Abstract

The direct and sequence-dependent investigation of photochemical processes in DNA on the way to cyclobutane pyrimidine dimers (CPDs) as DNA damage requires the probing by photochemically different photosensitizers. The C-nucleosides of xanthone, thioxanthone, 3-methoxyxanthone, and triphenylene as photosensitizers were synthesized by Heck couplings and incorporated into ternary photoactive DNA architectures. This structural approach allows the site-selective excitation of the DNA by UV light. Together with a single defined site for T–T dimerization, not only the direct CPD formation but also the distance-dependent CPD formation in DNA as well as the possibility for energy transport processes could be investigated. Direct CPD formation was observed with xanthone, 3-methoxyxanthone, and triphenylene as sensitizers but not with thioxanthone. Only xanthone was able to induce CPDs remotely by a triplet energy transfer over up to six intervening A-T base pairs. Taken together, more precise information on the sequence dependence of the DNA triplet photochemistry was obtained.
Keywords: cyclobutane pyridimidine dimers, triplet, energy transfer, DNA photodamage, DNA synthesis, oligonucleotide
Introduction
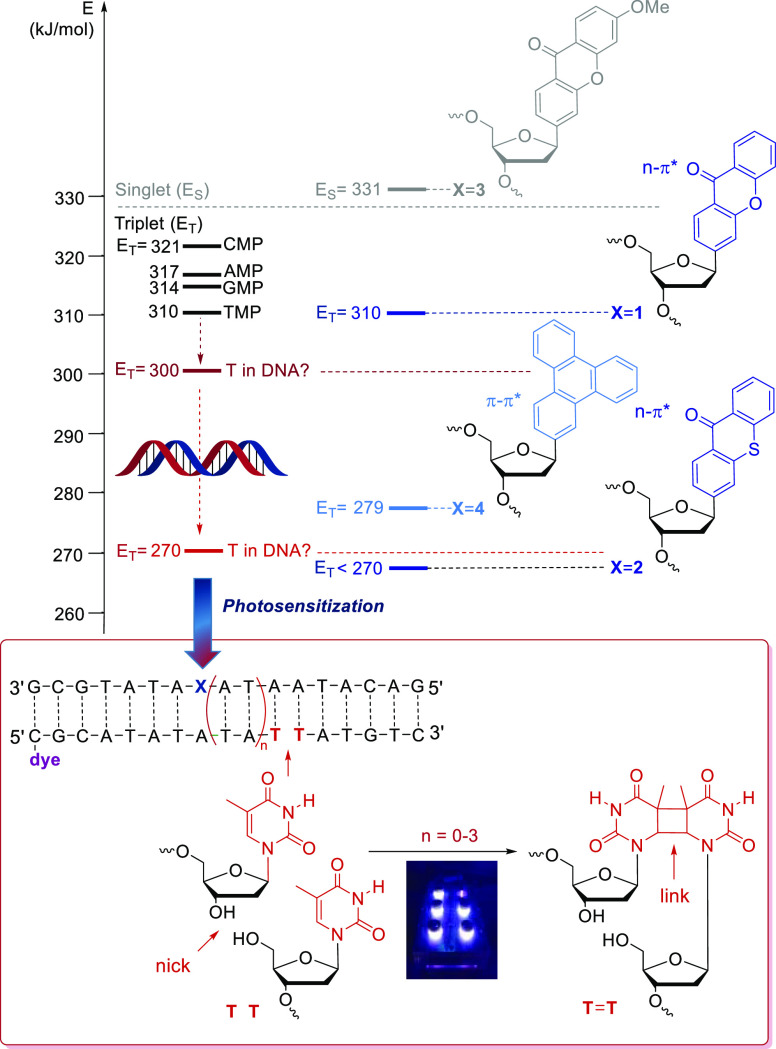
Absorption of sunlight can lead to cellular DNA damage which may cause skin cancer.1−3 It is important to elucidate the pathways of DNA photodamages4−6 to gain the best possible understanding of excited-state dynamics in DNA. UV-B directly excites DNA and yields charge separated states (excitons) with charges delocalized over several base pairs.7−11 The subsequent charge recombination in a few hundred ps protects DNA from most UV damages.12 However, UV-B light induces also the formation of cyclobutane pyrimidine dimers (CPDs) as primary DNA photodamages4−6 formed by a nearly barrier-free and thus extremely fast reaction.13 CPDs are considered as one of the molecular origins of skin cancer.2,3,14 These damages occur by singlet photochemistry and locally at the site of excitation by light. UV-A light is able to excite stacked ensembles of several nucleotides in double-stranded DNA which leads to the formation of CPDs, too.15 This observation indicates that DNA is able to transfer excitation energy, which may cause long-range damages, in particular CPDs. In contrast to these well-studied singlet photochemical processes, triplet photochemistry in DNA is less well explored because it is difficult to excite triplet states in DNA selectively and precisely at defined sites. These DNA triplet states are accessible through photosensitization by organic chromophores.16,17 The triplet energy of these triplet donor chromophores has to be higher than that of the acceptors, i.e., the DNA bases. The triplet state energies (T1) of isolated nucleotide monophosphates range between ET = 310 kJ/mol (for TMP) and ET = 321 kJ/mol (for CMP).18 Not every influence by stacking and base-pairing has been studied so far, but it is clear from the literature that stacked Ts in DNA have a reduced triplet state of ET = 270 kJ/mol (Figure 1).19 However, it is also reasonable to assume that the triplet energy of Ts might depend on the type of adjacent stacked base. Recent theoretical molecular dynamics simulations revealed that the triplet energy of T might be decreased only to ET = 300 kJ/mol, due to polarization effects, when stacked Ts up to the inclusion of four nucleobases are regarded.20 These are the critical values: The T1 state of the DNA photosensitizer has to be higher to selectively induce triplet states on Ts in DNA. If these photosensitizers are incorporated as C-nucleosides into DNA, the site of triplet excitation is well-defined. We recently introduced benzophenones, methoxyacetophenone, and methoxyxanthone as triplet photosensitizers for DNA and evidenced that photodamage at remote sites in DNA occurs by DNA-mediated energy migration.21,22 However, we did not yet report on the synthesis of the methoxyxanthone C-nucleoside. Herein, we show the synthesis of four different photosensitizers as C-nucleosides 1–4 and their synthetic incorporation into DNA. This structural approach places the photosensitizers at defined positions inside the base stack of DNA and allows their selective excitation by light with different types of probing of the DNA photochemistry. The C-nucleosides 1–3 bear xanthone, thioxanthone, and 3-methoxyxanthone as n−π*-type and UV-A-absorbing photosensitizers, whereas the C-nucleoside 4 bears triphenylene as a π–π*-type and UV-B-absorbing photosensitizer. Moreover, they differ by their photochemistry, which is characterized by their singlet (ES) or triplet energy (ET), respectively. We demonstrate and compare the DNA photosensitization of these structurally and also photochemically different C-nucleosides by the CPD formation in ternary DNA architectures that allow quantifying this photodamage simply by PAGE or HPLC.
