Abstract
Bacillus subtilis JH642 and a wild strain of B. subtilis called 22a both produce an antilisterial peptide that can be purified by anion-exchange and gel filtration chromatography. Amino acid analysis confirmed that the substance was the cyclic bacteriocin subtilosin. A mutant defective in production of the substance was isolated from a plasmid gene disruption library. The plasmid insertion conferring the antilisterial-peptide-negative phenotype was located in a seven-gene operon (alb, for antilisterial bacteriocin) residing immediately downstream from the sbo gene, which encodes the precursor of subtilosin. An insertion mutation in the sbo gene also conferred loss of antilisterial activity. Comparison of the presubtilosin and mature subtilosin sequences suggested that certain residues undergo unusual posttranslational modifications unlike those occurring during the synthesis of class I (lantibiotic) or some class II bacteriocins. The putative products of the genes of the operon identified show similarities to peptidases and transport proteins that may function in processing and export. Two alb gene products resemble proteins that function in pyrroloquinoline quinone biosynthesis. The use of lacZ-alb and lacZ-sbo gene fusions, along with primer extension analysis, revealed that the sbo-alb genes are transcribed from a major promoter, residing upstream of sbo, that is very likely utilized by the ςA form of RNA polymerase. The sbo and alb genes are negatively regulated by the global transition state regulator AbrB and are also under positive autoregulation that is not mediated by the subtilosin peptide but instead requires one or more of the alb gene products.
Polypeptide antibiotics possess bacteriocidal, fungicidal, metal-chelating, and immunomodulating activities. They are frequently found as secondary metabolites or small, secreted proteins produced by various microorganisms, such as the gram-positive bacteria of the genus Bacillus, lactic acid bacteria, and the genus Streptomyces (22, 23, 25, 27, 60). In Bacillus subtilis, some polypeptide antibiotics, such as bacteriocins, are gene encoded and are synthesized ribosomally while others are produced nonribosomally by the multienzyme thiotemplate mechanism (60). Bacteriocins such as nisin, produced by Lactococcus lactis, can be used in foods as antimicrobial agents to replace chemical preservatives, such as nitrite (35), that are potentially hazardous or carcinogenic. As demonstrated in studies of subtilin (B. subtilis), nisin (L. lactis), pediocin (Pediococcus acidilactici), and other known bacteriocins, the bacteriocins of gram-positive bacteria are typically first formed as precursors with reduced biological activity (6, 21, 38). The C-terminal ends of the precursors are then cleaved from the N-terminal leader sequences to yield the mature, active bacteriocins. In some cases, the precursor polypeptide undergoes posttranslational modifications. The formation of lanthionine and thiazole or oxazole adducts is characteristic of the maturation processes of class 1 bacteriocins (lantibiotics) and microcins of the gram-negative bacterium Escherichia coli, respectively (4, 32). Transport of the peptides to the external environment is carried out by ATP-dependent efflux protein complexes that are membrane associated (16). Genes involved in the biosynthesis of bacteriocins are typically organized into operons (21, 28) which include the bacteriocin structural gene and genes whose products function in bacteriocin maturation, export, immunity, and, in some cases, the regulation of operon expression. The operons that encode proteins that function in lantibiotic biosynthesis also contain genes encoding simple signal transduction systems composed of two-component regulatory proteins (6, 26, 29). These genes mediate a form of positive-feedback regulation that is induced in response to the presence of the lantibiotic (nisin or subtilin).
In B. subtilis, production of and resistance to antibiotics are regulated by the spo0-abrB system of control. The Spo0 phosphorelay is activated by conditions of nutritional stress and high cell density (2, 17, 18). The signals derived from these conditions are integrated into the phosphorelay and promote the accumulation of Spo0A phosphate, which activates sporulation gene transcription and represses the transcription of the transition state regulatory gene abrB (11, 48, 51). Mutations in spo0A render B. subtilis cells unable to produce certain antibiotics and confer sensitivity to those antibiotics. A mutation in abrB suppresses this spo0A phenotype (13, 14, 19, 54), indicating that AbrB exerts negative control of antibiotic production and resistance. AbrB is known to interact directly with the promoter regions of several genes that are normally induced in the transition from exponential growth to stationary phase (44, 48–50). The tycA operon, encoding the enzyme tyrocidine synthetase, which catalyzes the synthesis of a cyclic peptide antibiotic, is but one operon that is repressed by AbrB (8, 33).
Antimicrobial substances produced by a wild strain of B. subtilis isolated from an Oriental fermented food (57a, 58) are currently under investigation in our laboratory. One of these substances was initially identified as a bacteriocin endowed with activity against Listeria monocytogenes and Bacillus cereus. As detailed in this report, an operon required for the observed activity has been identified by insertion mutagenesis. The operon (alb, for antilisterial bacteriocin) consists of seven genes and is preceded by the gene sbo, encoding subtilosin, a modified antimicrobial peptide originally identified by Kurahashi and coworkers (1). The sbo gene resides in the vicinity of fnr and argS (encoding arginyl-tRNA synthetase) (30). The peptide product is composed of 32 common amino acids and some unusual residues that are likely the result of posttranslational modifications (Fig. 1). Comparison of the presubtilosin and mature subtilosin sequences suggests that the Sbo primary translation product may undergo novel modifications. The regulation of sbo-alb was also investigated by using alb- and sbo-lacZ fusions. A novel form of autogenous regulation that does not involve the product of sbo but instead requires an alb operon product(s) was uncovered.
FIG. 1.
