Abstract
Objective:
To train a deep learning (DL) algorithm to perform fully automated semantic segmentation of multiple autofluorescence lesion types in Stargardt disease.
Design:
Cross-sectional study with retrospective imaging data.
Subjects:
193 images from 193 eyes of 97 patients with Stargardt disease.
Methods:
Fundus autofluorescence (FAF) images obtained from patient visits between 2013 and 2020 were annotated with ground-truth labels. Model training and evaluation were performed with five-fold cross-validation.
Main Outcomes and Measures:
Dice similarity coefficients, intraclass correlation coefficients (ICCs), and Bland-Altman analyses comparing algorithm-predicted and grader-labeled segmentations.
Results:
The overall Dice similarity coefficient across all lesion classes was 0.78 (95%CI, 0.69–0.86). Dice coefficients were 0.90 (95%CI, 0.85–0.94) for areas of definitely decreased autofluorescence (DDAF), 0.55 (95%CI, 0.35–0.76) for areas of questionably decreased autofluorescence (QDAF), and 0.88 (95%CI, 0.73–1.00) for areas of abnormal background autofluorescence (ABAF). ICCs comparing the ground truth and automated methods were 0.997 (95%CI, 0.996–0.998) for DDAF, 0.863 (95%CI, 0.823–0.895) for QDAF, and 0.974 (95%CI, 0.966–0.980) for ABAF.
Conclusions:
A DL algorithm performed accurate segmentation of autofluorescence lesions in Stargardt disease, demonstrating the feasibility of fully automated segmentation as an alternative to manual or semi-automated labeling methods.
Precis
A ResNet-UNet convolutional neural network can accurately label multiple lesion types in autofluorescence images for Stargardt disease, facilitating automated monitoring of disease progression.
Introduction
Stargardt disease is the most common juvenile-onset macular dystrophy, affecting approximately 1 in 8,000–10,000 individuals.1 It is inherited in an autosomal recessive manner, and over 900 disease-causing variants have been identified in the gene ABCA4.2,3 The disease is clinically heterogenous, with significant variation in age of onset and presentation, imaging findings, and rate of progression.4
When disease-causing variants disrupt function of the ABCA4 transporter protein, toxic bisretinoids accumulate in the retinal pigment epithelium (RPE), leading to progressive changes on autofluorescence (AF) imaging. The AF lesions that arise in Stargardt disease have been well described.5–7 Lesions of definitely decreased autofluorescence (DDAF) are defined as areas where the level of AF darkness is at least 90% that of the optic disc. DDAF lesions signify substantial atrophy of the RPE-photoreceptor complex. Lesions of questionably decreased autofluorescence (QDAF) are defined as areas where the level of AF darkness is between 50% and 90% that of the optic disc. QDAF lesions represent areas of diseased outer retina that may convert into DDAF lesions. Other abnormal AF features in Stargardt disease include localized hypofluorescent and hyperfluorescent changes associated with pisciform flecks, diffuse background hyperfluorescence, and heterogenous hypofluorescent and hyperfluorescent background changes.8–10
The ProgStar natural history studies of Stargardt disease revealed that changes in AF lesions may be useful for monitoring disease progression and serving as an outcome measure for clinical trials.11 Automated segmentation of AF lesions may assist manual graders or serve as a basis for consensus in segmentation tasks for therapeutic trials. Additionally, automated segmentation could provide rapid or real-time rates of progression to physicians and patients. Previous studies have described using machine learning or deep learning methods to segment isolated AF features of Stargardt disease including atrophic lesions and flecks.12–14 However, simultaneous multi-label segmentation has not been previously achieved. Here we describe the training and validation of a DL algorithm for performing multi-label segmentation of DDAF and QDAF lesions, as well as regions of abnormal background AF (ABAF).