Figure 1.
Top: Structure of C-nucleosides 1–4, their singlet/triplet energies ES/ET in kJ/mol, and the triplet energies (ET) of the nucleoside monophosphates in comparison to the DNA photosensitizers xanthone (in 1), thioxanthone (in 2), and triphenylene (in 4). In contrast, the methoxyxanthone (in 3) shows only singlet photochemistry. Bottom: CPD formation in DNA requires a triplet energy ET higher than that of T in DNA (270 kJ/mol) and is studied in ternary photoactive DNA architectures. The designated site of CPD formation lacks the phosphodiester bond. Thus, the T–T dimerization links the two oligonucleotides which can be analyzed by PAGE or HPLC using the fluorescence of the 5′-terminal dye.
Results
Xanthone has a high T1 energy of ET = 310 kJ/mol,23 whereas thioxanthone has a lower one; the literature values range from 263 to 274 kJ/mol.23−27 Accordingly, C-nucleoside 1 should be clearly able to sensitize Ts, whereas C-nucleoside 2 is a borderline case, and it is not clear whether it could sensitize Ts or not.19 As structurally comparable, but a photochemically different singlet photosensitizer, we included C-nucleoside 3 with 3-methoxyxanthone, for which only a S1 energy of 332 kJ/mol is reported, but no triplet photochemistry.28 C-nucleoside 4 contains triphenylene with a T1 energy of ET = 279 kJ/mol23 as a different type of photosensitizer because triphenylene is excited by a π–π*-transition, whereas xanthones are excited by an n−π*-transition of their carbonyl groups. Thereby, we are able to compare how these different excitations and types of photochemistry influence the CPD formation in DNA. Although singlet and triplet energy levels of the photosensitizers might be affected by their stacking in the DNA, we assume that this is not significantly changing the photochemical scenario in our architectures.
All four C-nucleosides were synthesized in two central steps from the halogenated chromophore precursors. The first and key step is a Heck reaction (Scheme 1).29 The advantage of this type of C-nucleoside synthesis is the extremely high stereoselectivity to the β-anomers because the 3’-TBDMS group shields the downface of the glycal 6 and efficiently inhibits the formation of the α-anomer. The TBDMS-protected glycal 6 for these Heck reactions was obtained from TBDMS-protected thymidine by elimination of thymine as published.30 The brominated precursors 7 and 22 are commercially available, while precursor 12 had to be prepared according to a literature description.31 Precursor 17 was also prepared according to a different literature procedure.32 Based on our experience with benzophenone C-nucleosides,33 Pd(dppf)Cl2 was the best catalyst also for the central Heck-type coupling between the brominated chromophore precursors 12, 17, and 22 and the glycal 6. Yet, BrettPhos Pd G3 as the catalyst showed to be the most efficient for the coupling between precursor 7 and glycal 6. The yields lie in the range between 29% for Heck product 18 and 75% for 8, but it is important to keep in mind that this is a very straightforward approach to these C-nucleosides as pure β-anomers. The xanthones including their C-nucleosides are subject to sublimation, which complicates not only the workup and purification of products 8, 13, and 18 but also reduces their yields. The remaining TBDMS groups of the Heck products 8, 13, 18, and 23 were removed by treatment with Et3N·HF in quantitative yields. After this deprotection at their 3′- and 5′-positions, the resulting 3′-keto derivatives 9, 14, 19, and 24 were reduced with NaBH(OAc)3 as described in literature.34 This is another stereoselective reaction since the reducing agent NaBH(OAc)3 binds to the 5’-OH group that substitutes one of the OAc ligands at the boron, and the hydride attacks from the top to deliver the 3′-hydroxy functions in the desired ribo configuration. The C-nucleosides 1–4 were obtained in yields between 55 and 78%. For subsequent incorporation into oligonucleotide by solid-phase chemistry, the C-nucleosides 1–4 had to be converted into the corresponding phosphoramidites 11, 16, 21, and 26 by standard chemical methods. This includes first the protection of their 5′-groups by DMT-Cl in yields between 54 and 79% and subsequent phosphitylation of the remaining unprotected 3′-hydroxy groups of the products 10, 15, 20, and 25 in yields between 70 and 97%.
Scheme 1. Synthesis of C-Nucleosides 1–4 with the Photosensitizers (Ps) and Their Phosphoramidites 11, 16, 21, and 26 as DNA Building Blocks.
The architectures DNAX-0, DNAX-1, DNAX-2, and DNAX-3 were prepared by means of the corresponding phosphoramidites 11, 16, 21, and 26 to photochemically evaluate the sensitizing properties. These defined and ternary DNA architectures allow the site-selective photoexcitation and the site-selective formation of T–T dimers as CPD damages (Figure 2). The incorporated C-nucleoside (photosensitizer) X serves as the site of photoexcitation and energy injection. The sequences of the DNA architectures bear exactly one defined site where two adjacent Ts are located as designated sites for CPD formation. These adjacent Ts lack the phosphodiester bonds between them, and the T–T dimerization links the two oligonucleotides together. This linkage can be analyzed and quantified by PAGE or HPLC analysis because it changes both the electrophoretic mobility and the chromatographic retention time. For the analysis, the 5′-terminal oligonucleotides complementary to the template strand bearing X were marked by the atto550 dye (atto), whereas the third and 3′-terminal counterstrand does not need any marker. HPLC analysis in combination with LC–MS analysis was applied to verify and quantify the T–T dimerization. Additionally, PAGE analysis was performed according to previous studies with C-nucleosides and their potential to yield CPDs.21,22 The melting analysis of the DNA architectures DNAX-n revealed two melting temperatures, Tm1 in the range 11–15 °C and Tm2 in the range 43–49 °C (Table S4). The occurrence of two distinct melting temperatures proves the full hybridization of the ternary DNA architectures at temperatures below 10 °C. We assume that the presumably low amount of the dissociated 6-mer component at this temperature is similar in all irradiation experiments because the 6-mer oligonucleotide is the same in all DNA architectures. The CD spectra of representative DNA architectures (Figure S58) evidence the B-type helical conformation typical for double-stranded DNA.