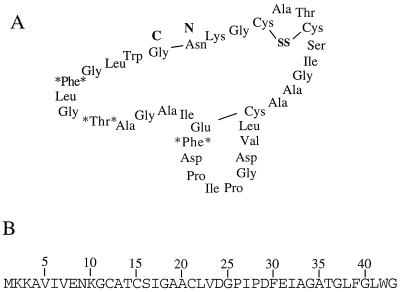
(A) Proposed structure of subtilosin (1). The boldfaced N and C mark the N and C termini of the prosubtilosin peptide (presubtilosin leader peptide removed), respectively. The proposed link between Glu31 and Cys21 (1) is shown. The asterisks on either side of an amino acid indicate residues that have likely undergone chemical modification. SS, disulfide link between Cys12 and Cys15. (B) Amino acid sequence of the presubtilosin peptide, deduced from the nucleotide sequence of the sbo coding region.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains used in this study are listed in Table 1. Strain 22a is a wild strain of B. subtilis isolated from an Oriental fermented soybean product (58). With the exception of wild strain 22a, all strains constructed are derivatives of B. subtilis JH642. The indicator bacterium, L. monocytogenes F4244, was provided by M. Slavik (University of Arkansas). The gene disruption plasmid library, in which 0.5- to 2-kb genome DNA fragments of JH642 were randomly inserted into the vector pJPM1 (31, 47), was obtained from A. L. Sonenshein (Tufts University). ZB449 is an SPβ-cured strain bearing a frameshift mutation in the abrB gene (59). TT71 (a gift from T. Tanaka) carries an insertion of a neomycin resistance (Neor) cassette (neo) in the abrB gene.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype (phenotype) | Reference or source |
|---|---|---|
| L. monocytogenes F4224 | M. Slavik | |
| B. subtilis | ||
| JH642 | trpC2 pheA1 | J. Hoch |
| 22a | Wild strain | 58 |
| ZB449 | trpC2 pheA1 abrB703 | 59 |
| LAB2136 | trpC2 pheA1 fnr::spc | 31 |
| TT71 | trpC2 abrB::neo | T. Tanaka |
| ORB3146 | trpC2 pheA1 alb::pE1 (Cmr) | This study |
| ORB3147 | trpC2 pheA1 alb::pMUPE1 (Ermr) | This study |
| ORB3148 | trpC2 pheA1 sbo::neo-1 | This study |
| ORB3149 | trpC2 pheA1 sbo::neo-2 | This study |
| ORB3152 | trpC2 pheA1 sbo::neo-1 albABC::pMUPE1 (Ermr) | This study |
| ORB3153 | trpC2 pheA1 sbo::neo-2 albABC::pMUPE1 (Ermr) | This study |
| ORB3154 | trpC2 pheA1 SPβc2del2::Tn917::pTK-lac (Ermr Cmr) | This study |
| ORB3158 | trpC2 pheA1 SPβc2del2::Tn917::pTK-sboΔEB (Ermr Cmr) | This study |
| ORB3159 | trpC2 pheA1 sbo::neo-1 SPβc2del2::Tn917::pTK-sboΔEB (Ermr Cmr) | This study |
| ORB3160 | trpC2 pheA1 sbo::neo-2 SPβc2del2::Tn917::pTK-sboΔEB (Ermr Cmr) | This study |
| ORB3161 | trpC2 pheA1 alb::pE1 SPβc2del2::Tn917::pTK-sboΔEB (Ermr Cmr) | This study |
| ORB3162 | trpC2 pheA1 SPβc2del2::Tn917::pTK-sboΔBH (Ermr Cmr) | This study |
| ORB3163 | trpC2 pheA1 sbo::neo-1 SPβc2del2::Tn917::pTK-sboΔBH (Ermr Cmr) | This study |
| ORB3164 | trpC2 pheA1 sbo::neo-2 SPβc2del2::Tn917::pTK-sboΔBH (Ermr Cmr) | This study |
| ORB3165 | trpC2 pheA1 alb::pE1 SPβc2del2::Tn917::pTK-sboΔBH (Ermr Cmr) | This study |
| ORB3166 | trpC2 pheA1 SPβc2del2::Tn917::pTK-sboEH (Ermr Cmr) | This study |
| ORB3167 | trpC2 pheA1 sbo::neo-1 SPβc2del2::Tn917::pTK-sboEH (Ermr Cmr) | This study |
| ORB3168 | trpC2 pheA1 sbo::neo-2 SPβc2del2::Tn917::pTK-sboEH (Ermr Cmr) | This study |
| ORB3169 | trpC2 pheA1 alb::pE1 SPβc2del2::Tn917::pTK-sboEH (Ermr Cmr) | This study |
| ORB3230 | trpC2 pheA1 sbo::neo-2 alb::pMUALBG (Ermr) | This study |
| ORB3284 | trpC2 pheA1 sbo::neo-1 alb::pMUALBG (Ermr) | This study |
| ORB3231 | trpC2 pheA1 alb::pMUALBG (Ermr) | This study |
| ORB3237 | trpC2 pheA1 abrB::neo | This study |
| ORB3238 | trpC2 pheA1 abrB::neo alb::pE1 (Cmr) | This study |
To construct the sbo::neo mutant, an EcoRI-HindIII fragment containing the sbo gene along with 512 bp of sequence upstream and 578 bp of sequence downstream of sbo was obtained by PCR with primers osboP1 and osboP2 (Table 2) and was then inserted into EcoRI- and HindIII-cleaved pUC18 (57). The resulting plasmid, pUC-sboEH, was used as template for a PCR using two partially complementary oligonucleotides (osboP3 and osboP4) specifying a BamHI site within the sbo coding sequence 43 bp downstream of the ATG start codon. The PCR DNA product was then cleaved with BamHI and subjected to intramolecular ligation, yielding pUC-sboEBH. A BglII-BamHI fragment bearing the neo gene from pDG782 (12) was inserted into the BamHI site, and the resulting ligation mixture was used to transform competent cells of E. coli, with selection for Neor (50 μg/ml). The plasmid was purified from the Neor transformants and used to transform cells of JH642 to create mutant strains ORB3148 (sbo::neo-1) and ORB 3149 (sbo::neo-2). The plasmid DNA recombined with the sbo gene DNA by a double-crossover mechanism by virtue of the homologous sbo DNA fragments flanking the neo insertion cassette. The insertion was confirmed by PCR analysis. The plasmid pE1 was acquired by a spontaneous loop-out recombination event that occurred in the isolate obtained in the plasmid disruption library mutagenesis. It contains a 1,178-bp fragment of the alb locus extending from 472 bp upstream to 646 bp downstream of albB (ywhR). Plasmid pE1 was used to transform competent cells of JH642 to yield the alb::pE1 insertion mutant ORB3146.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| osboP1 | CCTCATGACCAGGACTTCGCCTTCGCTTACTTT |
| osboP2 | CGGTGCCGAGCGCTTCAGGTAAGCTTTCCAAA |
| osboP3 | TGCTGGATCCGAGCCGCTTGTCTAGTGGACGGTCCTAT |
| osboP4 | CTCGGATCCAGCATGTTGCACAACCTTTGTTTTCTA |
| oywhm-U | GCACTGTCTTTAAGCTTTTATTGCTGCTTA |
| oywhm-L | GGATCAGTAGGATCCCAAGTCCCATTGAAA |
| ocalbA-L | CCCTCAGGAAGCTGGTGAACTCTTACACTTT |
To construct pMUPE1, a HindIII-BamHI fragment containing the 1,178-bp segment of alb DNA in pE1 was inserted into HindIII- and BamHI-cleaved pMUTIN2 (55). This plasmid was used to transform JH642 and mutant derivatives, with selection for erythromycin resistance. Plasmid pMUALBG, another derivative of pMUTIN2, was constructed by inserting a HindIII-BamHI PCR fragment (generated with primers oywhm-U and oywhm-L) containing bp 22 to 465 of the albG (ywhM) coding sequence into HindIII- and BamHI-cleaved pMUTIN2. The resulting recombinant plasmid was used to transform competent cells of JH642, with selection for erythromycin resistance, yielding strain ORB3231.
Plasmids pTK-sboEH, pTK-sboEBH, pTK-sboΔEB, and pTK-sboΔBH, all derivatives of pTKlac (24), were used to construct transcriptional lacZ fusions for analyses of promoter activity and regulation of the sbo gene and alb operon. The EcoRI-HindIII fragments released from pUC-sboEH (carrying the intact sbo gene) and pUC-sboEBH (containing the sbo BamHI allele [see below]) were inserted into EcoRI- and HindIII-cleaved pTKlac to generate pTK-sboEH and pTK-sboEBH, respectively. pTK-sboΔEB was a derivative of pTK-sboEBH with a deletion of the EcoRI-BamHI fragment, while pTK-sboΔBH was a derivative of pTK-sboEBH with a deletion of the BamHI-HindIII fragment. The plasmid pTK-sboΔEB contains the 3′ half of the sbo gene and the 5′ end of albA (to bp 442).
The sbo-lacZ fusion plasmids derived from pTKlac were introduced into prophage SPβc2del2::Tn917::pSK10Δ6 of strain ZB307 by transformation as previously described (59). Heat-induced lysates containing fusion-bearing phages were obtained and were used to transfer the sbo-lacZ fusions to mutant derivatives of JH642 by specialized transduction.
Isolation of an antilisteria-protein-negative mutant.
