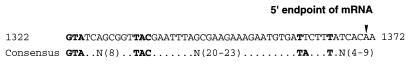Abstract
The nrtP and narB genes, encoding nitrate/nitrite permease and nitrate reductase, respectively, were isolated from the marine cyanobacterium Synechococcus sp. strain PCC 7002 and characterized. NrtP is a member of the major facilitator superfamily and is unrelated to the ATP-binding cassette-type nitrate transporters that previously have been described for freshwater strains of cyanobacteria. However, NrtP is similar to the NRT2-type nitrate transporters found in diverse organisms. An nrtP mutant strain consumes nitrate at a 4.5-fold-lower rate than the wild type, and this mutant grew exponentially on a medium containing 12 mM nitrate at a rate approximately 2-fold lower than that of the wild type. The nrtP mutant cells could not consume nitrite as rapidly as the wild type at pH 10, suggesting that NrtP also functions in nitrite uptake. A narB mutant was unable to grow on a medium containing nitrate as a nitrogen source, although this mutant could grow on media containing urea or nitrite with rates similar to those of the wild type. Exogenously added nitrite enhanced the in vivo activity of nitrite reductase in the narB mutant; this suggests that nitrite acts as a positive effector of nitrite reductase. Transcripts of the nrtP and narB genes were detected in cells grown on nitrate but were not detected in cells grown on urea or ammonia. Transcription of the nrtP and narB genes is probably controlled by the NtcA transcription factor for global nitrogen control. The discovery of a nitrate/nitrite permease in Synechococcus sp. strain PCC 7002 suggests that significant differences in nutrient transporters may occur in marine and freshwater cyanobacteria.
Cyanobacteria are photoautotrophic procaryotes that perform oxygen-evolving photosynthesis with carbon dioxide as the primary oxidizing agent and carbon source. Requiring only light, water, carbon dioxide, and inorganic salts, cyanobacteria have very simple nutrient requirements that allow these organisms to occupy highly diverse ecological niches. All cyanobacteria can utilize nitrate as their sole nitrogen source (30), although reduced nitrogen sources, such as ammonia and urea, can also be utilized (7). Since nitrogen is a major nutrient and accounts for about 11% of the dry weight of cyanobacterial cells (39), nitrate assimilation and reduction place a high demand for energy and electrons on the photosynthetic machinery. The estimated ratio of the maximal rates of carbon assimilation and nitrate assimilation is roughly 2 to 2.5 (6, 26) under optimized assay conditions. This suggests that up to 30% of the electrons generated by photosynthetic water oxidation are consumed during the reduction of nitrate to ammonia when cells are grown on nitrate.
Cyanobacterial cells preferentially utilize reduced nitrogen sources such as ammonia and urea. Nitrate consumption is completely inhibited within minutes of the addition of exogenous ammonia (5, 7, 19), and the nitrate transport system is thought to be the rate-limiting step for nitrate assimilation (7). It is believed that the nitrate transporter is rapidly inactivated in the presence of ammonia, although the molecular mechanism of this inhibition of nitrate assimilation by ammonia has not yet been elucidated (33).
It is important to know the structure of the nitrate transport system in cyanobacteria in order to analyze the regulation of nitrate assimilation by the availability of nitrogen sources as well as by photosynthesis and carbon assimilation. However, most recent molecular biological studies of the nitrate transport system have been limited to the freshwater cyanobacteria Synechococcus sp. strain PCC 7942 (21, 33) and Anabaena sp. strain PCC 7120 (2, 9). The genes for nitrite reductase (nirA), the nitrate transporter (nrtABCD) and nitrate reductase (narB) are transcriptionally regulated as an operon, nirA-nrtABCD-narB, in Synechococcus sp. strain PCC 7942 (for reviews, see references 20 and 21).
Nitrate assimilation is also very important in the low-temperature physiology of cyanobacteria. We have previously demonstrated that when cells are grown with nitrate as a nitrogen source, growth at a low temperature causes nitrogen limitation in the unicellular marine cyanobacterium Synechococcus sp. strain PCC 7002 (25). Cells of this cyanobacterium become chlorotic and grow arithmetically at 15°C in a medium containing nitrate as the sole nitrogen source. However, when cells are grown at 15°C on urea as the nitrogen source, cells grow exponentially and the symptoms of chlorosis are not observed (25). In the freshwater cyanobacterium Synechococcus sp. strain PCC 6301, nitrate transport also limits cell growth at low temperatures (26). The temperature-dependent decrease in the rate of nitrate consumption is similar to the temperature-dependent decrease in the growth rate, and both nitrate consumption and cell growth cease at 15°C in Synechococcus sp. strain PCC 6301. The marine cyanobacterium Synechococcus sp. strain PCC 7002 is somewhat more tolerant of low-temperature growth conditions, and both nitrate consumption and growth of this strain cease at 12°C (27). In order to understand better the important role of nitrate transport in defining the lower limits of cyanobacterial growth, we decided to characterize the nitrate transport system in the marine cyanobacterium Synechococcus sp. strain PCC 7002 in detail. These studies led to the discovery of a novel gene encoding a nitrate/nitrite permease, which was functionally characterized by reverse genetics and physiological studies.
MATERIALS AND METHODS
Organisms and culture conditions.
A laboratory wild-type strain of Synechococcus sp. strain PCC 7002, denoted PR6000, has been maintained at The Pennsylvania State University since 1981. Cells were grown photoautotrophically in medium A+ containing 12 mM NaNO3 (31), in medium AU10Ni containing 10 mM urea as a sole nitrogen source and supplemented with 5 μM NiSO4 (28), or in medium A-HEPES containing 5 mM NH4Cl as a nitrogen source and buffered with 25 mM HEPES-NaOH at pH 7.2 (28) under constant illumination from cool-white fluorescent lamps (250 μE m−2 s−1) with aeration from 1% (vol/vol) CO2 in air at 38°C. The photon flux density was measured with a model QSL-100 light meter (Biospherical Instruments, San Diego, Calif.). For the selection of kanamycin-resistant mutants, kanamycin (50 μg ml−1) was added to liquid media and agar plates. To change the growth media, cells were collected by centrifugation at room temperature, washed with nitrogen-free medium, and inoculated in fresh medium. Liquid cultures (25 ml) were incubated in 22- by 175-mm glass culture tubes, and the initial cell density for growth curve experiments was adjusted to an optical density at 550 nm (OD550) of 0.05 (90% light transmittance). Cell growth was monitored by the OD550 with a model Spectronic 20 spectrophotometer (Bausch & Lomb [now Milton Roy], Rochester, N.Y.). A cell suspension from an exponential-phase culture grown at 38°C in medium A+ at a light intensity of 250 μE m−2 s−1 with an OD550 of 1.0 contained 3.4 ± 0.3 μg of chlorophyll ml−1 and 1.0 × 108 ± 0.2 × 108 cells ml−1 as determined by microscopic count (25). To induce the nitrate consumption activity of cells grown with urea as a nitrogen source, cells grown overnight at 38°C in medium AU10Ni were collected by centrifugation and washed twice with fresh medium A+; the cells were then resuspended in the fresh medium A+ at the original cell density and incubated for 1 h under the same growth conditions. To inhibit the glutamine synthetase–glutamine(amide)-2-oxoglutarate aminotransferase (GOGAT) cycle in Synechococcus sp. strain PCC 7002, 6-diazo-5-oxo-l-norleucine (DON) (D-2141; Sigma Chemical Co., St. Louis, Mo.), which is a specific inhibitor for GOGAT, was used when indicated for some experiments. l-Methionine sulfoximine (M-5379; Sigma Chemical Co.) cannot be used with Synechococcus sp. strain PCC 7002, since l-methionine sulfoximine was not effective in releasing nitrate consumption from the repression effects of ammonia (data not shown). Wild-type cells of Synechococcus sp. strain PCC 7002 grow in medium A+ containing 0.5 mM l-methionine sulfoximine, although 0.5 mM DON completely inhibited cell growth under otherwise standard growth conditions (data not shown).
Cloning and interposon mutagenesis of nrtP and narB.