Methods
We included patients seen in the inherited retinal disease (IRD) clinic with a clinical diagnosis of Stargardt disease and molecular confirmation of disease with two or more pathogenic or likely pathogenic variants in ABCA4. The clinical diagnosis was established by an expert in IRDs (A.T.F or K.T.J.). Comprehensive evaluation consisted of family pedigree obtained by a genetic counselor (D.S. or K.B.), clinical examination, electroretinogram, Goldmann visual field testing, fundus photography, fundus AF, and macular optical coherence tomography. Molecular diagnosis was confirmed by a Clinical Laboratory Improvement Amendments certified laboratory. We excluded patients found to only have one pathogenic or likely pathogenic variant in ABCA4 or those found to have another likely cause of disease in a different IRD gene. Institutional Review Board Committee approval was obtained. This study adhered to the tenets of the Declaration of Helsinki.
AF images were captured using Optos 200Tx or Optos California Ultra-widefield (Optos Inc., Dunfermline, United Kingdom) imaging devices using an excitation wavelength of 532 nm. Each image was cropped to the central 512 × 512 pixels including the macula and optic disc. Labeling of DDAF and QDAF lesions was performed by a clinician with training in IRDs (P.Y.Z) using Photoshop (Adobe Inc., Mountain View, California) to create the multi-channel label masks according to previously established criteria for DDAF and QDAF lesions.5,6 A 20% random sampling of the images and ground truth labels were verified by an expert in IRDs (KTJ). Additionally, areas of abnormal background autofluorescence that did not meet criteria for DDAF or QDAF were labeled as ABAF lesions. ABAF lesions included hypofluorescent and hyperfluorescent changes surrounding flecks, patches of homogenous increased background hyperfluorescence, and areas of heterogenous hypofluorescent and hyperfluorescent background changes. Images were categorized into AF subtypes as previously described.10
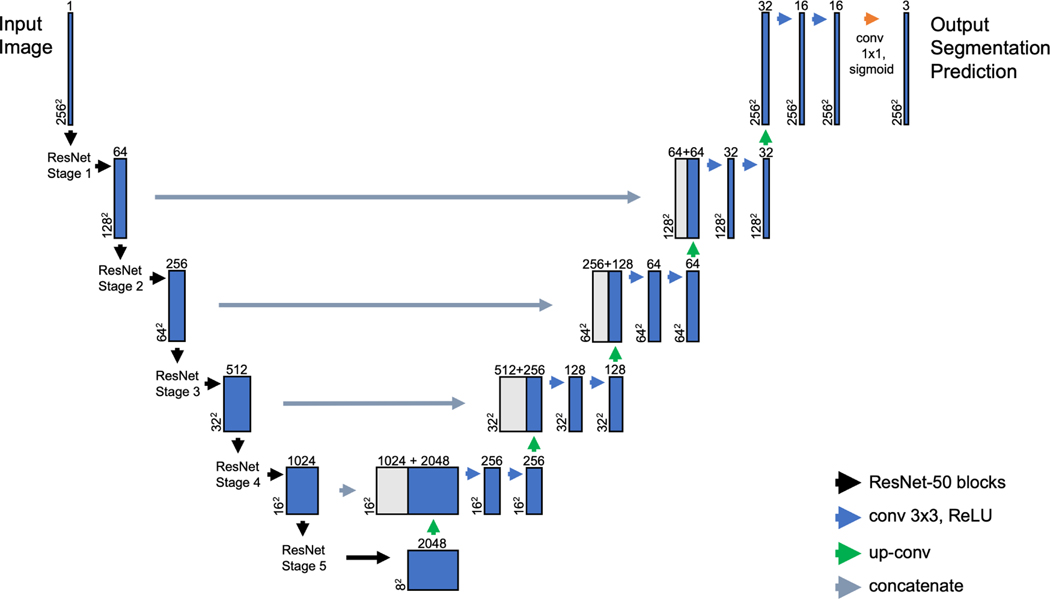
A modified UNet model was constructed using ResNet-50 encoder blocks (Figure 1) for the down-sampling portion of the model. The model is publicly available at (https://github.com/retina-deep-learning/StarSeg). We used this ResNet-UNet model because it contains features to optimize segmentation. The architecture of UNet contains an encoder and decoder portion.15 The encoder collects feature information, while the decoder combines the feature information from the encoder with spatial information passed through long skip connections. Additionally, we modified the base UNet architecture by specifying ResNet50 as the encoder portion of the model. Deeper neural networks have increased ability to extract higher-level features from images, but with deeper networks it was also paradoxically found that training errors increased. This issue, known as the degradation problem, was addressed by He and colleagues using short skip connections to pass identity mappings, which allow the network layers to fit residual mappings.16 This ResNet-UNet model was constructed to accept grayscale image input and generate output of multi-label classifications with different color channels for each feature (DDAF, blue; QDAF, green; ABAF, red).
