Abstract
A 320-nucleotide RNA with several characteristic features was expressed in Bacillus subtilis to study RNA processing. The RNA consisted of a 5′-proximal sequence from bacteriophage SP82 containing strong secondary structure, a Bs-RNase III cleavage site, and the 3′-proximal end of the ermC transcriptional unit. Comparison of RNA processing in a wild-type strain and a strain in which the pnpA gene, coding for polynucleotide phosphorylase (PNPase), was deleted, as well as in vitro assays of phosphate-dependent degradation, showed that PNPase activity could be stalled in vivo and in vitro. Analysis of mutations in the SP82 moiety mapped the block to PNPase processivity to a particular stem-loop structure. This structure did not provide a block to processivity in the pnpA strain, suggesting that it was specific for PNPase. An abundant RNA with a 3′ end located in the ermC coding sequence was detected in the pnpA strain but not in the wild type, indicating that this block is specific for a different 3′-to-5′ exonuclease. The finding of impediments to 3′-to-5′ degradation, with specificities for different exonucleases, suggests the existence of discrete intermediates in the mRNA decay pathway.
One goal of studies on RNA processing in Bacillus subtilis is to determine the function of various RNases. To date, the B. subtilis RNases shown to be involved in mRNA processing are Bs-RNase III, a narrow-specificity endonuclease (15, 19), and polynucleotide phosphorylase (PNPase), a 3′-to-5′ exonuclease (2, 11). Previously, we constructed a strain in which part of the PNPase gene (pnpA) is deleted and that contains no PNPase activity (18). Decay of mRNA encoded by ermC, a plasmid-borne erythromycin resistance gene, was studied in the pnpA deletion strain. We showed that 5′-proximal ermC RNA fragments were easily detectable in the pnpA strain but barely detectable in the wild-type strain (2). Since mRNA turnover is thought to be essential for viability (6), the lack of a strong growth phenotype in the strain deficient in PNPase indicated the presence of at least one other 3′-to-5′ exoribonuclease that can compensate for the missing PNPase activity. We proposed that the 5′-proximal ermC fragments were mRNA decay intermediates that are normally degraded rapidly by PNPase. The 3′-to-5′ exonuclease that is responsible for mRNA turnover in the pnpA deletion strain is less efficient at degrading these fragments, possibly due to particular structures at their 3′ ends. In addition to its function in mRNA turnover, PNPase processing may be required for the expression of particular genes. The absence of such processing events in the pnpA strain may be the cause of the phenotypes displayed by this strain: competence deficiency, cold sensitivity, tetracycline sensitivity, and filamentous growth (11, 18).
Although PNPase is apparently able to degrade ermC mRNA fragments efficiently, we have found that PNPase processivity can be impeded. In the course of assaying for purified Bs-RNase III, we discovered an RNA-processing product that was the result of PNPase stalling (14). A similar PNPase stall site was detected in vivo (2).
In this study, we examined in more detail the previously observed PNPase block by using a model RNA that contained 5′-proximal SP82 sequences and 3′-proximal ermC sequences. Experiments show that PNPase activity behaves similarly in vitro and in vivo, suggesting that the block to processivity is an inherent property of the RNA. Decay intermediates of the model RNA that were detected only in the pnpA strain suggest that other sequences, which are not a barrier to PNPase activity, function to block a different 3′-to-5′ exonuclease.
MATERIALS AND METHODS
Bacterial strains.
The wild-type B. subtilis host was BG1, which is trpC2 thr-5. The pnpA mutant host was BG119, a derivative of BG1 in which an internal portion of the pnpA gene was replaced with a kanamycin resistance cassette (18). B. subtilis growth media and competent B. subtilis cultures were prepared as described previously (7). E. coli DH5α (9) was the host for plasmid constructions.
Plasmids.
Plasmid pYH199 was a derivative of pYH188 (2). Plasmid pYH188 contains an SP82-ermC transcriptional unit which is similar to the one described here but has an altered transcription terminator sequence. It also contains a second (promoterless) copy of ermC downstream. We found that strains carrying pYH188 were erythromycin resistant, indicating substantial readthrough transcription from the SP82-ermC sequence into the downstream ermC sequence. This would give additional RNA products upon probing with an ermC-specific probe. To avoid this, we constructed plasmid pYH199, an HpaI deletion derivative of pYH188 which does not contain any ermC sequences other than that of the SP82-ermC RNA transcriptional unit and which has the native ermC transcription terminator.
Mutations in the SP82 sequence were constructed either by the method of Kunkel et al. (10) on M13 single-stranded phage DNA templates or by the QuikChange site-directed mutagenesis protocol (Stratagene) on plasmid DNA templates.
In vivo RNA analysis.
Growth of strains and isolation of RNA were as described previously (2). Northern blot analysis, using 6% denaturing polyacrylamide gels, was performed as described previously (19). Electroblotting from a high-resolution (sequencing) gel was performed in the same manner as for low-resolution gels, except that the blotting was done at 10 V overnight followed by 30 V for 30 min. Radioactivity in the bands on Northern blots was quantitated with a PhosphorImager instrument (Molecular Dynamics). Half-lives were determined by linear regression analysis on semilogarithmic plots of percent RNA remaining versus time. Reverse transcriptase products were generated with reagents from the Gibco-BRL Superscript Preamplification System. Riboprobes were synthesized by T7 RNA polymerase transcription, in the presence of [α-32P]UTP, with isolated PCR fragments as templates. For the PCR amplifications, the downstream primer contained at its 5′ end the T7 RNA polymerase promoter sequence (TAATACGACTCACTATA), allowing transcription of the PCR product.
