Abstract
Arginase of the Helicobacter pylori urea cycle hydrolyzes l-arginine to l-ornithine and urea. H. pylori urease hydrolyzes urea to carbon dioxide and ammonium, which neutralizes acid. Both enzymes are involved in H. pylori nitrogen metabolism. The roles of arginase in the physiology of H. pylori were investigated in vitro and in vivo, since arginase in H. pylori is metabolically upstream of urease and urease is known to be required for colonization of animal models by the bacterium. The H. pylori gene hp1399, which is orthologous to the Bacillus subtilis rocF gene encoding arginase, was cloned, and isogenic allelic exchange mutants of three H. pylori strains were made by using two different constructs: 236-2 and rocF::aphA3. In contrast to wild-type (WT) strains, all rocF mutants were devoid of arginase activity and had diminished serine dehydratase activity, an enzyme activity which generates ammonium. Compared with WT strain 26695 of H. pylori, the rocF::aphA3 mutant was ∼1,000-fold more sensitive to acid exposure. The acid sensitivity of the rocF::aphA3 mutant was not reversed by the addition of l-arginine, in contrast to the WT, and yielded a ∼10,000-fold difference in viability. Urease activity was similar in both strains and both survived acid exposure equally well when exogenous urea was added, indicating that rocF is not required for urease activity in vitro. Finally, H. pylori mouse-adapted strain SS1 and the 236-2 rocF isogenic mutant colonized mice equally well: 8 of 9 versus 9 of 11 mice, respectively. However, the rocF::aphA3 mutant of strain SS1 had moderately reduced colonization (4 of 10 mice). The geometric mean levels of H. pylori recovered from these mice (in log10 CFU) were 6.1, 5.5, and 4.1, respectively. Thus, H. pylori rocF is required for arginase activity and is crucial for acid protection in vitro but is not essential for in vivo colonization of mice or for urease activity.
Helicobacter pylori causes gastritis (2, 14, 35), is strongly associated with the development of peptic ulcers (3, 16), and constitutes a risk factor for gastric adenocarcinoma (9, 27). Although the mechanisms behind the development of these diseases are not well understood, urease, which catalyzes the hydrolysis of urea to carbon dioxide and ammonium, is clearly central to the pathogenesis of H. pylori infection since urease is absolutely essential for colonization of a variety of animal models (4, 5; for a review, see references 19, 24, and 25).
In situ studies have shown that H. pylori synthesizes urea and ornithine from the catabolism of arginine (23). This reaction is catalyzed by arginase, an enzyme of the H. pylori urea cycle (21). Thus, in the metabolism of nitrogenous end products in H. pylori, arginase activity is upstream of urease (21). H. pylori may also obtain some of its urea requirements through arginase activity of the host, since some mammalian cells have a complete urea cycle (21, 38). H. pylori does not have the enzymes for arginine biosynthesis and is therefore dependent on host-derived arginine (1, 20, 21, 26, 29, 34). It is thus possible that urease, arginine, arginase, and the other enzymes of the urea cycle play a fundamental role in the release and assimilation of ammonium, thereby contributing to maintaining the nitrogen balance in H. pylori (21, 23). Indeed, most arginases play a crucial biological role in regulating cytosolic arginine and ornithine levels, which are required for numerous metabolic processes, such as protein synthesis and production of polyamines and nitric oxide (1a).
H. pylori is an acid-sensitive organism but is protected from acid (pH <4.0) by the ammonia released from urea hydrolysis due to urease activity (3a, 10, 15, 31, 33). Thus, H. pylori exposed to acid in vitro survive in the presence of urea; survival in the presence of other metabolites has not been reported. H. pylori is also able to generate ammonium via the actions of other enzymes, including an aliphatic amidase (32), and various amino acid deaminases, such as asparaginase, aspartase, glutaminase, and serine dehydratase (20). The latter four enzymes provide the bacterium with intracellular ammonium derived from the catabolism of the respective amino acids. It is currently unclear what role, if any, these ammonium-generating enzymes play in the protection of H. pylori from acid exposure.
Partially purified H. pylori arginase is a homo-oligomer (100 to 300 kDa) consisting of ∼37-kDa subunits, each of which is predicted to contain two metal ions in the active site (23). H. pylori arginase is unique among the arginase enzyme family in that it has highest activity for cobalt rather than manganese, and the enzyme is associated with the cell wall fraction rather than the cytosol (23). H. pylori arginase has very high specificity for arginine and does not recognize the structurally similar compound agmatine. Previously, it was hypothesized that an open reading frame, hp1399, of H. pylori 26695 encoded the arginase since the deduced amino acid sequence of this protein showed 20 to 27% identity with other bacterial arginases, with greatest homology (27.1%) to Bacillus subtilis RocF (1, 8, 23, 34). In accordance with the B. subtilis gene encoding arginase, the putative arginase gene hp1399 of H. pylori was referred to as rocF (1, 34). However, there are no reports confirming that hp1399 actually encodes the H. pylori arginase, and it is not known whether H. pylori rocF is required either in vitro for growth or, given its close metabolic relationship with urease, in vivo for the colonization of mice.
To establish that rocF encodes arginase, the gene was cloned and rocF isogenic mutants of H. pylori were constructed by allelic exchange. To provide unambiguous functional data on H. pylori RocF, in vitro and in vivo studies were performed by using rocF mutants in which the rocF gene was either inactivated by gene disruption alone or, alternatively, by both gene disruption and deletion. The resulting rocF mutants and corresponding wild-type (WT) strains of H. pylori were tested for arginase activity by employing proton nuclear magnetic resonance (1H-NMR) spectroscopy. Since arginase may be metabolically coupled with other ammonium-generating enzymes, including urease, the WT and rocF mutants of H. pylori were also tested for urease and deaminase activities. Finally, the effects of rocF gene inactivation on the survival of H. pylori to acid exposure in vitro and on gastric colonization in the mouse model (7, 13) were investigated. The mouse model for H. pylori mimics the human disease in inflammation, in colonization location (i.e., antrum of the stomach), and in the degree of colonization.
MATERIALS AND METHODS
Bacterial strains, growth conditions, primers, and plasmids.
H. pylori strains 26695, N6, and SS1 were grown on brucella agar with 10% (vol/vol) sheep defibrinated blood or on blood agar base number 1 or 2 with 10% (vol/vol) horse blood in a microaerobic environment for 2 to 3 days by using the CampyPak Plus system (Becton Dickinson). Kanamycin (20 to 30 μg/ml) was added to the growth medium as appropriate. Escherichia coli strains were grown on Luria (L) agar and in L broth plus appropriate antibiotics (ampicillin, 100 μg/ml; kanamycin, 50 μg/ml). For growth of H. pylori in broth, bacteria were inoculated to a starting A600 of ∼0.05 in 100 ml of Mueller-Hinton broth with 3% heat-inactivated fetal bovie serum (Sigma Cell Culture, St. Louis, Mo.) and grown in a 250-ml flask under microaerobic conditions with aeration (200 rpm) for 2 days. The oligonucleotide primers, plasmids, and bacterial strains are listed in Table 1.
TABLE 1.