Figure 1.
Architecture of the deep learning model used to train on the Stargardt disease imaging data set. The ResNet-UNet encoder-decoder model, consists of a contracting (down-sampling) path of ResNet-50 blocks and an expanding (up-sampling) path, with feature maps passed from the contracting path to the expanding path. “conv” refers to a spatial convolution layer, “3×3” refers to the kernel size for the convolution operation, and “ReLU” refers to the rectified linear unit activation function. “max pool” refers to a max pooling layer. “up conv” refers to an up-sampling layer. “concatenate” refers to the joining of a feature map from the contracting path with an up-sampling layer from the expanding path. “sigmoid” refers to the sigmoid activation function used after the final convolutional layer to generate probabilities for predictions.
The model was evaluated using five-fold cross-validation. The RMSprop algorithm was used for optimization, and soft Dice loss was used for the loss function.17,18 The ResNet-50 part of the model was pre-trained on ImageNet. Learning rate was set to 0.001 for initial training, and then reduced to 0.0001 for fine-tuning. Training data batch size was set to 16. Training data was randomly augmented using rotation of up to 30 degrees, horizontal and vertical translation of up to one-quarter image length, shear of up to 15 degrees, and zoom between 75 and 125 percent of image size. Training of each fold was completed after 600 epochs. Computation was performed using Keras with TensorFlow version 2.4.1 as backend on the University of Michigan High Performance Computing Cluster (16GB NVIDIA Tesla V100; NVIDIA Corporation, Santa Clara, California). Intra-class correlation coefficient (ICC) calculations and Bland-Altman analyses were performed in R (The R Foundation, Vienna, Austria). ICC estimates and their 95% confidence intervals were calculated based on a two-way random-effects, absolute agreement, single-measurement model.
Results
The study included 193 images from 193 eyes of 97 patients seen in the IRD clinic between 2013 and 2020. We excluded 19 images from 10 patients due to media opacity or poor image quality precluding accurate image labeling. Clinical characteristics are shown in Table 1. Mean age was 35.0 (SD = 17.3), and mean logMAR visual acuity was 0.76 (SD = 0.53) or Snellen equivalent 20/115. Out of 97 patients, 79 (81%) had 2 disease-causing variants, 17 (18%) had 3 disease-causing variants, and 1 (1%) had 4 disease-causing variants. There were 77 unique disease-causing variants found in 97 patients. The most frequently found disease-causing variants were missense variant c.5882G>A (16 patients), missense variant c.3113C>T (14 patients), splice variant c.2588G>C (12 patients), missense variant c.5603A>T (12 patients), and missense variant c.1622T>C (11 patients). Image characteristics are shown in Table 2. Out of 193 images, 83 (43%) images were categorized as AF subtype 1 (localized low signal at the fovea surrounded by homogenous background), 77 (40%) as subtype 2 (localized low signal at the macula surrounded by heterogenous background), and 33 (17%) as subtype 3 (multiple low-signal areas in the posterior pole surrounded by heterogenous background).
Table 1.
Clinical Characteristics (N = 97)
| Mean | SD | |
|---|---|---|
| Age (years) | 35.0 | 17.3 |
| Visual Acuity (logMAR) | 0.76 | 0.53 |
| N (%) | ||
| Female Sex | 53 (55%) | |
| Race | ||
| White | 81 (84%) | |
| Black | 9 (9%) | |
| Asian | 4 (4%) | |
| Other | 3 (3%) | |
| No. of Disease-Causing Variants | ||
| 2 Variants | 79 (81%) | |
| 3 Variants | 17 (18%) | |
| 4 Variants | 1 (1%) | |
| Most Common Disease-Causing Variants | Frequency | |
| c.5882G>A, (p.Gly1961Glu), Missense | 16 | |
| c.3113C>T, (p.Ala1038Val), Missense | 14 | |
| c.2588G>C, (p.Gly863Ala), Splice | 12 | |
| c.5603A>T, (p.Asn1868Ile), Missense | 12 | |
| c.1622T>C, (p.Leu541Pro), Missense | 11 | |
| c.4139C>T, (p.Pro1380Leu), Missense | 9 | |
| c.5461–10T>C, Intronic | 7 | |
| c.6079C>T, (p.Leu2027Phe), Missense | 7 | |
| c.3322C>T, (p.Arg1108Cys), Missense | 5 | |
Table 2.