In vitro RNA analysis.
Replicative-form preparations of M13 DNA containing the SP82 sequence and mutant derivatives thereof were linearized by HindIII digestion for use as transcription templates. The M13 clone containing the SP82 sequence carries a T7 RNA polymerase promoter upstream of the SP82 sequence such that transcription begins as shown in Fig. 2 (see below). Transcription was performed in the presence of [α-32P]UTP, and the labeled RNA products were isolated from a 6% denaturing polyacrylamide gel as described previously (1). Labeled RNA was incubated in the presence of 10 mM Tris (pH 7.8)–100 mM NaCl–3 mM Mg2+, either with or without 0.5 mM Na2HPO4, and with approximately 4 μg of a B. subtilis protein extract. The extract was prepared by sonication of lysozyme-treated cells that were harvested from a late-logarithmic-phase B. subtilis culture grown in rich medium. After centrifugation at 15,000 × g for 20 min, the supernatant was dialyzed overnight against 20 mM Na-Tricine (pH 8.0)–100 mM KCl–10% glycerol–0.2 mM EDTA–0.1 mM phenylmethylsulfonyl fluoride, and 1.0 mM dithiothreitol. Aliquots were stored at −70°C. After incubation at 37°C with the extract, the reaction mixture was extracted with phenol-chloroform (1:1) and precipitated in 2 M ammonium acetate with 2.5 volumes of ethanol. The products were analyzed on a 6% denaturing polyacrylamide gel.
FIG. 2.
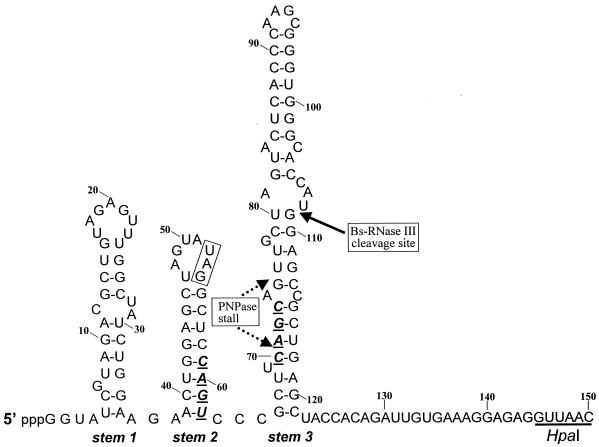
Secondary structure of the SP82 moiety of SP82-ermC RNA. The sequence shown is the one transcribed in vitro by T7 RNA polymerase. In vivo RNA contains an additional 2 nt (GU) at the 5′ end. Numbering is from the in vitro transcriptional start site. The 4-nt deletions in stems 2 and 3 are shown in boldface. The CAGC sequence that was deleted in stem 3 was replaced with an A to create an AflII site. The boxed UAG sequence in the loop portion of stem 2 is the stop codon for the putative SP82 gene 60 coding sequence.
RESULTS
Properties of the model RNA.
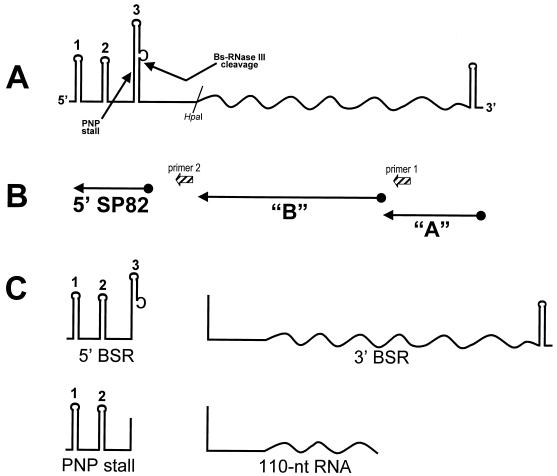
To study RNA processing in B. subtilis, a plasmid (pYH199) was constructed that contained a transcriptional unit with the following characteristics (Fig. 1): (i) transcription from the constitutive ermC promoter to give a 320-nucleotide (nt) RNA; (ii) 150 nt of bacteriophage SP82 sequence at the 5′ end and 170 nt of ermC sequence, including the transcription terminator, at the 3′ end; (iii) a Bs-RNase III cleavage site between nt 108 and 109 of the SP82 moiety; and (iv) a strong block to PNPase processivity, which was characterized in this study, located in the SP82 moiety. Thus, the expressed RNA contained elements (Bs-RNase III cleavage site, PNPase stall site) that had been found previously in vitro (13, 14), as well as a classical transcription terminator consisting of a strong stem-loop followed by a run of U residues. The proposed secondary structure for the SP82 moiety of this “SP82-ermC” RNA is shown in Fig. 2 and is based on a previous analysis by using structure-specific RNases (14). SP82 sequences are up to the HpaI site shown, followed by the 3′-terminal 170 nt beginning at the ermC HpaI site.
FIG. 1.