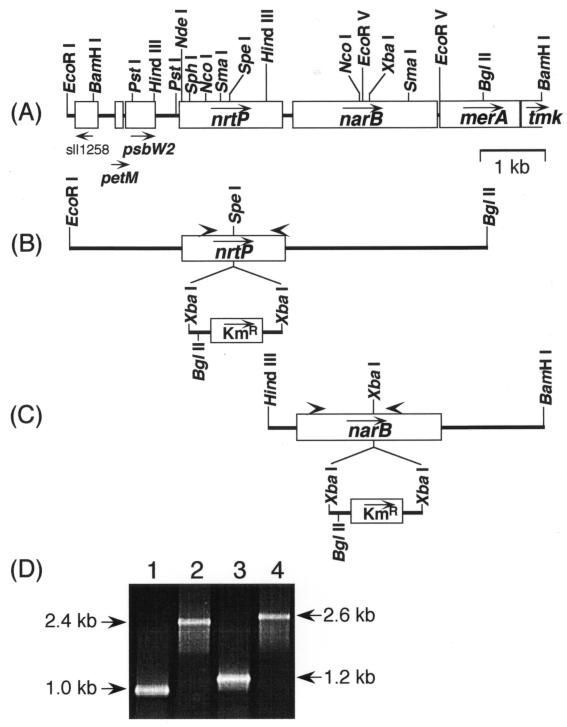
Genomic DNA of Synechococcus sp. strain PCC 7002 was partially digested with EcoRI and inserted into the EcoRI site of cosmid vector of pHC79 to construct a genomic DNA library as described previously (38). A cosmid clone was isolated from the genomic DNA library of Synechococcus sp. strain PCC 7002 by cross-hybridization with a 2.1-kb DNA fragment carrying the narB gene of Synechocystis sp. strain PCC 6803 (11). The screening probe was amplified by PCR with genomic DNA from Synechocystis sp. strain PCC 6803 as the template and the specific primers 5′-ATG GAT TCA CCA GCT ATT CTT (forward) and 5′-TTA AAC TGG TGT AAT ATT AAC (reverse). Heterologous hybridization experiments were performed as previously described (1). The complete nucleotide sequence of the 7.2-kb region from EcoRI to BamHI of the positive clone (Fig. 1A) was determined at the Nucleic Acid Facility of the Biotechnology Institute of the Pennsylvania State University. The nrtP and narB genes were insertionally inactivated by insertion of a 1.4-kb XbaI fragment carrying the aphII gene and derived from plasmid pRL170 (4) into a unique SpeI site within the nrtP gene (position 2432 in the nucleotide sequence of the 7.2-kb region (Fig. 1B) and into an XbaI site within the narB gene (position 4583) (Fig. 1C). Wild-type cells of Synechococcus sp. strain PCC 7002 (strain PR6000) were transformed with plasmid DNA containing the interrupted gene essentially as described by Stevens and Porter (32). Kanamycin-resistant transformants were selected on medium AU10Ni supplemented with 50 μg of kanamycin ml−1. The aphII gene insertion into each gene was confirmed by PCR with total genomic DNA as the template and the following specific primers: forward primer for the nrtP gene, 5′-AAC TTT GGC TCG GCA TTT TC; reverse primer for the nrtP gene, 5′-CAG CAC TTG GAA GAA GCC A; forward primer for the narB gene, 5′-ATT CCA GCG CAT CAG TTG; and reverse primer for the narB gene, 5′-CGG AAC TGG GGA CAA TAG (Fig. 1).
FIG. 1.
(A) Physical map of the 7.2-kb genomic region of the nrtP and narB genes in Synechococcus sp. strain PCC 7002. Arrows indicate the direction of transcription for the following open reading frames: sll1258, which is similar to hypothetical gene in Synechocystis sp. strain PCC 6803; petM, encoding a subunit of b6f complex; psbW2, encoding a subunit of photosystem II; merA, encoding mercuric reductase; and tmk, encoding thymidylate kinase. (B) The construct for the insertional inactivation of the nrtP gene in Synechococcus sp. strain PCC 7002. The kanamycin resistance gene cartridge (aphII gene) of 1.4 kb was inserted at the SpeI site in the nrtP gene with the same transcription orientation. The EcoRI-to-BglII DNA fragment was cloned between the EcoRI and BamHI sites of pUC19. Arrowheads indicate the positions of the primers for the PCR analysis. (C) The construct for insertional inactivation of the narB gene in Synechococcus sp. strain PCC 7002. The kanamycin resistance gene cartridge (aphII gene) of 1.4 kb was inserted into the XbaI site in the narB gene with the same transcription orientation. The HindIII-to-BamHI DNA fragment was cloned into the HindIII and BamHI sites of pUC19. Arrowheads indicate the positions of the primers for the PCR analysis. (D) Evaluation of gene replacement in the chromosomal DNAs of the nrtP mutant and the narB mutant by PCR analysis. Genomic DNAs from the wild type (lanes 1 and 3), the nrtP mutant (lane 2), and the narB mutant (lane 4) were used as the template for PCR with primers specific for the nrtP gene (lane 1 and 2) and for the narB gene (lanes 3 and 4). PCR products were separated by agarose gel electrophoresis and detected by ethidium bromide staining.
Nitrate consumption by whole cells (ammonia evolution).
The rate of nitrate consumption by whole cells was measured by ammonia release to the assay medium under illumination in the presence of the GOGAT inhibitor DON. Cells were collected by centrifugation and washed twice with 25 mM HEPES-NaOH buffer (pH 7.0) containing 1 mM KCl; the cells were resuspended at a chlorophyll concentration of 25 or 50 μg ml−1 in 25 mM HEPES-NaOH buffer (pH 7.0) containing 1 mM KCl, 10 mM NaHCO3, and 1 mM DON. The 2.5-ml sample in a 12- by 75-mm test tube was illuminated at 250 μE m−2 s−1 provided from a halogen floodlight after passage through a 5-cm layer of water as a heat filter, and the reaction temperature was maintained at 38°C by using a refrigerated, circulating water bath. The samples were continuously mixed with a magnetic stirrer. The reaction was initiated by the addition of 12 mM NaNO3 to the assay medium. Aliquots of 200 μl were taken from the reaction mixture at 15-min intervals up to 60 min, and the formation of ammonia was determined. Ammonia concentrations were determined by the indophenol method. After cells were removed by centrifugation, 120 μl of the supernatant was transferred to a test tube, and 200 μl of phenol-nitroprusside solution (640-1; Sigma Chemical Co.) and 200 μl of alkaline hypochlorite solution (640-3; Sigma Chemical Co.) were added to the sample. The sample mixture was incubated at room temperature for approximately 20 min to allow color development. Distilled water (1.0 ml) was added, and the absorbance at 570 nm was measured with a spectrophotometer (model 14R; Cary, San Fernando, Calif.) modified for computerized data acquisition by On-Line Instrument Systems (Bogart, Ga.). The ammonia concentration was determined from a standard curve constructed with known NH4Cl concentrations (0.1 to 1 mM) in the assay medium.
Nitrate consumption by whole cells (high-affinity uptake assay).
High-affinity nitrate uptake was measured by monitoring the disappearance of nitrate with a nitrate-specific electrode as described previously (26). Cells were collected by centrifugation and washed twice with 25 mM HEPES-NaOH buffer (pH 7.0) containing 1 mM KCl. The cells were resuspended at a chlorophyll concentration of 10 μg ml−1 in 25 mM HEPES-NaOH buffer (pH 7.0) containing 1 mM KCl, 10 mM NaHCO3, and 0.5 mM DON. Illumination (250 μE m−2 s−1) was provided from a halogen floodlight after passage through a 5-cm layer of water, and the temperature of the samples was maintained at 38°C by using a refrigerated, circulating water bath. The 25-ml sample was placed in a 50-ml jacketed beaker and was preilluminated for 15 min before the reaction was started. The assay was started by the addition of NaNO3 to give an initial concentration of 100 μM in the assay medium. The disappearance of nitrate from the medium was directly monitored in real time with a model PHM240 ion meter (Radiometer, Westlake, Ohio) equipped with a model 9746 BN combination nitrate electrode (Orion, Beverly, Mass.). Changes in the electrode potential, and thus in the nitrate concentration, were monitored with a chart recorder.
Nitrite consumption by whole cells.