Imaging Characteristics (193 images from 193 eyes)
| FAF Subtype | N (%) | |
|---|---|---|
| Type 1 | 83 (43%) |
|
| Type 2 | 77 (40%) | |
| Type 3 | 33 (17%) | |
| Heterogenous FAF Background | 107 (55%) | |
| Mean Lesion Area (mm2) | Mean | SD |
| DDAF | 9.9 | 20.0 |
| QDAF | 3.4 | 5.9 |
| ABAF | 36.1 | 29.4 |
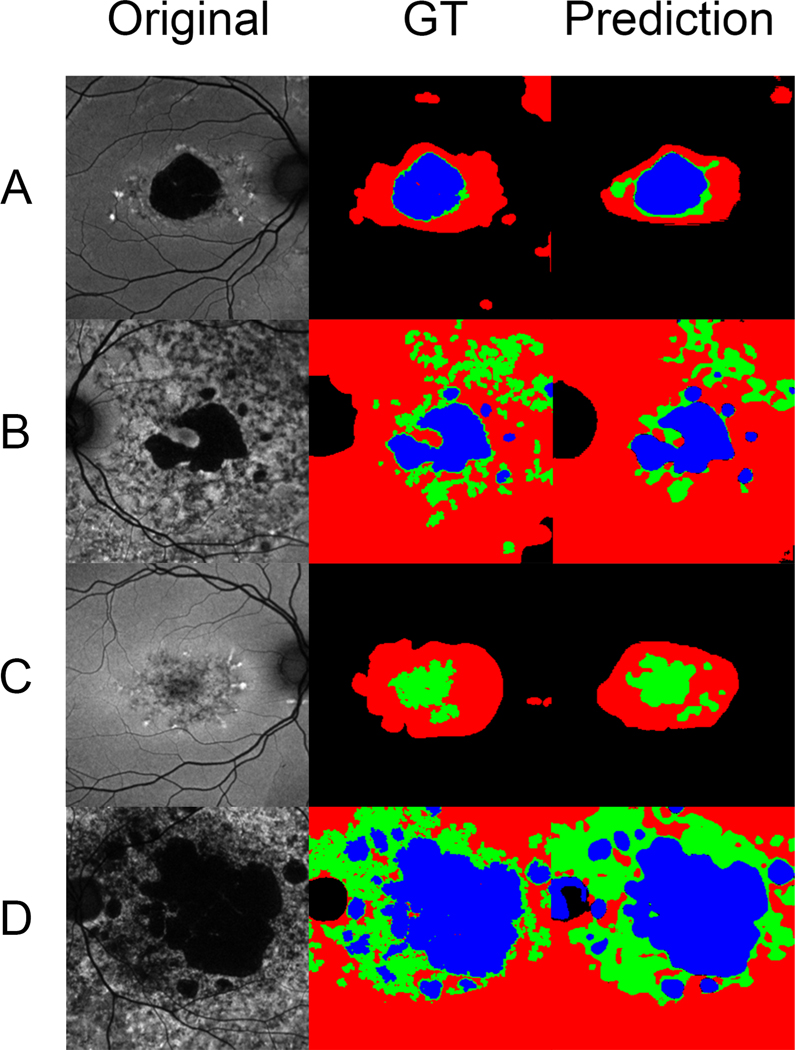
DL algorithm performance was evaluated using five-fold cross-validation. The Dice similarity coefficient over all classes of lesions was 0.78 (95%CI, 0.69–0.86). Dice coefficients varied among the different lesion classes. For DDAF lesions, the Dice coefficient was 0.90 (95%CI, 0.85–0.94). For QDAF lesions, the Dice coefficient was 0.55 (95%CI, 0.35–0.76). For ABAF, the Dice coefficient was 0.88 (95%CI, 0.73–1.00). Figure 2 shows images representative of the different AF subtypes found in Stargardt disease, their labeled ground truths, and the DL algorithm segmentation predictions. The algorithm was able to predict segmentations for DDAF, QDAF, and ABAF lesions across the three previously described AF subtypes (type 1, type 2, and type 3).10 Predicted segmentations for DDAF and ABAF were more consistent with the respective ground truth labels than the predicted segmentations for QDAF, consistent with the higher Dice coefficients for DDAF and ABAF.