Diagram of SP82-ermC RNA. Straight lines, SP82 sequences; wavy lines, ermC sequences. (A) Sites of PNPase stalling and Bs-RNase III cleavage are indicated. Stem structures in the SP82 moiety are numbered 1, 2, and 3. The ermC transcription terminator is indicated by the 3′-terminal stem-loop. (B) Location of riboprobes used in Northern blot analyses and oligonucleotide primers used for reverse transcriptase mapping. (C) Schematic diagram of major RNAs detected in Northern blot analyses. BSR, Bs-RNase III; PNP, PNPase.
Block to PNPase processivity.
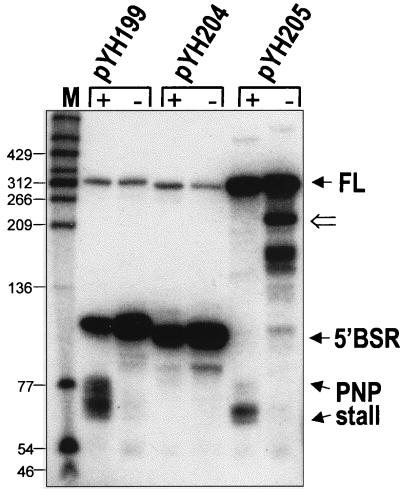
The presence of a PNPase-specific block in SP82-ermC RNA encoded by pYH199 was demonstrated by the Northern blot analysis in Fig. 3 (pYH199 lanes). The blot was probed with a riboprobe complementary to the SP82 moiety (Fig. 1B). Little of the full-length RNA was observed due to rapid Bs-RNase III cleavage. In addition to the 5′ Bs-RNase III cleavage product, a group of bands in the range of 60 to 80 nt, with a concentration of bands of approximately 65 to 70 nt, was observed in the wild-type strain but not in the pnpA deletion strain. Previous in vitro experiments, using 5′-end-labeled RNA, demonstrated that these products started at the native 5′ end and terminated with 3′ ends located upstream of the Bs-RNase III cleavage site (14). Since PNPase is a 3′-to-5′ exonuclease, we hypothesized that this set of bands was the result of an encounter by PNPase with a structural element in the SP82 moiety that stalled PNPase processivity. It was also observed that the 5′ Bs-RNase III cleavage product was present in somewhat greater abundance in the pnpA strain, indicating that PNPase is capable of degrading this RNA fragment.
FIG. 3.
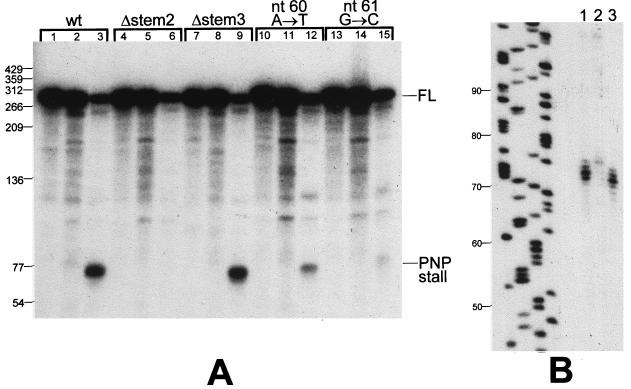
Northern blot analysis of SP82-ermC RNA, using the SP82 5′ riboprobe. RNA was isolated from wild-type (+) and pnpA (−) strains containing the wild-type SP82 sequence (pYH199), the 4-nt deletion in stem 2 (pYH204), and the 4-nt deletion in stem 3 (pYH205). The marker lane (M) contained 5′-end-labeled DNA fragments of TaqI-digested plasmid pSE420 (3). Values to the left are molecular sizes in nucleotides. The location of the 220-nt RNA detected in the strain carrying pYH205, which encodes the SP82-ermC RNA that is not cleaved by Bs-RNase III, is indicated by the open arrow to the right of the blot. FL, full-length RNA.
Stem 2 is the strongest predicted structure in the SP82 moiety; it is located upstream of the 3′ end of the PNPase-specific products detected in the wild-type strain. A mutation was made that deleted four nucleotides (CAGU) in stem 2 (Fig. 2), severely disrupting the potential to form secondary structure at this site. Northern blot analysis of RNA encoded by a pYH199 derivative containing this mutation (pYH204) showed an absence of PNPase-specific products, even in the wild type (Fig. 3, pYH204 lanes). Thus, stem 2 appeared to be important for formation of the PNPase block.
We wished to test whether the particular nucleotides at which PNPase stalls were important for the stalling effect. In addition, we wished to test whether prior Bs-RNase III cleavage was necessary for the attack and subsequent stalling of PNPase. In other words, is the 3′ end generated by Bs-RNase III cleavage necessary for attack by PNPase and subsequent stalling, or can PNPase initiate degradation at a distal 3′ end and stall at the same site? For this, a 4-nt deletion (CAGC) was made in the left side of stem 3 (Fig. 2), in the middle of the region at which PNPase processivity is stalled. Deletion of these nucleotides was expected to alter the secondary structure of stem 3 such that it may not serve as a target for Bs-RNase III cleavage. Previous in vitro experiments have shown that perturbations in the lower stem portion of the SP82 Bs-RNase III cleavage site affect Bs-RNase III activity (13). The plasmid carrying the stem 3 mutation was designated pYH205. Northern blot analysis of RNA encoded by pYH205 showed that, indeed, Bs-RNase III cleavage was not detectable (Fig. 3, pYH205 lanes). The stall to PNPase processivity was apparent in the wild-type strain, although the distribution of bands was more restricted than for pYH199 RNA. (The identity of the strong band [ca. 220 nt] below the full-length RNA, which is seen in the pnpA deletion strain carrying pYH205, is discussed below; the identity of the other bands [ca. 150 nt] detected in this strain has not been studied.) We concluded that PNPase degradation and subsequent stalling can initiate downstream of the Bs-RNase III cleavage site and that the specific nucleotides at the stall site are not necessary for stalling.