The rate of nitrite consumption by whole cells was measured by the rate of disappearance of nitrite from the assay medium under illumination. Since nitrite can be taken up by passive diffusion of nitrous acid at neutral pH (7), assays performed at pH 7 reflect the in vivo activity of nitrite reductase, while assays performed at pH 10 reflect the active uptake of nitrite by cells. Cells were collected by centrifugation and washed twice with 25 mM HEPES-NaOH buffer (pH 7.0) containing 1 mM KCl; the cells were then resuspended at a chlorophyll concentration of 5 or 10 μg ml−1 in 25 mM HEPES-NaOH buffer (pH 7.0) or 25 mM CAPS [3-(cyclohexylamino)-1-propanesulfonic acid]-NaOH buffer (pH 10.0) containing 1 mM KCl, 10 mM NaHCO3, and 0.5 mM DON. Illumination (250 μE m−2 s−1) was provided from a halogen floodlight, and the reaction temperature was maintained at 38°C by using a refrigerated, circulating water bath. The 5.0-ml samples in 13- by 100-mm test tubes were preilluminated for 15 min before the reaction was started. The assay was started by the addition of NaNO2 to give an initial concentration of 100 μM in the assay. Aliquots of 350 μl were taken from the reaction mixture at 5-min intervals up to 20 min, and nitrite concentration in the medium was determined by the diazo coupling method. After cells were removed by centrifugation, 250 μl of the supernatant was transferred to a test tube, and 250 μl of 1% (wt/vol) sulfanilamide (S-9251; Sigma Chemical Co.) in 3 M HCl, 250 μl of 0.02% (wt/vol) N-(1-naphthyl)ethylenediamine (N-5889; Sigma Chemical Co.), and 0.4 ml of distilled water were added to the sample. After incubation at room temperature for color development, the absorbance at 540 nm was measured. The concentration of nitrite was determined from a standard curve constructed with NaNO2 concentrations ranging from 1 to 100 μM.
RNA blot hybridization analyses, RT-PCR analysis, and 5′ end mapping of nrtP mRNA.
RNA blot hybridization analyses were carried out as described by Sakamoto and Bryant (24). Total RNA (10 μg) was fractionated by agarose gel electrophoresis and transferred onto a nylon membrane (Nytran+; Schleicher & Schuell, Keane, N.H.). A 1.3-kb DNA probe specific for the nrtP gene (positions 1641 to 2995 of the nucleotide sequence of the 7.2-kb region described above) was prepared by restriction digestion with NdeI and HindIII (Fig. 1A), and a 1.2-kb probe for the narB gene (positions 4468 to 5659) was prepared by restriction digestion with EcoRV (Fig. 1A). RNA blots were hybridized with the 32P-labeled probes. The membranes were exposed to Kodak X-Omat AR X-ray film with an intensifying screen (Cronex Lightning-Plus; DuPont, Wilmington, Del.) for 1 day to detect the nrtP transcripts and for 5 days to detect the narB transcripts. The sizes of the transcripts were estimated from the sizes of the rRNAs, i.e., 23S rRNA (2.8 kb), 16S rRNA (1.5 kb), and 23S rRNA in vivo cleavage products (2.3 and 0.5 kb). Genomic DNA Southern blot hybridization analyses were performed under the same stringency conditions as for the RNA blot hybridization analyses to verify the specificity of these probes; the probes hybridized only with the DNA fragments with the size expected from the restriction map (data not shown).
The cDNA templates for reverse transcription-PCR (RT-PCR) were synthesized by using 1 μg of total RNA from wild-type cells of Synechococcus sp. strain PCC 7002 grown on nitrate with 20 pmol of specific primer and Moloney murine leukemia virus reverse transcriptase (Promega, Madison, Wis.) in a reaction volume of 25 μl according to the manufacturer’s protocol. After digestion of excess RNA with RNase H and RNase A, the sample containing the synthesized cDNA was purified by using a microconcentrator (Nanosep 100; Pall Filtron Co., Northborough, Mass.) to remove the primer and the digested RNA. The cDNA fraction was recovered in 40 μl of 10 mM Tris-HCl–1 mM EDTA (pH 8.0), and 1/10 volume (4 μl) of the cDNA sample was used as the template for PCR to amplify the RT-PCR product. Three different primers were used for cDNA synthesis: primer 1, inside the nrtP gene, 5′-CAG CAC TTG GAA GAA GCC A (nucleotides 3065 to 3047); primer 2, inside the narB gene, 5′-CCA AAG CTC TGG ATA TAG (nucleotides 3865 to 3848); and primer 3, inside the narB gene, 5′-CGG AAC TGG GGA CAA TAG (nucleotides 4825 to 4808). To detect RT-PCR products containing the noncoding region between the nrtP and narB open reading frames, the forward primer 5′-TAG CCA AGT GGC TAT TTT AG (nucleotides 2534 to 2553) and primer 2 (as the reverse primer; described above) were used for the PCR amplification. As a positive control to detect RT-PCR products containing the nrtP and narB regions, PCR was performed with nrtP-specific primers 5′-AAC TTT GGC TCG GCA TTT TC (forward; nucleotides 2031 to 2050) and primer 1 (reverse) and narB-specific primers 5′-ATT CCA GCG CAT CAG TTG (forward; nucleotides 3632 to 3649) and primer 3 (reverse). The RT-PCR products derived from the nrtP and narB genes, and with the expected sizes, were detected after 25 cycles of PCR. However, RT-PCR products derived from the noncoding region between nrtP and narB (positions 3201 to 3407) were not detected after 40 cycles of PCR (data not shown).
The 5′ endpoint of the mRNA product of the nrtP gene was determined by RT-PCR by amplification of the 5′ region of the mRNA and subsequent sequence analysis of the RT-PCR product as follows. A cDNA template was synthesized by using the nrtP-specific primer 5′-CCA GGT CAT ATG CAG GAT CT (nucleotides 1652 to 1633) as described as above. A poly(C) tail was added to the 3′ end of the synthesized cDNA by using terminal transferase (New England Biolabs, Beverly, Mass.) and dCTP as described in manufacturer’s protocol. The first PCR was performed with the poly(C)-tailed cDNA with a poly(G)-anchored primer, 5′-ACC TGC AGG CAT GCA AGC TTG GGG GGG GGG GGG GGG, as the forward primer and the reverse primer 5′-CAA GCT GCA GGT CAG ACT GTC (nucleotides 1568 to 1548). With the first PCR product as the template DNA, a second PCR was performed to amplify a DNA fragment of 0.22 kb with the forward primer 5′-ACC TGC AGG CAT GCA AGC TTG and the reverse primer 5′-GTC AAT TTT CGA CCT TAT GG (nucleotides 1550 to 1531), and the PCR product of 0.22-kb was subcloned into the EcoRV site of Bluescript KS(−) by the AT-overhang ligation method. The nucleotide sequences of the insert DNAs of three independent clones were determined by using both the T7 and T3 sequencing primers; identical DNA sequence results for the 5′ end of the RT-PCR product were obtained from each plasmid clone.
Nucleotide sequence accession number.
The nucleotide sequence of a 7,210-bp region carrying the nrtP and narB genes in Synechococcus sp. strain PCC 7002 has been submitted to GenBank (accession no. AF089813).
RESULTS
Cloning and insertional inactivation of the nrtP and narB genes.
We initially tried to isolate the nrtA and nrtC genes, encoding subunits of an expected ABC-type nitrate transporter from Synechococcus sp. strain PCC 7002. However, probes for nrtA and nrtC from Synechococcus sp. strain PCC 7942 and from Synechocystis sp. strain PCC 6803 did not hybridize with genomic DNA fragments of Synechococcus sp. strain PCC 7002 under low-stringency conditions (data not shown). These results suggested either that the nrtA and nrtC genes of Synechococcus sp. strain PCC 7002 were unexpectedly divergent in sequence from those of these related cyanobacteria or that Synechococcus sp. strain PCC 7002 in fact does not possess the genes encoding an ATP-binding cassette-type nitrate transporter. As described below, the latter appears to be the case. Since nitrate permease/transporter genes are rather poorly conserved in sequence but are often linked to the gene encoding nitrate reductase, we thus used the highly conserved narB gene of Synechocystis sp. strain PCC 6803 as a heterologous hybridization probe to isolate a cosmid clone carrying the narB gene and its flanking genes from Synechococcus sp. strain PCC 7002. A 7.2-kb region of this cosmid was sequenced, and the physical map of the sequenced region is shown in Figure 1A.
An open reading frame that predicts a protein of 534 amino acids was found in the 5′ upstream flanking region of the narB gene in Synechococcus sp. strain PCC 7002. The deduced amino acid sequence of this open reading frame showed significant similarity to the sequences of a number of nitrate and nitrite transporters of the major facilitator superfamily, which includes the LacY permease. This gene was thus assumed to encode the nitrate permease of Synechococcus sp. strain PCC 7002 and was designated the nrtP gene. The deduced amino acid sequence of the NrtP protein of Synechococcus sp. strain PCC 7002 showed 31 to 32% sequence identity to the NRT2;1 protein of Chlamydomonas reinhardtii, the Nrt2 proteins of Oryza sativa and Arabidopsis thaliana, the Ynt1 protein of Pichia angusta (Hansenula polymorpha), and the CrnA protein of Aspergillus nidulans. The NrtP protein also showed sequence similarity to the nitrate permease NasA, as well as the nitrite extrusion protein NarK, of Bacillus subtilis. Figure 2 shows a sequence alignment of the NrtP protein of Synechococcus sp. strain PCC 7002 and these NRT2-type nitrate/nitrite transporters of other organisms. The NrtP protein is predicted to be a highly hydrophobic protein that probably contains 12 transmembrane regions. The topology of the NrtP protein is probably similar to that of the Ynt1 and CrnA proteins, since these proteins seem to contain extra hydrophobic domains (Fig. 2, transmembrane regions 7 and 8). Although we do not yet have experimental evidence concerning the localization of NrtP, the protein presumably is localized in the plasma membrane based on its function as a nitrate permease (see below).
