Abstract
Colicin-producing strains occur frequently in natural populations of Escherichia coli, and colicinogenicity seems to provide a competitive advantage in the natural habitat. A cka-lacZ fusion was used to study the regulation of expression of the colicin K structural gene. Expression is growth phase dependent, with high activity in the late stationary phase. Nutrient depletion induces the expression of cka due to an increase in ppGpp. Temperature is a strong signal for cka expression, since only basal-level activity was detected at 22°C. Mitomycin C induction demonstrates that cka expression is regulated to a lesser extent by the SOS response independently of ppGpp. Increased osmolarity induces a partial increase, while the global regulator integration host factor inhibits expression in the late stationary phase. Induction of cka was demonstrated to be independent of the cyclic AMP-Crp complex, carbon source, RpoS, Lrp, H-NS, pH, and short-chain fatty acids. In contrast to colicin E1, cka expression is independent of catabolite repression and is partially affected by anaerobiosis only upon SOS induction. These results indicate that while different colicins are expressed in response to some common signals such as nutrient depletion, the expression of individual colicins could be further influenced by specific environmental cues.
Colicins are bacteriocins synthesized by, and active against, Escherichia coli cells and sometimes cells of closely related species such as Shigella and Salmonella. Twenty-three colicin types have been characterized (24). Colicin-producing cells synthesize an immunity protein, which protects the producers against the colicin synthesized inside the same cell or by other cells.
Colicin K kills cells by forming channels in membranes, destroying the electrochemical potential of the cytoplasmic membrane. Uptake into sensitive cells requires the outer membrane proteins Tsx, OmpF, TolR, and TolB. The genes cka, encoding colicin activity, cki, encoding immunity, and ckl, encoding lysis, have been previously found on pColK-K49 (21) and pColK-K235 (23) of strain BZB2116.
Already studied in detail is the expression of the colicin E1 structural gene. Its regulation has been demonstrated to be complex, involving repression by LexA, stimulation of transcription by the cyclic AMP (cAMP) receptor protein (Crp)-cAMP complex (25), Fnr-mediated stimulation of expression in anaerobic conditions (7), and induction by depletion of nutrients (8).
The high frequency of synthesis of colicins in E. coli strains implies that their role could be the defense or invasion of an ecological niche. Expression of structural genes of different colicins could therefore involve common mechanisms of regulation. On the other hand, strains can produce more than one colicin, and even though production is induced by some common conditions, e.g., the SOS response, synthesis of individual colicins could be influenced by different signals. In the present study, we used a cka-lacZ fusion to study the regulation of cka expression with regard to growth phase, nutrient depletion, growth temperature, SOS induction, and osmotic shock. The role of certain global regulators of gene expression, i.e., integration host factor (IHF), Lrp, ppGpp, and Crp (cAMP-Crp), was also investigated.
Our results demonstrate that colicin K synthesis is growth phase dependent and induced by nutrient depletion due to an increase in ppGpp. cka expression is strongly influenced by temperature, with only basal-level activity being present at 22°C. The SOS response is not a strong signal for cka expression. Increased osmolarity partially increases expression, while the global regulator IHF modulates cka expression negatively with regard to growth phase. Compared with regulation of expression of the ColE1 structural gene cea (8), our results demonstrate that while nutrient depletion induces the synthesis of ColE1 and colicin K and while LexA is a common repressor, other global regulatory proteins and environmental conditions modulate synthesis differently. Such differential regulation could be of ecological significance, since the synthesis of individual colicins could be induced by different signals at different sites within the warm-blooded host.
MATERIALS AND METHODS
Bacteria and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| Strains | ||
| KS533 | Rough:K1:H7 | 2 |
| DH5α | thi-1 recA hsdR17 lac | A. Francky |
| GE2897 | MC4100 Δ82(himA)::tet | 8 |
| GE3653 | MC4100 lrp-201::Tn10 | 8 |
| MC4100 | araD139Δ(argF-lac)U169 rpsL150 relA1 flbB5301 ptsF25 deoC1 | 16 |
| SBS688 | MC4100 Δcrp39 | 12 |
| RH90 | MC4100 rpoS::Tn10 | R. Hengge-Aronis |
| RH98 | MC4100 relA251::kan spoT207::cat | 16 |
| MC4100 hns::kan | hns | 30 |
| RK5288 | MC4100 fnr-250 zcj-637::Tn | 7 |
| AB1133 | Sensitive to all colicins | B. Bachman |
| BZB2116 | Harboring pColK-K235 | 23 |
| Plasmids | ||
| pColK-JA533 | Colicin K | This study |
| pCB267 | AprlacZ promoter probe vector | 27 |
| pIK32 | pUC19 with the 7-kb Sau3AI fragment encoding cka, cki, ckl | This study |
| pIK100 | pUC19 with the 1-kb EcoRI fragment of pIK32 | This study |
| pIK471 | pCB267 cka-lacZ | This study |
| pUC19 | Apr cloning vector | 31 |
Media and chemicals.
Strains were grown in Luria-Bertani (LB) medium with aeration at 37 or at 22°C where indicated. Where indicated, LB medium was supplemented with 0.3 M NaCl to increase osmolarity. Ampicillin was used at 100 μg/ml, kanamycin was used at 30 μg/ml, tetracyline was used at 10 μg/ml, and streptomycin was used at 100 μg/ml. o-Nitrophenyl-β-d-galactopyranoside (ONPG) was used as substrate in β-galactosidase assays of cells treated with sodium dodecyl sulfate (SDS)–chloroform and washed with Z buffer. Enzyme activity is defined in units of optical density at 420 nm (OD420) per minute per unit of OD600 (20).