Single-nucleotide changes in stem 2.
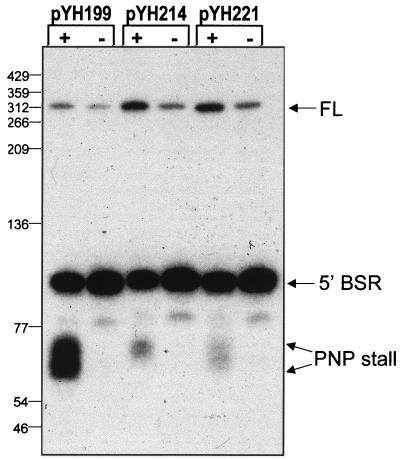
The 4-nt deletion in stem 2, contained on plasmid pYH204, was predicted to severely affect the stem structure. To determine the effect of less drastic structural alterations, we made single-nucleotide changes in stem 2. Plasmid pYH214 contained an SP82-ermC RNA with an A-to-T change at nt 60, while plasmid pYH221 had a G-to-C change at nt 61 (Fig. 2). Northern blot analysis of RNA isolated from strains carrying these plasmids showed that the PNPase-stalled products were detected but in sharply decreased amounts relative to the wild type (Fig. 4). Quantitation of the group of bands representing PNPase stalling revealed that the amount of these products in strains with the single-nucleotide mutations in stem 2 was 7- to 7.5-fold smaller than in the wild type.
FIG. 4.
Northern blot analysis of SP82-ermC RNA with single-nucleotide mutations. RNA was isolated from wild-type (+) and pnpA (−) strains carrying the A-to-T change in nt 60 (pYH214) and the G-to-C change in nt 61 (pYH221).
In vitro analysis of PNPase stalling.
Since the RNA products ending at the SP82 stall site were observed only in the wild-type strain and not in the pnpA deletion mutant, it could be assumed that these products were generated by PNPase activity that was stalled at this site. However, it could also be that the absence of PNPase disrupts the function of other RNA cleavage/recognition proteins that are involved in the formation of these RNA products. Furthermore, the evidence that stem 2 creates a block to PNPase processivity could be explained differently. For example, the observed products could be the result of endonucleolytic cleavage, followed by limited degradation or by 3′ polyadenylation. Perturbation of the stem 2 sequence might interfere with such endonucleolytic cleavage. We therefore used a B. subtilis protein extract to examine PNPase-dependent stalling in vitro. The substrates used were transcribed by T7 RNA polymerase from a template that contained the same 150 nt of SP82 sequence to the HpaI site as in pYH199 (except for the first 2 nt at the 5′ end), followed by an additional 145 nt of SP82 sequence. Mutant templates were prepared which contained the same changes as those present on the plasmids used for the in vivo analyses described above.
PNPase-specific activity was tested by the presence or absence of inorganic phosphate (Pi). To avoid the complication of Bs-RNase III activity, which is present in an extract from a wild-type strain, an extract from strain BG218 was used. BG218 contains a partial disruption of the rncS gene such that Bs-RNase III activity is virtually undetectable in an extract from this strain (19). The results with the wild-type substrate are shown in Fig. 5A, lanes 1 to 3. Incubation in the presence of Mg2+ alone (lane 2) resulted in little degradation of the full-length substrate. This is consistent with our previous findings of low-level Mg2+-dependent decay in a B. subtilis extract (18). When Pi was added (lane 3), the PNPase activity resulted in degradation of about 85% of the full-length substrate, with the appearance of an RNA product representing a stalling of PNPase processivity. The size of this RNA product was similar to that observed in vivo, but there was a tighter distribution of 3′ ends. Thus, the observed products were phosphate dependent, indicating strongly that they are generated by PNPase activity. Reducing the substrate concentration 10-fold did not affect the results (data not shown), suggesting that PNPase activity was in excess.
FIG. 5.
In vitro analysis of PNPase processing. (A) Low-resolution denaturing gel showing processing of labeled SP82 RNA incubated in the presence of a BG218 protein extract. The RNA substrate is indicated at the top of each set (Δ, deletion mutant). The three lanes in each set contained a control in which RNA processing was inactivated by the immediate addition of phenol-chloroform (lanes 1, 4, 7, 10, and 13), the mixture without Pi (lanes 2, 5, 8, 11, and 14), and the mixture with Pi (lanes 3, 6, 9, 12, and 15). (B) High-resolution denaturing gel showing the migration of the PNPase processing product from the wild-type (lane 1) and stem 3 deletion mutant (lane 3) substrates. The leftmost lanes show a sequencing reaction used as a molecular size marker (sizes in nucleotides are indicated at the left). Lane 2 contains RNA that was incubated with the extract in the presence of Pi and 3 mM ATP. The products of this incubation migrated near the top of the gel (not shown), suggesting the presence of polyadenylating activity in the extract. This result is not relevant to the subject of this paper.