FIG. 2.
Sequence alignment of the NrtP protein of Synechococcus sp. strain PCC 7002 to other Nrt2-type nitrate transporter sequences. Amino acids identical and similar to those in the NrtP protein of Synechococcus sp. strain PCC 7002 are marked by shading. Amino acids that are identical in more than five of the nine proteins are shown in boldface. Twelve putative hydrophobic transmembrane regions are indicated by the numbered lines. Osci, partial sequence of the open reading frame in the 5′ upstream region of the narB gene in Oscillatoria chalybea (accession number of the protein sequence, S57965); NrtP, Synechococcus sp. strain PCC 7002 (this study); Chlamy, Chlamydomonas reinhardtii (S40142); Rice, Oryza sativa (BAA33382); Arabi, Arabidopsis thaliana (CAB09794); Ynt1, Pichia angusta (CAA93631); CrnA, Aspergillus nidulans (P22152); NarK, Bacillus subtilis (P46907); NasA, B. subtilis (P42432).
The amino acid sequence deduced from the narB gene of Synechococcus sp. strain PCC 7002 showed a high degree of identity and similarity to the sequences of nitrate reductases of other organisms. NarB of Synechococcus sp. strain PCC 7002 is 58 to 68% identical to the NarB proteins of the cyanobacteria Synechococcus sp. strain PCC 7942, Synechocystis sp. strain PCC 6803, Anabaena sp. strain PCC 7120, and Oscillatoria chalybea. These data strongly suggest that the narB gene of Synechococcus sp. strain PCC 7002 encodes nitrate reductase. As shown in Fig. 1, homologs of petM, psbW2, merA, tmk, as well as a homolog of sll1258 of Synechocystis sp. strain PCC 6803, were also found in the sequences flanking nrtP and narB of Synechococcus sp. strain PCC 7002.
For the functional characterization of the nrtP and narB genes in Synechococcus sp. strain PCC 7002, each gene was insertionally inactivated by using an aphII gene cartridge that encodes aminoglycoside 3′-phosphotransferase II and confers kanamycin resistance (Fig. 1B and C). PCR analyses were performed to confirm that complete segregation of the nrtP and nrtP::aphII alleles had occurred in the transformed strain. The nrtP-specific PCR primers amplified a 1.0-kb DNA fragment when the template was total genomic DNA from the wild-type strain (Fig. 1D, lane 1). When genomic DNA from the nrtP mutant strain was used as template, no 1.0-kb DNA fragment from the wild-type gene was amplified, but a 2.4-kb PCR product corresponding to the nrtP gene plus the 1.4-kb aphII insertion was observed (Fig. 1D, lane 2). These results indicate that the nrtP mutant strain is homozygous and that the expected aphII insertion occurred within the nrtP gene as shown in Fig. 1B. Similarly, PCR analyses were performed to demonstrate complete segregation of the narB gene and the narB::aphII alleles. When the narB-specific primers were used with total genomic DNA from the wild-type strain, a single DNA fragment of 1.2 kb was amplified (Fig. 1D, lane 3). When the same primers were used with total DNA extracted from the narB mutant strain, no 1.2-kb DNA fragment was amplified, but instead a 2.6-kb product corresponding to the narB gene fragment with the 1.4-kb aphII gene insertion was amplified (Fig. 1D, lane 4). These results indicate that the narB mutant strain is homozygous and that it contains the expected insertion of the aphII gene into the narB gene.
Growth of the nrtP and narB mutants.
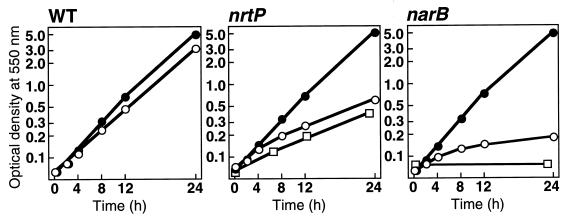
Figure 3 shows the growth profiles of wild-type cells, the nrtP mutant cells and the narB mutant cells on urea and after transfer to nitrate growth conditions. Wild-type cells of Synechococcus sp. strain PCC 7002 could grow on either nitrate or urea as the sole nitrogen source, with a doubling time of 4 h on nitrate and 3.5 h on urea at 38°C under saturating light and carbon dioxide conditions. The growth rates of the nrtP and narB mutant strains on urea were identical to that of the wild-type strain (Fig. 3). However, when the nrtP mutant cells grown on urea were transferred to fresh medium A+ containing 12 mM nitrate as the sole nitrogen source, the growth rate decreased and reached steady-state conditions with a doubling time of 9 h (Fig. 3) approximately 4 to 8 h after the nitrogen source shift from urea to nitrate. The nrtP mutant cells continued to grow exponentially on nitrate with a doubling time of 9 h in the second subculture (Fig. 3), but the mutant cells had a chlorotic yellow-green coloration (data not shown). When the nitrate concentration in the growth medium was increased to 50 mM NaNO3, the nrtP mutant cells still exhibited a chlorotic yellow-green coloration, and the doubling time (9 h) was identical to that of cells grown on 12 mM NaNO3. These results indicate that the nrtP mutant has a much lower capacity for nitrate utilization for cell growth and that this limited ability to use nitrate limits the growth of the nrtP mutant cells. In the narB mutant cells, the growth rate dramatically decreased after the nitrogen source was changed from urea to nitrate (Fig. 3). The narB mutant cells could not grow at all after subculturing of the narB mutant cells a second time into fresh medium A+ (Fig. 3). These results indicate that the narB mutant cells are unable to utilize nitrate and that cell growth ceases after some initial scavenging of nitrogenous storage materials has occurred in the cells. The narB mutant cells could grow with a rate similar to that of the wild type in a medium containing 5 mM NaNO2 as the sole nitrogen source. Both the wild-type and the narB mutant cells had a lower growth rate on nitrite than on nitrate, with a doubling time of about 7 h; moreover, a lag phase occurred after transfer of cells from urea growth conditions to nitrite growth conditions (data not shown). These results suggest that the wild-type and narB mutant cells of Synechococcus sp. strain PCC 7002 have similar capacities for nitrite utilization.
FIG. 3.
Growth curves for the wild-type strain, the nrtP mutant strain, and the narB mutant strain on urea and after transfer to nitrate growth conditions. Cells grown on medium AU10Ni containing urea as a nitrogen source were collected by centrifugation and washed with nitrogen-free medium. The cells were then inoculated into fresh medium A+ containing nitrate as a nitrogen source (open circles) and into medium AU10Ni containing urea as a nitrogen source (closed circles) and were cultured at 38°C under 250 μE m−2 s−1 with aeration with 1% CO2-enriched air. After growth in medium A+ for 24 h, the cells of the nrtP and narB mutants were diluted into fresh medium A+ and cultured under identical growth conditions (squares). The data shown are derived from a single experiment; however, this experiment was repeated three times, and essentially identical results were obtained in each case.
Nitrate and nitrite assimilation rates in the nrtP and narB mutants.