Oligonucleotide primers, plasmids, and bacterial strains used in this study
| Oligonucleotide primer, plasmid, or strain | Coordinates in rocFa | Relevant genotype or description | DNA sequence (5′ to 3′) | Source or reference |
|---|---|---|---|---|
| Primersb | ||||
| RocF-F3 (for) | −149–−130 | GCCTGCAGTATTGGGGTGTTTTTCTATC (PstI site underlined) | ||
| RocF-R13 (for) | 18–37 | AACTGCAGAAGCAGAGTTAGGAGCG (PstI site underlined) | ||
| RocF-F4 (for) | 204–223 | AAATCTGATCCCTTGCATGA | ||
| RocF-R14 (rev) | 339–360 | CGGGATCCGCATGCGCGTCTAAATAC (BamHI site underlined) | ||
| RocF-R6 (rev) | 554–573 | TTCGCTCTGTTCGGTGCTTC | ||
| RocF-R15 (for) | 562–580 | CGGGATCCGAACAGAGCGAAAGAGATG (BamHI site underlined) | ||
| RocF-R17 (rev) | 691–708 | GTCCAAATCCAAACTGAG | ||
| RocF-R16 (rev) | 898–915 | CGGAATTCGATGAGATCTAAGATCTC (EcoRI site underlined) | ||
| RocF-R5 (rev) | 987–1006 | GGATCGATCTTTTTCAACCTTTTATCGT (ClaI site underlined) | ||
| Kan-H16 (rev) | NA | CGGTATAATCTTACCTATCACCTCA | ||
| Kan-K4 (rev) | NA | TCCAATTCACTGTTCCTT | ||
| Kan-K5 (for) | NA | TATATTTAAAAATGACGG | ||
| Kan-K8 (for) | NA | TTTGACTTACTGGGGATCAAGCCTG | ||
| Plasmids (parent) | ||||
| pBluescript II SK (−) | Apr, cloning vector | Stratagene | ||
| pBS-rocF (pBluescript II SK) | Apr, 1,154-bp rocF PCR product (nucleotides −149 to 1006) cloned into the PstI and ClaI sites | This study | ||
| pBS-rocF::aphA3 (pBluescript II SK) | Apr Knr, 1.2-kb EcoRI aphA3 cassette from pHP1 ligated to the EcoRI site of rocF in pBS-rocF | This study | ||
| pHP1 (pUC19) | Apr Knr, source of aphA3 cassette | H. Kleanthous | ||
| pILL570 | Spr, cloning vector | 11 | ||
| pILL570 Not (pILL570) | Spr, NotI and EcoRI sites have been introduced between the ClaI and HindIII sites in the pILL570 polylinker | This study | ||
| pILL600 | Knr, source of aphA3 cassette | 12 | ||
| pILL235 (pILL570 Not) | Spr, 676-bp rocF spliced PCR products (nucleotides 18 to 339 and 562 to 915) | This study | ||
| pILL236-2 (pILL570) | Spr Knr, aphA3 cassette from pILL600 ligated to the BamHI site in rocF in pILL235 | This study | ||
| Strains | ||||
| E. coli DH5α | F−supE44 ΔlacU169 (φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories | ||
| H. pylori | ||||
| 26695 | WT, genome sequenced | 4, 34 | ||
| 26695 rocF::aphA3 | 26695 allelic exchange arginase (rocF) mutant by using pBS-rocF::aphA3 | This study | ||
| N6 | WT, naturally transformable laboratory strain | 6 | ||
| N6 236-2 | N6 allelic exchange arginase (rocF) mutant by using pILL236-2 | This study | ||
| N6 rocF::aphA3 | N6 allelic exchange arginase (rocF) mutant by using pBS-rocF::aphA3 | This study | ||
| SS1 | WT (mouse-adapted) | 13 | ||
| SS1 236-2 | SS1 allelic exchange arginase (rocF) mutant by using pILL236-2 | This study | ||
| SS1 rocF::aphA3 | SS1 allelic exchange arginase (rocF) mutant by using pBS-rocF::aphA3 | This study |
+1 refers to the A residue in the start codon of rocF. The coding region of rocF is +1 to +969. Negative numbers refer to upstream residues. NA, not applicable.
for, forward; rev, reverse.
Mice.
Eight-week-old specific-pathogen-free Swiss mice (Centre d’Elevage R. Janvier, Le-Genest-St.-Isle, France) were housed in polycarbonate cages in isolators and fed a commercial pellet diet with water ad libitum. All animal experimentation was performed in accordance with institutional guidelines.
Chemicals.
l-Arginine, l-asparagine, l-aspartate, l-arginine, l-glutamine, l-serine, l-agmatine, and urea were obtained from Sigma. All reagents were of analytical grade.
Molecular biology techniques.
Chromosomal DNA was isolated by a previously described method (17) or by the Qiamp technique (Qiagen, Inc., Valencia, Calif.). Plasmid DNA was isolated by the alkaline lysis method (30) or by column chromatography (Qiagen). Restriction endonuclease digestions, ligations, and other enzyme reactions were conducted according to the manufacturer’s instructions. PCR reactions (50 μl) contained 10 to 100 ng of DNA, PCR buffer, 2.0 to 2.5 mM MgCl2, deoxynucleoside triphosphates (each nucleotide at a concentration of 0.20 to 0.25 mM), 50 to 100 pmol of each primer, and 2.5 U of thermostable DNA polymerase.
Cloning of rocF from H. pylori by PCR.
PCR primers were synthesized (Gibco BRL or the University of Maryland Biopolymer Facility) based on the published DNA sequence of the rocF gene from H. pylori 26695 (GenBank accession number AE000639 [34]) (Table 1). H. pylori 26695 chromosomal DNA was digested with AvaI and the 1.5- to 2.5-kb fragments were gel purified (Qia-Quick; Qiagen, Inc.) and employed as template in PCR reactions by using the primers RocF-F3 and RocF-R5 with the following conditions: 30 cycles, with 1 cycle consisting of 94°C for 5 min, 51°C for 1 min 30 s, and 72°C for 2 min with Pfu DNA polymerase (Stratagene) in a thermocycler (MJ Research, Waterford, Mass.). The predicted 1,154-bp product (corresponding to nucleotides 1459777 to 1460930 of the H. pylori genome, GenBank accession number AE000511) was digested with PstI and ClaI and directionally ligated into pBluescript II SK(−) to yield pBS-rocF. The construct was confirmed by restriction analysis, PCR, and sequence analysis of the insert by using the primers T3, T7, RocF-F4, RocF-F5, and RocF-R6.
Construction of rocF mutants of H. pylori strains by allelic exchange.
The rocF gene in H. pylori was inactivated by either gene disruption (i) or by gene disruption and deletion strategies (ii) as described below. (i) An ∼1.2-kb EcoRI fragment containing a kanamycin cassette (aphA3) from pHP1 (kindly provided by H. Kleanthous, Oravax) was inserted into the unique EcoRI site within rocF (nucleotides 244 to 249) in pBS-rocF to yield pBS-rocF::aphA3 (Fig. 1). This construct was sequenced for confirmation. (ii) Two regions of the rocF gene of H. pylori 26695 were PCR amplified (an initial step of 94°C for 5 min, followed by 30 cycles, with 1 cycle consisting of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min) by using oligonucleotide primers R13 and R14 and primers R15 and R16 (Table 1) derived from the published genome sequence (34). The resulting ∼330 and ∼365 bp with incorporated PstI-BamHI and BamHI-EcoRI cloning sites, respectively, were digested and purified on Elutip columns (Schleicher & Schuell) and cloned into linearized pILL570 Not (Table 1). This generated a deletion of nucleotides 340 to 561 of the coding sequence of rocF (Fig. 1). Spectinomycin-resistant transformants were screened by colony hybridization by using 32P-labeled probes (Amersham Megaprime kit). Plasmid DNA (pILL235) from one of the probe-positive clones was linearized with BamHI and ligated to the BamHI-digested 1.4-kb kanamycin cassette from pILL600 (11). The resulting plasmid, pILL236-2, was sequenced for confirmation.