A plasmid gene disruption library was obtained from L. Sonenshein (47). The plasmid library was used to transform JH642, with selection for chloramphenicol resistance. Total chromosomal DNA was purified from the pool of transformants and used to transform competent cells of B. subtilis 22a. Chloramphenicol resistant (Cmr) transformants were then screened for loss of activity against L. monocytogenes F4244. After the transformants were patched onto yeast extract-glucose (YG) plates and incubated for 20 to 24 h at 30°C, the YG cultures were then overlaid with brain-heart infusion semisoft agar (0.8% agar; 25 μg of nalidixic acid/ml) containing 0.1 ml of an overnight culture of L. monocytogenes and incubated at 37°C for 18 to 24 h. Of 3,400 resultant colonies, 7 had no zone of inhibition when overlayed with a suspension of L. monocytogenes. To confirm the phenotypic linkage of antilisterial activity and Cmr, chromosomal DNA was prepared from the mutants and used to transform B. subtilis 22a. Mutants that showed 100% transformation linkage between the antilisterial phenotype and plasmid-associated drug resistance were chosen. One of the mutations was transferred to strain JH642 by transformation, using mutant chromosomal DNA, with selection for Cmr. The mutation also conferred an antilisterial-protein-negative phenotype in JH642.
To identify the gene(s) of the mutant locus affecting antilisterial activity in JH642, the integrated plasmid and flanking region were outcloned. Chromosomal DNA of a mutant was digested with EcoRI or HindIII, ligated at a low DNA concentration to facilitate intramolecular ligation, and then used to transform E. coli DH5α competent cells. However, the plasmid obtained, pE1, was not generated by restriction endonuclease digestion but rather was a product of a spontaneous loop-out recombination event. The pE1 insert was subjected to nucleotide sequence analysis.
Culture media.
YG (2% glucose, 0.5% yeast extract, and 0.1% trace-metal solution [2.2 g of ZnSO4 · 7H2O, 1.1 g of H3BO3, 0.5 g of MnCl2 · 4H2O, 0.5 g of FeSO4 · 7H2O, 0.16 g of CoCl2 · 5H2O, 0.16 g of CuSO4 · 5H2O, 0.11 g of (NH4)5Mo7O24 · 4H2O, and 5.0 g of disodium EDTA in 100 ml]) was used to culture strain JH642 for the purification of subtilosin. B. subtilis cells were routinely grown on agar plates containing Difco sporulation medium (DSM) (15). Cells of lacZ fusion-bearing strains were grown on DSM plus 0.5% glucose (DSM-G) for the time course β-galactosidase assay experiments. Solid TSS minimal medium (7) was used to examine auxotrophic phenotypes. E. coli cells were routinely grown in 2× YT (yeast extract-tryptone) medium.
Transformation.
Preparation of competent B. subtilis cells and genetic transformation were carried out as previously described (5). Preparation of E. coli competent cells and plasmid transformation were performed according to published procedures (45).
Assay of β-galactosidase activity.
All inocula were grown overnight at 37°C on solid DSM supplemented with the appropriate antibiotics. The cells, harvested by washing the plate surface with 2 ml of DSM, then were used to inoculate batch cultures containing either DSM or DSM-G to an initial optical density at 595 nm of about 0.17 or an initial Klett value (red filter) of about 8. The cultures were grown at 37°C in a shaking water bath. Collection of 1-ml samples was started when the culture reached an optical density at 595 nm of OD595 0.4 or had a Klett reading of around 20. Collection of samples for β-galactosidase activity assays continued at 30-min or 1-h intervals. Measurement of β-galactosidase activity has been described previously (36).
Purification of subtilosin.
Two hundred milliliters of YG broth was inoculated with a single colony of B. subtilis JH642 and incubated at 32°C with shaking (200 rpm) for 36 h. The supernatant was collected by centrifugation (21,252 × g, 10 min) and adjusted with 1 M Tris buffer (pH 7.5) to 20 mM Tris-HCl (final concentration). The buffered supernatant was filtered through a 0.45-μm-pore-size syringe filter and subjected to chromatography with an anion-exchange cartridge (5-ml High Q; Bio-Rad). After sample application, the cartridge was washed with 20 mM Tris (pH 7.5) for 20 min (3 ml/min) followed by elution with a linear gradient, starting with 20 mM Tris (pH 7.5) and ending with 1 M NaCl in 20 mM Tris (pH 7.5), over 50 min. Fractions were collected, and those showing inhibitory activity against L. monocytogenes F4244 (between 100 and 200 mM NaCl) were pooled. The pooled fractions were extracted with one-fourth volume of butanol at room temperature for 1 h and allowed to stand overnight at room temperature. The butanol layer was dried by evaporation at 55°C. The residue was dissolved in 1 ml of methanol and subjected to chromatography on a Sephadex LH-20 column (1.5 by 25 cm). Elution was performed with methanol at a rate of 3 ml/min. Fractions (3 ml each) of the first absorption peak (at 280 nm) were pooled, dried by evaporation, and resuspended in 400 μl of 20 mM sodium phosphate buffer (pH 7.0).
Subtilosin A activity assay and protein concentration determinations.
Antilisterial activity of subtilosin A was determined by the critical dilution assay (58). Protein concentrations were determined spectrophotometrically by using the Bio-Rad (Hercules, Calif.) protein assay with bovine serum albumin as a standard.
Bioautography of subtilosin by SDS-PAGE.
Samples of Sephadex LH-20 eluates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 16% gels with Tris-Tricine running buffer. Each sample (10 μl) was loaded in duplicate onto the gel. After electrophoresis was conducted at 100 V for 120 min, each lane of the gel was cut vertically. For each sample, one lane was stained with Coomassie blue to visualize the separated protein bands while the other lane was assayed for inhibitory activity against L. monocytogenes F4244 according to the method of Zheng and Slavik (58).
Sequencing and primer extension.
Total RNA was purified by the method of Nakano et al. (37) from JH642 and ZB449 cells collected at T2 (i.e., 2 h after the end of the exponential growth phase) from DSM-G cultures. Primer extension was performed with oligonucleotides osboP4 (hybridizing to nucleotides 20 to 55 of the sbo coding sequence) and ocalbA-L (hybridizing to a sequence within albA from 31 to 61 bp from the TTG start codon). Primer extension was carried out according to published protocols (37). DNA sequencing was conducted by using a Sequenase version 2.0 kit (U.S. Biochemical Corp.), [α-35S]dATP (ICN), and plasmid pTK-sboEH (as a template).
RESULTS
Studies of the nature of the antilisterial activity produced by the wild strain of B. subtilis known as 22a, isolated from Oriental fermented food (58), and the standard genetic strain JH642 were conducted. The sbo gene, encoding the bacteriocin subtilosin (Fig. 1) (1), and the genes of the alb operon (albA to -G are, respectively, genes ywiA, ywhR, ywhQ, ywhP, ywhO, ywhN, and ywhM, according to the B. subtilis genome sequencing consortium [30]), which function in the production of antilisterial activity, were identified in a search for mutants defective in production of the antilisterial activity.
Nucleotide sequence analysis of the sbo-alb locus.
Mutants of B. subtilis 22a (58) and JH642 that were defective in production of the antilisterial activity were identified by using a plasmid insertion library constructed by Serror and Sonenshein (47). The plasmid insertion clone pE1 contains an insert corresponding to the region from albA (ywiA) to albC (ywhQ) (30) and linked by transformation to an fnr::spc marker (Fig. 2). Sequence analysis revealed that the insertion took place within a cluster of seven genes putatively constituting an operon (Fig. 2) that we called alb (for antilisterial bacteriocin).
FIG. 2.
(A) Organization of the alb operon and flanking chromosomal regions of the B. subtilis chromosome. Rectangular boxes indicate the positions of genes in the alb region. The arrows above the boxes indicate the transcription units deduced by the Bacillus genome sequencing project (30). Also shown are the locations of the alb operon and the sbo gene, as well as the genetically linked fnr gene and the fnr::spc insertion mutation (used to demonstrate linkage) (31). The integrative plasmid pE1 and the site of its recombination with the chromosome, which causes disruption of the alb operon, are shown. T, putative transcription termination site. (B) Location of the sbo::neo-1 and sbo::neo-2 insertions. (C) The stem-loop structure predicted from inspection of the sbo-albA intergenic-region sequence that overlaps with the 3′ end of the sbo gene (indicated in boldface). Also shown is the free energy of secondary-structure formation, ΔG.