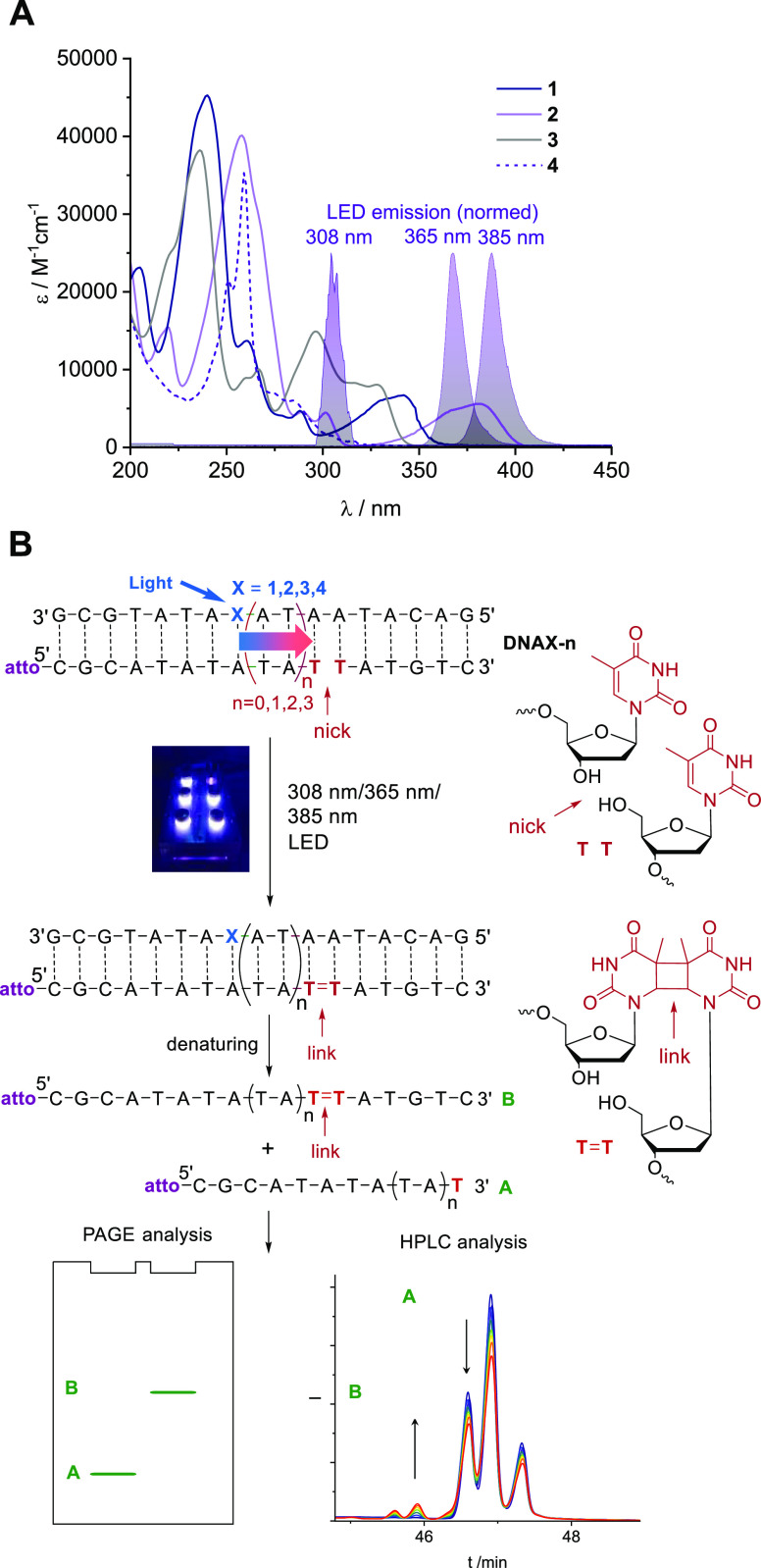
Figure 2.
(A) Absorption spectra of the nucleosides 1, 2, 3, and 4 (18.8, 15.0, 25.0, and 8.22 μM in MeCN) and corresponding normalized LED emission spectra. (B) DNA architectures DNAX-n (X = 1–4, n = 0–3) for the photochemical evaluation of the photosensitizers 1–4 by the yield of CPDs. The C-nucleosides 1–4 are excited by a 305 nm (UV-B), 369, or 385 nm (UV-A) LED. The counterstrand is marked at the 5′-terminus with atto550 (atto) and elongated by the T–T formation lacking the phosphodiester bonds between them. This extension is analyzed quantitatively by denaturing PAGE or HPLC.
The optimal wavelength to selectively excite the photosensitizer was determined by measuring the extinction coefficient as well as the absorption spectra of the nucleosides 1, 2, 3, and 4 (Figure 2). The irradiations were done (i) using a 369 nm LED (for DNA1-n and DNA3-n), a 385 nm LED (for DNA2-n), or a 305 nm LED (for DNA4-n), (ii) at a temperature of 10 °C to secure complete annealing of the ternary DNA architectures, and (iii) under an argon atmosphere to prevent oxidative DNA damages. All experiments were repeated at least three times. In the architectures DNAX-0, the T–T dimerization site is placed next to the photoexcitation site X allowing direct sensitization. In DNAX-1, DNAX-2, and DNAX-3, the T–T dimerization is separated by two, four, and six intervening A-T base pairs to evaluate if the photosensitizer X is able to induce remote T–T dimerization by energy transport through the DNA. It is assumed that the photobleaching of the atto550 dye occurs to the same extent in starting DNA material and in the CPD product because it occurs independently from the modification X and the CPD formation. Accordingly, the CPD yields were determined as ratios by means of the fluorescence readout (λem = 540 ± 10 nm) of the PAGE or the emission channel (λem = 554–576 nm) of the HPLC. Background reactions as negative control experiments were performed with the architectures DNAT-0 in which X has been replaced by T. Irradiations with the 365 and 385 nm LEDs gave neglectable amounts of background T–T dimers. The triphenylene photosensitizer in the architectures DNA4-n requires an irradiation at 305 nm, which, of course, induces a detectable background reaction because this wavelength is within the nucleic acid absorption range. Here, the control DNA hybrid DNAT-0 showed 13 ± 2% yield in CPDs over 6 h of irradiation (Figure S70). Samples were taken at distinct time intervals during each irradiation experiment to analyze the CPD formation in a time-dependent way.