Conditioned medium was prepared by diluting an overnight culture of MC4100 1:500 into 100 ml of fresh LB medium. After growth for 4 or 12 h with aeration at 37°C, the cells were centrifuged twice and the medium was filter sterilized. Conditioned medium was used within 24 h.
Strains to be tested for cka induction in conditioned medium were first grown overnight at 37°C in LB medium with the appropriate antibiotic. The bacterial cells were then diluted 1:500 into fresh LB medium and grown to an OD600 of 1. Subsequently, the cells were inoculated at a density corresponding to an OD600 of 0.015 into conditioned medium and further incubated with aeration at 37°C. Samples were periodically removed and assayed for β-galactosidase activity.
cka induction under anaerobic conditions was tested by first growing bacteria overnight with aeration at 37°C. The bacteria were grown under the same conditions to 2 × 108 to 4 × 108 cells/ml (OD600 = 1). They were subsequently diluted 1:100 and grown aerobically for 30 min. The bacterial culture was then divided into two halves; one was incubated further aerobically, while the other was incubated in tubes without aeration for the establishment of anaerobic conditions (7). However, this is not a strictly anaerobic but a semianaerobic environment. After 1 h, each culture was split into two parts, and one was induced with 0.5 μg of mitomycin C per ml.
General DNA manipulation techniques.
Plasmid DNA isolation, ligation, and transformation experiments were performed by standard methods (26). Restriction endonuclease digestions were carried out as specified by the supplier (Boehringer). DNA fragments were purified from agarose gels by using the GeneClean II system (Bio 101). DNA-labelling and hybridization experiments were carried out by using the DIG DNA labelling and detection kit (Boehringer). Hybridization experiments were performed to detect the cloned chromosomal colicin K genes by using the 1.15-kb EcoRI-DraI fragment of the cka structural gene. DNA sequencing was performed by a dye rhodamine terminator cycling reaction on an ABI 377 automated sequencer at the Department of Biochemistry, Colorado State University. The relative plasmid pIK471 content was densitometrically determined (13) in all the studied strains and compared at 22 and 37°C.
Cloning of the colicin K genes.
Colicin K in strain KS533 (1) is encoded by pColK-JA533. Partially Sau3A1-digested plasmid DNA was cloned into the BamHI site of vector pUC19. A colicinogenic clone was isolated, and the plasmid was designated pIK32. The presence of colicin K-specific genes on plasmid pIK32 was confirmed by DNA hybridization analysis. The location of the cka promoter region with part of the structural gene on an approximately 1-kb EcoRI fragment of plasmid pIK100 was deduced by comparison with the restriction maps of the cka structural, cki immunity, and ckl lysis genes (21). The nucleotide sequence of the cka promoter region was determined by DNA sequencing of the cloned 1-kb EcoRI fragment and is presented in Fig. 1.
FIG. 1.
Regulatory region of the cka gene including 397 nucleotides upstream of transcription initiation and 74 nucleotides of the cka gene. The predicted Shine-Dalgarno sequence (S.D.), the −10 region, the −35 region, and the two SOS boxes are indicated.
Construction of the cka-lacZ gene fusion.
PCR was carried out to amplify the 471-bp fragment encompassing 397 nucleotides upstream from the transcription initiation site and 74 nucleotides of the cka structural gene. Two primers were designed, one designated K1 (5′-TCGGATCCATGCGTCTTGCCTGGTATAC-3′pro1) on the basis of the determined promoter region and an added BamHI restriction sequence and the other designated K2 (5′-TCTCTAGATGATTCAGATTCGCCCCTG-3′), corresponding to colicin K nucleotides 74 to 55 (21) and an added XbaI restriction sequence. PCR was carried out in the following steps: heating at 93°C for 5 min followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 61°C for 1 min, and extension at 72°C for 1 min and one final extension for 10 min at 72°C. The PCR-generated fragment was recovered from a 0.9% agarose gel, cut with BamHI and XbaI, and cloned into the promoter probe plasmid pCB267 with the promoterless lacZ gene, also cut with BamHI and XbaI (27), generating pIK471.
Double-stranded nucleotide sequencing with K1 and K2 oligonucleotides as primers was carried out to confirm that no base changes had occurred in cloning the 471-bp amplified fragment in plasmid pIK471.
Osmotic pressure.
An overnight culture of the tested strain was diluted 1:500 into fresh LB medium and grown with aeration at 37°C to mid-exponential phase (OD600 = 0.2). The culture was then divided into two parts, and to one of these was added 0.3 M NaCl. Samples were periodically removed and assayed for β-galactosidase activity.
Colicin production.
Colicin K production was determined by the overlay method (23) by testing the sensitivity of the indicator strain AB1133 (sensitive to all colicins) and immunity of strain BZB2116 with plasmid pColK-K235 (23).
Induction of the SOS response with mitomycin C.
Cells of an overnight culture of strain MC4100 with plasmid pIK471 were diluted 1:500 in LB medium and grown with aeration at 37°C. At an OD600 of 0.3, the culture was divided into two parts and to one was added mitomycin C to a concentration of 0.5 μg/ml; the culture was then incubated further.
Temperature-dependent synthesis of colicin K.
Cells of an overnight culture of strain MC4100 with plasmid pColK-JA533 were diluted 1:500 in LB medium and grown with aeration at 37°C. At an OD600 of 0.3, the culture was divided into two parts, and to one was added mitomycin C to a concentration of 0.5 μg/ml; the culture was then incubated further for 3 h. Alternatively, to detect Cka at 22°C, an overnight culture of the pColK-JA533-carrying strain was diluted as above and incubated with aeration at 22°C to an OD600 of 2. The culture was subsequently divided into two parts, and to one was added mitomycin C to a concentration of 0.5 μg/ml; the culture was then incubated further for 8 h. A 1.5-ml portion of the cultures was removed, and whole-cell extracts were prepared (24). Samples were analyzed by SDS-polyacrylamide gel electrophoresis.