Table 1 shows nitrate and nitrite assimilation rates in whole cells of wild-type, nrtP mutant, and narB mutant cells of Synechococcus sp. strain PCC 7002. Wild-type cells grown on nitrate had the maximum activities for ammonia evolution from nitrate, high-affinity nitrate uptake, and nitrite consumption. When cells were grown on urea, no nitrate consumption occurred and the rate of nitrite consumption was very slow in wild-type cells of Synechococcus sp. strain PCC 7002. These results suggest that the expression of the enzymes for nitrate uptake and reduction are suppressed when cells are grown on urea. When urea-grown cells were treated in A+ medium for 1 h under 250 μE m−2 s−1 at 38°C with aeration of 1% CO2-enriched air, all activities of ammonia evolution, high-affinity nitrate uptake, and nitrite consumption were induced in the wild-type cells. In the nrtP mutant cells after a 1-h induction of these activities by incubation of cells under nitrate growth conditions, the reduction of nitrate to ammonia was detectable, but the rate of ammonia evolution was approximately 4.5-fold lower than that observed for wild-type cells. However, high-affinity nitrate uptake was not detectable in the nrtP mutant cells. These results indicate that the nrtP mutant cells cannot utilize nitrate as efficiently as the wild-type cells. Thus, the nrtP mutant can still take up and reduce nitrate when it is present at the concentration in the growth medium (12 mM), but the nrtP mutant does not exhibit nitrate uptake and consumption when the external concentration is only 100 μM. These data for the nitrate consumption rates in the nrtP mutant are consistent with the data for growth of this mutant on nitrate. The nitrite consumption rate of the nrtP mutant cells at neutral pH was similar to that of the wild-type cells, and this indicates that the in vivo activity of nitrite reductase in the mutant strain is similar to that in the wild type. In the narB mutant cells, no nitrate reduction occurred as assayed either by ammonia evolution or by the high-affinity uptake assay. These results again indicate that cells lacking nitrate reductase cannot utilize nitrate at all. The narB mutant cells did not consume nitrite as efficiently as the wild type, and the in vivo level of nitrite reductase in the narB mutant cells after a 1-h induction under nitrate growth conditions was similar to the level found in wild-type cells grown on urea. These results indicate that the narB mutant has a significantly lower nitrite reductase activity than the wild type or the nrtP mutant. Since the data for the nitrite consumption rate in the narB mutant was inconsistent with the data for growth of this mutant on nitrite, it was hypothesized that the product of nitrate reductase, nitrite, is a positive effector of the activity of nitrite reductase (see below).
TABLE 1.
Nitrate and nitrite assimilation rates in wild-type and nrtP and narB mutant Synechococcus sp. strain PCC 7002a
| Growth condition(s) and strain | Mean nitrate consumption ± SD (n)
|
Mean nitrite consumption ± SD (n) (μmol of NO2− mg of Chl−1 h−1) at pH 7 | |
|---|---|---|---|
| Ammonia evolutionb (NO3− to NH3) (μmol of NH3 mg of Chl−1 h−1) | High-affinity uptakec (μmol of NO3− mg of Chl−1 h−1) | ||
| Grown on nitrate (medium A+), wild type | 32 ± 5 (3) | 42 ± 4 (5) | 82 ± 4 (4) |
| Grown on urea (medium AU10Ni), wild type | NDd (3) | ND (3) | 9.5 ± 1e (3) |
| Grown on urea and then treated in medium A+ for 1 h | |||
| Wild type | 18 ± 3 (3) | 18 ± 2 (7) | 77 ± 8 (4) |
| nrtP mutant | 4 ± 0.5 (4) | ND (3) | 55 ± 6 (4) |
| narB mutant | ND (3) | ND (3) | 9.5 ± 4e (3) |
Cells were grown in medium A+ (12 mM NaNO3 as a nitrogen source) or in medium AU10Ni (10 mM urea plus 5 μM NiSO4) at 38°C under 250 μE m−2 s−1 in 1% CO2-enriched air. To induce the activity of nitrate assimilation, cells were harvested by centrifugation, washed twice with fresh medium A+, and then incubated in medium A+ for 1 h under growth conditions. Ammonia evolution, high-affinity nitrate uptake, and nitrite consumption at pH 7 (in vivo activity of nitrite reductase) were measured with whole cells as described in Materials and Methods.
The detection limit of this assay was approximately 2 μmol of NH3 mg of Chl−1 h−1.
The detection limit of this assay was approximately 0.5 μmol of NO3− mg of Chl−1 h−1.
ND, not detectable.
The rate of nitrite consumption increased during the assay. The initial rate of nitrite consumption is shown.
High-affinity nitrite uptake in the nrtP mutant.
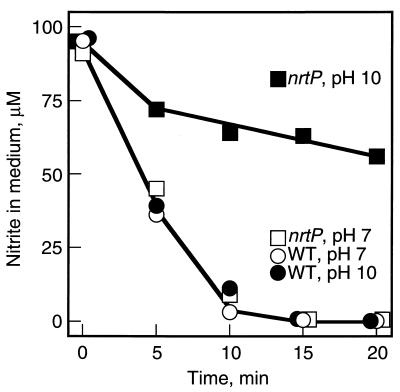
Since nitrite can be taken up passively by the diffusion of nitrous acid at neutral pH (7), high-affinity uptake of nitrite at pH 10 in the nrtP mutant was measured to test the possible role of NrtP in nitrite uptake (Fig. 4). Wild-type cells consumed nitrite at both pH 7 and pH 10, with essentially identical rates of approximately 70 μmol mg of chlorophyll (Chl)−1 h−1. The nrtP mutant cells could consume nitrite with a rate nearly identical to that of the wild-type cells at pH 7 (Fig. 4), but at pH 10 the nrtP mutant cells could consume nitrite only very slowly (Fig. 4). The rate of nitrite consumption at pH 10 in the nrtP mutant was approximately 10 μmol mg of Chl−1 h−1 and was sevenfold lower than that in the wild type under these conditions. Although the nrtP mutant cells had a much lower rate of nitrite consumption at pH 10, nitrite was eventually completely consumed (data not shown). These results suggest that NrtP plays a specific role in the uptake of nitrite as well as nitrate. Moreover, these results indicate that nitrite uptake by Synechococcus sp. strain PCC 7002 takes place (i) by diffusion of nitrous acid at neutral pH; (ii) by high-efficiency uptake via the NrtP permease, and (iii) by low-efficiency but high-affinity uptake through another, unidentified transport mechanism(s).
FIG. 4.
Nitrite consumption by whole cells of the wild-type strain and the nrtP mutant strain of Synechococcus sp. strain PCC 7002. The disappearance of nitrite from the assay medium was measured for wild-type cells at pH 7, wild-type cells at pH 10, nrtP mutant cells at pH 7, and nrtP mutant cells at pH 10. The samples were preilluminated for 15 min under 250 μE m−2 s−1 at 38°C in the presence of the GOGAT inhibitor DON. At time zero, 100 μM NaNO2 was added to initiate the reaction. The assay medium contained 25 mM HEPES-NaOH (pH 7.0) or 25 mM CAPS-NaOH (pH 10.0), 1 mM KCl, 10 mM NaHCO3, 0.5 mM DON, 100 μM NaNO2, and cells equivalent to 10 μg of chlorophyll ml−1. The data shown are the average values from two independent experiments.
Effect of nitrite on the in vivo activity of nitrite reductase in the narB mutant.
In the narB mutant cells, the in vivo activity of nitrite reductase was as low as that of wild-type cells grown on urea (Table 1). Since the narB mutant cells could grow on nitrite with a 7-h doubling time, similar to that of the wild-type cells, the narB mutant was expected to have a nitrite reductase activity equivalent to that of the wild-type cells. It has been reported that nitrite positively regulates the transcription of the nirA-nrtABCD-narB operon in Synechococcus sp. strain PCC 7942 (12). Thus, the effect of exogenously added nitrite on the level of the nitrite reductase activity was tested with the narB mutant strain of Synechococcus sp. strain PCC 7002, in which no endogenous nitrite can be produced from nitrate reduction. Table 2 shows the effect of 1 mM NaNO2 on the in vivo activity of nitrite reductase. When the narB mutant cells were grown on urea and incubated in a medium containing 1 mM NaNO2 in the absence of other reduced nitrogen sources, the activity of nitrite reductase in the narB mutant was enhanced. After a 1-h treatment in medium A+, the activity of nitrite reductase was slightly enhanced, but the activity level was lower than that in cells treated with 1 mM nitrite. Only a basal level of nitrite reductase activity was detected in the narB mutant cells when they were treated with AU10Ni medium for 1 h, and the addition of nitrite with urea did not increase the activity of nitrite reductase. These results indicate that nitrite plays a role as a positive effector of nitrite reductase in the absence of urea but that the presence of urea suppresses the activity of nitrite reductase.
TABLE 2.
Effect of nitrite on in vivo nitrite reductase activity in the Synechococcus sp. strain PCC 7002 narB mutanta
| Medium for treatment | Mean nitrite reductase (nitrite consumption at pH 7) ± SD (n) (μmol NO2− mg of Chl−1 h−1) |
|---|---|
| A− (nitrogen free) | 5 ± 1 (3) |
| A− with 1 mM NaNO2 | 29 ± 2 (3) |
| A+ (12 mM NaNO3) | 10 ± 2 (3) |
| A+ with 1 mM NaNO2 | 27 ± 2 (4) |
| AU10Ni (10 mM urea) | 2 ± 0.5 (3) |
| AU10Ni with 1 mM NaNO2 | 1.5 ± 0.3 (3) |
Cells of the narB mutant were grown in AU10Ni (10 mM urea plus 5 μM NiSO4) at 38°C under 250 μE m−2 s−1 in 1% CO2-enriched air. Cells were harvested by centrifugation, washed twice with fresh nitrogen-free medium A−, and then incubated in the specified medium with and without 1 mM NaNO2 for 1 h under growth conditions. After the 1-h treatment, the rate of in vivo nitrite reductase activity at pH 7 was measured with whole cells as described in Materials and Methods.