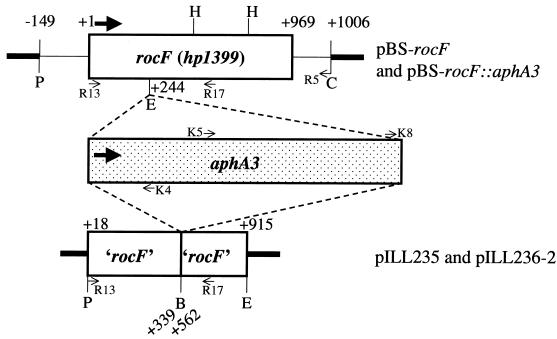
FIG. 1.
Schematic diagrams of rocF constructs used in this study. pBS-rocF::aphA3 and pILL236-2 contain the aphA3 kanamycin-resistant cassette. Thick arrows denote the direction of transcription. Restriction enzyme abbreviations: B, BamHI; C, ClaI; E, EcoRI; H, HindIII; P, PstI. Thick lines refer to vector sequences. The symbols ‘ and ’ refer to truncations at the 5′ or 3′ end, respectively. +1 refers to the first nucleotide in the coding region of rocF. Thin arrows denote primers used for PCR confirmation and sequencing of rocF mutants. See Table 1 for primer sequences, Table 2 for the PCR strategy, and Fig. 2B for the results.
H. pylori strains 26695, N6, and SS1 were transformed with pILL236-2 or pBS-rocF::aphA3 by either electroporation (2.5 kV, 25 μF, 800 Ω) or natural transformation (36).
Confirmation of H. pylori allelic exchange rocF mutants.
Kanamycin-resistant transformants for each of the two rocF disruption constructs in each strain of H. pylori were verified by Southern blot, PCR, and/or PCR-sequencing analysis. For Southern blotting, chromosomal DNA from H. pylori 26695 and the rocF::aphA3 mutant (∼6 μg) was digested with AvaI and electrophoresed through an agarose gel (30). DNA was transferred to Hybond-N+ nylon membrane and treated according to the manufacturer (Amersham Pharmacia Biotech). Hybridization was performed by using the ECL Direct Nucleic Acid Labeling and Detection System (Amersham) with labeled probes of the 1.2-kb gel-purified EcoRI fragment containing the kanamycin cassette from pHP1 and the ∼240-bp gel-purified HindIII rocF fragment (nucleotides 481 to 721) from pBS-rocF. Blots were washed stringently and treated according to the manufacturer’s specifications.
For PCR analyses, chromosomal DNA from each H. pylori mutant was subjected to PCR by using a pair of oligonucleotide primers in which one primer was specific for the rocF gene while the other primer was specific for the kanamycin cassette (Table 2). Primers specific to rocF sequences both upstream and downstream of the kanamycin insertion sites were used. DNA prepared from the parental strains (H. pylori N6 and SS1) served as negative controls.
TABLE 2.
Strategy used for verification of rocF mutants by PCR and sequence analyses
| Strain | PCR primer pairsa
|
PCR-sequencing
|
||
|---|---|---|---|---|
| Up, bpb | Down, bp | Up | Down | |
| N6 WT | R13+K4, none | R5+K8, none; R17+K8, none | NAc | NA |
| NA | NA | |||
| N6 rocF::aphA3 | R13+K4, 680 | R5+K8, 860 | NDd | R5+K8e |
| N6 236-2 | R13+K4, 890 | R17+K8, 250 | R13+H16 | R17+K8 |
| SS1 WT | R13+K4, none | R17+K5, none; R17+K8, none | NA | NA |
| NA | NA | |||
| SS1 rocF::aphA3 | R13+K4, 680 | R17+K5, 1160 | ND | R5+K8 |
| SS1 236-2 | R13+K4, 890 | R17+K8, 250 | R13+H16 | R17+K8 |
Primer pairs used for PCR are listed. Primers are schematically depicted in Fig. 1. The first primer of the pair is specific for rocF, whereas the second primer is specific for the kanamycin resistance gene. The primer sequences are given in Table 1. The results for the PCR reactions from columns 2 and 3 of the table are shown in Fig. 2B.
Up and down, rocF regions immediately up- or downstream, respectively, of insertion sites for kanamycin cassettes. The sizes of the PCR products (shown in Fig. 2B) are given in base pairs.
NA, not applicable.
ND, not determined.
Underlining indicates primers used for direct sequencing of PCR products.
For PCR sequence analysis of each H. pylori mutant, the Thermo Sequenase radiolabeled terminator cycle sequencing kit (Amersham) was used. Chromosomal DNA was PCR amplified to generate DNA products corresponding to sequences both upstream and downstream of the kanamycin insertions sites in H. pylori rocF. PCR products were treated with 10 U of exonuclease I and 2 U of shrimp alkaline phosphatase at 37°C for 15 min. The enzymes were inactivated by heating at 80°C for 15 min. DNA products were subjected to cycle sequencing by using oligonucleotides specific to sequences either internal to the rocF or kanamycin cassette genes, and α33P-labeled dideoxyribonucleotides. Cycle sequencing was performed as follows: 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min (30 cycles). Sequencing gels were dried under vacuum and exposed to Storage Phosphor Screens (Molecular Dynamics). Screens were scanned on a Storm Phosphor/Fluor imaging machine (Molecular Dynamics).
Urease extract preparations, protein determinations, and quantitative determination of urease activity by the phenol-hypochlorite method.
H. pylori sonicated extracts were prepared as described previously (18). Protein concentrations were determined by the Bicinchoninic Acid assay method (Pierce Chemical Company, Rockford, Ill.), according to the manufacturer’s 30-min protocol, with bovine serum albumin as the standard. Urease activity of extracts was determined by measuring the amount of ammonium release from urea in the phenol-hypochlorite urease assay as described previously (37), with the modifications described earlier (18). Data are presented as urease specific activity (nanomoles of ammonium per minute per milligram of protein). Statistical analysis of the data was conducted by use of the Alternative Welch’s t test with InStat 2.03 software (GraphPad Software, San Diego, Calif.).
Preparation of cells and lysates for NMR spectroscopy.
H. pylori cell suspensions were prepared by harvesting cells in sterile NaCl (150 mM) and centrifuging them at 17,000 × g (6°C, 8 min). The supernatant was discarded, and the pellet was collected and resuspended in 150 mM NaCl. The procedure was repeated three times. After the final wash, packed cells were resuspended to a concentration of approximately 108 to 109 cells/ml in sterile 150 mM NaCl. Cell lysates were prepared by resuspending packed cells in sterile NaCl (150 mM) and lysing by two freeze-thaw cycles in liquid nitrogen.
In situ quantitative determination of arginase, aspartase, asparaginase, glutaminase, and serine dehydratase activities of H. pylori by NMR spectroscopy.
For the NMR measurements, cells suspended in a phosphate (20 mM, pH 7.0)–NaCl (115 mM)–KCl (15 mM) buffer were placed into 5-mm tubes (Wilmad, Buena, N.J.), and l-arginine (100 mM), l-asparagine (80 mM), l-aspartate (80 mM), l-glutamine (80 mM), or l-serine (80 mM) was added to start the reactions. Measurements were carried out at 37°C. Substrates and reaction products were observed by NMR spectroscopy. 1H-NMR free induction decays were assessed by using a Bruker DMX-500 or DMX-600 spectrometer operating in the pulsed Fourier transform mode with quadrature detection with presaturation of the water resonance. The instrumental parameters for the DMX-500 instrument were as follows: operating frequency, 500.13 MHz; spectral width, 5,000 Hz; memory size, 16 K; acquisition time, 1.638 s; number of transients, 48 to 144; pulse angle, 50° (3 μs); and relaxation delay with solvent presaturation, 1.0 s. The instrumental parameters for the DMX-600 instrument were as follows: operating frequency, 600.13 MHz; spectral width, 6,009.61 Hz; memory size, 16 K; acquisition time, 1.363 s; number of transients, 48 to 144; pulse angle, 50° (3 μs); and relaxation delay with solvent presaturation, 1.6 s. Spectral resolution was enhanced by Gaussian multiplication with line broadening of −1 Hz and a Gaussian broadening factor of 0.19.