DNA1-n showed the T–T dimerization by an additional peak in the HPLC analysis with a shorter retention time (Figures S85–S88). The CPD adducts were verified by LC-ESI-MS (Figures S98, S104, and S111). For DNA1-1, DNA1-2, and DNA1-3, the HPLC analysis showed a third peak with even lower retention time. According to LC–MS (Figure S110), we assign these bands interstrand crosslinking products (ICLs) that were photochemically induced by the xanthone C-nucleoside in yields between 12 and 18% after 6 h of irradiation (Figures S86–S88). ICLs are most likely formed by H abstraction at the 2′-deoxyribofuranoside in the counterstrand. Interstrand T–T dimerization is excluded as a possible origin of ICLs since we know from previous work with 5-methyl C vs T22 that the Ts are stacked at the nicked position and not very flexible. DNA1-0 reaches a maximum of 73% of CPDs after 1 h of irradiation (Figure 3), and a decrease of the CPD yields was observed during the irradiation longer than 1 h. A repair mechanism of the CPD damage via photoinduced charge transfer between the T–T dimer and the xanthone moiety makes this observation plausible.35−37 Oxidative38 and reductive39 cleavage of T–T dimers has been evidenced in DNA. With a reduction potential of Ered = 1.57 V (vs SCE) and a triplet energy of ET = 3.22 V, xanthone is a strong photoreductant24,40 and should be able to open CPDs oxidatively by light based on the estimated oxidation potential for T, Eox = 1.84 V (vs SCE)41 and for T–T dimers, Eox = 1.4–1.8 V (vs SCE).42 In case of DNA1-1, DNA1-2, and DNA1-3, respectively, the yield of the T–T dimer reaches a plateau after the irradiation over 2 h. We assume that is due to a stationary state between photoinduced CPD formation and CPD cleavage remotely over distances of 2–6 A-T pairs. Overall, an exponential distance dependence was observed for the series DNA1-n which is consistent with our previous results for a Dexter-type triplet energy transport with a typical exponential dependence on the distance r.21 This is best described by the parameter β according to the fitting function y = constant·exp(−β·r). The β-value for DNA1-n is 0.05 Å–1 and even lower than the β-values observed for energy transport induced by benzophenones and methoxyacetophenone (β = 0.13–0.34 Å–1). Such a shallow distance dependence indicates an energy hopping process based on the similarly low values experimentally obtained for DNA-mediated electron transport by hopping in the late 1990s and early 2000s.43 While the architectures DNA1-n showed CPD formation over time, DNA2-n showed only ICL formation ranging from 9 to 17% yield and no CPD formation. This is due to the low triplet energy of thioxanthone (ET = 263–274 kJ/mol) which is neither sufficient to directly sensitize the T–T dimer formation in DNA2-0 nor the DNA-mediated energy transport in DNA2-1, DNA2-2, and DNA2-3.23−27 There is no distance dependence of the observed photoinduced product in the series DNA2-n, and the yields stay in the range of 9–17%, which additionally indicates that these are not the CPDs but the ICL products. LC–MS analysis could confirm this (Figure S121). The time-dependent product analysis of the irradiated DNA3-n (365 nm LED) and DNA4-n (305 nm LED) by PAGE reveals the formation of the T–T dimer by an additional gel band with slower mobility. The irradiation of the architectures in the series DNA3-n showed a slow and steady increase in CPD formation over time. In contrast to DNA1-n, no plateau of the CPD yield was reached even after 6 h of irradiation. This is due to the fact that 3-methoxyxanthone induces the CPD formation by singlet photochemistry. This gives a rather shallow dependence of the CPD yields over the small distances of 2–6 A-T base pairs according to the sigmoidal distance dependency shown (with R0 = 25 Å equal to 7.4 base pairs) from our previous work, which again is the result of the singlet photochemistry in these DNA architectures.22 The CPD yields obtained in the architectures in the series DNA4-n had to be corrected by the background reaction due to the irradiation wavelengths in the UV-B range (305 nm LED), as already mentioned above, which increases the experimental error. A detectable amount of CPD above the error was observed only in case of DNA4-0 but not in DNA4-1 and DNA4-2. Photosensitization was only successful in DNA4-0, in which the photosensitizer is located directly next to the T–T dimerization site. Obviously, triphenylene is not able to induce an energy transport through the DNA to yield CPDs remotely in DNA4-1 and DNA4-2, which might be due to the π–π*-type excitation of this chromophore, because the triplet energy is sufficiently high (ET = 279 kJ/mol).23 We did not prepare DNA4-3 because we did not expect any damage over the distance of 6 A-T pairs.
Figure 3.
Time-dependent yields of CPDs and ICLs for up to 6 h of irradiation (left, A–D) and distance dependence after 6 h of irradiations (right, a–d) for n = 0 to n = 3 (6 A-T-pairs); A/a: DNA1-n showing the exponential distance dependence; B/b: DNA2-n showing only the distant-independent formation of ICLs; C/c: DNA3-n showing the sigmoidal distance dependence published recently;22 D/d: DNA4-n showing only the direct CPDs in DNA4-0 but no remote CPD formation in the other DNA architectures.