RNA substrates containing the mutations described above were tested in the in vitro assay. The amount of degradation of the full-length substrate in the presence of Pi was approximately 85% in all cases. The stem 2 deletion mutant (Fig. 5A, lanes 4 to 6) showed no RNA resulting from PNPase stalling, as was observed in vivo. The stem 3 deletion mutant showed the same pattern as the wild-type substrate (lanes 7 to 9). On closer examination, using a higher resolution (sequencing) gel, the PNPase stalling products for the wild type and stem 3 deletion mutant were very similar (Fig. 5B). The amount of RNA resulting from PNPase stalling in the substrates that contained single-nucleotide changes in stem 2 was dramatically affected, as it was in vivo. The A-to-T change at nt 60 resulted in a weak PNPase stall (Fig. 5A, lanes 10 to 12), while the G-to-C change at nt 61 resulted in a barely detectable PNPase stall (lanes 13 to 15).
Since the in vitro system appeared to mirror the in vivo results, the in vitro assay was used to determine whether PNPase was completely blocked by stem 2 or whether PNPase was able to proceed past the block at some frequency. For this, uniformly labeled SP82 RNA that terminated at the HpaI site was incubated for 15 min at 37°C in the presence of extract with Mg2+ only or with Mg2+ plus Pi. The products were run on a 6% denaturing polyacrylamide gel, and the counts in the full-length and the PNPase-processed RNA bands were measured. It was assumed that the RNA substrate was uniformly labeled; thus, based on the number of U residues in each of the RNA species, quantitation of the full-length input RNA and the small RNA resulting from the PNPase block would give the percentage of RNA molecules on which PNPase was blocked and could not be further degraded. The percentage of RNA remaining in the PNPase-blocked form was 97.8 ± 10.8 (mean of five experiments; data not shown). Thus, the block to PNPase processivity on RNA molecules that had been attacked by PNPase was virtually complete. We tentatively concluded that the nature of the stem 2 structure provides an absolute block to PNPase processivity.
Alternatives to this conclusion were considered and were tested (data not shown). For example, it could be that the PNPase activity in the extract dies after a few minutes of incubation at 37°C. To test this, we analyzed the PNPase activity after preincubation of the extract in buffer containing Pi but without labeled RNA substrate (the extract contains cellular RNAs, including substantial amounts of rRNA). Preincubation for as long as 30 min showed no effect on the ability of PNPase to degrade the substrate up to the block. It was also possible that upon incubation with the extract, the substrate undergoes some change (e.g., protein binding), which would interfere with PNPase activity. To test this, the RNA was preincubated in the extract prior to addition of Pi. This treatment also did not affect the pattern of PNPase-dependent degradation. It appeared, therefore, that the block to PNPase processivity was solely due to the presence of stem 2.
Block to decay in the pnpA deletion strain.
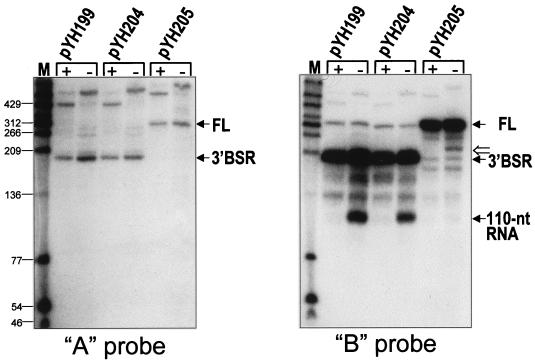
The data presented so far focused on the block to PNPase processivity, which occurs in the SP82 moiety of SP82-ermC RNA. We next examined the ermC moiety of SP82-ermC RNA by using probes directed at the upstream and downstream portions of the ermC sequence, as shown in Fig. 1B (probes A and B). RNA isolated from wild-type and pnpA mutant strains carrying pYH199 was examined by Northern blot analysis (Fig. 6). The A probe, which is complementary to 3′-proximal sequences, detected the 3′ Bs-RNase III cleavage product as well as a faint signal for full-length SP82-ermC RNA. (The A probe consistently gave weaker signals than other probes.) By using the A probe, only the 3′ Bs-RNase III cleavage product was detected, with the pnpA strain containing about twice the amount of this product as the wild-type strain. This suggested that PNPase is not required for decay of the 3′ Bs-RNase III cleavage product. On the other hand, the B probe, which also detected the 3′ Bs-RNase III cleavage product, gave a strong signal for a band of about 100 nt in the pnpA strain only (Fig. 1C). This band was also detected in the strain carrying pYH204, which contained the stem 2 deletion mutant.
FIG. 6.
Northern blot analysis of SP82-ermC RNA, using the A and B riboprobes. RNA was isolated from wild-type (+) and pnpA (−) strains containing the wild-type SP82 sequence (pYH199), the 4-nt deletion in stem 2 (pYH204), and the 4-nt deletion in stem 3 (pYH205). Values to the left are molecular sizes in nucleotides. The location of the 220-nt RNA detected in the strain carrying pYH205, which encodes the SP82-ermC RNA that is not cleaved by Bs-RNase III, is indicated by the open arrow to the right of the blot probed with the B riboprobe.