Transcripts of the nrtP and narB genes.
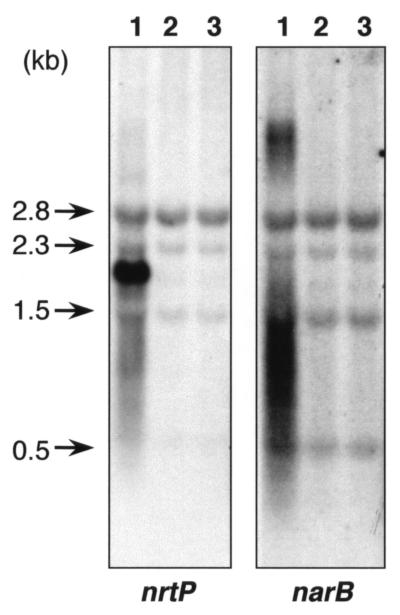
To investigate the nitrogen source dependence of expression of the nrtP and narB genes, total RNAs were isolated from wild-type cells of Synechococcus sp. strain PCC 7002 that had been grown on nitrate, urea, and ammonium, and these RNA samples were subjected to RNA blot hybridization analyses (Fig. 5). When a nrtP hybridization probe was used, a major and very strong hybridization signal corresponding to transcripts of about 2.0 kb was detected in RNA extracted from nitrate-grown cells (Fig. 5, panel nrtP, lane 1). However no transcripts hybridizing to the nrtP probe were detected in the RNA of cells grown on urea or ammonium (Fig. 5, panel nrtP, lanes 2 and 3). When the narB hybridization probe was used, a hybridization signal from transcripts of approximately 3.5 to 4.8 kb and a smear of hybridization to transcripts smaller than 2 kb (Fig. 5, panel narB, lane 1) were detected only in the RNA sample prepared from nitrate-grown cells and not in the RNAs of cells grown on urea or ammonium (Fig. 5, panel narB, lanes 2 and 3). Based on the signal intensities and the exposure times, it was estimated that the signal intensity of the 2.0-kb transcripts hybridizing to the nrtP probe was more than eightfold stronger than that of the 4.8-kb transcript hybridizing with the narB probe. To test further the possibility of dicistronic transcription of the nrtP and narB genes, an RT-PCR analysis was performed in an effort to detect RT-PCR products derived from the noncoding region between the nrtP and narB open reading frames. However, no amplification of this region was detected with an RNA template prepared from nitrate-grown cells (data not shown). The 5′ endpoint of the mRNA product of the nrtP gene was determined by sequencing the RT-PCR product from cDNA that was synthesized with a specific oligonucleotide primer for the nrtP gene and that was subsequently tailed with a poly(C) tract by terminal transferase as described in Material and Methods (Fig. 6). The 5′ end of the nrtP mRNA was mapped at position 1371 in the sequence under GenBank accession no. AF089813, which is located 226 nucleotides upstream of the putative initiation codon of the nrtP gene. The upstream flanking sequence of the 5′ mRNA endpoint closely resembles a consensus, NtcA-dependent promoter sequence (8). This result strongly suggests that the nrtP gene is transcribed from an NtcA-dependent promoter.
FIG. 5.
Nitrogen source-dependent expression of the nrtP and narB genes in Synechococcus sp. strain PCC 7002. Total RNA (10 μg per lane) was isolated from cells grown on nitrate (lane 1), urea (lane 2), or ammonium (lane 3) as a sole nitrogen source and transferred to nylon membrane filters. The membranes were hybridized with probes specific for the nrtP or narB genes. After washing to remove the excess probe DNA, the membranes were exposed to X-ray films for 1 day for the nrtP gene and for 5 days for the narB gene. The 23S (2.8-kb) and 16S (1.5-kb) rRNA bands and the in vivo cleavage products of the 23S rRNA (2.3 and 0.5 kb) were detected as nonspecific hybridization background (arrows). The data shown are from a single experiment which was repeated twice, and identical results were obtained in each experiment.
FIG. 6.
The 5′ endpoint of the nrtP mRNA and the putative NtcA-dependent promoter sequence of the nrtP gene in Synechococcus sp. strain PCC 7002. The 5′ endpoint of the mRNA was determined by the sequence of RT-PCR products as described in Materials and Methods. The numbering for the sequence corresponds to the nucleotide numbering of the sequence under GenBank accession no. AF089813. The consensus sequence for an NtcA-dependent promoter is that described by Flores et al. (8).
These results indicate that the nrtP and narB genes are transcribed as monocistronic mRNAs. The narB gene is probably transcribed from a weaker promoter than the nrtP gene, and the stability of the narB mRNA also seems to be lower than that for nrtP. The absence of transcripts for these genes when cells are grown on reduced nitrogen sources suggests that both genes are subject to regulation by NtcA (7, 8).
DISCUSSION
Three distinct nitrate transport systems have been identified so far (for reviews, see references 3 and 20): the CHL1-type (35), the NRT2-type (36), and the ABC-type (22) nitrate transport systems. Based on their primary structures, these different nitrate transport systems are not evolutionarily related to one another (3). The CHL1 (NRT1) gene was originally identified in a chlorate-resistant mutant of Arabidopsis thaliana and encodes the dual-affinity nitrate transporter in plants (14). Expression of the CHL1 protein in Xenopus oocytes directly demonstrated the function of this gene product in nitrate transport (35). The NRT2-type transporters are high-affinity nitrate transporters that occur in wide range of organisms, including bacteria (17), fungi (23, 36), algae (10), and plants (3, 34). One transporter of this group, the Nrt2;1 transporter of Chlamydomonas reinhardtii, has been reported to be bispecific for nitrate and nitrite (10). Until this study, the ABC-type nitrate transporter was the only known high-affinity nitrate transporter in cyanobacteria. In the study presented here, the nrtP gene of Synechococcus sp. strain PCC 7002 was shown to encode a bispecific nitrate/nitrite permease of the NRT2 class. Since a partial open reading frame with very high sequence similarity to NrtP was found in the 5′ upstream region of the narB gene in the cyanobacterium Oscillatoria chalybea (37), the NrtP permease will almost certainly prove to be more widely distributed among cyanobacteria. An important question that remains to be answered in future studies concerns the distribution of these two types of transporters and the possible physiological implications (if any) of this difference.
Based on the data presented here, the NrtP permease is involved in the active uptake of nitrate in Synechococcus sp. strain PCC 7002. Presumably, the energy for nitrate uptake can be provided by the proton gradient or proton motive force, as is the case in plants (3). It has been proposed that nitrate is taken up by a 2H+/NO3− symport in plants (3), but the energy source (e.g., H+ versus Na+) for the nitrate uptake in cyanobacteria has not yet been directly characterized. In the ABC-type nitrate transporter in cyanobacteria, hydrolysis of ATP is presumed to provide the energy for the active uptake of nitrate (20), although coupling of ATP hydrolysis to transport has not been directly demonstrated yet. The discovery of the NrtP permease in Synechococcus sp. strain PCC 7002 provides an excellent model system for a combined biochemical and genetic analysis of the molecular mechanism of nitrate transport by the NRT2-type permeases that occur in a broad range of organisms, including higher plants.
Based on the genomic Southern hybridization analysis and the insertional inactivation of the nrtP gene, the nrtP gene appears to be a single-copy gene in Synechococcus sp. strain PCC 7002. Additionally, the data presented here suggest that the NrtP permease is the only high-efficiency transport system for both nitrate (Fig. 4; Table 1) and nitrite (Fig. 4) in this cyanobacterium. Since the in vivo activity of nitrite reductase was identical in both the wild-type and the nrtP mutant cells (Table 1; Fig. 4, pH 7), the results in Fig. 4 clearly show that NrtP is involved in the high-efficiency and high-affinity transport of nitrite at alkaline pH. Since the nrtP mutant could reduce nitrate to ammonia (Table 1) and grow on nitrate (Fig. 3), both nitrate and nitrite can also be taken up by low-efficiency transport mechanisms that have not yet been identified. Nonetheless, the data presented here clearly demonstrate that the NrtP permease is the primary, bispecific transporter for nitrate and nitrite in Synechococcus sp. strain PCC 7002.