Immediately upstream of the alb operon is the sbo gene, encoding the 43-amino-acid precursor of the bacteriocin subtilosin (30), which was originally characterized by Kurahashi and coworkers (1) (Fig. 1). The subtilosin precursor contains no obvious leader peptide sequence, which is normally required for peptide export, nor are there the typical motifs associated with the processing of prelantibiotics. The peptide appears to undergo some unique modifications during maturation. Asn9 is linked to the C-terminal glycine, and Cys21 has been proposed to be linked to Glu31 (1) (Fig. 1A). Although codons specifying phenylalanine at positions 30 and 39 and Thr at position 36 are present in the nucleotide sequence (Fig. 1B), the corresponding amino acids are not found in the mature subtilosin peptide (1); this was confirmed by amino acid analysis (described below). It is possible that these residues play some role in intrachain cross-linking. As described further below, mutations in either the alb operon or sbo eliminated the antilisterial activity.
Overlapping with the C-terminal coding end of sbo and residing 93 bp upstream of the albA TTG start codon is a 55-bp sequence that potentially codes for a region of RNA secondary structure (location in the sbo-alb region denoted as T in Fig. 2A). The stem-loop depicted in Fig. 2C has a ΔG of −23.4 and could impede transcriptional readthrough from upstream. A similar structure, found between the mutA gene, encoding mutacin II, and the remainder of the mut operon, is thought to reduce transcriptional readthrough from the upstream mut operon promoter (42).
The putative coding sequences of the alb operon encode proteins that potentially function in the processing and export of peptides, such as an ATP-binding cassette transport complex (albC) and two processing peptidases. Interestingly, one of these peptidases, the product of albF (ywhN), shows significant sequence similarity to mitochondrial zinc-endoproteinase (3, 20, 39, 41) and to PqqF, a protein required for the synthesis of the cofactor pyrroloquinoline quinone (PQQ). PqqF is thought to cleave the PqqA peptide, thereby releasing glutamate and tyrosine, which are precursors of PQQ (34, 52, 53, 56). AlbE and the putative peptidase encoded by the gene albF (30) could function in the processing of subtilosin or a protein required for subtilosin maturation. The first gene of the alb operon, albA (ywiA) (30), encodes a protein with significant homology to those that function in cofactor heme, PQQ, and molybopterin cofactor synthesis (9, 34, 43, 52). The albA product is thought to function in the association of a metal ion with an enzyme-bound cofactor. It is possible that the product of albA (ywiA) activates a metalloenzyme that catalyzes modification of prosubtilosin.
A mutation in sbo confers loss of antilisterial activity.
We sought to determine if the sbo gene was required for the observed antilisterial activity. A BamHI-BglII fragment bearing a neomycin resistance cassette was inserted into the sbo gene at the BamHI site, thereby bisecting the sbo gene (see Materials and Methods). The plasmid was used to transform competent cells of JH642 and strain 22a, resulting in the sbo::neo insertion mutation. The colonies of the resulting sbo mutant derivative of JH642 did not exhibit the antilisterial phenotype on YG plates (Fig. 3). The antilisterial activity of strain 22a was reduced but not eliminated. This was due to the presence of other antilisterial activities produced by 22a. These other, low-level activities could be detected in supernatant fluid from liquid cultures of strain 22a (data not shown).
FIG. 3.
Antilisterial substance produced by B. subtilis JH642 and 22a. (A) The strains are shown as colonies that were overlayed with soft agar containing a suspension of L. monocytogenes cells. Growth of Listeria is inhibited by wild-type (WT) JH642 and by 22a but not by the sbo and alb mutants of these organisms. Some slight inhibition is observed around the colonies of the 22a sbo and alb mutant cells. (B) A Tricine-SDS-PAGE gel that is stained with Coomassie blue shows the antilisterial peptide produced in WT JH642 but not present in cultures of strains ORB3148 (sbo) and ORB3146 (alb). (C) A bioautograph (see Materials and Methods) of the gel in panel B, showing the antilisterial activity of the peptide and the absence of activity in the lanes containing the sbo and alb mutant culture extracts.
Purification of antilisterial activity and evidence that the bacteriocin subtilosin is the antilisterial agent.
Further purification of subtilosin was carried out from supernatant fluid collected from JH642 cultures, since the fluid from strain 22a cultures contained other antilisterial activities that might interfere with subtilosin purification. Supernatant fluid from YG cultures of JH642 was precipitated with 65% (NH4)2SO4. The precipitate was extracted twice with methanol, evaporated, and subjected to Tricine-SDS-PAGE analysis (46). A single band migrating at approximately 4,000 Da, which was absent from the sbo::neo-1 mutant and plasmid alb::pE1 insertion mutant cultures (Fig. 3), was detected. Bioautography was performed by overlaying the proteins of the Tricine-SDS gel with molten soft brain-heart infusion agar in which cells of an overnight culture of L. monocytogenes were suspended. A zone of lysis was observed over the area of the gel where the subtilosin band was located (Fig. 3C). At this same position in the lanes containing the supernatant of the mutant cultures, no band was evident in the Tricine-SDS gel and no zone of lysis occurred in the bioautograph.
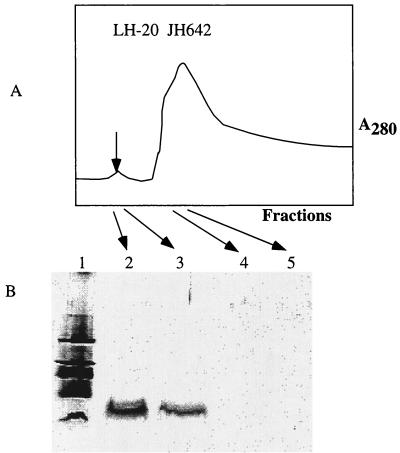
Culture fluid was also subjected to ion-exchange chromatography, and fractions exhibiting antilisterial activity were collected. This material was extracted with butanol, evaporated, dissolved in methanol, and resolved further by Sephadex LH-20 gel filtration column chromatography. The stepwise purification (7.1-fold) is outlined in Table 3. A single peak running in the early fractions of the void volume possessed the antilisterial activity and contained the 4,000-Da band evident on Tricine-SDS gels (Fig. 4). The purified substance, found to be greater than 90% pure by high-performance liquid chromatography, was subjected to amino acid analysis, which revealed that its composition was the same as that previously published for subtilosin (data not shown). The identification of the substance as subtilosin was confirmed by Edman degradation sequence analysis (data not shown) of peptide fragments generated by partial acid hydrolysis. Again, no Phe residues were detected despite the fact that there are two Phe codons in the sbo coding sequence. Additionally, only one Thr residue was detected, confirming previously published data (1).
TABLE 3.
Purification of subtilosin A
| Purification step | Vol (ml) | Total protein (μg) | AUa
|
Recovery (%) | Fold purification | |
|---|---|---|---|---|---|---|
| Total | Specific (AU/μg) | |||||
| Culture supernatant | 250 | 13,000 | 5,000 | 0.38 | 100 | 1 |
| Fractions from High Q column | 60 | NDb | 1,600 | ND | 32 | ND |
| Butanol extraction | 15 | 400 | 1,024 | 2.56 | 20.5 | 6.7 |
| Suspension after LH-20 chromatography | 0.4 | 240 | 640 | 2.7 | 12.8 | 7.1 |
AU, arbitrary units.