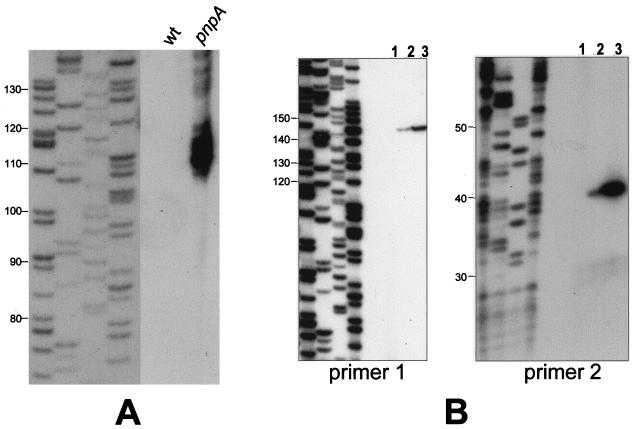
The nature of the band that was detected only in the pnpA strain was examined further by blotting from a sequencing gel and by reverse transcriptase analysis. The sequencing gel blot was used to more accurately determine the size of this RNA or group of RNAs. The data in Fig. 7A show a group of bands detected in the pnpA strain ranging from 108 to 120 nt, with the strongest signal at about 110 nt. Below, we call this group of RNAs the 110-nt RNA. To map the 5′ end of this RNA, reverse transcriptase analysis was performed (Fig. 7B). For this analysis, two oligonucleotide primers (primers 1 and 2) were used (Fig. 1B). Primer 1 was complementary to ermC sequences, with its 5′ end located 145 nt downstream of the Bs-RNase III cleavage site. A single 5′ end was mapped for the SP82-ermC RNA in the wild-type and the pnpA strains, and this corresponded precisely to the Bs-RNase III cleavage site. The amount of reverse transcriptase product in the pnpA strain was about three times that in the wild-type strain, which reflects the relative abundance of the 3′ Bs-RNase III cleavage product detected by the A probe (Fig. 6). Primer 2 was complementary to SP82 sequences, with its 5′ end located 39 nt downstream of the Bs-RNase III cleavage site. Again, a single 5′ end was mapped, which was at the Bs-RNase III cleavage site. The amount of reverse transcriptase product from the pnpA strain (in which there was abundant 110-nt RNA) was approximately 13-fold more than in the wild-type strain (which had no detectable 110-nt RNA). From these mapping data, we conclude that there is a block to 3′-to-5′ degradation in the ermC sequence, which is located about 110 nt downstream of the Bs-RNase III cleavage site.
FIG. 7.
Analysis of 110-nt RNA. (A) Northern blot analysis of RNA from wild-type and pnpA strains carrying pYH199, which were separated on a sequencing gel. The probe was the B riboprobe (Fig. 1B). The leftmost lanes show a sequencing reaction used as a molecular size marker (sizes in nucleotides are indicated at the left). The figure is a composite of two exposures of the same filter. (B) Reverse transcriptase analysis of RNA isolated from wild-type and pnpA strains carrying pYH199, using primer 1 (left) or primer 2 (right). The four leftmost lanes in each panel show sequencing reactions used as molecular size markers (sizes in nucleotides are indicated at the left). Lanes: 1, wild-type strain with no plasmid; 2, wild-type strain with pYH199; 3, pnpA strain with pYH199.
We would expect that in the strain carrying pYH205, which contains the stem 3 deletion mutant for which no Bs-RNase III cleavage occurs, the block to decay in the ermC sequence would produce an RNA that was 110 nt larger than the 110-nt RNA (since the Bs-RNase III cleavage site is at nt 110 of the in vivo-transcribed RNA [Fig. 2 legend]). In the Northern blot analysis probed with the B riboprobe, a band running at about 220 nt was detected in the lane for the pnpA strain carrying pYH205 (Fig. 6). The same result was observed for the pnpA strain carrying pYH205 when the SP82 5′ riboprobe was used (Fig. 3). This band was not detected in the lanes for the wild-type strain. These results indicate that blockage of 3′-to-5′ decay at the site in the ermC moiety is not dependent on Bs-RNase III cleavage.
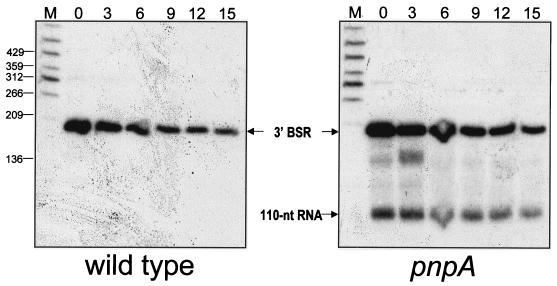
The stability of the 110-nt RNA detected in the pnpA strain carrying pYH199 was measured relative to the stability of the 3′ Bs-RNase III cleavage product (Fig. 8). The stability of the 3′ Bs-RNase III cleavage product in the wild-type and pnpA strains was determined to be 5.7 and 6.5 min, respectively. However, the 110-nt RNA was about three times as stable, with a half-life longer than 15 min (and projected to be 19.5 min). Since it is likely that the 110-nt RNA is generated by processing of the 3′ Bs-RNase III cleavage product, the half-life measurement of the 110-nt RNA probably reflects both the production and degradation of the 110-nt RNA.
FIG. 8.
Northern blot analysis of processed RNAs after the addition of rifampin. Total RNA from wild-type and pnpA strains was probed with the B riboprobe. Above each lane is the time (minutes) after rifampin addition.