Nucleotide sequence accession number.
The sequence reported in this study has been deposited in the EMBL nucleotide sequence database under accession no. Y18549.
RESULTS
cka expression is growth phase dependent.
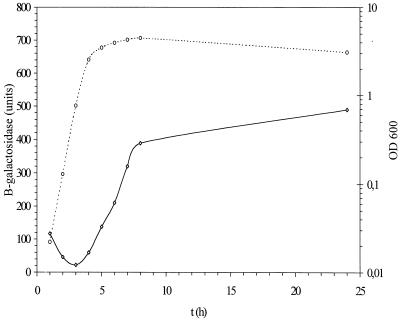
Overnight cells of strain MC4100 carrying plasmid pIK471 were diluted 1:500 in LB medium and grown with aeration at 37°C (Fig. 2). Dilution of residual β-galactosidase activity from the overnight culture was observed in the mid-exponential phase, when a basal level of 22 U of β-galactosidase was produced. Expression of β-galactosidase activity subsequently increased in a linear manner, with a 17-fold increase in the stationary phase (8 h after inoculation) and a 22-fold increase, amounting to 490 U, in the late stationary phase.
FIG. 2.
Growth phase-dependent expression of cka-lacZ. Strain MC4100 with pIK471 was grown in LB medium. Expression in Miller units of the β-galactosidase level (diamonds) and growth (OD600) (circles) were determined. The experiment was repeated five times, and the results of one experiment closest to the average were used.
Depletion of nutrients induces cka expression.
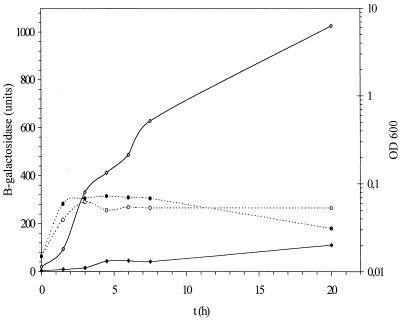
Induction of colicin production in the stationary phase could be due to a variety of changes in the medium, for example, anaerobiosis, pH, depletion of metabolites, excretion of compounds, and high cell density (8). To study the effect of depletion of nutrients, cells were inoculated, at low density (OD600 = 0.015), into medium conditioned for 12 h as described in Materials and Methods. As is evident from Fig. 3, from a basal level a rapid 16-fold increase in activity was established, while growth stopped 3 h after inoculation at an OD600 of 0.062. Subsequently (24 h after inoculation), an approximately 46-fold increase, resulting in 1,026 U, was observed. Activity was thus greater in conditioned medium than in the late-stationary-phase culture which is also depleted of nutrients.
FIG. 3.
Expression of the cka-lacZ fusion in conditioned medium. Expression of MC4100 (open symbols) and in the relA spoT double mutant (solid symbols) in medium conditioned for 12 h is shown. Levels of β-galactosidase activity in Miller units (diamonds) and growth OD600 (circles) are depicted. Experiments were carried out in duplicate, and a representive result is shown.
To determine whether cka expression is induced by depletion of nutrients in conditioned medium or by an inducer released by cells used to deplete the medium, expression was also tested in media containing different proportions of conditioned and fresh medium. Differentially conditioned media were used; one had sustained growth for 4 h, and the other had sustained growth for 12 h (Table 2). After 4 h of growth, the medium is not completely devoid of nutrients, the OD600 reaches 1 to 2, and at this time the increase in expression is greatest. If nutrient depletion induces expression, addition of fresh medium should inhibit expression. Alternatively, the increase in expression at the beginning of the stationary phase could be due to an inducer present in the medium whose concentration is dependent upon cell density. In this case, expression should not be affected by the addition of fresh medium. In both differentially depleted media, the addition of fresh LB medium resulted in a 0.5-fold reduction in β-galactosidase activity for up to 3 h after inoculation.
TABLE 2.
Expression of the cka-lacZ fusion in conditioned and diluted mediaa
| Time (h) | Activityb in medium containing LB at:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0%
|
25%
|
50%
|
75%
|
100%
|
||||||
| OD600 | Units | OD600 | Units | OD600 | Units | OD600 | Units | OD600 | Units | |
| 4-h conditioned medium | ||||||||||
| 1.5 | 0.066 | 150 | 0.24 | 50 | 0.34 | 32 | 0.399 | 27 | 0.46 | 24 |
| 3 | 0.136 | 307 | 0.69 | 155 | 1.32 | 84 | 1.85 | 79 | 2.43 | 60 |
| 4.5 | 0.239 | 484 | 1.25 | 381 | 2.16 | 268 | 2.85 | 250 | 3.53 | 201 |
| 6 | 0.450 | 486 | 1.65 | 651 | 2.46 | 597 | 3.04 | 547 | 3.56 | 439 |
| 7.5 | 0.740 | 543 | 1.96 | 760 | 2.82 | 619 | 2.71 | 597 | 3.32 | 458 |
| 20 | 0.90 | 1,820 | 1.56 | 1,080 | 2.57 | 711 | 2.35 | 714 | 2.90 | 504 |
| 12-h conditioned medium | ||||||||||
| 1.5 | 0.039 | 94 | 0.21 | 31 | 0.33 | 24 | 0.498 | 18 | 0.53 | 17 |
| 3 | 0.062 | 330 | 0.638 | 170 | 1.50 | 168 | 2.71 | 89 | 3.30 | 102 |
| 4.5 | 0.050 | 412 | 1.14 | 245 | 1.95 | 474 | 3.28 | 107 | 3.96 | 131 |
| 6 | 0.054 | 487 | 0.88 | 1,165 | 2.02 | 652 | 3.16 | 393 | 4.10 | 411 |
| 7.5 | 0.053 | 628 | 0.99 | 1,200 | 2.23 | 593 | 3.34 | 340 | 4.39 | 376 |
| 20 | 0.053 | 1,026 | 0.73 | 1,447 | 1.72 | 674 | 2.82 | 366 | 3.42 | 390 |
MC4100 carrying pIK471 was grown in media conditioned for 4 or 12 h and in media with 25, 50, and 75% fresh LB as well as in 100% LB.