The time-evolution data for substrates and products were obtained by acquiring sequential spectra of the reactions. Progress curves were obtained by measuring the integrals of substrate resonances at each time point. Maximal rates were calculated from good fits (correlation coefficients of >0.99) of the data to straight lines for 30 to 80 min of incubation. Calibrations of the peaks arising from substrates were performed by extrapolating the resonance intensity data to zero time and assigning to this intensity the appropriate concentration value. Data are presented as specific activity (nanomoles of amino acid hydrolyzed per minute per milligram of protein).
Determination of cellular viability of H. pylori cells exposed to various pH buffers and metabolites.
Suspensions of H. pylori 26695 and its isogenic rocF::aphA3 mutant were adjusted to an A600 of ∼1.5 (108 to 109 viable cells/ml) in prewarmed 0.9% NaCl. Bacteria (1-ml suspension) were washed and resuspended in prewarmed 0.1 M citrate-phosphate buffer at a pH of 2.3 or 6.0 with or without urea (5 to 20 mM), l-arginine (10 mM), or l-agmatine (10 mM). Bacteria were incubated at 37°C for 30 min in a microaerobic environment with a Campy Pouch (Becton Dickinson). The buffers did not change pH after this incubation period. In control experiments at neutral pH, H. pylori did not lose viability under these conditions. After incubation, H. pylori cells were washed and resuspended in 0.9% NaCl, and the samples were serially diluted in 0.9% NaCl and plated for enumeration of viable CFU on blood agar. Plates were incubated under microaerobic conditions for 4 to 5 days. Viable CFU were plated prior to start of the assay (T0) and were used as the baseline to compare the viability after 30 min. Data are presented as log10 CFU killed by 30 min under the conditions indicated in Fig. 3 and 4.
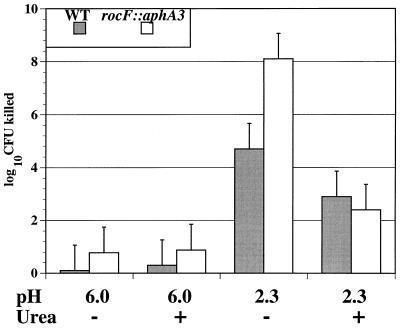
FIG. 3.
Exposure of WT and the rocF::aphA3 mutant of H. pylori 26695 to acid. H. pylori strains were treated with 0.1 M citrate-phosphate buffer at the pHs indicated in the presence or absence of 5 mM urea. After 30 min, bacteria were washed, diluted, and plated for viable CFU. A representative experiment of six experiments, each conducted in duplicate, is shown. Data are presented as the log10 CFU killed, which is the log10 CFU that survived at time = 0 min (T0, 108 to 109) minus the log10 CFU that survived at 30 min under the condition indicated.
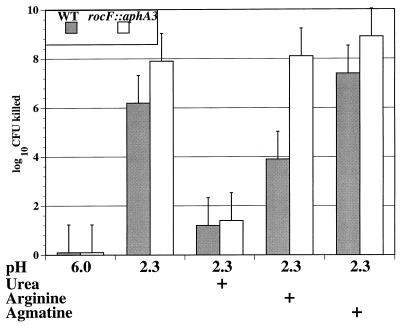
FIG. 4.
Arginine rescues WT H. pylori 26695 but not the isogenic rocF::aphA3 mutant from acid exposure. H. pylori strains were treated with 0.1 M citrate-phosphate buffer at the pHs indicated in the presence or absence of 10 mM l-arginine, l-agmatine, or 20 mM urea. After 30 min, bacteria were washed, diluted, and plated to determine viable CFU. A representative experiment of three experiments, each conducted in duplicate, is shown. The log10 CFU killed is as described in the legend to Fig. 3.
Mouse colonization experiments with the mouse-adapted H. pylori SS1 strain.
H. pylori inocula for mouse colonization experiments were prepared from 36- to 48-h plate cultures of H. pylori mouse-adapted strain SS1 WT, and isogenic rocF mutants SS1 rocF::aphA3 and SS1 236-2. All strains had undergone exactly 24 subcultures in vitro. Inocula were examined under phase-contrast microscopy for the presence of predominantly spiral-shaped, motile bacteria, prior to infection of animals. Mice (n = 9 to 11/group) were each inoculated orogastrically by using polyethylene catheters with 100-μl suspensions containing 3 × 105 to 5.0 × 105 CFU, which is ∼100 times the 100% infectious dose in this model (7).
Mice were sacrificed at 1 month postinoculation. The presence of H. pylori infection in mice was determined by biopsy urease and quantitative culture analyses as previously described (7). Briefly, stomachs were washed in 0.85% (wt/vol) NaCl and dissected longitudinally into two sections. Fragments of each stomach were placed into aliquots (400 μl) of urea-broth medium and peptone trypsin broth (Organotéchnique, La Courneuve, France). The presence of urease activity in tissue fragments was determined by monitoring the urea-broth at room temperature for a color indicator shift to an alkaline pH as a result of the production of ammonium from urea hydrolysis. To perform quantitative bacterial cultures on stomach samples, tissue fragments were homogenized by using disposable plastic grinders and tubes (PolyLabo, Strasbourg, France). Quantitative culture was performed on homogenized samples, as previously described (7). The homogenates were serially diluted in sterile 0.85% NaCl and plated onto blood agar plates, supplemented with 200 μg of bacitracin and 10 μg of naladixic acid (Sigma Chemical Co.) per ml. After 3 to 4 days of incubation, colonies with an H. pylori morphology were counted. The CFU data were statistically analyzed by the Alternative Welch’s t test, whereas colonization data were analyzed by using chi-squared analysis with the Fisher’s exact test.
RESULTS
Features of the rocF gene and the RocF protein of H. pylori.
Previous results indicated that H. pylori has arginase activity and a urea cycle (21, 23). The H. pylori sequenced genomes revealed genes hp1399 and jhp1427 in strains 26695 and J99, respectively, as predicted orthologs of arginase (1, 34). To investigate the functions of rocF in vitro and in vivo, the rocF gene was cloned by PCR, sequenced, and found to be identical to the previously published sequence of strain 26695 (34). The 969-bp coding region of rocF does not appear to be cotranscribed with any other gene based on the finding that the upstream gene (hp1398) is in the opposite orientation to that of rocF, and the 5′ end of the downstream gene (hp1400) is more than 500 bp away from the 3′ end of rocF (1, 34; see also below). Additionally, rocF has a predicted near consensus rho-independent transcriptional terminator (CUUUUCAAACCN11GGUUGAAAAAG), followed by an AU-rich region. The rocF gene has a predicted Shine-Dalgarno sequence of TAAGGAGGTG that closely matches the consensus (TAAAGGAGT) and two strong predicted ς70-like promoters.