Discussion
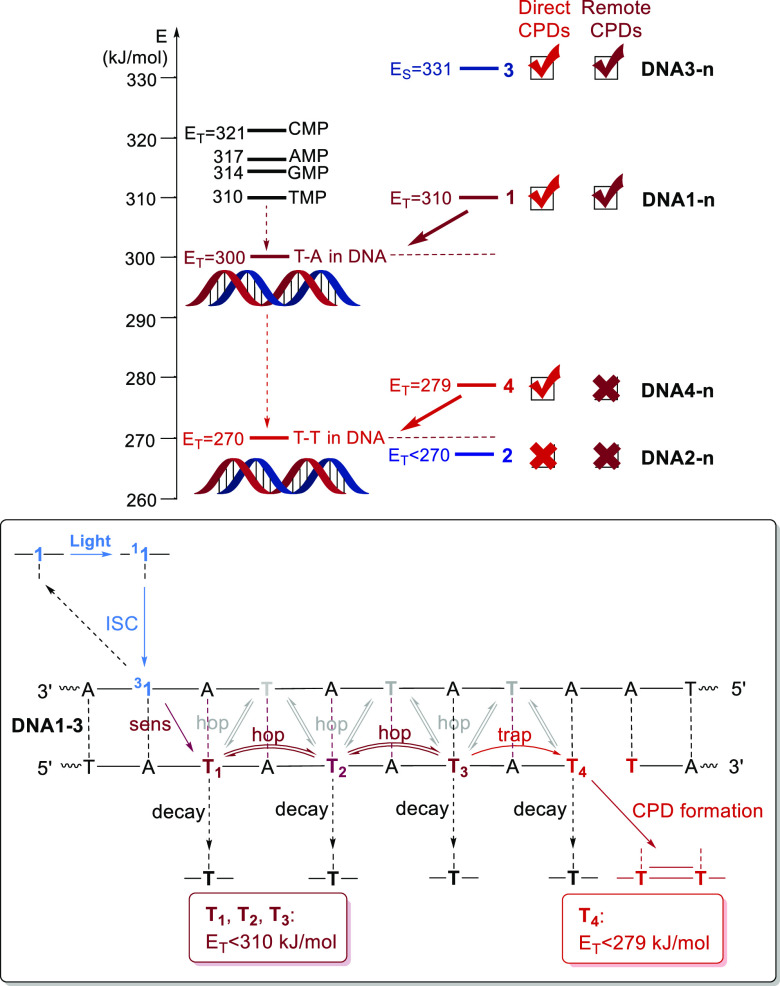
The comparison of the four different photosensitizers 1–4 in the corresponding DNA architectures DNAX-n reveals important mechanistic insights about the photochemical pathways to CPDs as typical DNA photodamage. As discussed recently,22 3-methoxyxanthone in DNA3-n is a special case. Although this photosensitizer is able to induce both direct and remote CPD formation, it shows no characteristic triplet photochemistry based on the shallow distance dependence for remote CPD yields in the range DNA3-1, DNA3-2, and DNA3-3. These results together with the synthesis of the C-nucleoside 3 (which was not yet published) complement the results for the other photoactive DNA architectures, where the triplet energy of the photosensitizer is the most critical value for CPD formation and for energy transport. In DNA1-n, DNA2-n, and DNA4-n, the triplet donors xanthone, thioxanthone, and triphenylene, respectively, were used to probe the triplet photochemistry of DNA. In double-stranded DNA, the triplet state energies of the isolated nucleotide monophosphates (in the range of ET = 310–321 kJ/mol) are significantly influenced mainly by stacking interactions. The lowest value of ET = 270 kJ/mol for Ts in DNA was determined by Miranda et al. using the experimentally but indirectly determined CPDs yields with different photosensitizers. It is clear that such a low triplet energy makes Ts to preferred energy traps resulting in the chemical formation of CPDs. However, this value is under debate, and theoretical molecular dynamics simulations revealed that the triplet energy of T might be decreased only to ET = 300 kJ/mol.20 It is reasonable to assume that the local triplet energy of Ts inside the DNA base stack depends on which type of DNA bases are stacked on the 3′- and 5′-side of the T. Our experiments support this assumption and give a more precise scenario by probing the different triplet energy levels in DNA (Figure 4). The triplet energy of xanthone (ET = 310 kJ/mol23), but more importantly of triphenylene (ET = 279 kJ/mol23), is higher than ET = 270 kJ/mol and thus able to photoinduce the direct CPD formation in the architectures DNA1-0 and DNA4-0, respectively, in which the photosensitizer was placed next to the T–T dimerization site. In contrast, thioxanthone is not even able to photoinduce direct CPD formation in DNA2-0 because the triplet energy is obviously too low (ET = 263–274 kJ/mol). Taken together, these results confirm the Miranda value of ET = 270 kJ/mol in DNA. It is also important to note here that direct CPD formation can be induced by both n−π* excitation in DNA1-0 and π–π* excitation in DNA4-0. When it comes to the photoinduction of the DNA-mediated energy transport to form remote CPDs over the distance of 2–6 AT-pairs, the results differ. Only xanthone as the photosensitizer in the architectures DNA1-1, DNA1-2, and DNA1-3 is able to induce the remote CPD formation, and the distance dependence of the CPD yields drops exponentially. We previously observed such exponential distance dependencies with benzophenones and acetophenones in DNA over distances of up to 37 Å (10 A-T pairs).21 Blancafort and Voityuk et al.44 suggested that the energy is first injected into the next T (interstrand), and the experimentally observed exponential distance dependence can be best explained by a stepwise intrastrand energy hopping between the Ts via the alternating A-T sequence between energy donor (X) and energy acceptor (T T dimerization site). The T–T coupling for the interstrand energy hopping that would involve every T in the A-T pairs is too low. Accordingly, we propose an energy transport by intrastrand hopping from T to T until the energy is trapped by the CPD formation at the T T site. In such a mechanistic scenario, the triplet energy of the Ts in the alternating A-T sequence of the bridge between the donor and acceptor must be significantly higher than 270 kJ/mol, possibly near the calculated value of ET = 300 kJ/mol,20 because only xanthone (ET = 310 kJ/mol23) in DNA1-n is able to induce the hopping process. Finally, the energy is trapped at the T–T dimerization site because the local triplet energy there is lower (ET < 279 kJ/mol). An alternative mechanism could be that all Ts have the same approximate ET, and only the final T has one additional decay mode, the dimerization, that is not available to the other Ts in the sequence. However, the results with the DNA architectures DNA4-n contradict this possible explanation if we assume that the type of transition, π–π* vs n−π*, does not influence the result. The ET of triphenylene is higher than 270 kJ/mol; accordingly, CPDs are formed in DNA4-0, but there is no remote photodamage observed in the architectures DNA4-1 and DNA4-2 which would be observed if all Ts have the same ET.
Figure 4.
Mechanistic scenario for direct and remote CPD formation in the photoactive DNA architectures with local triplet energies for Ts: Left: Xanthone in DNA1-0 and triphenylene in DNA4-0, but not thioxanthone in DNA2-0, are able to photosensitize direct CPD formation. Thus, the triplet energy of the T–T dimerization site in DNA must be lower than 279 kJ/mol. Right: Only xanthone is able to induce the triplet energy transport and remote CPD formation in DNA1-n, and it is proposed that the Ts in the A-T base pairs serve as hopping steps for the triplet energy. Thus, the triplet energy of the alternating A-T sequence must be lower than 310 kJ/mol, but higher than 279 kJ/mol. 3-Methoxyxanthone in DNA3-n induces singlet photochemistry which forms also remote CPDs.
