. eLife 2021, 10, e69091 10.7554/eLife.69091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan B.; Wang H.; Huynh T. Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS Lett. 2021, 595, 1454–1461. 10.1002/1873-3468.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief M.; Gautel M.; Oesterhelt F.; Fernandez J. M.; Gaub H. E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 1997, 276, 1109–1112. 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- Yu H.; Siewny M. G.; Edwards D. T.; Sanders A. W.; Perkins T. T. Hidden dynamics in the unfolding of individual bacteriorhodopsin proteins. Science 2017, 355, 945–950. 10.1126/science.aah7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.; Li H. B. Engineered elastomeric proteins with dual elasticity can be controlled by a molecular regulator. Nat. Nanotechnol. 2008, 3, 512–516. 10.1038/nnano.2008.168. [DOI] [PubMed] [Google Scholar]
- Zhuang X.; Rief M. Single-molecule folding. Curr. Opin. Struct. Biol. 2003, 13, 88–97. 10.1016/S0959-440X(03)00011-3. [DOI] [PubMed] [Google Scholar]
- Alonso-Caballero A.; Echelman D. J.; Tapia-Rojo R.; et al. Protein folding modulates the chemical reactivity of a Gram-positive adhesin. Nat. Chem. 2021, 13, 172–181. 10.1038/s41557-020-00586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl Z. N.; Yang W. T.; Marszalek P. E. Direct Observation of Multimer Stabilization in the Mechanical Unfolding Pathway of a Protein Undergoing Oligomerization. ACS Nano 2015, 9, 1189–1197. 10.1021/nn504686f. [DOI] [PubMed] [Google Scholar]
- Perales-Calvo J.; Giganti D.; Stirnemann G.; Garcia-Manyes S. The force-dependent mechanism of DnaK-mediated mechanical folding. Sci. Adv. 2018, 4, eaaq0243 10.1126/sciadv.aaq0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Liu Z.; Yang B.; Lopez Morales J.; Nash M. A. Optimal Sacrificial Domains in Mechanical Polyproteins: S. epidermidis Adhesins Are Tuned for Work Dissipation. JACS Au 2022, 2, 1417–1427. 10.1021/jacsau.2c00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl S. W.; Nash M. A.; Fried D. B.; et al. Single-molecule dissection of the high-affinity cohesin-dockerin complex. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 20431–20436. 10.1073/pnas.1211929109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc M. A.; Fink M. R.; Perkins T. T.; Sousa M. C. Type III secretion system effector proteins are mechanically labile. Proc. Natl. Acad. Sci. U.S.A. 2021, 118, e2019566118 10.1073/pnas.2019566118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y.; Wu T.; Wang M.; et al. Enzymatic biosynthesis and immobilization of polyprotein verified at the single-molecule level. Nat. Commun. 2019, 10, 2775 10.1038/s41467-019-10696-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.; Lam C.; Wang M.; Li H. Nonmechanical protein can have significant mechanical stability. Angew. Chem., Int. Ed. 2006, 45, 642–645. 10.1002/anie.200502623. [DOI] [PubMed] [Google Scholar]
- Alegre-Cebollada J.; Kosuri P.; Giganti D.; et al. S-Glutathionylation of Cryptic Cysteines Enhances Titin Elasticity by Blocking Protein Folding. Cell 2014, 156, 1235–1246. 10.1016/j.cell.2014.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R. C.; Durner E.; Schoeler C.; et al. Mechanisms of Nanonewton Mechanostability in a Protein Complex Revealed by Molecular Dynamics Simulations and Single-Molecule Force Spectroscopy. J. Am. Chem. Soc. 2019, 141, 14752–14763. 10.1021/jacs.9b06776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel R.; Nassoy P.; Leung A.; Ritchie K.; Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 1999, 397, 50–53. 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- Dudko O. K.; Hummer G.; Szabo A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 15755–15760. 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C.; Chemla Y. R.; Forde N. R.; Izhaky D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004, 73, 705–748. 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- Zhou W.; Fiorin G.; Anselmi C.; et al. Large-scale state-dependent membrane remodeling by a transporter protein. eLife 2019, 8, e50576 10.7554/eLife.50576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova A.; Zhang Z.; Acharya A.; et al. Machine Learning Reveals the Critical Interactions for SARS-CoV-2 Spike Protein Binding to ACE2. J. Phys. Chem. Lett. 2021, 12, 5494–5502. 10.1021/acs.jpclett.1c01494. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Xiang R.; Huo S.; et al. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduction Targeted Ther. 2021, 6, 233 10.1038/s41392-021-00653-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.; Kolawole E. M.; Chan W.; et al. Tuning T cell receptor sensitivity through catch bond engineering. Science 2022, 376, eabl5282 10.1126/science.abl5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Radford S. E.; Brockwell D. J. Force-induced remodelling of proteins and their complexes. Curr. Opin. Struct. Biol. 2015, 30, 89–99. 10.1016/j.sbi.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruszka D. T.; Whelan F.; Farrance O. E.; et al. Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein. Nat. Commun. 2015, 6, 7271 10.1038/ncomms8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M.; Woodside M. T. Mechanical strength of RNA knot in Zika virus protects against cellular defenses. Nat. Chem. Biol. 2021, 17, 975–981. 10.1038/s41589-021-00829-z. [DOI] [PubMed] [Google Scholar]
- Dong X.; Zhao B.; Iacob R. E.; et al. Force interacts with macromolecular structure in activation of TGF-beta. Nature 2017, 542, 55–59. 10.1038/nature21035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra S.; Lin C.-T.; Feng X. A.; Basu A.; Ha T. Single-Molecule Analysis and Engineering of DNA Motors. Chem. Rev. 2020, 120, 36–78. 10.1021/acs.chemrev.9b00361. [DOI] [PubMed] [Google Scholar]
- Le S.; Yu M.; Yan J. Direct single-molecule quantification reveals unexpectedly high mechanical stability of vinculin-talin/alpha-catenin linkages. Sci. Adv. 2019, 5, eaav2720 10.1126/sciadv.aav2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasoba D.; Kimura I.; Nasser H.; et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell 2022, 185, 2103–2115. 10.1016/j.cell.2022.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P.; Zhang L.; Rocha C.; et al. Comparable neutralisation evasion of SARS-CoV-2 omicron subvariants BA.1, BA.2, and BA.3. Lancet Infect. Dis. 2022, 22, 766–767. 10.1016/S1473-3099(22)00224-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangavarapu K.; Latif A. A.; Mullen J. L.; et al. Outbreak. info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Nat. Methods 2023, 20, 512–522. 10.1038/s41592-023-01769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I.; Yamasoba D.; Tamura T.; et al. Virological characteristics of the novel SARS-CoV-2 Omicron variants including BA. 2.12. 1, BA. 4 and BA. 5. Cell 2022, 185, 3992–4007. 10.1016/j.cell.2022.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the Supporting Information.