ND, not done.
FIG. 4.
(A) Profile of fractions collected from an LH-20 size exclusion column onto which a methanol extract of concentrated JH642 culture supernatant was applied. A small absorbance peak (λ = 280) (arrow) contains the putative subtilosin peptide, as shown by Tricine-SDS-PAGE (B).
Expression of alb-lacZ is observed in DSM-G and in sbo::neo mutants.
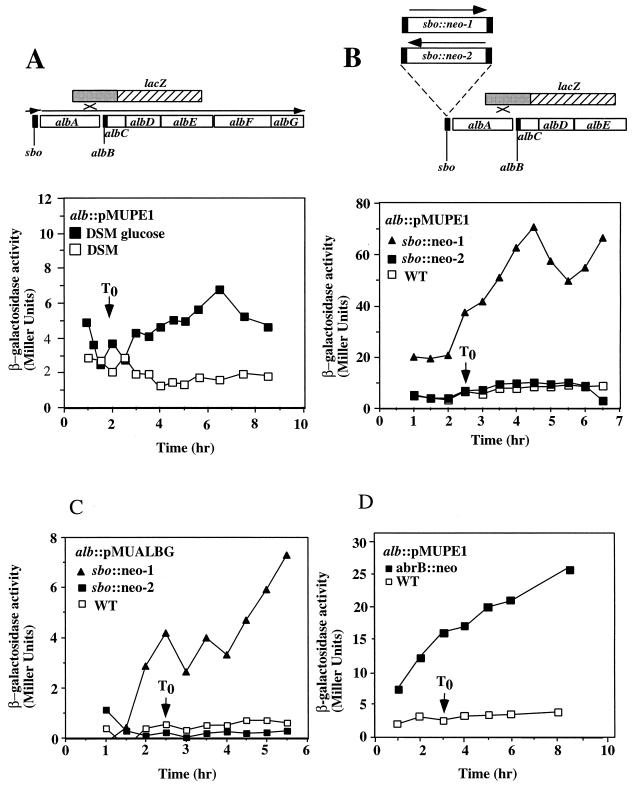
To begin to understand the organization and regulation of the sbo-alb genes, the expression of sbo- and alb-lacZ gene fusions was analyzed. The insert of plasmid pE1 was cloned into the gene disruption vector pMUTIN2 (55). The resulting plasmid (pMUPE1) was introduced by transformation into cells of JH642, thus creating a strain containing a disruption of the alb operon and a transcriptional fusion of the 5′ end of the operon with the vector-borne lacZ gene. Expression of the fusion was very near background levels in TSS minimal medium with either ammonium or glutamate as the nitrogen source (data not shown). Expression was also at background levels in DSM medium (Fig. 5A) but was observed to be higher in DSM-G, with activity accumulating in stationary phase. The introduction of the sbo::neo mutation (sbo::neo-1) resulted in high-level expression of alb-lacZ throughout growth (Fig. 5B), but this only occurred when the neomycin resistance gene was oriented in the same direction as the alb transcription unit. In the reverse orientation, the sbo::neo insertion (sbo::neo-2) showed much-reduced activity.
FIG. 5.
Expression of an alb-lacZ fusion constructed by creation of the alb::pMUPE1 insertion. Cultures were grown in DSM or DSM-G, and 1-ml samples were collected at either 30-min or 1-h intervals. β-Galactosidase activity was determined and plotted versus time. T0 indicates the end of the exponential growth phase. (A) Map of the alb operon and location of the lacZ fusion generated by recombination between the DNA of the alb locus and pMUPE1. Below the map is the expression profile of alb-lacZ in cells of strain ORB3147 grown in DSM and DSM-G. (B) Location of the sbo::neo-1 and sbo::neo-2 insertions with respect to the alb::pMUPE1 lacZ fusion. Also shown below the map is the expression profile of alb-lacZ in strains ORB3152 (sbo::neo-2) and ORB3153 (sbo::neo-1). (C) Expression profile of the albG-lacZ of strain ORB3231 (alb::pMUALBG) and its sbo::neo-1 (ORB3284) and sbo::neo-2 (ORB3230) mutant derivatives. (D) Effect of an abrB::neo insertion on the expression profile of alb-lacZ (alb::pMUPE1).
A pMUTIN disruption of the last gene of the operon, albG, using plasmid pMUALBG resulted in reduced antilisterial activity, as judged by the size of the zone of inhibition of albG::pMUALBG colonies on lawns of L. monocytogenes. The level of expression of the albG::pMUALBG fusion was low in DSM-G but was elevated when the sbo::neo-1 mutation was introduced (Fig. 5C). The sbo::neo-2 mutation did not elevate the level of expression of albG::pMUALBG, indicating that the derepression observed in the sbo::neo-1 mutant was not due to the sbo mutation per se but was likely due to transcription from the neo promoter. The similar responses to the presence of the sbo::neo-1 insertion observed for albG::pMUALBG and alb::pMUPE1 indicate that the two fusions reside in the same transcription unit. The observed transcriptional activity could originate from a promoter residing between sbo and alb, as well as from within the neo gene. The putative transcription termination sequence downstream of the sbo gene would limit transcription from a promoter upstream of sbo, but the fact that transcription from the neo gene is observed to traverse the termination sequence suggests that alb expression in wild-type cells could also be the result of transcription initiation upstream of sbo.
The expression of alb-lacZ is regulated by abrB.
The production of and resistance to antibiotics observed for B. subtilis are known to be under the control of the abrB gene (13, 14, 19), encoding the transition state regulator of late-growth gene transcription (40, 48). The effect of abrB mutations on the expression of alb::pMUPE1 was examined. The abrB::neo insertion mutation was introduced into the alb::pMUPE1 mutant by transforming competent cells of strain ORB3147 with DNA from strain TT71. The level of expression of alb::pMUPE1 was observed to be nearly 10-fold higher in the abrB mutant (Fig. 5D). These data and data presented below indicate that transcription of the sbo-alb genes is under the negative control of the abrB gene product.
Identification of the sbo promoter region.
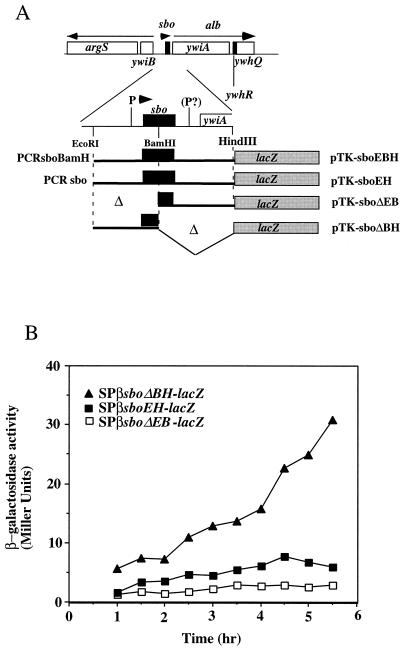
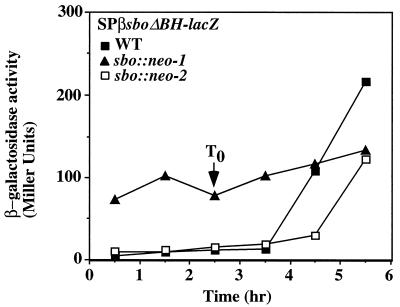
The BamHI site in the sbo coding sequence of plasmid pUC-sboEBH was used to isolate two fragments, one containing the 5′ end of sbo and the putative sbo promoter region and the other containing the 3′ end of sbo and the sbo-albA intergenic region. Both were inserted into plasmid pTKlac (24), yielding pTK-sboΔBH and pTK-sboΔEB, respectively (Fig. 6A). The EcoRI-HindIII fragment of pUC-sboEH containing the entire wild-type sbo gene and flanking DNA was also inserted into pTKlac, as was the EcoRI-HindIII fragment of plasmid pUC-sboEBH. The resulting plasmids were introduced into the SPβ prophage of strain ZB307 (59). Transducing phages carrying the fusions were generated and used to lysogenize cells of strain JH642. The level of expression of the SPβ–sboEH-lacZ fusion peaked at 6 Miller units in stationary phase (Fig. 6B), while the level of expression of SPβ–sboΔEB-lacZ was the same as that of the SPβ-pTKlac negative control. The level of expression of the SPβ–sboEBH-lacZ fusion was similar to that observed for the SPβ–sboEH-lacZ construct. The level of expression of the SPβ–sboΔBH-lacZ fusion began at 10 Miller units and increased to 30 Miller units in stationary phase. The results suggest that the major promoter of the sbo and alb genes resides upstream of the sbo gene.