Activity (given in Miller units) was assayed at different times after inoculation. The experiments were performed in duplicate, and representative results are given.
However, subsequently, at 6 h following inoculation, the activity was up to twofold higher, particularly when 25% fresh LB medium was added to both conditioned media. In medium which had sustained growth for 4 h, the addition of 25% fresh LB medium resulted in higher activity 7.5 h after inoculation, while in medium depleted for 12 h, a pronounced increase was observed up to 20 h after inoculation.
To rule out the possibility that addition of fresh LB medium results in dilution of an inducer, tryptone and yeast extract were added to the conditioned medium to levels present in LB medium; the results were essentially the same (data not shown).
The decreased activity following the addition of fresh medium demonstrates that depletion of nutrients induces cka expression. However, while our results demonstrate initially lower activity of the cka-lacZ fusion in conditioned medium with added nutrients, the subsequently higher activity, which can exceed the activity in 100% depleted medium, indicates that cka expression is not induced solely by depletion of nutrients.
cka expression is positively affected by ppGpp.
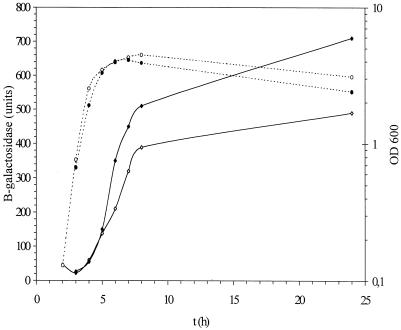
As cells exhaust nutrients, inbalances occur in amino acid biosynthetic pathways, resulting in empty acceptor sites in ribosomes and leading to an increase in guanosine tetraphosphate synthesis. To determine whether ppGpp is involved in cka expression, the β-galactosidase activity of the cka-lacZ fusion was studied in strain RH98 with mutations in relA and spoT. Strains with relA spoT double mutations produce no ppGpp. Approximately eightfold-lower β-galactosidase activity was found for the entire growth cycle in strain RH98 carrying plasmid pIK471 (Fig. 4). These results demonstrate that ppGpp is a positive effector of cka expression.
FIG. 4.
Expression of the cka-lacZ fusion in the double mutant relA spoT (solid symbols) and in MC4100 (open symbols). β-Galactosidase activity in Miller units (diamonds) and growth OD600 (circles) are presented. The experiments were repeated four times, and the results of one experiment closest to the average were used.
cka activity in the relA spoT mutant was also tested in medium conditioned for 12 h (Fig. 3). In conditioned medium, the β-galactosidase activity of the cka-lacZ fusion in the relA spoT mutant was approximately 10-fold lower than that in strain MC4100. cka expression in the relA spoT double mutant was similar in fresh and conditioned media during the first 8 h. Subsequently, the activity was approximately twofold higher in conditioned medium, again indicating that another factor besides ppGpp could be involved in induction of cka expression.
To rule out the possibility that β-galactosidase activity is affected by changes in the pIK471 copy number, the relative plasmid content was determined as described in Materials and Methods. In all the studied strains, the pIK471 copy number was altered only in the relA spoT mutant. The plasmid content was higher than for strain MC4100 (data not shown). The negative influence of ppGpp on the replication of a number of plasmids has been previously demonstrated (33). Our results demonstrate that ppGpp is a positive activator for induction of cka expression due to depletion of nutrients.
Mitomycin C induces cka expression.
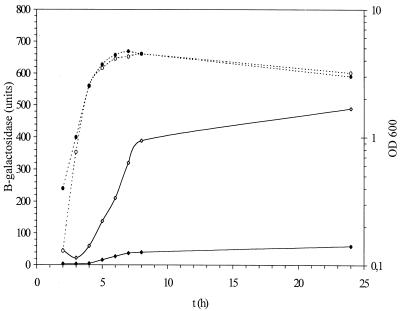
Expression of colicin activity genes is subject to SOS control with a LexA repressor binding region located upstream of all colicin activity genes. All ColE operons and the cka gene have two overlapping SOS boxes in their regulatory regions (Fig. 1). Induction of cka expression upon DNA damage was tested in the presence of mitomycin C at 0.5 μg/ml (Table 3). This dose gave the highest induction with the least killing. Induction with mitomycin C was observed in the exponential and stationary phases, with approximately threefold and twofold increases in β-galactosidase activity, respectively. The relA spoT mutant was also treated with mitomycin C, and the same level of induction was observed as in the wild-type MC4100 strain (Fig. 5).
TABLE 3.
Influence of mitomycin C and IHF on cka-lacZ expression
| Time (h) | Expression ina:
|
|||||
|---|---|---|---|---|---|---|
| MC4100
|
Mitomycin Cb
|
himAc or HimA−
|
||||
| OD600 | Units | OD600 | Units | OD600 | Units | |
| 3 | 0.76 | 22 | 0.42 | 60 | 0.52 | 62 |
| 5 | 3.46 | 138 | 2.81 | 343 | 2.92 | 137 |
| 8 | 4.47 | 389 | 3.53 | 807 | 4.23 | 394 |
| 24 | 3.09 | 490 | 1.70 | 980 | 4.28 | 695 |
The expression in Miller units is indicated at four different times (during the growth cycle) of cell growth and compared with the expression in strain MC4100.