DISCUSSION
That stem 2 of the SP82 moiety afforded the block to PNPase processivity was confirmed by showing an absence of this block when 4 nt of stem 2 was deleted and a strong reduction in PNPase stalling when single-nucleotide changes were introduced. The site of stalling was downstream of stem 2, but the distribution of RNA bands detected in vitro was narrower than that detected by Northern blotting of in vivo RNA (compare Fig. 5A to Fig. 3). This may reflect further processing events in vivo that occur after the block to PNPase; e.g., it is possible that the 3′ ends generated by PNPase degradation at this site are polyadenylated by a poly(A) polymerase, thus generating a more extensive ladder of bands in vivo.
The block to PNPase activity was independent of the 3′ end at which PNPase initiated degradation. The in vitro results shown here were obtained with a protein extract from BG218, which does not have detectable Bs-RNase III activity. Thus, PNPase degradation initiated at the 3′ end of the substrate located approximately 190 nt downstream of the Bs-RNase III cleavage site. Similar results were obtained with RNA substrates whose 3′ end was at the HpaI site (Fig. 2) or whose 3′ end coincided with the site of Bs-RNase III cleavage (data not shown). The in vivo results with the stem 3 deletion mutant, which was not recognized as a substrate for Bs-RNase III cleavage, demonstrated that the same PNPase block was observed even though degradation must be initiating at a site downstream of the Bs-RNase III cleavage site. Further experiments are required to determine whether PNPase can initiate degradation in vivo at the native 3′ terminus, which is immediately downstream of a strong secondary structure, or whether degradation of SP82-ermC RNA by PNPase requires a prior endonucleolytic cleavage upstream of the terminator.
We found that the block to PNPase processivity in vitro could not be overcome by extended incubation. In earlier studies of 3′-to-5′ exonuclease processivity (4, 12), blocks to E. coli PNPase or RNase II were found to be transient. It is likely that the transient nature of a block is the result of alternative conformations that can be assumed by the blocking secondary structure. If an alternate structure, which does not block exonuclease processivity, can be assumed, then at some frequency during the incubation period this structure will form and will allow passage of the exonuclease. The complete block to PNPase activity offered by stem 2 may be due to the assumption of an extraordinarily stable structure or may be a special feature of B. subtilis PNPase.
In terms of the biological significance of the SP82 sequence, we note that in the native SP82 sequence, stem 2 is located at the end of an open reading frame that for most of its coding sequence is identical to gene 60 of phage SPO1 (16). The stop codon for the SP82 gene 60 is located in the loop region of stem 2 (Fig. 2). Since gene 60 is undoubtedly expressed as an early SP82 phage gene, processing by PNPase may be an important step in gene expression.
Although stem 2 provided a strong block to PNPase processivity, RNA bands that had 3′ ends at the site of PNPase stalling were not observed in the pnpA strain. Thus, although we assume that there are other 3′-to-5′ exonuclease activities present in the pnpA strain that can accomplish mRNA turnover, such activities are not blocked at the site of PNPase stalling. This may indicate a difference in the processivity mechanism of PNPase and other exonucleases, such that the stem 2 block is specific for PNPase.
An interesting parallel to the case of the stem 2 block was the finding, in the pnpA strain, of a strong RNA band with its 3′ end located in the ermC moiety—the 110-nt RNA. It has not been possible, so far, to recreate this processing event in vitro (unpublished results). We have not resolved whether the 110-nt RNA is formed by a block to exonuclease degradation or by endonuclease cleavage. The evidence arguing against endonuclease cleavage is that we have been unable to detect the 3′ RNA product that would result from endonucleolytic cleavage at this site. Neither the A riboprobe nor several different oligonucleotide probes directed at ermC sequences downstream of the putative cleavage site could detect such an RNA fragment (unpublished results). It is possible, however, that the 3′ cleavage product is extremely unstable and therefore undetectable even in the pnpA strain. An argument in favor of the 110-nt RNA representing an endonuclease cleavage site and not a block to exonuclease processivity is that there is no apparent upstream RNA structure in the vicinity of the 110-nt RNA 3′ end that would be predicted to block processivity. Tests for computer-predicted secondary structure show little potential for even moderately strong stem-loops in this region of the ermC sequence.
Strikingly, the sequence at the site of the 3′ end of the 110-nt RNA is GAUUU, which conforms precisely to the consensus sequence originally described for RNase E endonuclease cleavage in E. coli (8). RNase E is thought to be the major RNase involved in initiation of mRNA decay in E. coli, and evidence for a B. subtilis RNase E activity has been obtained by Condon et al. (5); however, a homologue to RNase E is not identifiable from the B. subtilis genome sequence. It is tempting to speculate that the 3′ end of the 110-nt RNA represents a B. subtilis “RNase E” cleavage site. Experiments involving site-directed mutagenesis of this site in the ermC sequence may reveal the requirements for formation of the 110-nt RNA and may resolve the issue of exonucleolytic versus endonucleolytic processing.