Figure 2.
Rows of representative images (A, B, C) from the data set with corresponding ground truth labeling (GT) and algorithm segmentation predictions. DDAF is labeled in blue, QDAF in green, and ABAF in red. Image A is a subtype 1 AF pattern with localized low signal in the macula (DDAF and QDAF) surrounded by homogenous background. Image B is a subtype 2 AF pattern with localized low signal in the macula (DDAF and QDAF) surrounded by heterogenous background. Image C shows a subtype 1 AF pattern with localized low signal (QDAF only) at the macula surrounded by homogenous background. Image D is a subtype 3 AF pattern with multiple low-signal areas (DDAF and QDAF) in the posterior pole surrounded by heterogenous background.
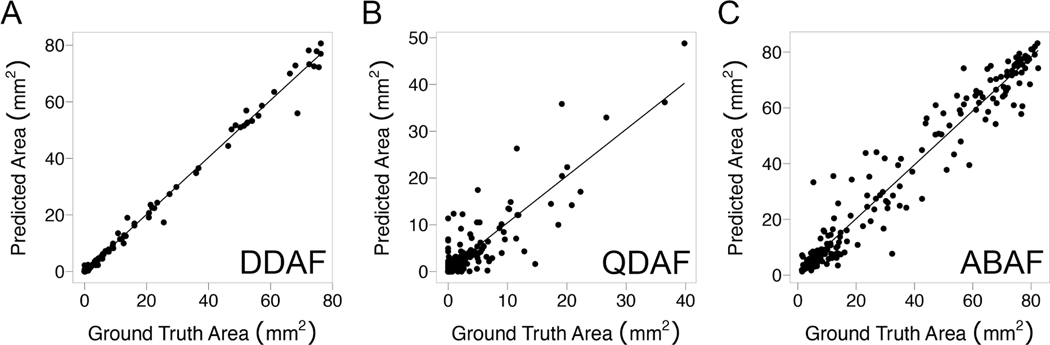
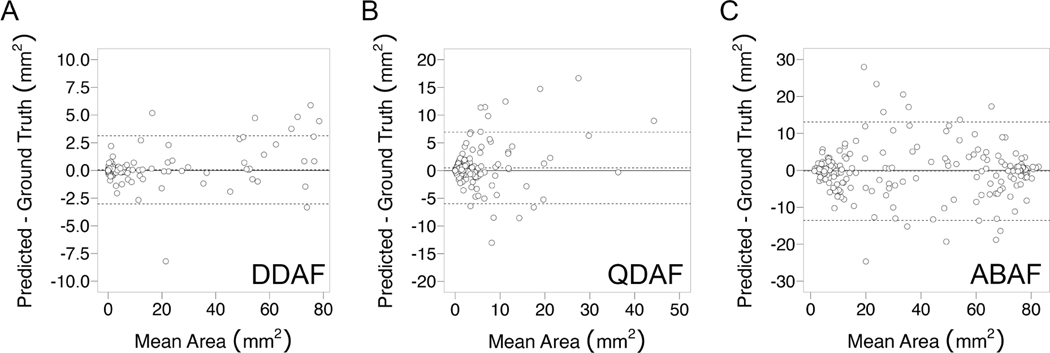
Correlation plots of ground truth versus predicted lesion area are shown in Figure 3. The highest correlation was observed for DDAF, with an ICC of 0.997 (95%CI, 0.996–0.998). For QDAF, the ICC was 0.863 (95% CI, 0.823–0.895). For ABAF, the ICC was 0.974 (95%CI, 0.966–0.980). Bland-Altman analyses were performed for each lesion type (Figure 4). For DDAF, there was no statistically significant mean bias, and the limits of agreement between methods were −3.0 to +3.1 mm2. For QDAF, there was a statistically significant mean bias of +0.5 mm2, and the limits of agreement were −6.0 to +6.9 mm2. For ABAF, there was no statistically significant mean bias, and the limits of agreement were −13.6 to +13.1 mm2.