Conclusions
The DNA photochemistry was probed by four photochemically different C-nucleosides as sensitizers of CPDs. The synthesis of four different photosensitizers as C-nucleosides 1–4 has been achieved by stereoselective Heck couplings to the TBDMS-protected glycal 6 and subsequent stereoselective reduction to regain the 2′-deoxyribofuranoside configuration. The C-nucleosides 1–3 bear xanthone, thioxanthone, and 3-methoxyxanthone as n−π*-type and UV-A absorbing photosensitizers, whereas the C-nucleoside 4 bears triphenylene as a π–π*-type and UV-B-absorbing photosensitizer. The synthesis of the phosphoramidites 11, 16, 21, and 26 allowed their synthetic incorporation into the photoactive architectures DNAX-n, with X being the type of photosensitizer 1, 2, 3, and 4, and n counting the number of intervening A-T pairs, 0 (none), 1 (2 A-T), 2 (4 A-T), and 3 (6 A-T). Thus, in these ternary DNA architectures, the photosensitizer X as the energy donor is placed at defined positions in the sequence and inside the base stack of double-stranded DNA and allows site-selective excitation by light. We assume that the stacking interactions in the DNA do not alter the singlet and triplet energies of the photosensitizers 1–4 to a significant extent. The sequences of the two counterstrands to the photosensitizer-modified strand in the DNA architectures bear a single site in the sequence where two adjacent Ts are located as designated sites for CPD formation. These adjacent Ts lack the phosphodiester bonds between them. Therefore, the T–T dimerization links the two oligonucleotides together which can be analyzed and quantified by PAGE or HPLC analysis using the emission of a fluorescent 5′-terminal atto550 marker. Only xanthone and triphenylene, but not thioxanthone, are able to photosensitize direct (adjacent) CPD formation. Based on the literature values for the triplet energies for these photosensitizers, the triplet energy of the T–T dimerization site in DNA must be lower than 274 kJ/mol. Moreover, only xanthone is able to induce the triplet energy transport and remote CPD formation over up to six intervening A-T base pairs. The yields of remote CPDs drop exponentially with increasing distance in the xanthone-modified DNA architectures. Thus, it is proposed that the Ts in the A-T base pairs serve as hopping steps for the triplet energy. Again, based on the literature values for the triplet energies for the xanthone, the triplet energy of the T in the alternating A-T sequence must be lower than 310 kJ/mol, but higher than 279 kJ/mol. 3-Methoxyxanthone is able to induce both direct and remote CPD formation but shows no characteristic triplet photochemistry based on the shallow distance dependence for remote CPD yields. Overall, we were able to obtain experimentally precise information on the sequence dependence of the DNA triplet photochemistry. The photoinduced DNA-mediated energy transport to remote sites in our photoactive DNA architectures will add an important extension to our understanding of DNA photodamaging and mutations.
Methods
Materials
All chemicals were purchased from Abovchem, Aldrich, Alfa Aesar, ABCR, Fluka, TCI. The unmodified and Atto550-modified DNA were purchased from metabion. Thin layer chromatography was performed on a Fluka silica gel 60 F254-coated aluminum foil. Flash chromatography was carried out on silica gel 60 from Aldrich (43–60 μm).
NMR Spectroscopy
1H NMR (400 MHz), 13C NMR (101 MHz), and 31P NMR spectra (162 MHz) were measured on a Bruker Advance DRX 500. 1H NMR (500 MHz), 13C NMR (126 MHz), and 31P NMR spectra (202 MHz) were measured on a Bruker Advance 500. The chemical shifts in the 1H and 13C NMR spectra are reported in parts per million (ppm) relative to tetramethylsilane as an internal standard. The chemical shifts in the 31P NMR spectra are also reported in ppm relative to the deuterated solvent. The coupling constant (J) is given in Hertz (Hz), and the multiplicity of signals are reported as follows: s (singlet), d (doublet), t (triplet), q (quadruplet), quint (quintet), sext (sextet), m (multiplet), br. s (broad singlet), dt (doublet of triplets), and td (triplet of doublets).
Mass Spectrometry
Mass spectrometry was performed on a Finnigan MAT 95 with FAB and EI as ionization methods. ESI mass spectrometry was performed on a Thermo Fisher Scientific Q Exactive (Orbitrap). Oligonucleotides were identified by MALDI mass spectrometry on an AXIMA Confidence spectrometer from Shimadzu.
DNA Synthesis and Purification
Reagents and CPG (1 μmol) were purchased from ABI and GlenResearch. The oligonucleotides were synthesized under an argon atmosphere on an H-6 DNA/RNA synthesizer from K&A LABORGERÄTE. For the incorporation of the xanthone-, thioxanthone-, 3-methoxyxanthone-, and triphenylene-phosphoramidite, the coupling time was increased (Table S1). After preparation, the trityl-off oligonucleotide was cleaved from the resin and deprotected by incubation with conc. NH4OH at 55 °C for 16 h. The oligonucleotide was dried and purified by high-performance liquid chromatography (HPLC) at a Thermo Scientific Dionex UltiMate on a RP-C18 column using the following conditions: A = NH4OAc buffer (50 mM; pH = 7.0); B = acetonitrile, flow rate 2.5 mL/min, UV/vis detection at 260, 280 and 340 nm. Xanthone- and thioxanthon-modified DNA strands were prepared as trityl-on oligonucleotide and were pre-purified using Glen-Pak DNA Purification Cartridges prior to the HPLC purification. The oligonucleotides were lyophilized, and the concentration was determined by their absorbance at 260 nm on a Nanodrop ND-1000 spectrophotometer.
Optical Spectroscopy
Absorption spectra and melting temperatures (2.5 μM DNA, 250 mM NaCl, 10 mM Na-Pi buffer, 10–90 °C, 0.7 °C/min, step width 0.5 °C) were recorded with a Cary 100 Bio Varian UV/vis spectrometer and a Cary.3500 UV/vis Multicell Peltier spectrophotometer.
Irradiation Experiments and Analysis
Irradiation experiments were performed with two Nichia UVA-LEDs (NCSU033B) at 365, 385, and 305 nm under the following conditions: 2.50 μM DNA, 10 mM Na-Pi buffer, 250 mM NaCl in ddH2O (1 mL) at 10 °C after degassing under an argon atmosphere. High-resolution polyacrylamid gels (PAGE) were carried out with a Sequi-Gen DT Sequencing Cell (21 × 40 cm) with a PowerPac HV by Bio-Rad (12.5% Rotiporese gel, T = 50 °C, t = 1.5 h, U = 3000 V, P = 50 W). Fluorescence was measured with a Stella 8300 Raytest spectrofluorometer containing LEDs radiating with a wavelength of 540 ± 10 nm. HPLC analysis was performed at Thermo Scientific Dionex UltiMate on a RP VDSpher OptiBio PUR 300 C18-SE 250 × 4.6 mm column using the following conditions (Table S2). Solution A = NH4OAc buffer (50 mM; pH = 6.5) and B = acetonitrile.
Acknowledgments
General financial support by KIT is gratefully acknowledged.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.3c00167.