The culture of strain MC4100 was divided into two parts in the early log phase at an OD600 of 0.3. Mitomycin C was added at 0.5 μg/ml to one part. The data shown represent the average of three independent experiments.
β-Galactosidase activity (in Miller units) was measured in the himA mutant strain GE2897, which is defective in IHF. The data shown represent the average of two independent experiments.
FIG. 5.
Expression of the cka-lacZ fusion in the relA spoT mutant induced with mitomycin C. β-Galactosidase activity in Miller units (diamonds) and growth OD600 (circles) are presented. Solid symbols depict induction with mitomycin C, and open symbols indicate no induction. The experiment was carried out in duplicate, and a representative result is shown.
cka expression is repressed by IHF and independent of ςS, H-NS, and Lrp.
To determine whether other global regulator proteins influence cka expression, the β-galactosidase activity of the cka-lacZ fusion was studied in strains defective in rpoS (RH90), himA (GE2897), and hns and lrp (GE3653). The results of our study showed that synthesis and the induction of cka expression in the stationary phase is H-NS independent (data not shown). β-Galactosidase activity of the cka-lacZ fusion was even somewhat higher in the rpoS mutant (data not shown), most probably due to competition between ς70 and ςS for RNAP core. In the absence of ςS, the cellular concentration of the ς70-containing holoenzyme is higher and can result in higher activity of some promoters (9). Our results show that induction of cka expression is ςS and Lrp independent. On the other hand the somewhat higher β-galactosidase activity in the late stationary phase in the himA mutant strain GE2897 indicates that IHF inhibits cka expression in this phase of the growth cycle (Table 3).
cka expression is temperature dependent.
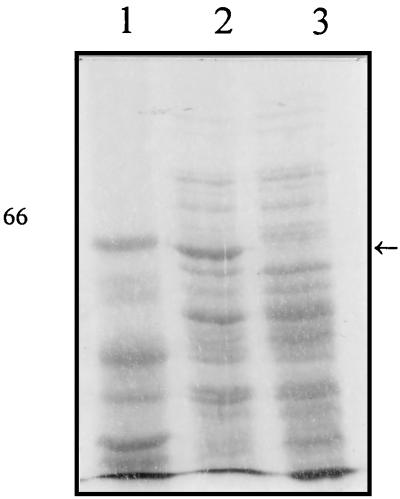
To determine whether the transcription of cka is thermoregulated, the β-galactosidase activity of the cka-lacZ fusion was monitored when cells were grown at 22°C. cka gene expression is influenced by temperature, since only 52 U of β-galactosidase activity was detected at 22°C (approximately 10-fold lower than at 37°C). When mitomycin C was added to the growth medium, fivefold-lower activity at 22°C than at 37°C was observed. Lower activity at 22°C was found not to be due to lower plasmid copy number (data not shown). These results were confirmed by SDS-PAGE of the Cka protein, as described in Materials and Methods (Fig. 6).
FIG. 6.
Temperature and colicin K synthesis. Strain MC4100 carrying pColK-JA533 grown in LB medium at either 37 or 22°C was treated with mitomycin C. Whole-cell extracts were prepared as described in Materials and Methods and run on an SDS–7% polyacrylamide gel. The gel was stained with Coomassie blue to visualize proteins. Lanes: 1, marker; 2, culture grown at 37°C; 3, culture grown at 22°C. The position of colicin K is indicated by an arrow. The number on the left represents the molecular mass (in kilodaltons) of the standard protein.
Since the global regulator H-NS is involved in thermoregulation of Pap pili (14), cka expression of the cka-lacZ fusion was tested in the hns, relA spoT, lrp, and himA mutant strains, and no influence on expression at 22°C was demonstrated. The level of osmotic induction in the wild-type MC4100 strain at 22°C was the same as at 37°C.
Increased osmolarity affects the expression of cka.
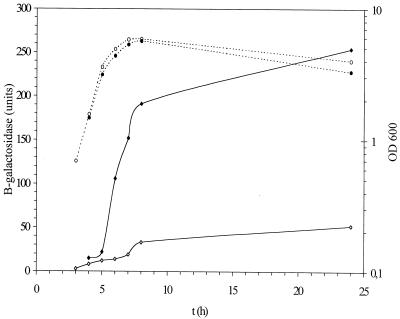
Bacterial cells are frequently subjected to fluctuations in the osmolarity of the environment. Osmolarity has been noted to be a signal controlling several virulence genes (19). Under conditions of increased osmolarity, in LB medium supplemented with 0.3 M NaCl, cka-lacZ expression in strain MC4100 was partially increased (Fig. 7).
FIG. 7.
Osmotic induction of expression of the cka-lacZ fusion in MC4100. β-Galactosidase activity in Miller units (diamonds) and growth OD600 (circles) are presented. Solid symbols depict osmotic induction, and open symbols indicate no osmotic induction. The experiment was repeated four times, and the results of one experiment closest to the average were used.
Colicin K synthesis is not affected by the cAMP-Crp complex.
To determine whether the cAMP-Crp complex and thus catabolite repression influences the expression of cka, β-galactosidase activity was monitored in strain SBS688 (defective in crp, encoding the cAMP receptor protein Crp). cka expression was shown to be independent of the cAMP-Crp complex (data not shown).
β-Galactosidase activity of strain MC4100 with pIK471 was also tested in M63 medium supplemented with Casamino Acids and glucose or glycerol as the carbon source. No differences in β-galactosidase activity were observed, confirming that cka expression is independent of catabolite repression (data not shown).
Effect of anaerobiosis, pH, and short-chain fatty acids on cka expression.