The predicted pI for H. pylori RocF is 6.4, and Leu, Lys, and Gln residues comprise 30% of the protein. The predicted RocF protein has the three conserved histidine residues (H91, H118, and H133), including the one within the putative divalent cation-binding motif D-A-H-X-D (amino acids 116 to 122) found in all agmatinases and arginases.
Construction and confirmation of rocF allelic exchange mutants of H. pylori.
Allelic exchange rocF mutants of H. pylori were constructed (Fig. 1; Table 1) by using two different strategies and three different H. pylori strains. To confirm the construction of rocF mutants of H. pylori, chromosomal DNA from WT and mutants was analyzed by Southern blot, PCR, and PCR sequencing.
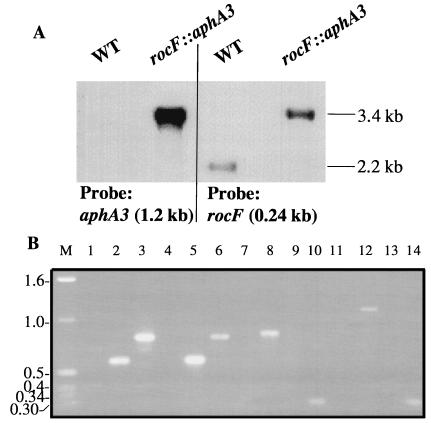
Southern blot analyses of AvaI-digested chromosomal DNA from H. pylori 26695 WT and rocF::aphA3 mutant strains, when probed with a 240-bp fragment of the rocF gene, resulted in probe-positive fragments with sizes of 2.2 and 3.4 kb, respectively (Fig. 2A). The aphA3 probe also hybridized to a 3.4-kb AvaI fragment from chromosomal DNA from the mutant but did not hybridize with WT chromosomal DNA (Fig. 2A), thus confirming the insertion of the kanamycin cassette in the rocF gene of H. pylori 26695.
FIG. 2.
Confirmation of rocF allelic exchange mutants of H. pylori. (A) Southern blot analysis of H. pylori WT and the rocF::aphA3 isogenic mutant. Chromosomal DNA (∼6 μg) from H. pylori 26695 and 26695 rocF::aphA3 was digested with AvaI and processed as described in Materials and Methods. The blot was probed with either the kanamycin cassette (1.2 kb) or a portion of the rocF gene (240-bp HindIII fragment). Note that when probed with rocF, the mutant has an increase in molecular weight by ∼1.2 kb, as expected. (B) PCR analysis of rocF mutants of H. pylori. The primer pair strategies used are shown in Table 2. Lanes (number, H. pylori strain, primer pair): 1, N6 WT, R13 and K4; 2, N6 rocF::aphA3, R13 and K4; 3, N6 236-2, R13 and K4; 4, SS1 WT, R13 and K4; 5, SS1 rocF::aphA3, R13 and K4; 6, SS1 236-2, R13 and K4; 7, N6 WT, R5 and K8; 8, N6 rocF::aphA3, R5 and K8; 9, N6 WT, R17 and K8; 10, N6 236-2, R17 and K8; 11, SS1 WT, R17 and K5; 12, SS1 rocF::aphA3, R17 and K5; 13, SS1 WT, R17 and K8; 14, SS1 236-2, R17 and K8; and M, 1-kb marker (Gibco BRL).
The rocF mutants of H. pylori strains N6 and SS1 were confirmed by PCR and PCR-sequencing analyses by using the strategies outlined in Table 2. Primers were designed to amplify DNA fragments corresponding to the sequences both immediately upstream and downstream of the insertion sites of the kanamycin cassettes (Fig. 1). The expected PCR product sizes were obtained in all cases (Fig. 2B; Table 2 shows the fragment sizes). As expected, no PCR products were obtained with any primer pair when using chromosomal DNA from any of the WT H. pylori strains (Fig. 2B, lanes 1, 4, 7, 9, 11, and 13). Additionally, PCR analysis of the hp1400 gene downstream of rocF in the rocF mutants with an hp1400 (fecA)-specific primer plus a rocF primer yielded products with the appropriate size in all of the strains tested (data not shown). Direct sequence analysis of selected PCR products (Table 2) provided unequivocal evidence that the junctions of rocF and aphA3 were correct in these H. pylori mutant strains.
H. pylori rocF mutants retain WT levels of urease activity.
The rocF mutants of H. pylori had similar rates of growth in broth to those of the WT strains (data not shown). To ascertain whether arginase affects urease activity in H. pylori, we measured this enzyme activity in the WT and rocF::aphA3 mutant of H. pylori 26695. No significant difference was observed in the urease activities, with averages of 4,400 ± 200 and 4,500 ± 500 nmol of ammonium/min/mg of protein for the WT and rocF::aphA3 mutant, respectively (n = 3; P = 0.69). Similarly, no differences in urease activity were noted between the parent and the corresponding isogenic rocF mutants of H. pylori strains N6 and SS1 (data not shown).
H. pylori rocF mutants lack arginase activity.
Catabolism of l-arginine by H. pylori was investigated in cell suspensions of the WT strains 26695, N6, and SS1 and the corresponding rocF mutants by employing the 1H-NMR spectroscopy method described previously (23). All three WT strains of H. pylori expressed arginase activity (Table 3). All five rocF mutants constructed were completely devoid of arginase activity (Table 3).
TABLE 3.
Rates of amino acid catabolism determined for H. pylori cells of WT and rocF mutant strains (n = 4)a
| Strain | Mean rate of enzyme activity (nmol/min/mg of protein) ± SD
|
||||
|---|---|---|---|---|---|
| Arginase | Asparaginase | Aspartase | Glutaminase | Serine dehydratase | |
| 26695 | 33 ± 4 | 19 ± 3 | 8 ± 2 | 66 ± 6 | 42 ± 7 |
| 26695 rocF::aphA3 | UDb | 19 ± 3 | 8 ± 2 | 62 ± 8 | 19 ± 3c |
| N6 | 51 ± 5 | 35 ± 6 | 11 ± 3 | 91 ± 10 | 37 ± 8 |
| N6 rocF::aphA3 | UD | 20 ± 4 | 12 ± 3 | 80 ± 11 | 14 ± 3c |
| N6 236-2 | UD | 25 ± 4 | 11 ± 2 | 85 ± 10 | 25 ± 5d |
| SS1 | 54 ± 5 | 32 ± 3 | 17 ± 3 | 81 ± 8 | 50 ± 7 |
| SS1 rocF::aphA3 | UD | 28 ± 5 | 11 ± 3 | 75 ± 8 | 17 ± 3e |
| SS1 236-2 | UD | 24 ± 3 | 15 ± 2 | 75 ± 6 | 32 ± 6e |
Bacteria were suspended in a phosphate (20 mM; pH 7)–NaCl (115 mM)–KCl (15 mM) buffer plus one of the following amino acids: l-arginine (100 mM), l-asparagine (80 mM), l-aspartate (80 mM), l-glutamine (80 mM), or l-serine (80 mM) and were measured for enzyme activity by NMR (see Materials and Methods).
UD, undetectable. P < 0.0001 compared with its isogenic WT strain.
P < 0.005 compared with its isogenic WT strain.
P < 0.05 compared with its isogenic WT strain.
P < 0.01 compared with its isogenic WT strain.
Asparaginase, aspartase, glutaminase, and serine dehydratase activities in WT and rocF mutants of H. pylori.