FIG. 6.
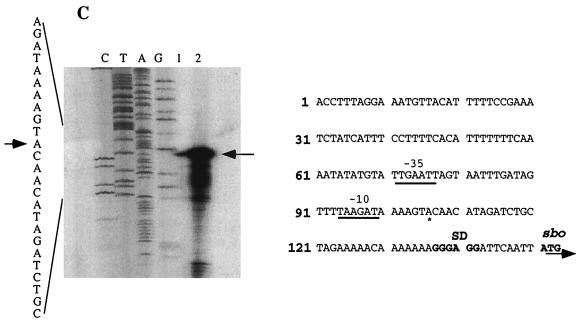
(A) A fragment containing the sbo gene and the 5′ end of the alb operon was obtained by PCR and inserted into the promoter probe plasmid pTKlac (24). The EcoRI-BamHI fragment bearing the putative sbo promoter region was deleted to create an alb-lacZ (sboΔEB-lacZ) fusion, and the BamHI-HindIII fragment was deleted to create an sbo-lacZ (sboΔBH-lacZ) fusion. Additionally, the EcoRI-HindIII fragment containing the entire sbo gene and flanking DNA was inserted into pTKlac to create sboEH-lacZ, while the fragment containing the PCR-generated sboEBH allele was inserted into pTKlac to create the sboEBH-lacZ construct. The fusions thus constructed were inserted into the SPβ prophage of B. subtilis, using a published protocol. (B) Expression of the phage-borne fusions in cells of strains ORB3158 (SPβsboΔEB-lacZ), ORB3162 (SPβ–sboΔBH-lacZ), and ORB3166 (SPβsboEH-lacZ) grown in DSM-G. T0, the time at which exponential growth ceases, is indicated by an arrow. (C) Primer extension analysis of RNA from JH642 and ZB449 (abrB703) cells. RNA was purified from cells of cultures grown to T2 in DSM-G. On the left is the autoradiograph showing the sequence pattern of a dideoxynucleotide sequencing reaction containing primer osboP4 and pTK-sboEH DNA. Lane 1, primer extension product from a reaction containing JH642 RNA and the osboP4 oligonucleotide; lane 2, primer extension product of the reaction of ZB449 RNA and osboP4 primer. The arrow indicates the primer extension products and the proposed start site of transcription in the sequence at the left. On the right is the nucleotide sequence of the sbo promoter region, with the ATG start codon and Shine-Delgarno (SD) sequence shown. The putative −10 and −35 regions of the sbo promoter are underlined. An asterisk marks the transcriptional start site.
This conclusion was supported by the identification of the transcriptional start site of the sbo gene by primer extension analysis. RNA was purified from JH642 cells and from cells of the ZB449 (abrB703) (59) mutant strain collected from cultures grown to T2 (2 h after the end of the exponential growth phase). Two oligonucleotides were used to generate primer extension products, one (osboP4) hybridizing with the sbo coding sequence beginning at bp 57 and the other, ocalbA-L, hybridizing to bp 35 to 65 of the albA coding region. Using osboP4, a primer extension product was generated whose length indicated the presence of a transcriptional start site residing 45 bp upstream of the sbo ATG (Fig. 6C). No primer extension product was detected with the ocalbA-L oligonucleotide. The RNA from abrB mutant cells yielded a primer extension product of the same size as that obtained from a reaction containing JH642 RNA and the osboP4 primer, but the product was much more abundant. This result is further evidence that sbo-alb transcription is under the negative control of abrB.
Expression of SPβ–sbo-lacZ is not regulated by subtilosin but is stimulated by alb operon expression.
We next investigated the possibility that the sbo gene and alb operon are autoregulated via the subtilosin peptide. DSM-G cultures of the SPβ–sboΔBH-lacZ lysogen ORB3162 were treated with subtilosin at a concentration of 7 μg/ml before the end of the exponential growth phase. No increase in sbo-lacZ expression was observed other than the normal post-exponential-phase induction of expression (data not shown). The existence of autoregulation was tested again by examining the expression of sbo-lacZ in sbo and alb mutant strains. A heat-induced lysate of SPβ–sboΔBH-lacZ was used to lysogenize cells of strains ORB3146 (alb::pE1), ORB3148 (sbo::neo-1), and ORB3149 (sbo::neo-2). sbo-directed β-galactosidase activity was measured in cells collected throughout growth and stationary phase in DSM-G. Post-exponential-phase induction of lacZ expression was observed in the ORB3162 (SPβ–sboΔBH-lacZ) cultures (Fig. 7B). The sbo::neo-1 mutant ORB3163, which cannot produce subtilosin but constitutively expresses alb, shows high-level constitutive expression of SPβ–sboΔBH-lacZ. The sbo:neo-2 mutant ORB3164 does not show high-level constitutive expression but exhibits a low level of expression that increases in stationary phase. Disruption of the alb operon with pE1 did not consistently affect SPβ–sboΔBH-lacZ expression, suggesting that the defect conferred by the insertion, while eliminating subtilosin production, may not be severe enough to affect sbo transcription. It is also possible that the albA and/or albB gene, the disposition of which is not affected by the pE1 insertion, is responsible for stimulating the expression of SPβ–sboΔBH-lacZ. The constitutive expression of phage-borne sbo-lacZ in the sbo::neo-1 mutant suggests that the transcription of sbo-alb is positively regulated by one or more of the alb operon products, which constitute an autoregulatory loop controlling sbo-alb transcription.
FIG. 7.
alb-dependent stimulation of sbo transcription. Strains ORB3162 (SPβ–sboΔBH-lacZ), ORB3163 (sbo::neo-1 SPβ–sboΔBH-lacZ), and ORB3164 (sbo::neo-2 SPβ–sboΔBH-lacZ) were grown in DSM-G. The profiles of sboΔBH-lacZ expression in the three strains over the exponential and stationary phases of growth are shown. WT, wild type.
DISCUSSION
The production of an antilisterial peptide by B. subtilis is dependent on the sbo and alb genes, as judged by the phenotypes of three mutant strains, the alb::pE1 operon disruption mutant, the sbo::neo mutant, and the albG::pMUALBG insertion mutant. The albABCDEFG genes are believed to constitute an operon that encodes the proteins that function in presubtilosin processing and subtilosin export. The sbo gene encodes presubtilosin, which is a 43-amino-acid peptide that likely undergoes processing steps that include proteolytic cleavage at the Asn9 residue, modification at the two Phe residues at positions 30 and 39, and modification of Thr36. A cross-link of unusual composition is thought to connect Cys21 and Glu31 (adjacent to the modified Phe residue). Finally, the cyclization of the presubtilosin peptide involves covalent linking of the C-terminal Gly with the N-terminal Asn. The involvement of modified Phe and Glu, along with the similarities revealed by aligning the amino acid sequences of the AlbA and AlbE products with those of proteins that function in PQQ synthesis (which is initiated by a condensation of Glu and Tyr [10]), suggests that PQQ synthesis and subtilosin processing may proceed through some common reaction mechanisms.