The influence of anaerobiosis on cka expression was monitored under anaerobic (7) and aerobic conditions in the wild-type MC4100 strain and in the fnr mutant. We compared the β-galactosidase activity of uninduced cultures and cultures induced with 0.5 μg of mitomycin C per ml.
It is evident that the basal level of cka expression was not affected by anaerobic conditions (Table 4). In the fnr mutant, expression under uninduced and induced anaerobic conditions was twofold lower than in strain MC4100. In the wild-type strain MC4100, the expression in induced cultures was higher under anaerobic conditions than under aerobic conditions. These results show that anaerobiosis partially affects cka expression only upon SOS induction.
TABLE 4.
Aerobic and anaerobic regulation of the cka-lacZ fusiona
| Strain | Activity under aerobiosis
|
Activity under anaerobiosis
|
||||||
|---|---|---|---|---|---|---|---|---|
| No induction
|
Induction
|
No induction
|
Induction
|
|||||
| OD600 | Units | OD600 | Units | OD600 | Units | OD600 | Units | |
| fnr+b | 3.83 | 79 | 2.91 | 225 | 1.59 | 30 | 1.48 | 428 |
| fnrc | 4.05 | 40 | 2.49 | 240 | 0.49 | 16 | 0.9 | 280 |
After inoculation, the strains were grown in LB medium aerobically, split into two parts, and further grown aerobically and anaerobically for 1 h, after which they were split again and one part was induced with mitomycin C. β-Galactosidase activity (in Miller units) was assayed 4 h after inoculation. The data represent the average of two independent experiments.
Strain MC4100 with pIK471.
Strain RK5288 with pIK471.
The pH of the LB medium was 7.0 initially and 8.5 after sustaining growth for 12 h. To test whether the pH influences cka expression, the β-galactosidase activity of the cka-lacZ fusion was tested in fresh LB medium adjusted to pH 8.5 with NaOH. Expression was the same as in LB medium at pH 7 (data not shown), demonstrating that the increase in pH in the stationary-phase culture did not affect cka-lacZ expression.
The effects of two short-chain fatty acids, acetic and propionic acids, on cka-lacZ expression were studied. Sodium salts of both acids were added at 50 mM to early-log-phase cultures (OD600 = 0.1) of MC4100 carrying pIK471 growing in LB medium. LB medium supplemented with 50 mM NaCl served as a control. β-Galactosidase activity was determined after 1 h of growth, and no differences in expression were found between the control medium and medium containing short-chain fatty acids (data not shown), demonstrating that accumulation of metabolic products plays no role in colicin K expression.
DISCUSSION
Natural populations of E. coli frequently produce colicins. Recently, the rapid invasion of established colicinogenic populations by another colicinogenic strain expressing novel immunity functions has been demonstrated (29). Studies of regulation of expression of colicin genes and the environmental conditions influencing expression could further elucidate colicin action in the natural environment.
The results presented in this study demonstrate that regulation of colicin K expression is growth phase dependent, with production increasing as cells proceed through the growth cycle. Expression of cka increases in a linear manner as the cells proceed from the exponential to the stationary growth phase. A less pronounced increase in expression is characteristically observed in the late stationary phase. Expression of another pore-forming colicin, colicin E1, has also been shown to be growth phase dependent; however, cea expression is low and constant in the early log phase and then dramatically increases (8).
As cells proceed through the growth cycle, changes in the medium, such as nutrient depletion, pH changes, anaerobiosis, or the production of metabolites or inducers, occur and regulate gene expression. Depletion of nutrients was shown to induce cka expression. Surprisingly, activity in conditioned medium exceeded activity in the stationary phase when the medium was also depleted of nutrients. Complete derepression of cka expression in stationary-phase and late-stationary-phase cultures might be inhibited by cellular remains, which can be used as nutrients for growth.
Bacterial cells of various species sense and respond to population densities by the production and detection of small signalling substances (pheromones), which regulate a number of processes including quorum sensing, competence, sporulation, and antibiotic as well as exoenzyme production (reviewed in references 10, 15, and 28). Since cka induction in conditioned medium could be due to the presence of an inducer, the β-galactosidase activity of the cka-lacZ fusion was tested in conditioned medium complemented by the addition of fresh LB medium. Expression was initially (in the first 4 h after inoculation) lower, demonstrating that cka expression is induced in response to starvation. However, subsequently (after 6 h), particularly in conditioned medium complemented with 25% fresh LB medium, expression was significantly greater than in 100% depleted medium. Our results demonstrated that the addition of low concentrations of nutrients to conditioned medium induces cka expression to the greatest extent. This effect could be due to a specific low level of metabolic activity, evidenced as slow growth to an OD600 of 1 to 2. It thus seems that cka expression is not solely induced by depletion of nutrients but that another unidentified regulator could also be acting at a specific physiological state. Since nutrient availability in the natural bacterial environment is scarce and starvation is common, the fine-tuning of high levels of cka expression with low metabolic activity could offer a population of colicin-producing cells a significant competitive advantage during invasion and establishment in an ecological niche.
Nutrient depletion in growing bacteria induces the production of guanosine tetraphosphate. ppGpp exerts pleiotropic effects collectively known as the stringent response. Positive stringent control is exerted on processes acting in overcoming amino acid starvation or preparatory for surviving prolonged starvation. ppGpp has been reported to be a positive regulator of rpoS expression (11, 16) and the his operon (5). Our data demonstrate that ppGpp is the main positive effector of cka expression, since only basal-level activity with only an approximately twofold increase in expression occurred in the relA spoT mutant.
The expression of several important virulence determinants is temperature regulated in a number of bacterial genera (3, 4, 14, 17, 22, 32). By monitoring the β-galactosidase activity of the cka-lacZ fusion at 22 and at 37°C, we demonstrated that temperature is another important signal influencing cka expression. At 22°C, only a basal level of β-galactosidase activity was produced in the late stationary phase. Our results thus indicate that expression of cka is induced in the warm-blooded host.