Synthesis of 5–24, images of 1H, 13C, 31P NMR and ESI-MS spectra, DNA analyses and melting temperatures, images of MS and HPLC analyses of the DNA, gel images and data, HPLC images and data, and images of LC–MS (PDF)
Author Contributions
S.H. synthesized and investigated the DNA modified with 1 and 2, A.K. the DNA modified with 3, and M.S. the DNA modified with 4. H.-A.W. supervised the research and wrote major parts of the manuscript. CRediT: Sebastian Häcker investigation, writing-original draft; Maren Schrödter investigation, methodology, writing-review & editing; Arthur Kuhlmann investigation, methodology; Hans-Achim Wagenknecht funding acquisition, project administration, resources, supervision, writing-review & editing.
Deutsche Forschungsgemeinschaft (grant Wa-1386/16-2).
The authors declare no competing financial interest.
Supplementary Material
References
- Pfeifer G. P.; Besaratinia A. UV wavelength-dependent DNA damage and human non-melanoma and melanoma skin cancer. Photochem. Photobiol. Sci. 2012, 11, 90–97. 10.1039/c1pp05144j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J.; Douki T. Formation of UV-induced DNA damaghe contributing to skin cancer development. Photochem. Photobiol. Sci. 2018, 17, 1816–1841. 10.1039/c7pp00395a. [DOI] [PubMed] [Google Scholar]
- Taylor J.-S. Unraveling the Molecular Pathway from Sunlight to Skin Cancer. Acc. Chem. Res. 1994, 27, 76–82. 10.1021/ar00039a003. [DOI] [Google Scholar]
- Heil K.; Pearson D.; Carell T. Chemical investigation of light induced DNA bipyrimidine damage and repair. Chem. Soc. Rev. 2011, 40, 4271–4278. 10.1039/C000407N. [DOI] [PubMed] [Google Scholar]
- Sage E.; Girard P.-M.; Francesconi S. Unravelling UVA-induced mutagenesis. Photochem. Photobiol. Sci. 2012, 11, 74–80. 10.1039/c1pp05219e. [DOI] [PubMed] [Google Scholar]
- Cadet J.; Grand A.; Douki T. Solar UV Radiation-Induced DNA Bipyrimidine Photoproducts: Formation and Mechanistic Insights. Top. Curr. Chem. 2014, 356, 249–276. 10.1007/128_2014_553. [DOI] [PubMed] [Google Scholar]
- Gustavsson T.; Markovitsi D. Fundamentals of the Intrinsic DNA Fluorescence. Acc. Chem. Res. 2021, 54, 1226–1235. 10.1021/acs.accounts.0c00603. [DOI] [PubMed] [Google Scholar]
- Schwalb N. K.; Temps F. Base Sequence and Higher-Order Structure Induce the Complex Excited-State Dynamics in DNA. Science 2008, 322, 243–245. 10.1126/science.1161651. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Harpe K. d. L.; Beckstead A. A.; Improta R.; Kohler B. UV-Induced Proton Transfer between DNA Strands. J. Am. Chem. Soc. 2015, 137, 7059–7062. 10.1021/jacs.5b03914. [DOI] [PubMed] [Google Scholar]
- Bucher D. B.; Pilles B. M.; Carell T.; Zinth W. Charge separation and charge delocalization identified in long-lived states of photoexcited DNA. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 4369–4374. 10.1073/pnas.1323700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchvarov I.; Wang Q.; Raytchev M.; Trifonov A.; Fiebig T. Electronic energy delocalization and dissipation in single- and double-stranded DNA. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 4794–4797. 10.1073/pnas.0606757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton C. T.; Harpe K. d. L.; Su C.; Law Y. K.; Crespo-Hernandez C. E.; Kohler B. DNA Excited-State Dynamics: From Single Bases to the Double Helix. Annu. Rev. Phys. Chem. 2009, 60, 217–239. 10.1146/annurev.physchem.59.032607.093719. [DOI] [PubMed] [Google Scholar]
- Schreier W. J.; Schrader T. E.; Koller F. O.; Gilch P.; Crespo-Hernandez C. E.; Swaminathan V. N.; Carell T.; Zinth W.; Kohler B. Thymine Dimerization in DNA Is an Ultrafast Photoreaction. Science 2007, 315, 625–629. 10.1126/science.1135428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouret S.; Baudouin C.; Charveron M.; Favier A.; Cadet J.; Douki T. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 13765–13770. 10.1073/pnas.0604213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouret S.; Philippe C.; Gracia-Chantegrel J.; Banyasz A.; Karpati S.; Markovitsi D.; Douki T. UVA-Induced cyclobutane pyrimidine dimers in DNA: a direct photochemical mechanism. Org. Biomol. Chem. 2010, 8, 1706–1711. 10.1039/b924712b. [DOI] [PubMed] [Google Scholar]
- Cadet J.; Douki T.; Ravanat J.-L.; Mascio P. D. Sensitized formation of oxidatively generated damage to cellular DNA by UVA radiation. Photochem. Photobiol. Sci. 2009, 8, 903–911. 10.1039/b905343n. [DOI] [PubMed] [Google Scholar]
- Cuquerella M. C.; Lhiaubet-Vallet V.; Cadet J.; Miranda M. A. Benzophenone Photosensitized DNA Damage. Acc. Chem. Res. 2012, 45, 1558–1570. 10.1021/ar300054e. [DOI] [PubMed] [Google Scholar]
- Wood P. D.; Redmond R. W. Triplet State Interactions between Nucleic Acid Bases in Solution at Room Temperature: Intermolecular Energy and Electron Transfer. J. Am. Chem. Soc. 1996, 118, 4256–4263. 10.1021/ja954340+. [DOI] [Google Scholar]
- Bosca F.; Lhiaubet-Vallet V.; Cuquerella M. C.; Castell J. V.; Miranda M. A. The Triplet Energy of Thymine in DNA. J. Am. Chem. Soc. 2006, 128, 6318–6319. 10.1021/ja060651g. [DOI] [PubMed] [Google Scholar]
- Allahkaram L.; Monari A.; Dumont E. The Behaviour of Triplet Thymine in a Model B-DNA Strand. Energetics and Spin Density Localization Revealed by ab initio Molecular Dynamics Simulations. Photochem. Photobiol. 2022, 98, 633–639. 10.1111/php.13549. [DOI] [PubMed] [Google Scholar]