Figure 3.
Correlation plots comparing algorithm-predicted to ground-truth segmentation area by lesion type. For DDAF lesions (A), the intra-class correlation coefficient (ICC) was 0.997 (95%CI, 0.996–0.998). For QDAF lesions (B), the ICC was 0.863 (95%CI, 0.823–0.895). For ABAF lesions (C), the ICC was 0.974 (95%CI, 0.966–0.980).
Figure 4.
Bland-Altman plots comparing algorithm-predicted to ground-truth segmentation area by lesion type. For DDAF lesions (A), the mean bias was not statistically significant, and the limits of agreement were −3.0 to +3.1 mm2. For QDAF lesions (B), there was a statistically significant mean bias of +0.5 mm2, and the limits of agreement were −6.0 to +6.9 mm2. For ABAF lesions (C), the mean bias was not statistically significant, and the limits of agreement were −13.6 to +13.1 mm2.
Discussion
Convolutional neural networks based on the UNet encoder-decoder architecture have been previously used in ophthalmology to perform automated segmentation of geographic atrophy on color photos and detect referable retinal disease on OCT.15,19,20 We used a modified architecture in which the encoder portion of the network consisted of a ResNet-50 network to identify and segment multiple AF lesions of interest in Stargardt disease.
DDAF and QDAF lesions have been identified as important features for establishing severity and monitoring progression of Stargardt disease, as well as serving as potential outcome measures for therapeutic trials.5,7,11 The DL algorithm we trained achieved a high Dice coefficient of 0.90 (95%CI, 0.85–0.94) for segmentation of DDAF, exceeding the Dice coefficient of 0.78 achieved for atrophic DDAF lesions in a previous study.13 The ICC for algorithm DDAF predictions (0.997) was similar to the inter-grader ICC between human graders calculated for DDAF for both manual (0.981) and semiautomated (0.993) methods.5 Bland-Altman analysis showed agreement between algorithm predictions and ground truth labels, with limits of agreement comparable to those between human graders.21
The DL algorithm achieved a lower Dice coefficient for QDAF lesions. Automated segmentation of QDAF lesions has not previously been described. It is challenging for algorithms to reliably reproduce QDAF lesions due to disagreement on lesion borders. Even for human graders, the inter-grader agreement for QDAF lesions is lower, especially for the subset of poorly-demarcated QDAF lesions (PDQDAF). The ICC for QDAF (0.863) was lower than that for DDAF, and comparable to the ICC previously achieved for semi-automated segmentation of PDQDAF (0.715).5 Further research is needed to better define QDAF lesions to improve reproducibility.
The DL algorithm was also trained to label ABAF lesions, which included various other AF features of Stargardt disease such as hypofluorescence and hyperfluorescence near flecks, homogenously increased background hyperfluorescence, and areas with heterogenous hypofluorescent and hyperfluorescent background. The natural history of these various other lesion types is not well understood, but the DL algorithm could reproduce a ground truth labeling of these regions. Bland-Altman analysis of ABAF regions revealed that the algorithm under-predicted ABAF in a minority of images with large mean area. Further adjustments to the DL model or ground truth labeling methodology may be needed to make accurate predictions for ABAF lesions.