Repression of colicin synthesis by the LexA protein is a well-established fact. However, mitomycin C induces only a two- to threefold increase in the β-galactosidase activity of cka-lacZ fusion, indicating that SOS induction is not a strong regulatory signal for cka expression. Mitomycin C induction of the cka-lacZ fusion in the relA spoT mutant, together with induction in the wild-type strain during exponential growth, demonstrates that ppGpp and the SOS response act independently in regulating cka expression.
Increased osmolarity induces a partial increase in expression of the cka-lacZ fusion, as determined in medium with 0.3 M NaCl. A partial increase is observed particularly in the stationary phase. As glucose concentrations differ in different parts of the intestinal tract, so do concentrations of salts. Higher concentrations of salts are present in the proximal than in the distal part of the intestinal tract, again indicating that expression of cka could be induced by specific signals in the host.
Our data also demonstrate that in addition to repression by LexA, cka expression is inhibited in the late stationary phase by IHF. In the himA mutant strain, cka expression as determined by β-galactosidase activity of the cka-lacZ fusion is higher in the late stationary phase, when IHF concentrations increase sixfold. IHF has been proposed to act as an architectural element and could play an indirect physiological role in gene expression when other proteins responding to environmental signals influence IHF binding or activity (2).
Expression of the cka-lacZ fusion is shown to be independent of the carbon source and the cAMP-Crp complex. It has been reported that virulence factors important for colonization of the proximal part of the small intestine are expressed independent of the carbon source and the cAMP-Crp complex, since glucose concentrations are high in this part of the intestinal tract. On the other hand, the expression of genes encoding factors crucial in the distal part of the small intestine is dependent upon the carbon source and the cAMP-Crp complex, since glucose concentrations are low in this part of the intestinal tract (reviewed in reference 6). Expression of the colicin K cka gene is independent of the cAMP-Crp complex and, in contrast to colicin E1, not significantly affected by anaerobiosis. On the other hand, the colicin E1 cea gene is dependent on both conditions (7, 25). This implies that different colicins, even though responding to some common signals, also respond to specific cues. A comparison of the factors affecting the expression of the colicin K cka gene and the E1 cea gene (7, 8) is presented in Table 5. With regard to nutrient availability and colicin production, lower levels of colicin E3, E5, E6, E8, and E9 production in minimal media than in LB medium have been reported (29), further demonstrating the differential expression of various colicins.
TABLE 5.
Factors regulating the expression of the colicin E1 (8) and colicin K activity genes
| Factor | Colicin E1 expressiona | Colicin K expressiona |
|---|---|---|
| SOS response | + | + |
| Induction in stationary phase | + | + |
| Kinetics of induction | Rapid | Slow |
| Expression in conditioned medium versus fresh LB medium | The same | Higher |
| Stringent response | + | + |
| Catabolite repression | + | − |
| Anaerobic expression | + | ± |
| IHF inhibition | − | + |
| RpoS-dependent expression | − | − |
| Lrp-dependent expression | − | − |
+, the tested factor influenced expression; −, no influence was found; ±, no increase in expression under anaerobic conditions was found without SOS induction.
Our results, together with results from other groups, demonstrate that regulation of colicin synthesis is complex and that environmental signals in the host could play an important role in regulation of expression. Two SOS boxes are present in the promoter region of colicin K (Fig. 1) and of ColE operons. It has been proposed that LexA binding to the two overlapping SOS boxes protects colicin-producing cells from lysis under conditions that do not induce the SOS response, since lysis is lethal to the producing cell (18). Growth phase-dependent colicin induction has been reported to be independent of LexA cleavage (8). Intracellular LexA concentrations vary following DNA damage and not with regard to growth phase and environmental cues, like the concentrations of other regulators. Thus, LexA represses colicin expression, preventing cell lysis, until specific environmental signals, e.g., nutrient depletion, temperature, and osmolarity, induce colicin synthesis in the host. The differential induction of at least some colicins by specific environmental signals suggests that individual colicins could be acting and imposing a competitive advantage at different sites within the natural environment. Colicinogenic strains producing more than one colicin could have a competitive advantage under several different environmental conditions.
ACKNOWLEDGMENTS
This work was supported by a grant from the Slovene Ministry of Science and Technology to Irena Kuhar.
We thank J. Blazquez, R. Hengge-Aronis, A. P. Pugsley, H. Yamada, and G. M. Weinstock for providing bacterial strains.