To determine whether the inactivation of rocF could result in alteration of other enzyme activities involved in nitrogen metabolism, the deamination rates of l-asparagine, l-aspartate, l-glutamine, and l-serine were determined in H. pylori cell suspensions of the WT and rocF mutants by employing 1H-NMR spectroscopy (20). Fast rates of asparaginase, aspartase, glutaminase, and serine dehydratase enzyme activities were observed for the WT strains (Table 3). No significant differences were observed between WT H. pylori and their corresponding rocF mutants in asparaginase, aspartase, and glutaminase activities (Table 3). Most notably, however, there was a significant decrease in serine dehydratase activities in all rocF mutants relative to the WT strains (Table 3). Additionally, the activities of the rocF::aphA3 mutants were lower than those of the 236-2 mutants (Table 3). To determine whether these differences arose from modulation of enzyme activity by ammonium ions, serine dehydratase activities were measured in lysates containing NH4Cl at concentrations of up to 15 mM. The presence of NH4Cl did not alter the rates of enzyme activity (data not shown).
Effect of RocF on survival of H. pylori from acid exposure.
Urease mutants of H. pylori are much more sensitive to acid in vitro than are WT strains due to loss of hydrolysis of urea to acid-neutralizing ammonium (3a, 15, 31, 33). Since arginine is a precursor source of urea for H. pylori through arginase activity (21), we investigated the role of H. pylori arginase activity in sensitivity of H. pylori to acid. H. pylori 26695 and its isogenic rocF::aphA3 mutant were incubated for 30 min in citrate-phosphate buffer at pH 2.3 or 6.0 under microaerobic conditions in the presence or absence of urea, arginine, or agmatine. At pH 2.3, although the WT was sensitive to acid (viability was reduced by about 5 orders of magnitude), the rocF mutant was ∼1,000-fold more sensitive to acid (viability reduced by about 8 orders of magnitude) (Fig. 3). Urea at a concentration of 5 mM added to citrate-phosphate buffer at pH 2.3 increased the viability of both strains, with a reduction in viable bacteria (relative to T0 counts) of only 2.4 and 2.9 orders of magnitude, respectively (Fig. 3). Urea at a concentration of 20 mM added to citrate-phosphate buffer at pH 2.3 further increased the viability of both strains (Fig. 4). As a control, addition of urea to citrate-phosphate buffer at pH 6.0 had no effect on the cell viability of either strain (Fig. 3). These results indicated that the rocF::aphA3 mutant was remarkably more susceptible to acid than the WT and that both strains survived equally well when exogenous urea was added.
The survival of the WT and rocF::aphA3 mutant in the presence of arginine or agmatine (10 mM) was subsequently investigated. Arginine dramatically increased the viability of the WT strain at pH 2.3, with a reduction of viable bacteria (relative to T0 counts) of approximately 6 and 4 orders of magnitude in the absence versus the presence of arginine, respectively. In contrast, the addition of arginine did not increase the viability of the rocF::aphA3 mutant in acidic conditions, with similar reductions in viable bacteria, irrespective of the absence or presence of arginine (Fig. 4). Thus, there was an ∼10,000-fold decrease in the viability of the rocF::aphA3 mutant compared with the WT under acid conditions in the presence of arginine. This decrease in the viability of the rocF::aphA3 mutant compared with the WT under acid conditions plus arginine was maintained in a dose-dependent fashion (data not shown), and maximal protection from acid of both strains was observed at 50 mM arginine, a concentration known to be near saturating for arginase activity (21). As a control, the addition of arginine to citrate-phosphate buffer (pH 6.0) had no effect on the viability of either the WT or rocF::aphA3 mutant (data not shown). Agmatine did not protect either the WT or mutant from acid exposure (Fig. 4). At an intermediate pH (4.2), the viabilities of the WT and the rocF::aphA3 mutant were similar, as expected: in two experiments, each conducted in duplicate, reductions of viable bacteria by 2.8 and 3.5 orders of magnitude for the WT and by 2.7 and 3.6 orders of magnitude for the mutant were observed (data not shown). This viability was intermediate to that of strains tested at pH 2.3 and 6.0.
H. pylori rocF is not required for colonization of mice.
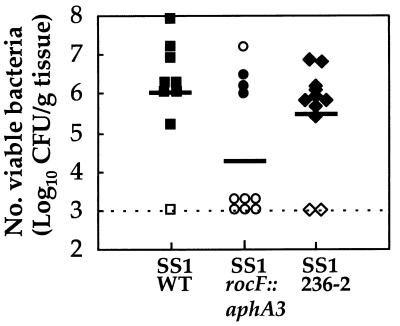
Since urease and arginase mutants both exhibit increased acid sensitivity in vitro (this study and references 3a, 15, 31, and 33) and urease is required for H. pylori colonization in vivo (4, 5), we investigated whether H. pylori arginase is required for colonization. Mice were challenged with equivalent inocula of WT H. pylori SS1 and the two rocF isogenic mutants. In mutant SS1 rocF::aphA3, rocF was disrupted by insertion of a kanamycin resistance cassette. In mutant SS1 236-2, rocF was disrupted by deletion of an internal rocF fragment and replaced with a kanamycin resistance cassette (Fig. 1 and 2). At 1 month postinoculation, 89% (8 of 9) of mice administered the H. pylori SS1 WT isolate were colonized with the organism, whereas 40% (4 of 10) and 82% (9 of 11) of animals that had been challenged with either H. pylori rocF::aphA3 and 236-2 mutants, respectively, were infected (P = 0.06 and P = 1.0, respectively, compared with the WT) (Fig. 5). Quantitative culturing on nonselective plates revealed similar numbers of viable H. pylori CFU per gram of tissue for the WT and mutant 236-2, with a mean log10 CFUs per gram of tissue ± standard deviation of 6.1 ± 1.4 and 5.5 ± 1.3, respectively (P = 0.36). In contrast, bacterial loads in mice challenged with the rocF::aphA3 mutant were significantly lower, with a 4.4 ± 1.8 log10 CFU/g of tissue (p = 0.03) (Fig. 5), a finding in agreement with its reduced colonization. Quantitative culturing on kanamycin-containing plates of gastric tissues from mice that had been administered either of the H. pylori SS1 rocF mutants revealed results similar to those obtained on nonselective plates, thus demonstrating the stability of the kanamycin cassettes in vivo (data not shown). Urease activities of the rocF mutants of H. pylori recovered from mice were similar to those of the WT strain and to those of the rocF mutant used to inoculate the mice (data not shown).
FIG. 5.
In vivo colonization of mice by WT and rocF mutants of H. pylori SS1. Bacterial loads were enumerated by quantitative culture on samples obtained 1 month postinoculation. The limit of detection is indicated by the dotted line. Symbols along the dotted line indicate that the strain failed to colonize the mouse. Urease-positive (solid symbols) and -negative (open symbols) gastric biopsy samples are also shown. Each point corresponds to a determination for a single mouse. Solid lines indicate the mean log10 of viable CFU per gram of tissue. In a second experiment (quantitative data not shown in the figure), the numbers of mice colonized with the WT, 236-2 rocF mutant, and rocF::aphA3 mutant were 64% (7 of 11), 100% (10 of 10), and 56% (5 of 9), respectively.
In a second independent experiment (data not shown in Fig. 5), the numbers of mice colonized with the WT, 236-2 rocF mutant, and rocF::aphA3 mutant were 64% (7 of 11), 100% (10 of 10), and 56% (5 of 9), respectively, a result in good agreement with the results of the first mouse experiment.