This study has limitations. The retrospective nature of image capture and use of a single AF imaging platform at a single institution. We specifically chose not to include longitudinal imaging data, since longitudinal images from the same patient are correlated. By excluding longitudinal imaging data, we sought to maximize algorithm validity and generalizability, but other factors may still limit generalizability. For example, other studies of FAF have used short-wave reduced-illuminance autofluorescence to assess areas of DDAF.22 There may be differences in the size of DDAF, QDAF, and ABAF lesion size depending on the imaging protocol. Images were labeled by a single grader, and future applications using multiple graders with varying levels of expertise would help further understanding of intra-class correlations. A larger data set containing images from multiple AF protocols from different institutions could train a DL algorithm capable of accurately reproducing segmentations across a variety of clinical and research applications. We hope that multi-center collaboration through an institution such as the Foundation Fighting Blindness Consortium will create opportunities to validate and generalize applications of deep learning to impact the care of patients with Stargardt disease worldwide.23
Acknowledgments
The authors would like to thank Cagri G. Besirli for supporting this work. Grant support from the Foundation Fighting Blindness (Clinical/Research Fellowship Award No. CD-CL-0619-0758 to P.Y.Z.), Heed Ophthalmic Foundation (P.Y.Z.) and from the National Eye Institute, National Institutes of Health (K12 Vision Clinician-Scientist Development Program Award No. 2K12EY022299 to A.T.F, and K23 Mentored Clinician Scientist Award No. 5K23EY026985 to K.T.J.).
Financial Support:
Foundation Fighting Blindness (CD-CL-0619-0758 to P.Y.Z.)
Heed Ophthalmic Foundation (P.Y.Z.)
National Eye Institute, National Institutes of Health (1K08EY032991 to A.T.F)
National Eye Institute, National Institutes of Health (5K23EY026985 to K.T.J.)
The funding organizations had no role in the design or conduct of this research.
Abbreviations:
- ABAF
abnormal background autofluorescence
- DDAF
definitely decreased autofluorescence
- DL
deep learning
- FAF
fundus autofluorescence
- ICC
intraclass correlation coefficient
- IRD
inherited retinal disease
- QDAF
questionably decreased autofluorescence
- RPE
retinal pigment epithelium
Footnotes
Conflict of Interest:
The authors have no proprietary or commercial interest in any materials discussed in this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Newsome DA, ed. Retinal Dystrophies and Degenerations. Raven Press; 1988. [Google Scholar]
- 2.Stargardt K.Über familiäre, progressive degeneration in der maculagegend des auges. Albrecht von Graefes Arch Klin Ophthalmol. Published online 1909:534–550. [Google Scholar]
- 3.Allikmets R, Singh N, Sun H, et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nature genetics. 1997;15:236–246. doi: 10.1038/ng0397-236 [DOI] [PubMed] [Google Scholar]
- 4.Tanna P, Strauss RW, Fujinami K, Michaelides M. Stargardt disease: clinical features, molecular genetics, animal models and therapeutic options. The British journal of ophthalmology. 2017;101:25–30. doi: 10.1136/bjophthalmol-2016-308823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuehlewein L, Hariri AH, Ho A, et al. COMPARISON OF MANUAL AND SEMIAUTOMATED FUNDUS AUTOFLUORESCENCE ANALYSIS OF MACULAR ATROPHY IN STARGARDT DISEASE PHENOTYPE. Retina. 2016;36(6):1216–1221. doi: 10.1097/IAE.0000000000000870 [DOI] [PubMed] [Google Scholar]
- 6.Strauss RW, Ho A, Munoz B, et al. The Natural History of the Progression of Atrophy Secondary to Stargardt Disease (ProgStar) Studies: Design and Baseline Characteristics: ProgStar Report No. 1. Ophthalmology. 2016;123:817–828. doi: 10.1016/j.ophtha.2015.12.009 [DOI] [PubMed] [Google Scholar]