REFERENCES
- 1.Ambrožič J, Ostroveršnik A, Starčič M, Kuhar I, Grabnar M, Žgur-Bertok D. Escherichia coli ColV plasmid pRK100: genetic organization, stability and conjugal transfer. Microbiology. 1998;144:343–352. doi: 10.1099/00221287-144-2-343. [DOI] [PubMed] [Google Scholar]
- 2.Aviv M, Giladi H, Schreider G, Oppenheim A B, Glaser G. Expression of the genes coding for the Escherichia coli integration host factor are controlled by growth phase, rpoS, ppGpp and by autoregulation. Mol Microbiol. 1994;14:1021–1031. doi: 10.1111/j.1365-2958.1994.tb01336.x. [DOI] [PubMed] [Google Scholar]
- 3.Coote J G. Antigenic switching and pathogenicity: environmental effects on virulence gene expression in Bordetella pertussis. J Gen Microbiol. 1991;137:2493–2503. doi: 10.1099/00221287-137-11-2493. [DOI] [PubMed] [Google Scholar]
- 4.Cornelis G R, Biot T, Lambert de Rouvroit C, Michelies T, Mulder B, Sluiters C, et al. The Yersinia yop regulon. Mol Microbiol. 1989;3:1455–1459. doi: 10.1111/j.1365-2958.1989.tb00129.x. [DOI] [PubMed] [Google Scholar]
- 5.Da Costa X J, Artz S W. Mutations that render the promoter of the histidine operon of Salmonella typhimurium insensitive to nutrient-rich medium repression and amino acid downshift. J Bacteriol. 1997;179:5211–5217. doi: 10.1128/jb.179.16.5211-5217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards R A, Puente J L. Fimbrial expression in enteric bacteria: a critical step in intestinal pathogenesis. Trends Microbiol. 1998;6:282–287. doi: 10.1016/s0966-842x(98)01288-8. [DOI] [PubMed] [Google Scholar]
- 7.Eraso J M, Weinstock G M. Anaerobic control of colicin E1 production. J Bacteriol. 1992;174:5101–5109. doi: 10.1128/jb.174.15.5101-5109.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eraso J M, Chidambaram M, Weinstock G M. Increased production of colicin E1 in stationary phase. J Bacteriol. 1996;178:1928–1935. doi: 10.1128/jb.178.7.1928-1935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farewell A, Kvint K, Nystrom T. Negative regulation by RpoS: a case of sigma factor competition. Mol Microbiol. 1998;29:1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- 10.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 11.Gentry D R, Hernandez V J, Nguyen L H, Jensen D B, Cashel M. Synthesis of stationary-phase ςS is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Gomez J M, Baquero F, Blazquez J. Cyclic AMP receptor protein positively controls gyrA transcription and alters DNA topology after nutritional upshift in Escherichia coli. J Bacteriol. 1996;178:3331–3334. doi: 10.1128/jb.178.11.3331-3334.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiszcynska-Sawicka E, Kur J. Effect of Escherichia coli IHF mutations on plasmid p15A copy number. Plasmid. 1997;38:174–179. doi: 10.1006/plas.1997.1307. [DOI] [PubMed] [Google Scholar]
- 14.Jordi B J A M, Dagberg B, deHaan L A M, Hamers A M, van de Zeijst B A M, Gaastra W. The positive regulator cfaD overcomes the repression mediated by the histone-like protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J. 1992;11:2627–2632. doi: 10.1002/j.1460-2075.1992.tb05328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleerebezem M, Quadri L E N, Kuipers O P, de Vos W M. Quorum sensing by peptide pheromones and two-component signal transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 16.Lange R, Fischer D, Hengge-Aronis R. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the ςS subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1995;177:4676–4680. doi: 10.1128/jb.177.16.4676-4680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leimester M, Domann E, Chakraborty T. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J Bacteriol. 1992;174:947–952. doi: 10.1128/jb.174.3.947-952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu F-M, Chak K-F. Two overlapping SOS-boxes in ColE operons are responsible for the viability of cells harboring the Col plasmid. Mol Gen Genet. 1996;251:407–411. doi: 10.1007/BF02172368. [DOI] [PubMed] [Google Scholar]
- 19.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 21.Pilsl H, Braun V. Strong function-related homology between the pore-forming colicins K and 5. J Bacteriol. 1995;177:6973–6977. doi: 10.1128/jb.177.23.6973-6977.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puente J L, Bieber D, Ramer S W, Murray W, Schoolnik G K. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol Microbiol. 1996;20:87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 23.Pugsley A P. Escherichia coli K12 strains for use in the identification and characterisation of colicins. J Gen Microbiol. 1985;131:369–376. doi: 10.1099/00221287-131-2-369. [DOI] [PubMed] [Google Scholar]
- 24.Pugsley A P, Oudega B. Methods for studying colicins and their plasmids. In: Hardy K G, editor. Plasmids, a practical approach. Oxford, United Kingdom: IRL Press; 1987. pp. 105–161. [Google Scholar]
- 25.Salles B, Weinstock G M. Interaction of the CRP-cAMP complex with the cea regulatory region. Mol Gen Genet. 1989;215:537–542. doi: 10.1007/BF00427053. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Schneider K, Beck C F. Promoter-probe vectors for the analysis of divergently arranged promoters. Gene. 1986;42:37–48. doi: 10.1016/0378-1119(86)90148-4. [DOI] [PubMed] [Google Scholar]
- 28.Sitnikov D M, Schineller J B, Baldwin T O. Transcriptional regulation of bioluminescence genes from Vibrio fischeri. Mol Microbiol. 1995;17:801–812. doi: 10.1111/j.1365-2958.1995.mmi_17050801.x. [DOI] [PubMed] [Google Scholar]
- 29.Tan Y, Riley M A. Rapid invasion of colicinogenic Escherichia coli with novel immunity functions. Microbiology. 1996;142:2175–2180. doi: 10.1099/13500872-142-8-2175. [DOI] [PubMed] [Google Scholar]
- 30.Yamamada H, Yoshida T, Tanaka K, Sasakawa C, Mizuno T. Molecular analysis of the E. coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol Gen Genet. 1991;230:332–336. doi: 10.1007/BF00290685. [DOI] [PubMed] [Google Scholar]
- 31.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–109. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 32.White-Zeigler C A, Blyn L B, Braaten B A, Low D A. Identification of a genetic locus in Escherichia coli involved in the thermoregulation of the pap operon. J Bacteriol. 1990;172:1775–1782. doi: 10.1128/jb.172.4.1775-1782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wrobel B, Wegrzyn G. Replication of plasmids derived from P1, F, R1, R6K, RK2 replicons in amino acid-starved Escherichia coli stringent and relaxed strains. J Basic Microbiol. 1997;37:451–463. doi: 10.1002/jobm.3620370614. [DOI] [PubMed] [Google Scholar]