DISCUSSION
The H. pylori rocF gene (34) was cloned, and five isogenic rocF mutants with disruptions in this gene were constructed in three strains and were shown to completely lack arginase activity and to have reduced serine dehydratase activity (Table 3). Additionally, the rocF::aphA3 mutant of strain 26695 was dramatically more sensitive to acid and could not be rescued from acid exposure by addition of exogenous arginine, in comparison with the WT. Based on these results and the homology of RocF with the arginase family (1, 23, 34), it is concluded that rocF encodes the H. pylori arginase of the bacterium. H. pylori rocF mutants colonized mice, albeit to different degrees depending on the rocF disruption strategy.
H. pylori rocF mutants had urease activities similar to those of WT H. pylori in vitro. Exogenous urea protected the WT and the rocF::aphA3 mutant of strain 26695 equally well from acid, indicating that H. pylori arginase has no effect on urease activity under the conditions tested. Conversely, a urease mutant of H. pylori N6 was previously shown to have the same arginase activity as the WT strain (23). Notwithstanding the apparent close metabolic relationship between arginase and urease, in H. pylori these enzymes did not appear to influence the activity of each other in vitro.
It was demonstrated that arginase activity plays a critically important role in the survival of H. pylori from acid exposure in vitro because the WT, but not the rocF::aphA3 mutant, can be rescued from acid exposure by exogenous addition of arginine (Fig. 3 and 4). Agmatine had no effect on the survival of H. pylori from acid exposure, indicating that the activity being observed is arginase, not agmatinase. The rocF::aphA3 mutant of H. pylori 26695 was ∼1,000-fold more sensitive to acid exposure in vitro than the WT strain. The increased sensitivity of the rocF mutant to acid may arise from its inability to synthesize urea from arginine, which would result in a deficiency of acid-neutralizing ammonia available from urease activity. However, other urease-independent mechanisms of ammonia generation also could be involved in acid protection of H. pylori, since H. pylori exhibits several active amino acid deaminases (Table 3 and reference 20) and one or more aliphatic amidases (32). Preliminary experiments, however, suggested that the amino acid deaminases play little or no role in protecting H. pylori from acid exposure (19a). The reason for the increased viability of WT H. pylori treated in acid conditions plus urea compared with those treated with arginine could be due to differences in affinities of specific transporters for urea versus arginine or to the WT strain’s ability to synthesize its own urea by arginase activity but an inability to synthesize its own arginine.
H. pylori rocF mutants showed pleiotropic effects: in addition to the loss of arginase activity, serine dehydratase activities were markedly reduced in all mutants compared with the corresponding WT H. pylori, with stronger effects observed for the rocF::aphA3 mutants than for the 236-2 mutants (Table 3). Serine dehydratase deaminates serine to pyruvate plus ammonium; pyruvate has been shown to play an important role in the energy metabolism of H. pylori (22, 28). H. pylori rocF is at a locus distinct from the serine dehydratase locus (genes sdaA, hp0132, and jhp0120 in H. pylori 26695 and J99, respectively) (1, 34). Although reduced serine dehydratase activities could be due to impaired systems of ammonium excretion in the rocF mutants, the mechanism of this effect and the differences in serine dehydratase activities in the rocF::aphA3 and 236-2 mutants remain to be elucidated, possibly with the help of H. pylori isogenic serine dehydratase mutants. However, the differences between the serine dehydratase activities of the SS1, SS1 236-2, and SS1 rocF::aphA3 strains suggested a relationship between the metabolic and mouse colonization data; the lower serine dehydratase enzyme rates of the SS1 rocF::aphA3 mutant may correlate with its less-effective colonization than the SS1 236-2 mutant. The in vivo data indicated that H. pylori arginase activity and the rocF gene are not essential for colonization of mice, since the 236-2 rocF mutant of H. pylori SS1 colonized mice as well as the WT strain. Interestingly, however, the rocF::aphA3 H. pylori mutant had a moderately reduced colonization efficiency, as demonstrated by the finding that only 40% of mice that were challenged with this mutant were still infected at 1 month postinoculation (Fig. 5). Arginase may only be necessary in vivo in situations such as limiting urea concentrations in the gastric mucosa, lowered host arginase levels, or lowered stomach pH. H. pylori rocF mutants may be able to colonize mice because the murine stomach has a slightly higher pH than that of humans, and in this changed environment arginase may not be essential to protect H. pylori from acid. Another reason for colonization by rocF mutants could be that host arginase activity compensated for arginase deficiency in the rocF mutants. Host arginase could provide the required urea concentrations to rocF mutants of H. pylori in vivo to generate acid-neutralizing ammonium from urease. Unfortunately, this hypothesis cannot be readily tested because host arginase activity cannot be inhibited without deleterious effects (2a).
It is unlikely that insertion of the kanamycin-resistance cassette into rocF causes polar effects on other genes adjacent to rocF, since there is a strong predicted transcriptional terminator downstream of the coding region of rocF and the translation start site for the gene downstream of rocF, hp1400, is more than 500 bp away from the 3′ end of rocF (1, 34). Also, the kanamycin cassettes used for construction of the rocF mutants lack a transcriptional terminator, thereby minimizing premature termination of rocF transcription. Additionally, the rocF mutants were vigorously confirmed by multiple techniques to rule out potential artifacts (Table 2 and Fig. 2). Thus, polar effects are probably not the reason for the colonization and serine dehydratase activity differences observed between the two rocF mutants. These differences, as well as the pleiotropic phenotypes of the rocF mutants, suggest that caution should be exercised in interpreting results of gene knockout experiments when only one allelic exchange mutant is used. Finally, it is highly unlikely that the decrease in serine dehydratase activity is due to spontaneous mutation of this locus because we have constructed five different arginase mutants in three different strains of H. pylori. The chances of a spontaneous mutation in this specific locus happening five independent times would be lower than 1 in 1016 (1,600 genes raised to the fifth power, assuming one phenotypic mutation per genome). Also the WT and mutant strains were passaged an equal number of times in vitro. All of the corresponding isogenic WT strains have WT rates of serine dehydratase activity even after several in vitro passages.
In summary, the H. pylori rocF gene encodes the urea cycle enzyme arginase and is required for arginase activity. A rocF mutant of H. pylori has increased susceptibility to acid treatment in vitro, and thus arginase activity dramatically helps H. pylori survive acid exposure in an arginine-dependent manner. rocF mutants also have decreased serine dehydratase activity, retain WT levels of urease activity, and are able to colonize mice but to different degrees. The data suggest that H. pylori and/or the host have a mechanism for in vivo compensation of the loss of the bacterial arginase.
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health Public Health Service grants AI25567 (to H.L.T.M.) and AI10098 (a postdoctoral fellowship to D.J.M.), by Pasteur-Mérieux-Connaught (Lyon, France) and OraVax, Inc. (Boston) (to R.L.F.), by Conseil Régional d’Ile de France (an INSERM Poste Vert postdoctoral scholarship to F.J.R.), and by the Australian Research Council and the National Health and Medical Research Council of Australia (to G.L.M.).
We thank Stephen M. Boyle for experimental suggestions and Keith T. Wilson for insightful discussions.