- 7.Strauss RW, Kong X, Ho A, et al. Progression of Stargardt Disease as Determined by Fundus Autofluorescence Over a 12-Month Period: ProgStar Report No. 11. JAMA Ophthalmol. 2019;137(10):1134–1145. doi: 10.1001/jamaophthalmol.2019.2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sparrow JR, Marsiglia M, Allikmets R, et al. Flecks in Recessive Stargardt Disease: Short-Wavelength Autofluorescence, Near-Infrared Autofluorescence, and Optical Coherence Tomography. Invest Ophthalmol Vis Sci. 2015;56(8):5029–5039. doi: 10.1167/iovs.15-16763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar V.Insights into autofluorescence patterns in Stargardt macular dystrophy using ultra-wide-field imaging. Graefes Arch Clin Exp Ophthalmol. 2017;255(10):1917–1922. doi: 10.1007/s00417-017-3736-4 [DOI] [PubMed] [Google Scholar]
- 10.Fujinami K, Lois N, Mukherjee R, et al. A longitudinal study of Stargardt disease: quantitative assessment of fundus autofluorescence, progression, and genotype correlations. Investigative ophthalmology & visual science. 2013;54:8181–8190. doi: 10.1167/iovs.13-12104 [DOI] [PubMed] [Google Scholar]
- 11.Strauss RW, Munoz B, Ho A, et al. Progression of Stargardt Disease as Determined by Fundus Autofluorescence in the Retrospective Progression of Stargardt Disease Study (ProgStar Report No. 9). JAMA Ophthalmol. 2017;135:1232–1241. doi: 10.1001/jamaophthalmol.2017.4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quellec G, Russell SR, Scheetz TE, Stone EM, Abràmoff MD. Computational quantification of complex fundus phenotypes in age-related macular degeneration and Stargardt disease. Invest Ophthalmol Vis Sci. 2011;52(6):2976–2981. doi: 10.1167/iovs.10-6232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Sadda SR, Hu Z. Deep learning for automated screening and semantic segmentation of age-related and juvenile atrophic macular degeneration. In: Hahn HK, Mori K, eds. Medical Imaging 2019: Computer-Aided Diagnosis. SPIE; 2019:62. doi: 10.1117/12.2511538 [DOI] [Google Scholar]
- 14.Charng J, Xiao D, Mehdizadeh M, et al. Deep learning segmentation of hyperautofluorescent fleck lesions in Stargardt disease. Sci Rep. 2020;10(1):16491. doi: 10.1038/s41598-020-73339-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv:150504597 [cs]. Published online May 18, 2015. Accessed August 9, 2019. http://arxiv.org/abs/1505.04597 [Google Scholar]
- 16.He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. arXiv:151203385 [cs]. Published online December 10, 2015. Accessed February 13, 2022. http://arxiv.org/abs/1512.03385 [Google Scholar]
- 17.Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. arXiv:141206980 null. Published online January 29, 2017. Accessed May 1, 2021. http://arxiv.org/abs/1412.6980 [Google Scholar]
- 18.Sudre CH, Li W, Vercauteren T, Ourselin S, Cardoso MJ. Generalised Dice overlap as a deep learning loss function for highly unbalanced segmentations. arXiv:170703237 [cs]. 2017;10553:240–248. doi: 10.1007/978-3-319-67558-9_28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liefers B, Colijn JM, González-Gonzalo C, et al. A Deep Learning Model for Segmentation of Geographic Atrophy to Study Its Long-Term Natural History. Ophthalmology. 2020;127(8):1086–1096. doi: 10.1016/j.ophtha.2020.02.009 [DOI] [PubMed] [Google Scholar]
- 20.De Fauw J, Ledsam JR, Romera-Paredes B, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24(9):13421350. doi: 10.1038/s41591-018-0107-6 [DOI] [PubMed] [Google Scholar]
- 21.Heath Jeffery RC, Thompson JA, Lo J, et al. Atrophy Expansion Rates in Stargardt Disease Using Ultra-Widefield Fundus Autofluorescence. Ophthalmology Science. 2021;1(1):100005. doi: 10.1016/j.xops.2021.100005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strauss RW, Muñoz B, Jha A, et al. Comparison of Short-Wavelength Reduced-Illuminance and Conventional Autofluorescence Imaging in Stargardt Macular Dystrophy. Am J Ophthalmol. 2016;168:269–278. doi: 10.1016/j.ajo.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durham TA, Duncan JL, Ayala AR, et al. Tackling the Challenges of Product Development Through a Collaborative Rare Disease Network: The Foundation Fighting Blindness Consortium. Transl Vis Sci Technol. 2021;10(4):23. doi: 10.1167/tvst.10.4.23 [DOI] [PMC free article] [PubMed] [Google Scholar]