Contradictory results have been reported concerning the specificity and mechanism of nitrite transport in Synechococcus sp. strain PCC 7942. Luque et al. (15) reported that insertional inactivation of the nrtD gene completely abolished active nitrite transport at pH 9.6. However, Maeda and Omata (16) reported that mutant cells harboring a deletion of the entire nrtABCD coding region could still consume nitrite at pH 9.6, albeit at a lower rate than wild-type cells. Our results show that a low rate of nitrite uptake (approximately 15% of the uptake rate observed for wild-type cells under the same assay conditions [Fig. 4]) occurs in the nrtP mutant cells of Synechococcus sp. strain PCC 7002 at pH 10.
The nrtP mutant cells could grow exponentially on nitrate but with a significantly lower rate than wild-type cells (Fig. 3), and whole cells of the nrtP mutant exhibited a lower rate of nitrate reduction to ammonia (Table 1). Although no direct experimental data have thus far been reported, it is likely that both nitrite and nitrate can also be taken up slowly by a transport system(s) for other ions (e.g., the bicarbonate transport system). Similar to the results presented here, Synechococcus sp. strain PCC 7942 mutants lacking a functional NrtABCD nitrate transporter could still grow on nitrate as the sole nitrogen source, and at high nitrate concentrations and under relatively low light intensity conditions, the mutants actually grew as rapidly as the wild-type strain (22). For a Synechococcus sp. strain PCC 7942 nrtD mutant, the growth rate increased as the concentration of nitrate was increased in the medium over the range of 2 to 40 mM KNO3 (22). However, the growth rate of the Synechococcus sp. strain PCC 7002 nrtP mutant did not increase when the nitrate concentration in the growth medium was increased from 12 to 50 mM (at saturating light and CO2 conditions) (data not shown). These results suggest that for Synechococcus sp. strain PCC 7002 the nitrate concentration (12 mM) in standard A+ medium is sufficient to saturate whatever transporter(s) are used. Moreover, this level of nitrate is sufficient, even in the absence of NrtP, to support exponential growth of the mutant cells that are otherwise supplied with sufficient light and other nutrients, although the nrtP mutant cells were chlorotic and had a lower growth rate under such conditions. The differences in the cell growth and saturation behavior in the various nitrate uptake mutants suggest that Synechococcus sp. strain PCC 7942 and Synechococcus sp. strain PCC 7002 may have different types of nitrate and nitrite transport systems for this slower, lower-efficiency uptake.
In Synechococcus sp. strain PCC 7942, the nirA-nrtABCD-narB genes are linked and regulated as a transcriptional operon, and the nirB-ntcB genes are linked to but divergently transcribed from the nirA gene (21). In Synechocystis sp. strain PCC 6803, the nrtABCD and narB genes are linked, but the nirA, ntcA, and ntcB genes are scattered about the genome (11). In Synechococcus sp. strain PCC 7002, the genes for nitrate assimilation are arranged quite differently. The nrtP and narB genes are linked, although the flanking genes (Fig. 1) are unlike those reported for Synechococcus sp. strain PCC 7942 (20) or Synechocystis sp. strain PCC 6803 (11). Although the genes involved in nitrate or nitrite uptake and assimilation seem to be scattered in the Synechococcus sp. strain PCC 7002 genome (29), the regulation of expression of these genes is apparently similar to that demonstrated in Synechococcus sp. strain PCC 7942 (21). The nrtP and narB genes were transcribed in nitrate-grown cells but not in urea- or ammonium-grown cells of Synechococcus sp. strain PCC 7002 (Fig. 5). These results strongly suggest that the nrtP and narB genes are probably coordinately controlled at the transcriptional level by the DNA-binding protein NtcA, which is the global transcriptional activator for the regulation of the genes for nitrogen metabolism in cyanobacteria (7). Consistent with the assumption of transcriptional regulation via NtcA, potential NtcA-dependent promoter sequence is found in the 5′ flanking regions of the 5′ end of the mRNA product of the nrtP gene (Fig. 6) and also in the 5′ flanking regions of the narB gene (GTAATGGCGTTTTAC; nucleotides 3250 to 3264 in the sequence under GenBank accession no., AF089813). The presence of these two putative NtcA binding sites is consistent with our suggestion above that the nrtP and narB genes are independently transcribed from separate promoters. The activity of nitrite reductase is subject to dual-mode control. The absence of reduced nitrogen sources causes increased expression of nitrite reductase, but nitrite appears to enhance the expression level of nitrite reductase specifically as well (Table 2). These observations are consistent with a model that has been proposed for Synechococcus sp. strain PCC 7942 (12). We are currently isolating the genes involved in nitrate and nitrite assimilation, as well as the genes encoding the control elements for the expression of those genes involved in global nitrogen metabolism, from Synechococcus sp. strain PCC 7002 (29). Since the genes encoding the nitrate permease, nitrate reductase, urease (27, 28), and glutamine synthetase (38) are transcribed from independent promoters, it may be easier to analyze the transcriptional regulation of these genes than has been the case with some other cyanobacteria. Such comparative studies of the transcriptional regulatory circuitry in different cyanobacteria should provide insights into those mechanisms which are common to all cyanobacteria and those which may be distinctive for a given species.
It is thought that nitrate uptake is the rate-limiting step in the overall rate of nitrate assimilation, which includes nitrate uptake, nitrate reduction, and nitrite reduction to ammonia (7). This has specifically been shown to be true when Synechococcus sp. strain PCC 6301 is grown at low temperature (26). It has been shown that exogenously added ammonium ion arrests nitrate consumption within minutes in Anabaena cylindrica (19), Synechococcus sp. strain PCC 7942 (21), Synechococcus sp. strain PCC 6301 (5, 26), and Synechococcus sp. strain PCC 7002 (27). Thus, although the nitrate transport system for Synechococcus sp. strain PCC 7002 differs significantly from those of these other cyanobacteria, exogenously added ammonia appears to control its activity in a very similar fashion. Ohmori and Hattori (18) also demonstrated that the addition of ammonium ions caused a rapid and transient decrease in the ATP level within a few minutes, and those authors suggested that this could be due to the rapid consumption of ATP by glutamine synthetase because of the increased substrate level for this enzyme. Although this rapid decrease in the ATP pool could be a circumstantial event induced by the exogenously added ammonia, the resulting decrease in the ATP level could nevertheless have a major impact on decreasing active transport of nitrate and in the early events in the repression of nitrate assimilation by ammonia (18). No biochemical model that satisfactorily explains the rapid effect of exogenous ammonia in inhibiting nitrate consumption has yet been proposed (33). However, it was recently shown that the glnB gene product, the PII protein, is involved in the regulation circuitry for the inhibition of nitrate or nitrite uptake by ammonium (13). It is assumed that such regulation of the nitrate transport system takes place to facilitate rapid responses to changes in nitrogen sources that are available in the environment. The discovery of the NrtP nitrate permease provides an important new tool for molecular biological approaches to analyses of the control mechanism(s) of the nitrate transport and reduction systems of cyanobacteria.
ACKNOWLEDGMENTS
We thank Veronica L. Stirewalt for her negative hybridization screening experiments for nrt and cmp genes, since these negative results ultimately provided the inspiration for finding the novel nitrate transporter. We also thank Joel Graham (Juniata College) and Julie Frey (Bloomsburg University), who were summer students supported by the NSF-RTG program in Microbial Structural Biology at Penn State University in 1998, for their technical assistance in DNA restriction mapping, sequencing, and gene disruption.
This work was supported by Public Health Service grant GM-31625 to D.A.B.