REFERENCES
- 1.Alm R, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–80. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 1a.Bewley M C, Jeffrey P D, Patchett M L, Kanyo Z F, Baker E N. Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily. Structure. 1999;7:435–448. doi: 10.1016/s0969-2126(99)80056-2. [DOI] [PubMed] [Google Scholar]
- 2.Blaser M J. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J Infect Dis. 1990;161:626–633. doi: 10.1093/infdis/161.4.626. [DOI] [PubMed] [Google Scholar]
- 2a.Braga A C, Vilarinho L, Ferreira E, Rocha H. Hyperargininemia presenting as persistent neonatal jaundice and hepatic cirrhosis. J Pediatr Gastroenterol Nutr. 1997;24:218–221. doi: 10.1097/00005176-199702000-00018. [DOI] [PubMed] [Google Scholar]
- 3.Buck G E, Gourley W K, Lee W K, Subramanyan J M. Relation of Campylobacter pyloridis to gastritis and peptic ulcers. J Infect Dis. 1986;153:664–669. doi: 10.1093/infdis/153.4.664. [DOI] [PubMed] [Google Scholar]
- 3a.Clyne M, Labigne A, Drumm B. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infect Immun. 1995;63:1669–1673. doi: 10.1128/iai.63.5.1669-1673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eaton K A, Brooks C L, Morgan D R, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59:2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eaton K A, Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect Immun. 1994;62:3604–3607. doi: 10.1128/iai.62.9.3604-3607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrero R L, Cussac V, Courcoux P, Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J Bacteriol. 1992;174:4212–4217. doi: 10.1128/jb.174.13.4212-4217.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrero R L, Thiberge J M, Huerre M, Labigne A. Immune responses of specific-pathogen-free mice to chronic Helicobacter pylori (strain SS1) infection. Infect Immun. 1998;66:1349–55. doi: 10.1128/iai.66.4.1349-1355.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardan R, Rapoport G, Debarbouille M. Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J Mol Biol. 1995;249:843–856. doi: 10.1006/jmbi.1995.0342. [DOI] [PubMed] [Google Scholar]
- 9.International Agency for Cancer Research. Monographs on the Evaluations of Cancer Risks to Humans. Vol. 61. Lyon, France: World Health Organization; 1994. pp. 177–240. [Google Scholar]
- 10.Krishnamurthy P, Parlow M, Zitzer J B, Vakil N B, Mobley H L T, Levy M, Phadnis S S H, Dunn B E. Helicobacter pylori containing only cytoplasmic urease is susceptible to acid. Infect Immun. 1998;66:5060–5066. doi: 10.1128/iai.66.11.5060-5066.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labigne A, Cussac V, Courcoux P. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J Bacteriol. 1991;173:1920–1931. doi: 10.1128/jb.173.6.1920-1931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labigne-Roussel A, Courcoux P, Tompkins L. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J Bacteriol. 1988;170:1704–1708. doi: 10.1128/jb.170.4.1704-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee A, O’Rourke J, Corazon de Ungria M, Robertson B, Daskalopoulos G, Dixon M F. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 14.Marshall B J. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;i:1273–1275. [PubMed] [Google Scholar]
- 15.Marshall B J, Barrett L J, Prakash C, McCallum R W, Guerrant R L. Urea protects Helicobacter (Campylobacter) pylori from the bactericidal effect of acid. Gastroenterology. 1990;99:697–702. doi: 10.1016/0016-5085(90)90957-3. [DOI] [PubMed] [Google Scholar]
- 16.Marshall B J, McGechie D B, Rogers P A, Glancy R J. Pyloric campylobacter infection and gastroduodenal disease. Med J Aust. 1985;142:439–444. doi: 10.5694/j.1326-5377.1985.tb113444.x. [DOI] [PubMed] [Google Scholar]
- 17.McAllister C F, Stephens D S. Analysis in Neisseria meningitidis and other Neisseria species of genes homologous to the FKBP immunophilin family. Mol Microbiol. 1993;10:13–23. doi: 10.1111/j.1365-2958.1993.tb00899.x. [DOI] [PubMed] [Google Scholar]
- 18.McGee D J, May C A, Garner R M, Himpsl J M, Mobley H L T. Isolation of Helicobacter pylori genes that modulate urease activity. J Bacteriol. 1999;181:2477–2484. doi: 10.1128/jb.181.8.2477-2484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGee D J, Mobley H L T. Mechanisms of Helicobacter pylori infection: bacterial factors. Curr Top Microbiol Immunol. 1999;241:155–180. doi: 10.1007/978-3-642-60013-5_9. [DOI] [PubMed] [Google Scholar]
- 19a.McGee, D. J., and H. L. T. Mobley. Unpublished observations.
- 20.Mendz G L, Hazell S L. Amino acid utilization by Helicobacter pylori. Int J Biochem Cell Biol. 1995;27:1085–1093. doi: 10.1016/1357-2725(95)00069-2. [DOI] [PubMed] [Google Scholar]
- 21.Mendz G L, Hazell S L. The urea cycle of Helicobacter pylori. Microbiology. 1996;142:2959–2967. doi: 10.1099/13500872-142-10-2959. [DOI] [PubMed] [Google Scholar]
- 22.Mendz G L, Hazell S L, van Gorkom L. Pyruvate metabolism in Helicobacter pylori. Arch Microbiol. 1994;162:187–192. doi: 10.1007/BF00314473. [DOI] [PubMed] [Google Scholar]
- 23.Mendz G L, Holmes E M, Ferrero R L. In situ characterisation of Helicobacter pylori arginase. Biochim Biophys Acta. 1998;1338:465–477. doi: 10.1016/s0167-4838(98)00207-6. [DOI] [PubMed] [Google Scholar]
- 24.Mobley H L T. Structure and function of Helicobacter pylori urease. In: Ernst P B, Michetti P, Smith P D, editors. Immunobiology of H. pylori from pathogenesis to prevention. New York, N.Y: Lippincott-Raven; 1997. pp. 59–73. [Google Scholar]
- 25.Mobley H L T, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nedenskov P. Nutritional requirements for growth of Helicobacter pylori. Appl Environ Microbiol. 1994;60:3450–3453. doi: 10.1128/aem.60.9.3450-3453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsonnet J, Friedman G D, Vandersteen D P, Chang Y, Vogelman J H, Orentreich N, Sibley R K. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 28.Pitson S M, Mendz G L, Srinivasan S, Hazell S L. The tricarboxylic acid cycle of Helicobacter pylori. Eur J Biochem. 1999;260:258–267. doi: 10.1046/j.1432-1327.1999.00153.x. [DOI] [PubMed] [Google Scholar]
- 29.Reynolds D J, Penn C W. Characteristics of Helicobacter pylori growth in a defined medium and determination of its amino acid requirements. Microbiology. 1994;140:2649–2656. doi: 10.1099/00221287-140-10-2649. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Segal E D, Shon J, Tompkins L S. Characterization of Helicobacter pylori urease mutants. Infect Immun. 1992;60:1883–1889. doi: 10.1128/iai.60.5.1883-1889.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skouloubris S, Labigne A, De Reuse H. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol Microbiol. 1997;25:989–998. doi: 10.1111/j.1365-2958.1997.mmi536.x. [DOI] [PubMed] [Google Scholar]
- 33.Sjöström J E, Larsson H. Factors affecting growth and antibiotic susceptibility of Helicobacter pylori: effect of pH and urea on the survival of a wild-type strain and a urease-deficient mutant. J Med Microbiol. 1996;44:425–433. doi: 10.1099/00222615-44-6-425. [DOI] [PubMed] [Google Scholar]
- 34.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 35.Warren J R. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;i:1273–1275. [PubMed] [Google Scholar]
- 36.Wang Y, Roos K P, Taylor D E. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J Gen Microbiol. 1993;139:2485–2493. doi: 10.1099/00221287-139-10-2485. [DOI] [PubMed] [Google Scholar]
- 37.Weatherburn M W. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39:971–974. [Google Scholar]
- 38.Wu C W, Chung W W, Chi C W, Kao H L, Lui W Y, Peng F K, Wang S R. Immunohistochemical study of arginase in cancer of the stomach. Virchows Arch. 1996;428:325–31. doi: 10.1007/BF00202199. [DOI] [PubMed] [Google Scholar]