- Antusch L.; Gaß N.; Wagenknecht H.-A. Elucidation of the Dexter-Type Energy Transfer in DNA by Thymine-Thymine Dimer Formation Using Photosensitizers as Artificial Nucleosides. Angew. Chem., Int. Ed. 2017, 56, 1385–1389. 10.1002/anie.201610065. [DOI] [PubMed] [Google Scholar]
- Kuhlmann A.; Bihr L.; Wagenknecht H.-A. How Far Does Energy Migrate in DNA and Cause Damage? Evidence for Long-Range Photodamage to DNA. Angew. Chem., Int. Ed. 2020, 59, 17378–17382. 10.1002/anie.202009216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkstroeter W. G.; Lamola A. A.; Hammond G. S. Mechanisms of Photochemical Reactions in Solution. XXVIII. Values of Triplet Excitation Energy of Selected Sensitizers. J. Am. Chem. Soc. 1964, 86, 4537–4540. 10.1021/ja01075a005. [DOI] [Google Scholar]
- Nikitas N. F.; Gkizis P. L.; Kokotos C. G. Thioxanthone: a powerful photocatalyst for organic reactions. Org. Biomol. Chem. 2021, 19, 5237–5253. 10.1039/D1OB00221J. [DOI] [PubMed] [Google Scholar]
- Meier K.; Zweifel H. Thioxanthone ester derivatives: efficient triplet sensitizers for photopolymer applications. J. Photochem. 1986, 35, 353–366. 10.1016/0047-2670(86)87066-6. [DOI] [Google Scholar]
- Elliott L. D.; Kayal S.; George M. W.; Booker-Milburn K. Rational design of triplet sensitizers for the transfer of excited state photochemistry from UV to visible. J. Am. Chem. Soc. 2020, 142, 14947–14956. 10.1021/jacs.0c05069. [DOI] [PubMed] [Google Scholar]
- Armesto D.; Ortiz M. J.; Agarrabeitia A. R.; El-Boulifi N. The Effects of Triplet Sensitizers’ Energies on the Photoreactivity of β, γ-Unsaturated Methyl Ketones. Angew. Chem., Int. Ed. 2005, 117, 7917–7919. 10.1002/ange.200502619. [DOI] [PubMed] [Google Scholar]
- Wolfbeis O. S.; Fuerlinger E. pH-dependent fluorescence spectroscopy. 15. Detection of an unusual excited-state species of 3-hydroxyxanthone. J. Am. Chem. Soc. 1982, 104, 4069–4072. 10.1021/ja00379a005. [DOI] [Google Scholar]
- Doyle Daves G. Jr. C-Glycoside Synthesis by Palladium-Mediated Glycal-Aglycon Coupling Reactions. Acc. Chem. Res. 1990, 23, 201–206. 10.1021/ar00174a006. [DOI] [Google Scholar]
- Cameron M. A.; Cush S. B.; Hammer R. P. Facile Preparation of Protected Furanoid Glycals from Thymidine. J. Org. Chem. 1997, 62, 9065–9069. 10.1021/jo970947s. [DOI] [Google Scholar]
- Zhang L.; Zhang J. Y. Microwave-Assisted Boron Trichloride Mediated Acylation of Phenols-Synthesis of (o-Hydroxyaryl)(Aryl)methanones and Xanthones. J. Comb. Chem. 2006, 8, 361–367. 10.1021/cc0501007. [DOI] [PubMed] [Google Scholar]
- Lumbomir V.; Foster H. M.. Process for preparing thioxanthones. US4101558A, 1978.
- Gaß N.; Wagenknecht H.-A. Synthesis of Benzophenone Nucleosides and Their Photocatalytic Evaluation for [2+2] Cycloaddition in Aqeous Media. Eur. J. Org. Chem. 2015, 2015, 6661–6668. 10.1002/ejoc.201500885. [DOI] [Google Scholar]
- Joubert N.; Pohl R.; Klepetárová B.; Hocek M. Modular and Practical Synthesis of 6-Substituted Pyridin-3-yl C-Nucleosides. J. Org. Chem. 2007, 72, 6797–6805. 10.1021/jo0709504. [DOI] [PubMed] [Google Scholar]
- Bucher D. B.; Kufner C. L.; Schlueter A.; Carell T.; Zinth W. UV-Induced Charge Transfer States in DNA Promote Sequence Selective Self-Repair. J. Am. Chem. Soc. 2016, 138, 186–190. 10.1021/jacs.5b09753. [DOI] [PubMed] [Google Scholar]
- Holman M. R.; Ito T.; Rokita S. E. Self-Repair of Thymine Dimer in Duplex DNA. J. Am. Chem. Soc. 2007, 129, 6–7. 10.1021/ja0668365. [DOI] [PubMed] [Google Scholar]
- Szabla R.; Kruse H.; Stadlbauer P.; Šponer J.; Sobolewski A. L. Sequential electron transfer governs the UV-induced self-repair of DNA photolesions. Chem. Sci. 2018, 9, 3131–3140. 10.1039/C8SC00024G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandliker P. J.; Holmlin R. E.; Barton J. K. Oxidative Thymine Dimer Repair in the DNA Helix. Science 1997, 275, 1465–1468. 10.1126/science.275.5305.1465. [DOI] [PubMed] [Google Scholar]
- Schwögler A.; Burgdorf L. T.; Carell T. Self-Repairing DNA Based on a Reductive Electron Transfer through the Base Stack. Angew. Chem., Int. Ed. 2000, 39, 3918–3920. . [DOI] [PubMed] [Google Scholar]
- Dantas J. A.; Correia J. T. M.; Paixao M. W.; Correa A. G. Photochemistry of Carbonyl Compounds: Application in Metal-Free Reactions. ChemPhotoChem 2019, 3, 506–520. 10.1002/cptc.201900044. [DOI] [Google Scholar]
- Seidel C. A. M.; Schulz A.; Sauer M. H. M. Nucleobase-Specific Quenching of Fluorescent Dyes. 1. Nucleobase One-Electron Redox Potentials and Their Correlation with Static and Dynamic Quenching Efficiencies. J. Phys. Chem. 1996, 100, 5541–5553. 10.1021/jp951507c. [DOI] [Google Scholar]
- Vicic D. A.; Odom D. T.; Nunez M. E.; Gianolio D. A.; McLaughlin L. W.; Barton J. K. Oxidative Repair of a Thymine Dimer in DNA from a Distance by a Covalently Linked Organic Intercalator. J. Am. Chem. Soc. 2000, 122, 8603–8611. 10.1021/ja000280i. [DOI] [Google Scholar]
- Wagenknecht H.-A. Electron Transfer Processes in DNA: Mechanisms, Biological Relevance and Applications in DNA Analytics. Nat. Prod. Rep. 2006, 23, 973–1006. 10.1039/B504754B. [DOI] [PubMed] [Google Scholar]
- Blancafort L.; Voityuk A. A. Thermally induced hopping model for long-range triplet excitation energy transfer in DNA. Phys. Chem. Chem. Phys. 2018, 20, 4997–5000. 10.1039/C7CP07811K. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.