The fact that the 110-nt band was observed only in the pnpA strain and not in the wild type suggests that PNPase is capable of degrading this portion of SP82-ermC RNA, while the exonuclease(s) that is active in the pnpA strain cannot proceed easily through this region. Thus, we have mapped two sites on SP82-ermC RNA that appear to represent blocks to 3′-to-5′ processivity: one which is specific to PNPase (stem 2) and one (in the ermC moiety) which is specific to exonucleases other than PNPase. These results indicate that the RNA products that form as a result of exonuclease activity may depend on particular degradative activities. Different RNA sites may act as blocks to different exonucleases, thus increasing the potential number of processed products. A study on the decay of the 3.7-kb cry1Aa mRNA in B. subtilis came to the conclusion that decay occurs through multiple endonuclease cleavages, whose sites are clustered near the 5′ and 3′ ends of the mRNA (17). It was pointed out, however, that the same mapped decay intermediates could result from limited exonuclease digestion. Our results with a much smaller RNA demonstrate that such limited exonuclease digestion does in fact occur. Further experiments to define the processing pathway of model RNA substrates, such as SP82-ermC RNA, are needed before we can better understand the role that RNA processing plays in mRNA decay and gene expression.
TABLE 1.
Plasmids used in this study
| Plasmid | Description |
|---|---|
| pYH199 | Wild-type SP82-ermC RNA sequence |
| pYH204 | 4-nt deletion in stem 2 |
| pYH205 | 4-nt deletion in stem 3 |
| pYH214 | A→T change of SP82 nt 60 |
| pYH221 | G→C change of SP82 nt 61 |
ACKNOWLEDGMENT
This work was supported by Public Health Service grant GM-48804 from the National Institutes of Health.
REFERENCES
- 1.Alberta J A, Rundell K, Stiles C D. Identification of an activity that interacts with the 3′ untranslated region of c-myc mRNA and the role of its target sequence in mediating rapid mRNA degradation. J Biol Chem. 1994;269:4532–4538. [PubMed] [Google Scholar]
- 2.Bechhofer D H, Wang W. Decay of ermC messenger RNA in a polynucleotide phosphorylase mutant of Bacillus subtilis. J Bacteriol. 1998;180:5968–5977. doi: 10.1128/jb.180.22.5968-5977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosius J. Compilation of superlinker vectors. Methods Enzymol. 1992;216:469–483. doi: 10.1016/0076-6879(92)16043-j. [DOI] [PubMed] [Google Scholar]
- 4.Coburn G A, Mackie G A. Differential sensitivities of portions of the mRNA for ribosomal protein S20 to 3′-exonucleases dependent on oligoadenylation and RNA secondary structure. J Biol Chem. 1996;271:15776–15781. doi: 10.1074/jbc.271.26.15776. [DOI] [PubMed] [Google Scholar]
- 5.Condon C, Putzer H, Luo D, Grunberg-Manago M. Processing of the Bacillus subtilis thrS leader mRNA is RNase E-dependent in Escherichia coli. J Mol Biol. 1997;268:235–242. doi: 10.1006/jmbi.1997.0971. [DOI] [PubMed] [Google Scholar]
- 6.Donovan W P, Kushner S R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci USA. 1986;83:120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubnau D, Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971;56:209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- 8.Ehretsmann C P, Carpousis A J, Krisch H M. Specificity of Escherichia coli endoribonuclease RNase E: in vivo and in vitro analysis of mutants in a bacteriophage T4 mRNA processing site. Genes Dev. 1992;6:149–159. doi: 10.1101/gad.6.1.149. [DOI] [PubMed] [Google Scholar]
- 9.Grant S G N, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunkel T A, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 11.Luttinger A, Hahn J, Dubnau D. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol Microbiol. 1996;19:343–356. doi: 10.1046/j.1365-2958.1996.380907.x. [DOI] [PubMed] [Google Scholar]
- 12.McLaren R S, Newbury S F, Dance G S C, Causton H C, Higgins C F. mRNA degradation by processive 3′-to-5′ exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J Mol Biol. 1991;221:81–95. [PubMed] [Google Scholar]
- 13.Mitra S, Bechhofer D H. Substrate specificity of an RNase III-like activity from Bacillus subtilis. J Biol Chem. 1994;269:31450–31456. [PubMed] [Google Scholar]
- 14.Mitra S, Bechhofer D H. In vitro processing activity of Bacillus subtilis polynucleotide phosphorylase. Mol Microbiol. 1996;19:329–342. doi: 10.1046/j.1365-2958.1996.378906.x. [DOI] [PubMed] [Google Scholar]
- 15.Panganiban A T, Whiteley H R. Bacillus subtilis RNase III cleavage sites in phage SP82 early mRNA. Cell. 1983;33:907–913. doi: 10.1016/0092-8674(83)90033-8. [DOI] [PubMed] [Google Scholar]
- 16.Stewart C R, Gaslightwala I, Hinata K, Krolikowski K A, Needleman D S, Peng A S-Y, Peterman M A, Tobias A, Wei P. Genes and regulatory sites of the “host-takeover module” in the terminal redundancy of Bacillus subtilis bacteriophage SPO1. Virology. 1998;246:329–340. doi: 10.1006/viro.1998.9197. [DOI] [PubMed] [Google Scholar]
- 17.Vázquez-Cruz C, Olmedo-Alvarez G. Mechanism of decay of the cry1Aa mRNA in Bacillus subtilis. J Bacteriol. 1997;179:6341–6348. doi: 10.1128/jb.179.20.6341-6348.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Bechhofer D H. Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J Bacteriol. 1996;178:2375–2382. doi: 10.1128/jb.178.8.2375-2382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Bechhofer D H. Bacillus subtilis RNase III gene: cloning, function of the gene in Escherichia coli, and construction of Bacillus subtilis strains with altered rnc loci. J Bacteriol. 1997;179:7379–7385. doi: 10.1128/jb.179.23.7379-7385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]