The sbo gene is followed by a sequence resembling a factor-independent transcriptional termination sequence. Yet, transcription can proceed through the sequence, as shown by the introduction of the neo gene upstream of the terminator that drives constitutive expression of the alb::pMUPE1 lacZ fusion. Other bacteriocin biosynthesis operons have a similar organization (21, 42). The positioning of the terminator (Fig. 2C) between the bacteriocin structural gene and the genes required for processing and export is thought to ensure that the proteins that carry out peptide modification and processing are present in small, catalytic amounts while the peptide substrate is produced in larger quantities. Transcription from the neo fragment can drive expression of the alb genes, including the last gene of the operon, albG. No promoter activity could be detected in the sbo-alb intergenic region by measuring lacZ activity of the SPβ–sboΔEB-lacZ cells or by primer extension analysis. However, we cannot rule out the possibility that within this region there is a weak promoter that is utilized under specific growth conditions. The major transcriptional start site for sbo-alb lies upstream of the sbo gene and is associated with a sequence resembling promoters utilized by the ςA form of B. subtilis RNA polymerase (−35 TTGAAT [17bp]−10 TAAGAT [Fig. 6C]). Strong evidence that sbo-alb is under the negative control of the global transition state transcriptional regulatory protein AbrB is presented here. In support of this conclusion is the observation that spo0A mutant cells, in which the AbrB protein is overproduced, do not exhibit antilisterial activity (data not shown). As with other AbrB-controlled genes, we would expect to find that AbrB protein directly interacts with the sbo promoter, thereby preventing RNA polymerase from establishing contacts with the −35 and −10 sequences.
Another form of regulation was revealed by examining the expression of SPβ–sboΔBH-lacZ in sbo and alb mutant strains. The sbo::neo-1 insertion was observed to drive the high-level constitutive expression of alb-lacZ (alb::pMUPE1). This mutation also caused constitutive expression of the sboΔBH-lacZ fusion positioned in the SPβ prophage, while the sbo::neo-2 insertion did not. Disruption of the sbo gene did not cause a decrease in the level of sbo-lacZ expression, and addition of subtilosin to cultures of low cell density did not stimulate expression. From these observations, we conclude that unlike the situation in the case of the nis or spa operon, the exogenous presence of the bacteriocin encoded by sbo does not stimulate expression of sbo. However, a form of positive autoregulation appears to exist, and it involves one or more of the alb gene products. The alb::pMUPE1 lacZ fusion has very low activity, which is likely due to the fact that the alb operon is disrupted by the fusion and there is no positive autoregulation. This can be suppressed by introduction of an abrB mutation (Fig. 5D), suggesting a link between the regulatory function of alb and AbrB activity.
The regulation of sbo-alb is more complex, however, since it is subject to control exerted by factors that regulate anaerobic gene expression, including ResDE and Fnr (37a). How these factors interact with those functioning in autoregulation and AbrB-dependent control presents an interesting problem for further investigation.
ACKNOWLEDGMENTS
Research reported herein was supported by grant GM45898 from the National Institutes of Health, a grant from the Oregon Medical Research Foundation, and funds from the Natural Sciences and Engineering Council of Canada.
REFERENCES
- 1.Babasaki K, Takao T, Shimonishi Y, Kurahashi K. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J Biochem. 1985;98:583–603. doi: 10.1093/oxfordjournals.jbchem.a135315. [DOI] [PubMed] [Google Scholar]
- 2.Burbulys D, Trach K A, Hoch J A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 3.Clary D O, Wahleithner J A, Wolstenholme D R. Sequence and arrangement of the genes for cytochrome b, URF1, URF4L, URF4, URF5, URF6 and five tRNAs in Drosophila mitochondrial DNA. Nucleic Acids Res. 1984;12:3747–3762. doi: 10.1093/nar/12.9.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Vos W M, Kuipers O P, Roelof van der Meer J, Siezen R J. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by Gram-positive bacteria. Mol Microbiol. 1995;17:427–437. doi: 10.1111/j.1365-2958.1995.mmi_17030427.x. [DOI] [PubMed] [Google Scholar]
- 5.Dubnau D, Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971;56:209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- 6.Entian K D, de Vos W M. Genetics of subtilin and nisin biosyntheses: biosynthesis of lantibiotics. Antonie Leeuwenhoek. 1996;69:109–117. doi: 10.1007/BF00399416. [DOI] [PubMed] [Google Scholar]
- 7.Fouet A, Sonenshein A L. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J Bacteriol. 1990;172:835–844. doi: 10.1128/jb.172.2.835-844.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furbaß R, Gocht M, Zuber P, Marahiel M A. Interaction of AbrB, a transcriptional regulator from Bacillus subtilis, with the promoters of the transition state-activated genes tycA and spoVG. Mol Gen Genet. 1991;225:347–354. doi: 10.1007/BF00261673. [DOI] [PubMed] [Google Scholar]
- 9.Glaser P, Danchin A, Kunst F, Zuber P, Nakano M M. Identification and isolation of a gene required for nitrate assimilation and anaerobic growth of Bacillus subtilis. J Bacteriol. 1995;177:1112–1115. doi: 10.1128/jb.177.4.1112-1115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodwin P M, Anthony C. The biochemistry, physiology and genetics of PQQ and PQQ-containing enzymes. Adv Microb Physiol. 1998;40:1–80. doi: 10.1016/s0065-2911(08)60129-0. [DOI] [PubMed] [Google Scholar]
- 11.Grossman A D. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 12.Guerout-Fleury A M, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 13.Guespin-Michel J F. Phenotypic reversion in some early blocked sporulation mutants of Bacillus subtilis: isolation and phenotype identification of partial revertants. J Bacteriol. 1971;108:241–247. doi: 10.1128/jb.108.1.241-247.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guespin-Michel J F. Phenotypic reversion in some early blocked sporulation mutants of Bacillus subtilis. Genetic study of polymyxin resistant partial revertants. Mol Gen Genet. 1971;112:243–254. [PubMed] [Google Scholar]
- 15.Harwood C R, Cutting S M. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons; 1990. [Google Scholar]
- 16.Havarstein L S, Diep D B, Nes I F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 17.Hoch J A. Control of cellular development in sporulating bacteria by the phosphorelay two-component signal transduction system. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: ASM Press; 1995. pp. 129–144. [Google Scholar]
- 18.Ireton K, Rudner D Z, Siranosian K J, Grossman A D. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- 19.Ito J, Mildner G, Spizizen J. Early blocked asporogenous mutants of Bacillus subtilis 168. I. Isolation and characterization of mutants resistant to antibiotic(s) produced by sporulating Bacillus subtilis 168. Mol Gen Genet. 1971;112:104–109. doi: 10.1007/BF00267488. [DOI] [PubMed] [Google Scholar]
- 20.Iwata S, Lee J W, Okada K, Lee J K, Iwata M, Rasmussen B, Link T A, Ramaswamy S, Jap B K. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 21.Jack R, Bierbaum B, Heidrich C, Sahl H-G. The genetics of lantibiotic biosynthesis. Bioessays. 1995;17:793–802. doi: 10.1002/bies.950170909. [DOI] [PubMed] [Google Scholar]