REFERENCES
- 1.Bryant D A, Tandeau de Marsac N. Isolation of genes encoding components of photosynthetic apparatus. Methods Enzymol. 1988;167:755–765. [Google Scholar]
- 2.Cai Y, Wolk C P. Nitrogen deprivation of Anabaena sp. strain PCC 7120 elicits rapid activation of a gene cluster that is essential for uptake and utilization of nitrate. J Bacteriol. 1997;179:258–266. doi: 10.1128/jb.179.1.258-266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford N M, Glass A D M. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 1998;3:389–395. [Google Scholar]
- 4.Elhai J, Wolk C P. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allow cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 5.Flores E, Guerrero M G, Losada M. Short-term ammonium inhibition of nitrate utilization by Anacystis nidulans and other cyanobacteria. Arch Microbiol. 1980;128:137–144. [Google Scholar]
- 6.Flores E, Guerrero M G, Losada M. Photosynthetic nature of nitrate uptake and reduction in the cyanobacterium Anacystis nidulans. Biochim Biophys Acta. 1983;722:408–416. [Google Scholar]
- 7.Flores E, Herrero A. Assimilatory nitrogen metabolism and its regulation. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 487–517. [Google Scholar]
- 8.Flores E, Muro-Paster A M, Herrero A. Cyanobacterial nitrogen assimilation genes and NtcA-dependent control of gene expression. In: Peshek G A, Löffelhardt W, Schmetterer G, editors. The phototrophic prokaryotes. New York, N.Y: Kluwer Academic Publishers; 1999. pp. 463–477. [Google Scholar]
- 9.Frias J E, Flores E, Herrero A. Nitrate assimilation gene cluster from heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:477–486. doi: 10.1128/jb.179.2.477-486.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galvan A, Quesada A, Fernandez E. Nitrate and nitrite are transported by different specific transport systems and by a bispecific transporter in Chlamydomonas reinhardtii. J Biol Chem. 1996;271:2088–2092. doi: 10.1074/jbc.271.4.2088. [DOI] [PubMed] [Google Scholar]
- 11.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 12.Kikuchi H, Aichi M, Suzuki I, Omata T. Positive regulation by nitrite of the nitrate assimilation operon in the cyanobacteria Synechococcus sp. strain PCC 7942 and Plectonema boryanum. J Bacteriol. 1996;178:5822–5825. doi: 10.1128/jb.178.19.5822-5825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H-M, Flores E, Herrero A, Houmard J, Tandeau de Marsac N. A role for the signal transduction protein PII in the control of nitrate/nitrite uptake in a cyanobacterium. FEBS Lett. 1998;427:291–295. doi: 10.1016/s0014-5793(98)00451-7. [DOI] [PubMed] [Google Scholar]
- 14.Liu K H, Huang C Y, Tsay Y F. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell. 1999;11:865–874. doi: 10.1105/tpc.11.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luque I, Flores E, Herrero A. Nitrate and nitrite transport in the cyanobacterium Synechococcus sp. PCC 7942 are mediated by the same permease. Biochim Biophys Acta. 1994;1184:296–298. [Google Scholar]
- 16.Maeda S-I, Omata T. Substrate-binding lipoprotein of the cyanobacterium Synechococcus sp. strain PCC 7942 involved in the transport of nitrate and nitrite. J Biol Chem. 1997;272:3036–3041. doi: 10.1074/jbc.272.5.3036. [DOI] [PubMed] [Google Scholar]
- 17.Ogawa K-I, Akagawa E, Yamane K, Sun Z-W, LaCelle M, Zuber P, Nakano M M. The narB operon and nasA gene are required for nitrate/nitrite assimilation in Bacillus subtilis. J Bacteriol. 1995;177:1409–1413. doi: 10.1128/jb.177.5.1409-1413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohmori M, Hattori A. Transient change in the ATP pool of Anabaena cylindrica associated with ammonia assimilation. Arch Microbiol. 1978;117:17–20. doi: 10.1007/BF00689345. [DOI] [PubMed] [Google Scholar]
- 19.Ohmori M, Ohmori K, Strotmann H. Inhibition of nitrate uptake by ammonia in a blue-green alga, Anabaena cylindrica. Arch Microbiol. 1977;114:225–229. [Google Scholar]
- 20.Omata T. Structure, function and regulation of the nitrate transport system of the cyanobacterium Synechococcus sp. PCC 7942. Plant Cell Physiol. 1995;36:207–213. doi: 10.1093/oxfordjournals.pcp.a078751. [DOI] [PubMed] [Google Scholar]
- 21.Omata T. Transcriptional and post-translational regulation of nitrate utilization in the cyanobacterium Synechococcus sp. strain PCC 7942. In: Satoh K, Murata N, editors. Stress responses of photosynthetic organisms. Amsterdam, The Netherlands: Elsevier Science, B. V.; 1998. pp. 197–214. [Google Scholar]
- 22.Omata T, Andriesse X, Hirano A. Identification and characterization of a gene cluster involved in nitrate transport in the cyanobacterium Synechococcus sp. PCC 7942. Mol Gen Genet. 1993;236:193–202. doi: 10.1007/BF00277112. [DOI] [PubMed] [Google Scholar]
- 23.Perez M D, Gonzalez C, Avila J, Brito N, Siverio J M. The YNT1 gene encoding the nitrate transporter in the yeast Hansenula polymorpha is clustered with genes YNI1 and YNR1 encoding nitrite reductase and nitrate reductase, and its disruption causes inability to grow in nitrate. Biochem J. 1997;321:397–403. doi: 10.1042/bj3210397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakamoto T, Bryant D A. Temperature-regulated mRNA accumulation and stabilization for fatty acid desaturase genes in the cyanobacterium Synechococcus sp. PCC 7002. Mol Microbiol. 1997;23:1281–1292. doi: 10.1046/j.1365-2958.1997.3071676.x. [DOI] [PubMed] [Google Scholar]
- 25.Sakamoto T, Bryant D A. Growth at low temperature causes nitrogen limitation in the cyanobacterium Synechococcus sp. PCC 7002. Arch Microbiol. 1998;169:10–19. doi: 10.1007/s002030050535. [DOI] [PubMed] [Google Scholar]
- 26.Sakamoto T, Bryant D A. Nitrate transport and not photoinhibition limits growth of the fresh water cyanobacterium Synechococcus species PCC 6301 at low temperature. Plant Physiol. 1999;119:785–794. doi: 10.1104/pp.119.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakamoto, T., and D. A. Bryant. Unpublished data.
- 28.Sakamoto T, Delgaizo V B, Bryant D A. Growth on urea can trigger death and peroxidation of the cyanobacterium Synechococcus sp. strain PCC 7002. Appl Environ Microbiol. 1998;64:2361–2366. doi: 10.1128/aem.64.7.2361-2366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schone, K., S. Persson, T. Sakamoto, V. L. Stirewalt, and D. A. Bryant. Unpublished data.
- 30.Stanier R Y, Cohen-Bazire G. Phototrophic prokaryotes: the cyanobacteria. Annu Rev Microbiol. 1977;31:225–274. doi: 10.1146/annurev.mi.31.100177.001301. [DOI] [PubMed] [Google Scholar]
- 31.Stevens S E, Jr, Patterson C O P, Myers J. The production of hydrogen peroxide by blue-green algae: a survey. J Phycol. 1973;9:427–430. [Google Scholar]
- 32.Stevens S E, Jr, Porter R D. Transformation in Agmenellum quadruplicatum. Proc Natl Acad Sci USA. 1980;77:6052–6056. doi: 10.1073/pnas.77.10.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tandeau de Marsac N, Lee H-M. Regulation of carbon and nitrogen metabolism in the unicellular cyanobacteria Synechococcus spp. In: Peshek G A, Löffelhardt W, Schmetterer G, editors. The phototrophic prokaryotes. New York, N.Y: Kluwer Academic Publishers; 1999. pp. 539–548. [Google Scholar]
- 34.Trueman L J, Richardson A, Forde B G. Molecular cloning of higher plant homologues of the high-affinity nitrate transporters of Chlamydomonas reinhardtii and Aspergillus nidulans. Gene. 1996;175:223–231. doi: 10.1016/0378-1119(96)00154-0. [DOI] [PubMed] [Google Scholar]
- 35.Tsay T F, Schroeder J I, Feldmann K A, Crawford N M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell. 1993;72:705–713. doi: 10.1016/0092-8674(93)90399-b. [DOI] [PubMed] [Google Scholar]
- 36.Unkles S E, Hawker K L, Grieve C, Campbell E I, Montague P, Kinghorn J R. crnA encodes a nitrate transporter in Aspergillus nidulans. Proc Natl Acad Sci USA. 1991;88:204–208. doi: 10.1073/pnas.88.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Unthan M, Klipp W, Schmid G H. Nucleotide sequence of the narB gene encoding assimilatory nitrate reductase from cyanobacterium Oscillatoria chalybea. Biochim Biophys Acta. 1996;1305:19–24. doi: 10.1016/0167-4781(95)00210-3. [DOI] [PubMed] [Google Scholar]
- 38.Wagner S J, Thomas S P, Kaufman R I, Nixon B T, Stevens S E., Jr The glnA gene of the cyanobacterium Agmenellum quadruplicatum PR-6 is nonessential for ammonium assimilation. J Bacteriol. 1993;175:604–612. doi: 10.1128/jb.175.3.604-612.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolk C P. Physiology and cytological chemistry of blue-green algae. Bacteriol Rev. 1973;37:32–101. doi: 10.1128/br.37.1.32-101.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]