- 22.Jack R W, Tagg F R, Ray B. Bacteriocins of gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katz E, Demain A L. The peptide antibiotics of Bacillus: chemistry, biogenesis, and possible functions. Bacteriol Rev. 1977;41:449–474. doi: 10.1128/br.41.2.449-474.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenney T J, Moran C P., Jr Genetic evidence for interaction of ςA with two promoters in Bacillus subtilis. J Bacteriol. 1991;173:3282–3290. doi: 10.1128/jb.173.11.3282-3290.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klaenhammer T R. Bacteriocins of lactic acid bacteria. Biochemie. 1988;70:337–349. doi: 10.1016/0300-9084(88)90206-4. [DOI] [PubMed] [Google Scholar]
- 26.Klein C, Kaletta C, Entian K-D. Biosynthesis of the lantibiotic subtilin is regulated by a histidine kinase/response regulator system. Appl Environ Microbiol. 1993;59:296–303. doi: 10.1128/aem.59.1.296-303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinkauf H, von Dohren H. Peptide antibiotics. In: Pape H, Rehm H-J, editors. Biotechnology, vol. 4. Microbiol products II. Weinheim, Germany: VCH Berlagsgesellschaft GmbH; 1984. pp. 284–307. [Google Scholar]
- 28.Kolter R, Moreno F. Genetics of ribosomally synthesized peptide antibiotics. Annu Rev Microbiol. 1992;46:141–164. doi: 10.1146/annurev.mi.46.100192.001041. [DOI] [PubMed] [Google Scholar]
- 29.Kuipers O P, Beerthuyzen M M, de Ruyter P G G A, Luesink E J, de Vos W M. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem. 1995;270:27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 30.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 31.LaCelle M, Kumano M, Kurita K, Yamane K, Zuber P, Nakano M M. Oxygen-controlled regulation of the flavohemoglobin gene in Bacillus subtilis. J Bacteriol. 1996;178:3803–3808. doi: 10.1128/jb.178.13.3803-3808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y M, Milne J C, Madison L L, Kolter R, Walsh C T. From peptide precursors to oxazole- and thiazole-containing peptide antibiotics: microcin B17 synthase. Science. 1996;274:1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 33.Marahiel M A, Zuber P, Czekay G, Losick R. Identification of the promoter for a peptide antibiotic biosynthesis gene from Bacillus brevis and its regulation in Bacillus subtilis. J Bacteriol. 1987;169:2215–2222. doi: 10.1128/jb.169.5.2215-2222.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meulenberg J J, Sellink E, Riegman N H, Postma P W. Nucleotide sequence and structure of the Klebsiella pneumoniae pqq operon. Mol Gen Genet. 1992;232:284–294. doi: 10.1007/BF00280008. [DOI] [PubMed] [Google Scholar]
- 35.Montville T J, Winkowski K. Biologically based preservation systems and probiotic bacteria. In: Doyle M P, Beuchat L R, Montville T J, editors. Food microbiology: fundamentals and frontiers. Washington, D.C.: ASM Press; 1997. pp. 557–577. [Google Scholar]
- 36.Nakano M M, Marahiel M A, Zuber P. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J Bacteriol. 1988;170:5662–5668. doi: 10.1128/jb.170.12.5662-5668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakano M M, Xia L, Zuber P. Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J Bacteriol. 1991;173:5487–5493. doi: 10.1128/jb.173.17.5487-5493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Nakano, M. M., G. Zheng, and P. Zuber. Unpublished results.
- 38.Nes I F, Diep D B, Havarstein L S, Brurberg M B, Eijsink V, Holo H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 39.Paces V, Rosenberg L E, Fenton W A, Kalousek F. The beta subunit of the mitochondrial processing peptidase from rat liver: cloning and sequencing of a cDNA and comparison with a proposed family of metallopeptidases. Proc Natl Acad Sci USA. 1993;90:5355–5358. doi: 10.1073/pnas.90.11.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perego M, Spiegelman G B, Hoch J A. Structure of the gene for the transition state regulator abrB: regulator synthesis is controlled by the Spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988;2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 41.Pollock R A, Hartl F U, Cheng M Y, Ostermann J, Horwich A, Neupert W. The processing peptidase of yeast mitochondria: the two co-operating components MPP and PEP are structurally related. EMBO J. 1988;7:3493–3500. doi: 10.1002/j.1460-2075.1988.tb03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi F, Chen P, Caufield P W. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl Environ Microbiol. 1999;65:652–658. doi: 10.1128/aem.65.2.652-658.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rivers S L, McNairn E, Blasco F, Giordano G, Boxer D H. Molecular genetic analysis of the moa operon of Escherichia coli K-12 required for molybdenum cofactor biosynthesis. Mol Microbiol. 1993;8:1071–1081. doi: 10.1111/j.1365-2958.1993.tb01652.x. [DOI] [PubMed] [Google Scholar]
- 44.Robertson J R, Gocht M, Marahiel M A, Zuber P. AbrB, a regulator of gene expression in Bacillus, interacts with the transcription initiation regions of a sporulation and an antibiotic biosynthesis gene. Proc Natl Acad Sci USA. 1989;86:8457–8461. doi: 10.1073/pnas.86.21.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 47.Serror, P., and A. L. Sonenshein. Unpublished data.
- 48.Strauch M A. AbrB, a transition state regulator. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 757–764. [Google Scholar]
- 49.Strauch M A. Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promoter. J Bacteriol. 1995;177:6999–7002. doi: 10.1128/jb.177.23.6999-7002.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strauch M A, Spiegelman G B, Perego M, Johnson W C, Burbulys D, Hoch J A. The transition state transcription regulator AbrB of Bacillus subtilis is a DNA binding protein. EMBO J. 1989;8:1615–1621. doi: 10.1002/j.1460-2075.1989.tb03546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strauch M A, Webb V, Spiegelman B, Hoch J A. The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci USA. 1990;87:1801–1805. doi: 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toyama H, Chistoserdova L, Lidstrom M E. Sequence analysis of pqq genes required for biosynthesis of pyrroloquinoline quinone in Methylobacterium extorquens AMI and the purification of a biosynthetic intermediate. Microbiology. 1997;143:595–602. doi: 10.1099/00221287-143-2-595. [DOI] [PubMed] [Google Scholar]
- 53.Toyama H, Lidstrom M E. pqqA is not required for biosynthesis of pyrroloquinoline quinone in Methylobacterium extorquens AM1. Microbiology. 1998;144:183–191. doi: 10.1099/00221287-144-1-183. [DOI] [PubMed] [Google Scholar]
- 54.Trowsdale J, Chen S M, Hoch J A. Genetic analysis of a class of polymyxin resistant partial revertants of stage 0 sporulation mutants of Bacillus subtilis: map of the chromosome region near the origin of replication. Mol Gen Genet. 1979;173:61–70. doi: 10.1007/BF00267691. [DOI] [PubMed] [Google Scholar]
- 55.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 56.Velterop J S, Sellink E, Meulenberg J J M, David S, Bulder I, Postma P W. Synthesis of pyrroloquinoline quinone in vivo and in vitro and detection of an intermediate in the biosynthetic pathway. J Bacteriol. 1995;177:5088–5098. doi: 10.1128/jb.177.17.5088-5098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 57a.Zheng G. Ph.D. thesis. Fayetteville: University of Arkansas; 1997. [Google Scholar]
- 58.Zheng G, Slavik M F. Isolation, partial purification and characterization of a bacteriocin produced by a newly isolated Bacillus subtilis strain. Lett Appl Microbiol. 1999;28:363–367. doi: 10.1046/j.1365-2672.1999.00545.x. [DOI] [PubMed] [Google Scholar]
- 59.Zuber P, Losick R. Role of AbrB in the Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol. 1987;169:2223–2230. doi: 10.1128/jb.169.5.2223-2230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuber P, Nakano M M, Marahiel M A. Peptide antibiotics. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 897–916. [Google Scholar]